Summary
Generating engraftable human hematopoietic cells from autologous tissues promises new therapies for blood diseases. Directed differentiation of pluripotent stem cells yields hematopoietic cells that poorly engraft. Here, we devised a method to phenocopy the vascular-niche microenvironment of hemogenic cells, thereby enabling reprogramming of human endothelial cells (ECs) into engraftable hematopoietic cells without transition through a pluripotent intermediate. Highly purified non-hemogenic human umbilical vein-ECs (HUVECs) or adult dermal microvascular ECs (hDMECs) were transduced with transcription factors (TFs), FOSB, GFI1, RUNX1, and SPI1 (FGRS), and then propagated on serum-free instructive vascular niche monolayers to induce outgrowth of hematopoietic colonies containing cells with functional and immunophenotypic features of multipotent progenitor cells (MPP). These reprogrammed ECs- into human-MPPs (rEC-hMPPs) acquire colony-forming cell (CFC) potential and durably engraft in immune-deficient mice after primary and secondary transplantation, producing long-term rEC-hMPP-derived myeloid (granulocytic/monocytic, erythroid, megakaryocytic) and lymphoid (NK, B) progeny. Conditional expression of FGRS transgenes, combined with vascular-induction, activates endogenous FGRS genes endowing rEC-hMPPs with a transcriptional and functional profile similar to self-renewing MPPs. Our approach underscores the role of inductive cues from vascular-niche in orchestrating and sustaining hematopoietic specification and may prove useful for engineering autologous hematopoietic grafts to treat inherited and acquired blood disorders.
Manufacture of autologous engraftable hematopoietic stem and progenitor cells (HSPC) offers tremendous therapeutic potential. Using in vitro cultures, human pluripotent stem cells can be differentiated into hematopoietic progenitors, which often have limited expansion potential and do not engraft myeloablated recipients1–3. Enforced expression of transcription factors (TFs) has also been used to reprogram somatic cells, into hematopoietic lineages4–6. Employing cellular fusion, we have shown that direct conversion of somatic cells into fetal HSPCs is also feasible7. Yet, these prior efforts have been unable to produce human hematopoietic cells capable of long-term multilineage engraftment4–7. We hypothesized that in addition to TF expression, hematopoietic specification and long-term engraftment may require inductive signals from the microenvironment. Indeed, the central instructive role of tissue-specific endothelial cells (EC)8in supporting organ regeneration9,10, including hematopoietic stem cell (HSC) self-renewal and reconstitution of multilineage hematopoiesis, has recently come to light11–18.
In mammals, definitive HSCs originate in the vascular microenvironment of the aorta-gonad-mesonephros (AGM)19–24, placenta25 and arterial vessels26. Putative HSCs bud off from hemogenic vascular cells lining the dorsal aorta floor and umbilical arteries, where they are in cellular contact with non-hemogenic ECs27. This ontological endothelial to hematopoietic transition (EHT) is mediated in part through expression of the TF RUNX121, its non-DNA binding partner core binding factor-β28, GFI1 and GFI1b29,30. However, the contribution of micro-environmental inductive signals provided by anatomically distinct niches and tissue-specific vascular niches8 within the AGM, fetal liver and placenta remain poorly defined.
We have identified a minimal set of four TFs—FOSB, GFI1, RUNX1, and SPI1 (FGRS)—that reprogram full-term human umbilical vein ECs (HUVECs) and human adult dermal microvascular ECs (hDMEC) into hematopoietic cells with long-term multipotent progenitor cell (MPP) activity (rEC-hMPP). The reprograming was successful only when a unique serum-free vascular niche platform was used. Subsets of rEC-hMPPs were immunophenotypically marked as HSCs and were capable of long-term primary and secondary multilineage engraftment in immunodeficient mice. We demonstrate that enforced or transient expression of FGRS-TFs combined with inductive signals from specialized vascular niche Calls1,11,31 are essential for efficient conversion of ECs into rEC-hMPPs.
Results
FGRS-TFs and vascular-induction reprogramming
Primitive HSCs emerge on a vascular bed during development. Thus, we hypothesized that the vascular niche could play an important role during reprogramming by inducing and maintaining nascent hematopoietic cells. Since serum impairs vascular function and interferes with expansion of HSCs and MPPs, we devised a vascular niche model, in which ECs transduced with the adenoviral E4ORF1 gene (E4ECs, VeraVecs) could be cultured without serum1,11,12,31. E4ORF1 activates survival pathways in ECs without provoking proliferation or cellular transformation and thereby maintain tissue-specific functional and metabolic attributes of ECs. E4ECs-derived from HUVECs1,11,12,31 or ECs purified and propagated from hematopoietic organs32,33 balance self-renewal and differentiation of human and mouse long-term HSCs and MPPs by production of physiological levels of Notch-ligands, Kit-ligand, BMPs, Wnts and other angiocrine factors14.
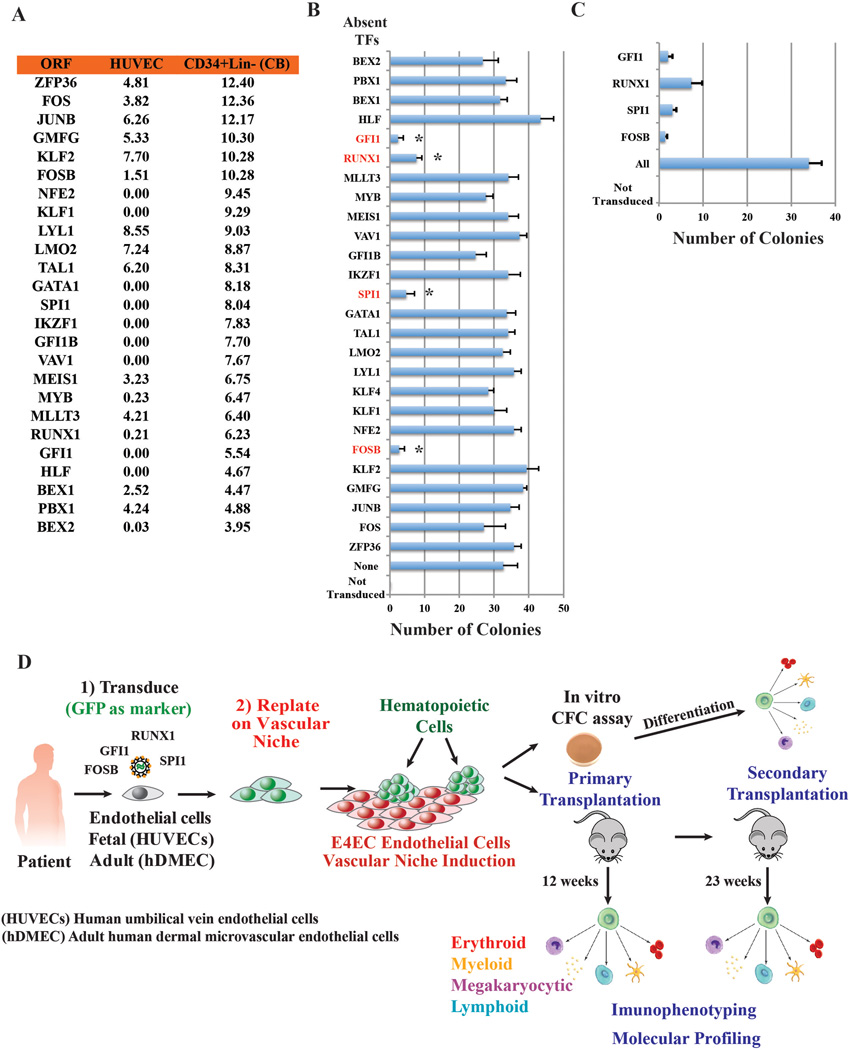
To identify TFs that drive EHT, we first identified TFs differentially expressed by Lin−CD34+ umbilical cord HSPCs, but not by HUVECs (Extended Data Figure 1A–D). We then cultivated CD45−CD133−cKit− CD31+ HUVECs that were devoid of hemogenic potential34 (Figure 1A) and transduced them with lentiviral-vectors expressing various combinations of differentially-expressed TF transcripts using GFP as a marker. Three days after transduction, HUVECs were replated on subconfluent serum-free E4EC-monolayers. Within2 weeks of co-culture with E4ECs, round GFP+CD45+ cells began to bud from transduced HUVECs and morph into grape-like colonies (Figure 1B). Systematic one-by-one dropout of candidate TFs demonstrated that expression of FOSB, GFI1, RUNX1, and SPI1 (FGRS) was necessary and sufficient for hematopoietic reprogramming of HUVECs (Extended Data Figure 1B, C). Co-culture of FGRS transduced-ECs (FGRS-EC) with E4EC monolayers augmented the yield and stability of the hematopoietic-like colonies, which manifested morphological features of hematopoietic progenitors (Figure 1C). Within 4 weeks of co-culture with E4ECs, FGRS-ECs began to proliferate and form GFP+CD45+ colonies (Figure 1A,C). Serum suppressed colony formation and naïve HUVECs could not survive without serum and failed to support the emergence of CD45+cells (Figure 1D). FGRS-transduction of 5×104 HUVECs followed by 3-weeks of serum-free co-culture with E4ECs yielded 32.3±10.5 colonies (Figure 1D) (efficiency of reprogramming 1.5% see methods), occasionally forming multi-colony structures (Extended Data Figure 2A). Once colonies formed, proliferation of GFP+ cells increased and after 5 weeks of co-culture with E4ECs, up to 20×106 GFP+CD45+ cells were produced, a ~400-fold expansion of the input FGRS-ECs (Figure 1D). Since clonal CD45+cells, but not CD45− cells, form colonies it is unlikely that E4ECs are mistakenly identified as hematopoietic cells (Extended Data Figure 2B,C). Thus, FGRS-ECs required sustained inductive and supportive signals from E4EC-vascular niche for efficient hematopoietic reprogramming.
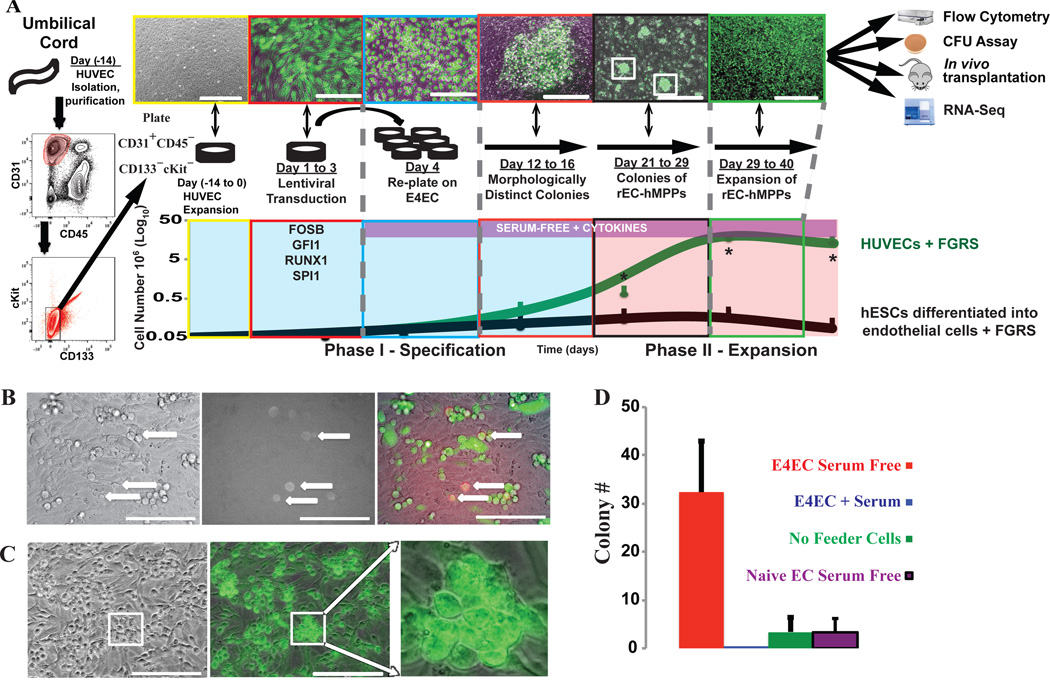
Figure 1. Reprogramming of HUVECs and hES-ECs into hematopoietic cells by FGRS TFs-transduction and vascular-induction.
A. Schema of reprogramming platform of HUVECs into hematopoietic cells. CD45−CD31+CD133−cKit− cells were sorted from freshly purified HUVECs and expanded (days −14 to 0). Sorted cells were transduced with FGRS (FGRS-ECs)(days 1–3) and grown in EC-media. On day 4, transduced cells were replated on E4ECs in serum-free hematopoietic media (days 12–40). Distinct GFP+ flat colonies were observed at days 12–16, which by days 21–29 remodeled into three-dimensional grape-like colonies. After a month (days 29–40) GFP+ cells expanded ~400-fold (n=4). CD144+VEGFR2+ECs derived from hES-ECs35 were also transduced with FGRS. The process of reprogramming is subdivided into two phases: Phase I–Specification (Day 1–20) and Phase II–Expansion (Day 21–40). The expanding cultures were assayed for morphological change, cell number, and CD45. Kinetics of reprogramming of HUVECs (Green trace) and expansion of reprogrammed hES-ECs cells (Black trace).
B. Emergence of rounded hematopoietic-like GFP+CD45+ cells two to three weeks after HUVECs were transduced with FGRS (white arrows).
C. Formation of GFP+ hematopoietic-like colonies on the E4ECs 3–4 weeks after FGRS transduction.
D. Generation of GFP+CD45+ hematopoietic-like colonies (C) from FGRS-ECs is enhanced by co-culturing with serum-free E4ECs and blocked by presence of serum (n=8, p<0.05). Scale bar: 200 µm.
To date, efforts to differentiate pluripotent stem cells into repopulating hematopoietic cells have had limited success1–3. We hypothesized that FGRS-TFs could augment hematopoietic differentiation of human embryonic stem cells (hES). To test this, we first differentiated hESCs into ECs (hES-EC)35,36 and then transduced purified VEGFR2+CD144+ hES-ECs with FGRS. Although this approach generated CD45+CD144− progeny (Extended Data Figure 2D), these cells did not form stable hematopoietic-like colonies and did not proliferate (Figure 1A black line). Thus, hES-ECs are not as permissive as HUVECs for reprogramming into hematopoietic cells.
rEC-MPPs have features of multilineage progenitors
During reprogramming, GFP+ FGRS-ECs and vascular-induced hematopoietic-like colonies lost CD31, but gained the expression of hematopoietic markers CD45, CD43, CD90(Thy-1) and CD14 (Figure 2A and Extended Data Figure 2E). In contrast, the GFPE4ECs remained CD31+CD34+CD45− Importantly, a subset of GFP+CD45+FGRS-EC progeny manifested the immunophenotype of human MPPs (CD45+Lin− CD45RA−CD38−CD90+CD34+) and HSCs (CD45+Lin−CD45RA−CD38−CD90−CD34+)37,38(Figure 2B). To assess the function of various populations of these ECs reprogrammed into human MPPs (rEC-hMPPs), we sorted four week-old GFP+CD45+CD34+ rEC-hMPPs and seeded them in colony-forming cells (CFC) assays to enumerate progenitor cells. The rEC-MPPs gave rise to GFP+ colonies with CFU-GEMM (granulocytic/erythroid/megakaryocytic/monocytic), CFU-GM (granulocytic/macrophage), and hemoglobinized BFU-E (erythroid) morphologies (Figure 2C). FACS and cytospin analysis documented presence of cells with morphological (Figure 2D) and immunophenotypic features of granulocyte/macrophage (CD11b+, CD14+), erythroid (CD235+), dendritic (CD83+) and megakaryocyte (CD41a+) progenies (Extended Data Figure 2F). The function of rEC-hMPP-derived macrophages was corroborated using a phagocytosis assay (Extended Data Figure 2G). Thus, rEC-hMPPs contain functional multilineage progenitor cells.
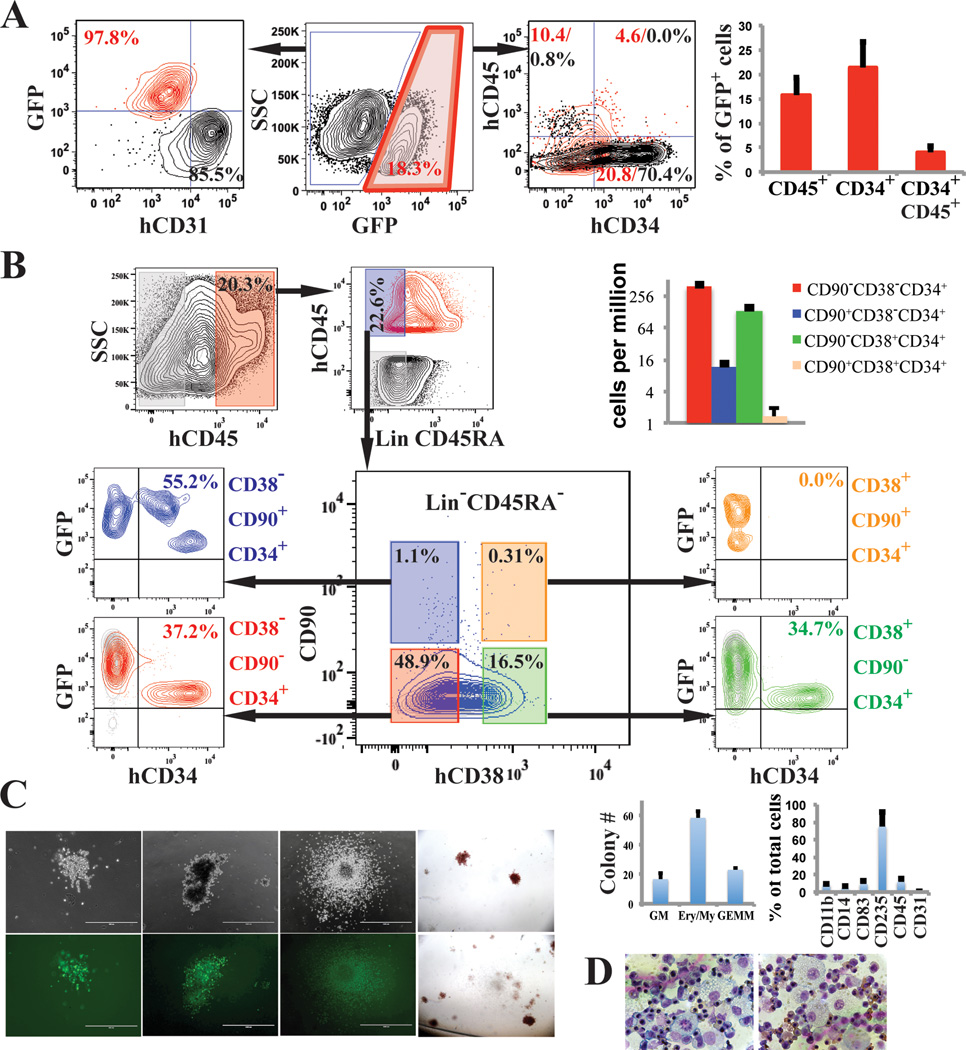
Figure 2. rEC-hMPPs phenotypically and functionally resemble multilineage HSPCs.
A. FACS analysis of cocultured GFP−E4ECs vascular niche along with GFP+ FGRS transduced HUVECs (FGRS-ECs) four weeks post-transduction (n=9).
B. Immunophenotypic analysis of FGRS reprogrammed HUVECs (red and blue; n=3).
C. Four weeks after FGRS-transduction and E4EC-induction, human GFP+CD45+CD34+ cells were sorted and seeded for CFC assay (n=3). Hematopoietic colonies arose in the CFC assay (magnificationx4); wide field (upper row) and corresponding fluorescent images (bottom row). Left to right: granulocytic-erythroid-monocytic-megakaryocytic (GEMM), Erythroid/Myeloid, and granulocytic-macrophage (GM) colonies, and hemoglobinized colonies. Graph shows CFC assay quantification.
D. Wright-Giemsa stain of cells obtained from the CFC assay colonies. 60x magnification.
rEC-hMPPs engraft long-term into primary recipients
To assess the engraftment potential of rEC-hMPPs, we transplanted 1.5×106 CD45+GFP+rEC-hMPPs into adult sub-lethally irradiated (275 Rad) immunocompromised-NSG mice (Figure 3A). We detected circulating human CD45+ cells in the peripheral blood (PB) of recipient engrafted mice from 2 to 44 weeks (Figure 3B) and found CD45−CD235+ erythroid cells 16 weeks, post-transplantation (Figure 3C). We then sorted hCD45+ cells from bone marrow (BM) of recipient mice 22 to 24 weeks after transplantation and cultured them for 24 hours. The hCD45+hCD34+ cells were resorted and seeded into CFC-assays. They formed CFU-GM, CFU-GEMM, and BFU-E hematopoietic colonies with typical myeloid progeny morphologies (Figure 3D). Hence, rEC-hMPPs are capable of robust multilineage engraftment.
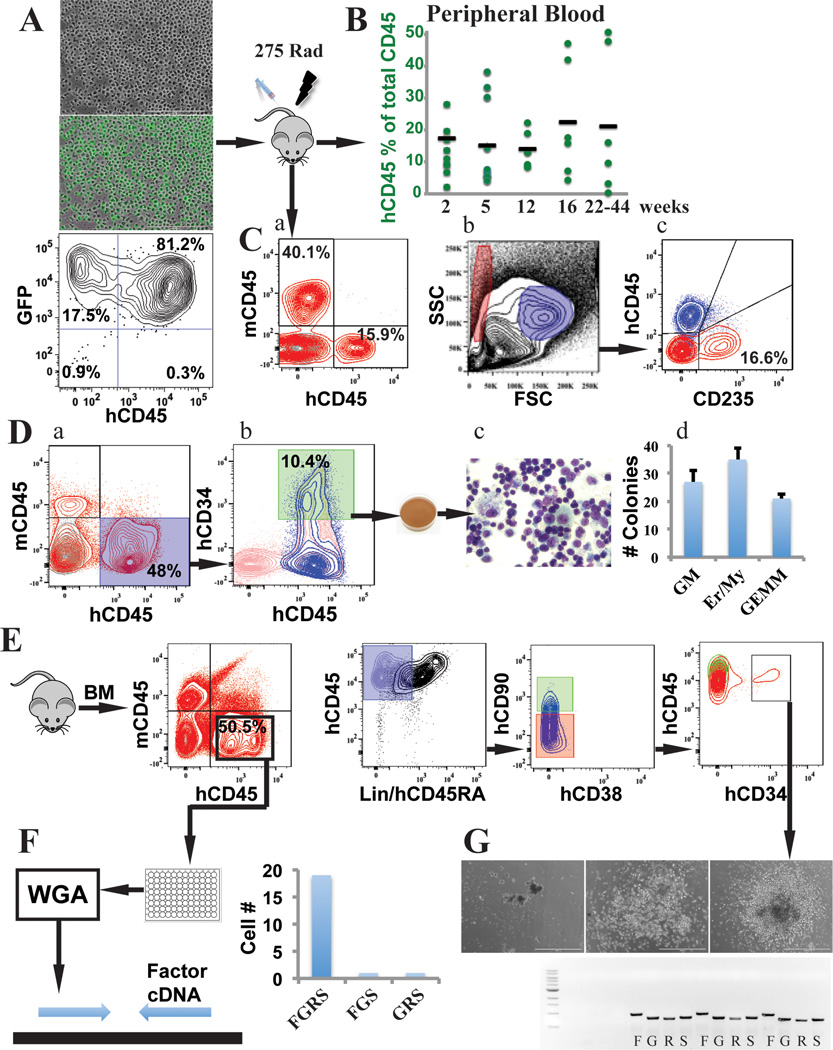
Figure 3. rEC-hMPPs are capable of in vivo erythro-myeloid-megakaryocytic multilineage engraftment.
A. Reprogrammed HUVECs into rEC-hMPPs were transplanted into sub-lethally irradiated (275 Rad) mice (n=9).
B. Circulating human CD45+ (hCD45+ ) cells were detected at 2 (n=7; 17.38±7.73%), 5 (n=6; 15.1±13.39%), 12 (n=6; 14.14±5.44%), 16 (n=6; 22.36±17.95%) and 22 to 44 (22–44) (n=6; 21.23±22.27%) weeks post transplantation. The 22–44 weeks engrafted mice were used for further analyses of the myelodysplasia and fibrotic changes (Extended Data Figure 8,9,10A).
C. Analysis of the total mononuclear peripheral blood cells at 16 weeks post-transplantation of hCD45+and mouse CD45+(mCD45+) cells, revealed presence of hCD45+(15 9%) and human non-erythroid circulating cells (a). We gated on the FSC/SSC hCD45− erythroid compartment (Red gate) and typical human non-erythroid hCD45+ compartment (Blue gate)(b,c).
D. rEC-hMPPs isolated from the host retained their multilineage potential in vitro; secondary CFC-assay. Engraftment of mouse bone marrow (BM) 22 weeks post-transplantation is shown (a). The cells were expanded in vitro for 24 hours (b) and FACS resorted for hCD45+hCD34+ cells for CFC assay. Wright-Giemsa stain (c) of the cytospin of the cells from CFC assay, (magnificationx100 right). Quantification of the CFC assay (d).
E. Phenotypic analysis of an in vivo 22 weeks engrafted rEC-hMPPs in BM shows human CD45+Lin− CD45RA−CD38−CD90−CD34+ MPPs.
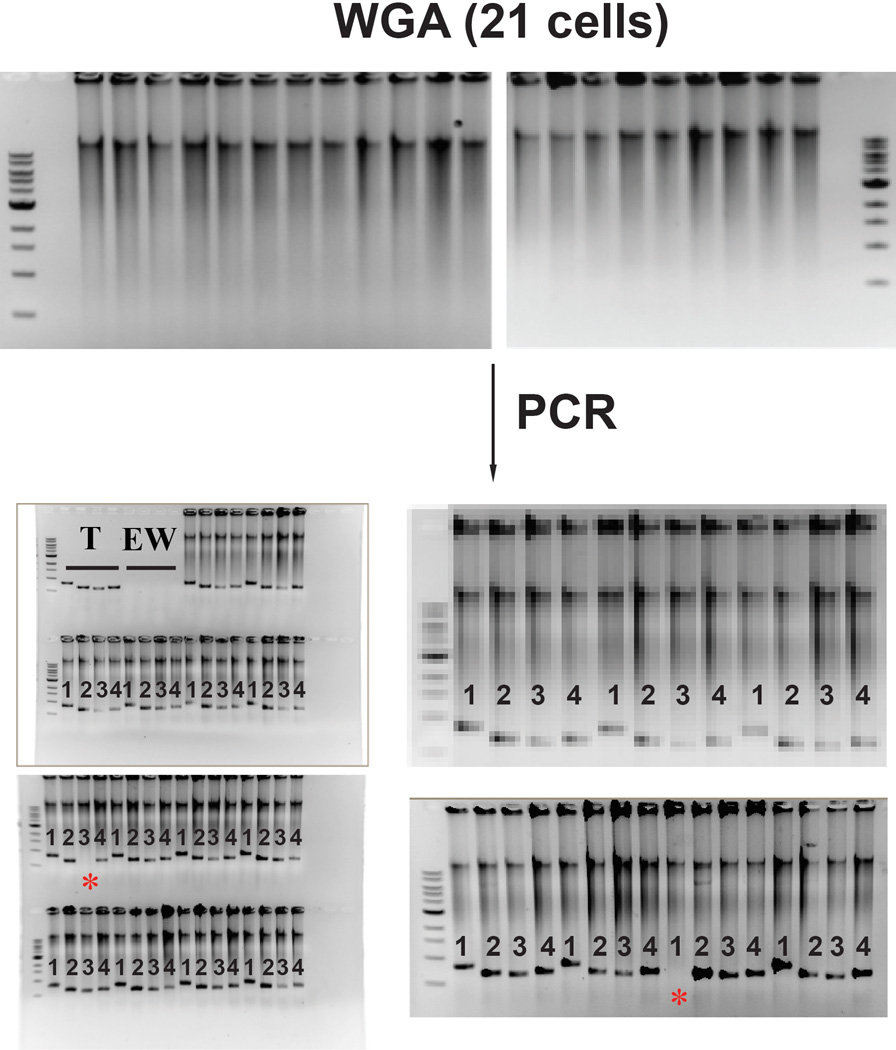
F. Identification of viral integration on a single-cell level. Whole genome amplification (WGA) of 21 hCD45+cells isolated from a host mouse (E). Quantification of the analysis is shown in the right graph.
G. Identification of viral integration on a single-colony level. Lin−CD45RA−CD38−CD90− CD34+cells (10 cells) were used for a CFC assay. We detected all four FGRS viral vectors in all CFC colonies tested (bottom image; Letters FOSB (F), GFI1 (G), RUNX1 (R), SPI1 (S) show PCR products specific for each of these factors in the first colony).
rEC-hMPPs are derived from non-hemogenic ECs
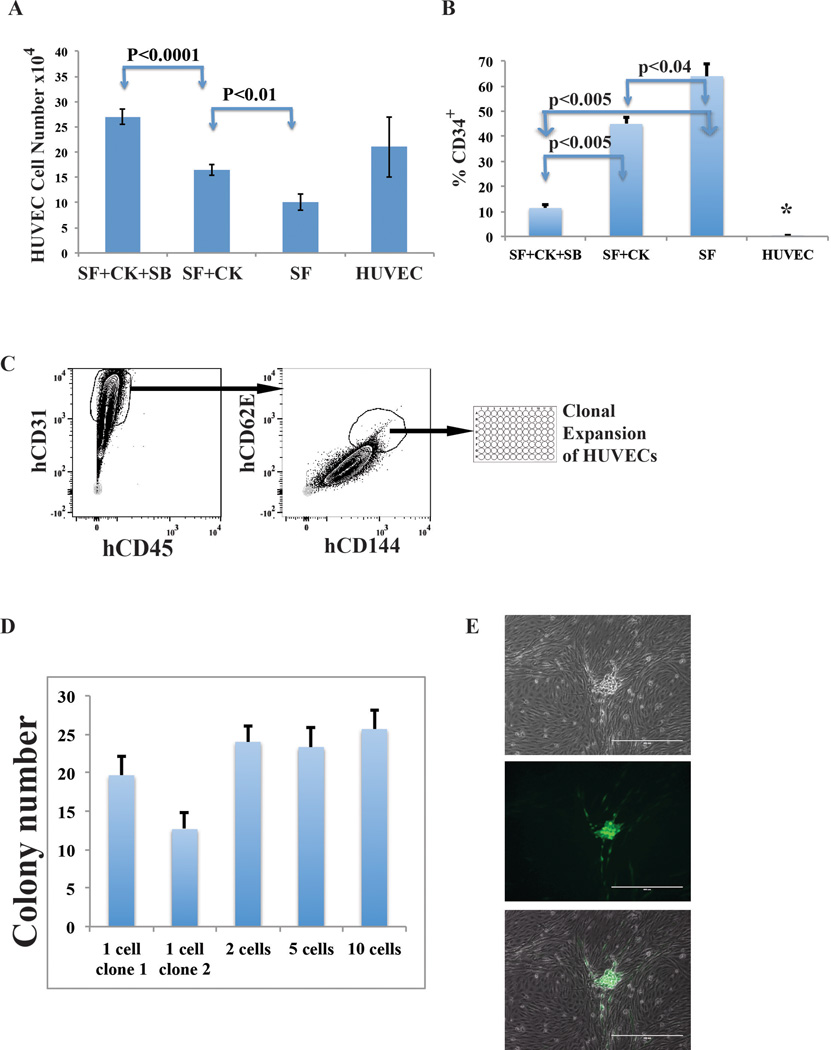
To rule out the possibility that rEC-hMPPs are derived from rare contaminating hemogenic cells25,34, we cultured naïve ECs in optimal pro-hematopoietic media or performed clonal reprogramming of ECs. Neither serum-withdrawal, nor addition of hematopoietic cytokines induced formation of CD45+CD34+cells from naïve HUVECs (Extended Data Figure 3A,B). Furthermore, clonal cultures of mature CD45−CD144+CD31+CD62E(E-selectin)+ ECs32,33 did not form CD45+rEC-MPPs (Extended Data Figure 3C,D and Extended Data Figure 4A,B,C). Thus, rEC-hMPPs were not derived from scarce population of spontaneously differentiating ECs with preexisting hemogenic potential.
The BM of robustly engrafted recipient NSG mice contained a small population of cells with Lin−CD45RA−CD38−CD90−CD34+ immunophenotype of human MPPs37 (Figure 3E). To ensure that engrafted cells were derived from FGRS-ECs, we purified hCD45+ cells from recipient BM (Figure 3F) and seeded single-cells into 96-well plates for whole genome amplification (WGA) and detection of viral vector integration. All hCD45+ cells had been transduced by lentiviral vectors, and 19/21 cells showed integration of all four FGRS-TFs (Figure 3F, Extended Data Figure 5). To verify that these cells were progeny of rEC-hMPPs, we seeded hCD45+ cells for CFC-assays to examine viral integration in individual colonies (Figure 3G). We demonstrated that all tested colonies were derived from cells that had integrated the lentiviral vectors expressing FGRS (Figure 3G). Therefore, engrafted human hematopoietic cells were derived from transplanted rEC-hMPPs.
To test whether FGRS-induced reprogramming triggered expression of endogenous human FGRS genes24, we expressed genetically distinct murine TFs (mFGRS) using inducible-lentiviral vectors to reprogram HUVECs into rEC-hMPPs and then assessed for endogenous human FGRS-gene expression. Transient expression of mFGRS with E4EC-coculture for 3-weeks induced a 100-fold greater expression of endogenous genes than that of switched-off mFGRS transcripts (Extended Data Figure 6A,B,C). Therefore, rEC-hMPPs do not require continuous expression of exogenous FGRS-TFs to sustain their hematopoietic cell fates.
Furthermore, we speculated that enforced SPI1 expression might prevent rEC-hMPPs from differentiating into T-cells39,40 Therefore, we constitutively expressed FGR-TFs with a Tet-inducible SPI1 and E4EC-induction, which resulted in generation of small but significant population of CD3+ T-cells (Extended Data Figure 6D,E). Thus, generation of lymphoid cells from rEC-hMPPs could be optimized by transient expression of specific TFs.
Adult ECs are reprogrammable to rEC-hMPPs
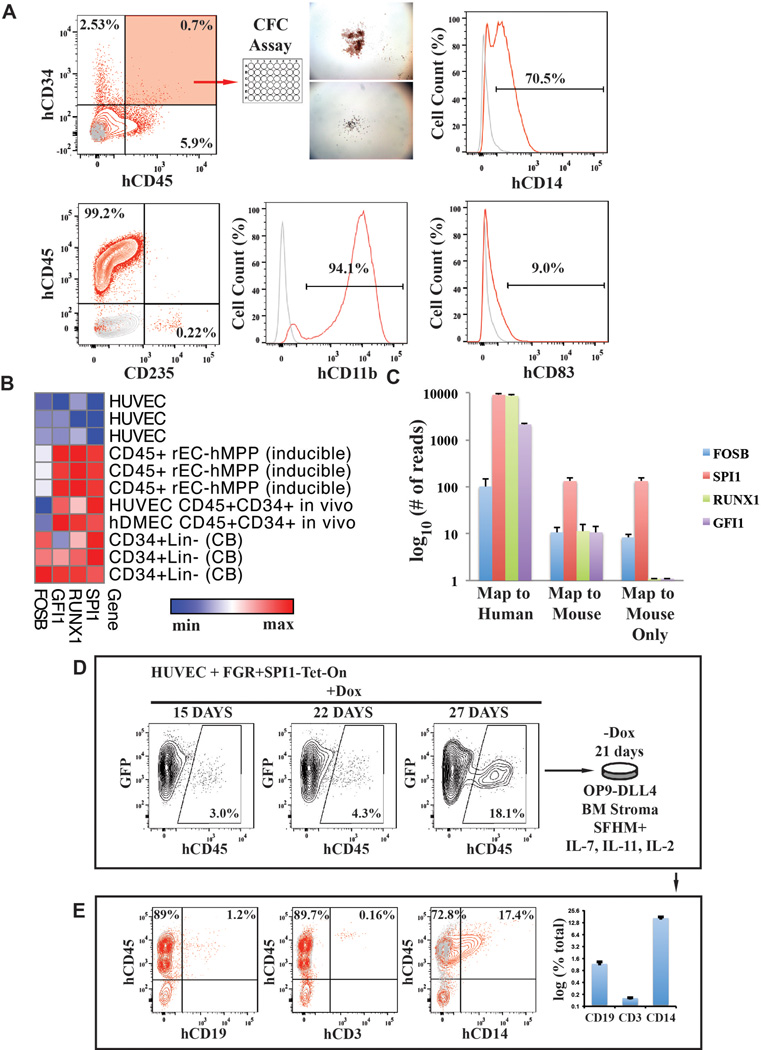
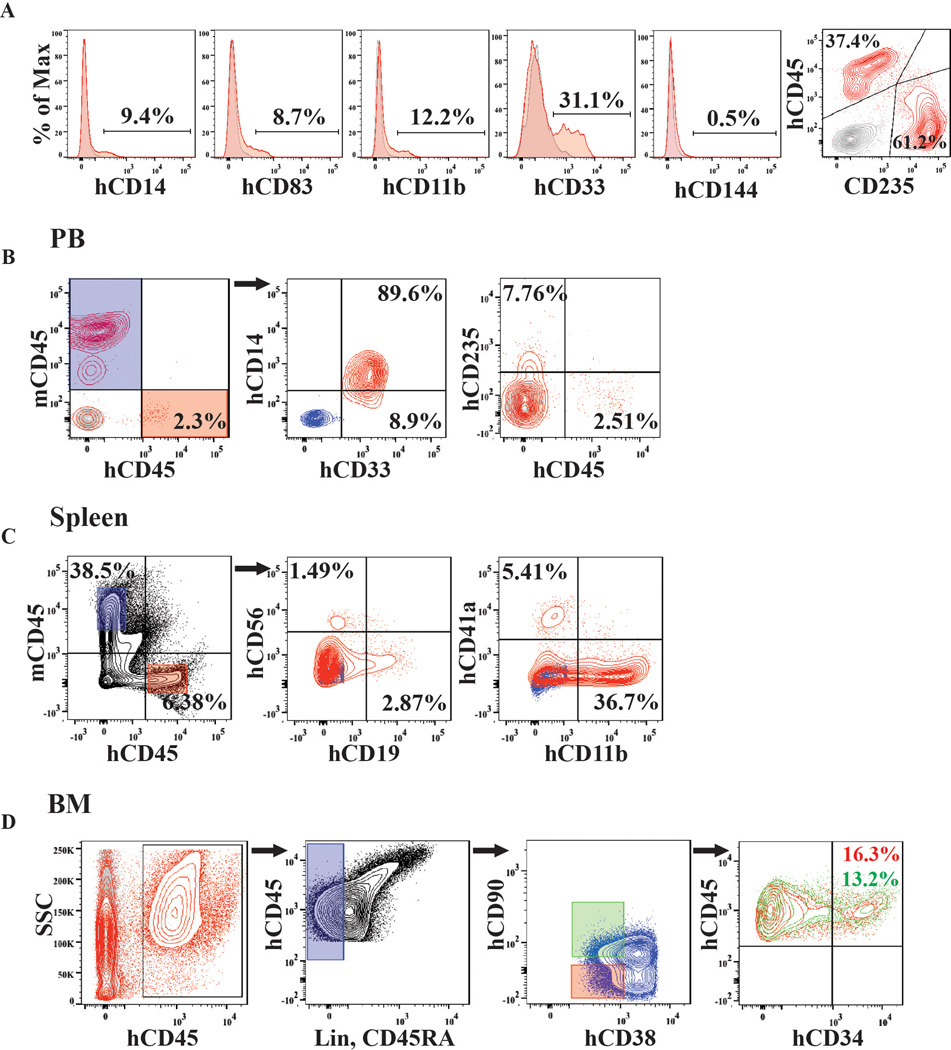
To test whether our approach could reprogram adult human ECs, we transduced hDMECs with FGRS-TFs and propagated them on serum-free E4EC-monolayers (Figure 4A). After 4 weeks GFP+hCD45+CD34+cells were sorted for CFC-assay. The rEC-hMPPs yielded cells with morphological features of hematopoietic (Figure 4Ba) and functional myeloid CFU-GM, CFU-GEMM, and BFU-Es (Figure 4Bb), containing CD235+erythroid, CD33+/CD14+/CD11b+macrophage/monocyte, and CD83+dendritic progenies (Figure 4Bb and Extended Data Figure 7A).
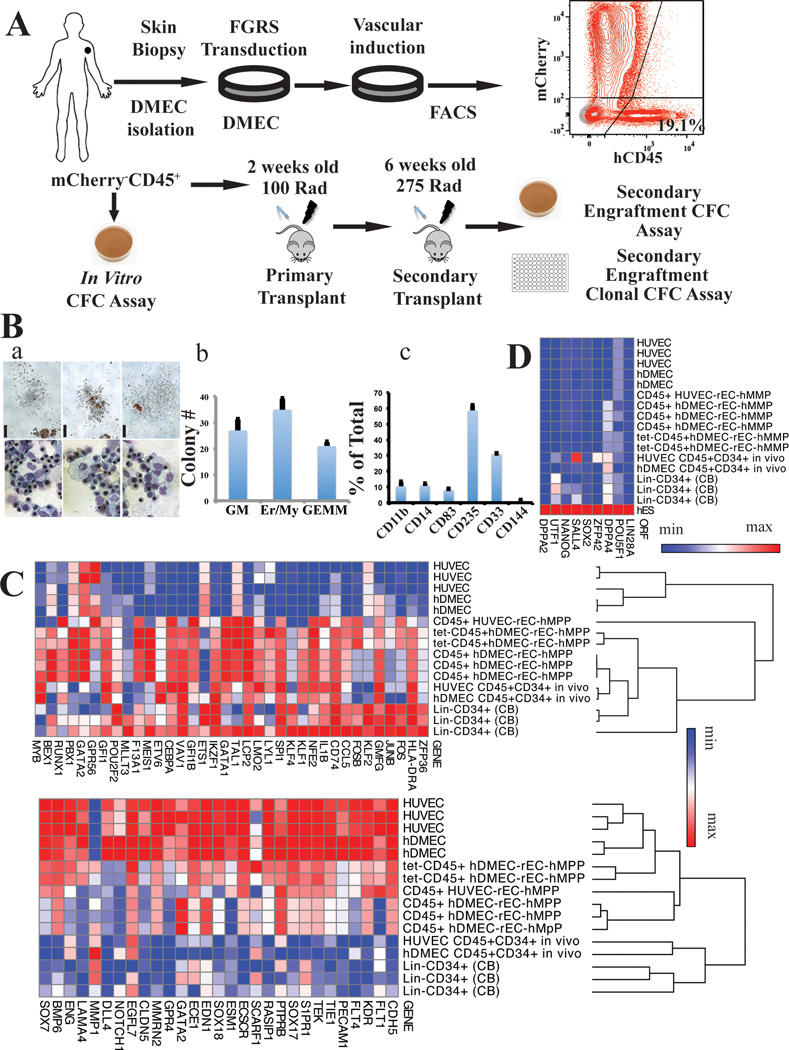
Figure 4. Functional and transcriptional analysis of adult hDMEC-derived rEC-hMPPs.
A. Schematic representation of in vitro and in vivo functional tests of hDMEC-derived rEC-hMPPs. B. (a) Hematopoietic colonies observed in CFC assay (scale bar: 200µm); wide field (upper row). Wright-Giemsa stain of cells from CFC colonies (magnification x60) (bottom row). (b) Quantification of the CFC assay(n=3). (c) Immunophenotypic quantification of surface marker expression in CFU colonies(n=3).
C. Global gene transcription profiling of in vitro generated CD45+ rEC-hMPPs (derived from hDMEC and HUVECs) and in vivo engrafted HUVEC-derived CD45+CD34+rEC-hMPPs, 22 weeks post-transplantation and hDMEC-derived CD45+CD34+rEC-hMPPs, 15 weeks post-secondary engraftment (4A). EC-endothelial cells, CB–cord blood cells.
D. Comparison of expression of prototypical pluripotency genes shown in (4C). Human embryonic stem cells (hES). The data in C and D are presented as log2 (transcription level).
Next, we compared the transcription profiles of rEC-hMPPs, before and after NSG engraftment, to those of naive HUVEC, hDMEC and purified Lin-CD34+ cord blood HSPCs (Figure 4C). FGRS-transduction plus E4EC-induction activated hematopoietic genes and down-regulated vascular gene signatures (Figure 4C). Importantly, 22-weeks post-transplantation, CD45+CD34+ rEC-hMPPs had a transcription pattern similar to normal CD34+cord blood HSPCs and distinct from the ECs from which they were derived (Figure 4C). Notably, pluripotency genes were not induced in rEC-hMPPs indicating that reprogramming does not require transition through a destabilizing pluripotent intermediate (Figure 4D).
rEC-hMPPs engraft primary & secondary recipients
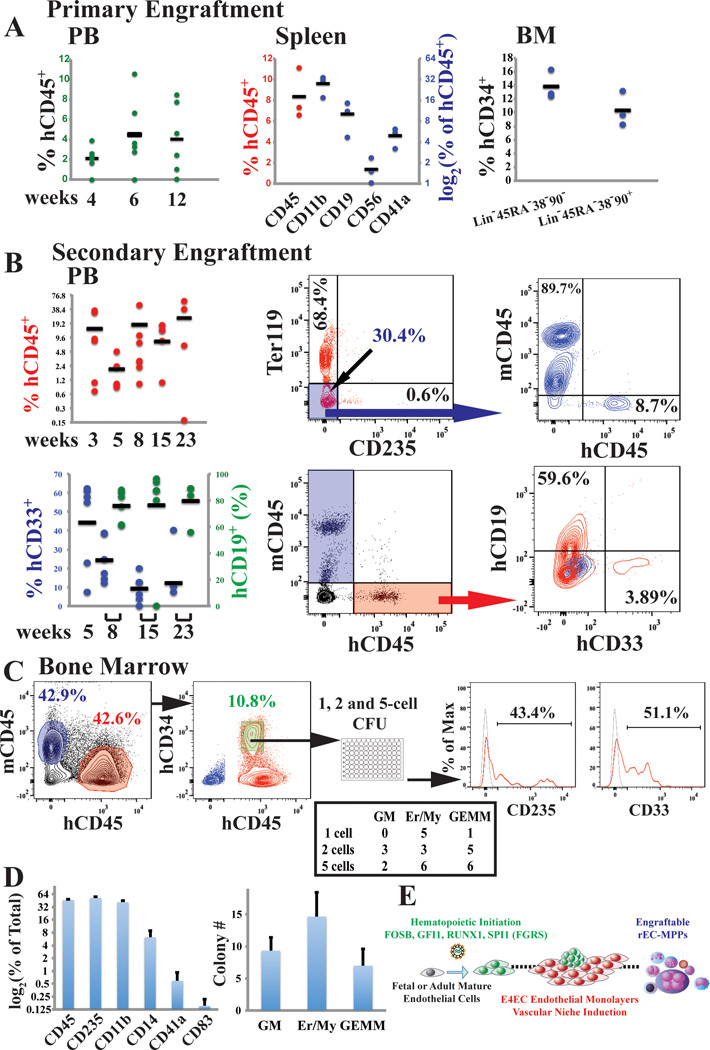
To assess engraftment potential of hDMEC-derived rEC-hMPP, we transplanted 1×105 CD45+GFP+rEC-hMPP into sub-lethally irradiated (100 Rads) 2-weeks old neonatal NSG mice (Figure 5A). Circulating hCD45+cells were detected in the PB of recipient animals 4 (2.09±1.27%), 6 (4.46±3.66%), and 12 (4.05±3.50%) weeks post-transplantation (Figure 5A). Fourteen weeks post-transplantation, human hematopoietic cells were found in PB, BM and spleen (Figure 5A and Extended Data Figure 5B,C, D). Notably, these recipient animals harbored myeloid and lymphoid populations, including CD19+ B-cells (10.13±4.98%), CD56+ NK-cells (1.62±0.67%), CD11b+ monocyte/macrophages (27.66±8.92%;) and CD41a+ megakaryocytes (4.90±1.51%;) in their spleens (Figure 5A and Extended Data Figure 7B,C,D). Hence, rEC-hMPPs are capable of prolonged multilineage hematopoietic engraftment. The BM of primary recipient mice (week 12–14) contained populations with the immunophenotype of human HSCs (hCD45+Lin− CD45RA−CD38−CD90+CD34+, 10.37±2.55%) and MPPs ((hCD45+Lin− CD45RA−CD38−CD90−CD34+, 13.83±2.14%)(Extended Figure 7D)37,38. Because these populations can self-renew, we tested whether BM cells of mice engrafted by primary rEC-hMPPs (12 weeks post-transplant) could engraft secondary NSG recipient mice. Indeed, the PB of secondary recipients was engrafted by human myeloid and lymphoid progenies 3(14.61±15.7%), 5(2.01±1.5%), 8(17.78±16.23%), 15(7.99±7.36%) and 23(26.3±25.7%) weeks post-transplantation (Figure 5B). Thus, subpopulations of rEC-hMPP scan self-renew and are capable of durable myeloid and lymphoid engraftment in NSG mice: characteristics similar to true hMPPs.
Figure 5. Adult human hDMECs-derived rEC-hMPPs are capable of in vivo primary and secondary multilineage engraftment.
A. Analysis of PB of mice at 4, 6, and 12 weeks post-primary transplantation(n=6). Analysis of spleen and BM of mice at 14 weeks post-primary transplantation(n=3).
B. Analysis of the PB of mice at 3, 5, 8 (n=6), 15 (n=4) and 23 (n=4) weeks post-secondary transplantation (n=6). FACS plots on the right show representative analysis of rEC-hMPP secondary engraftment. Mouse Ter119+ and human CD235+ erythroid populations were excluded to obtain an accurate estimation of hCD45+ and mCD45+cells.
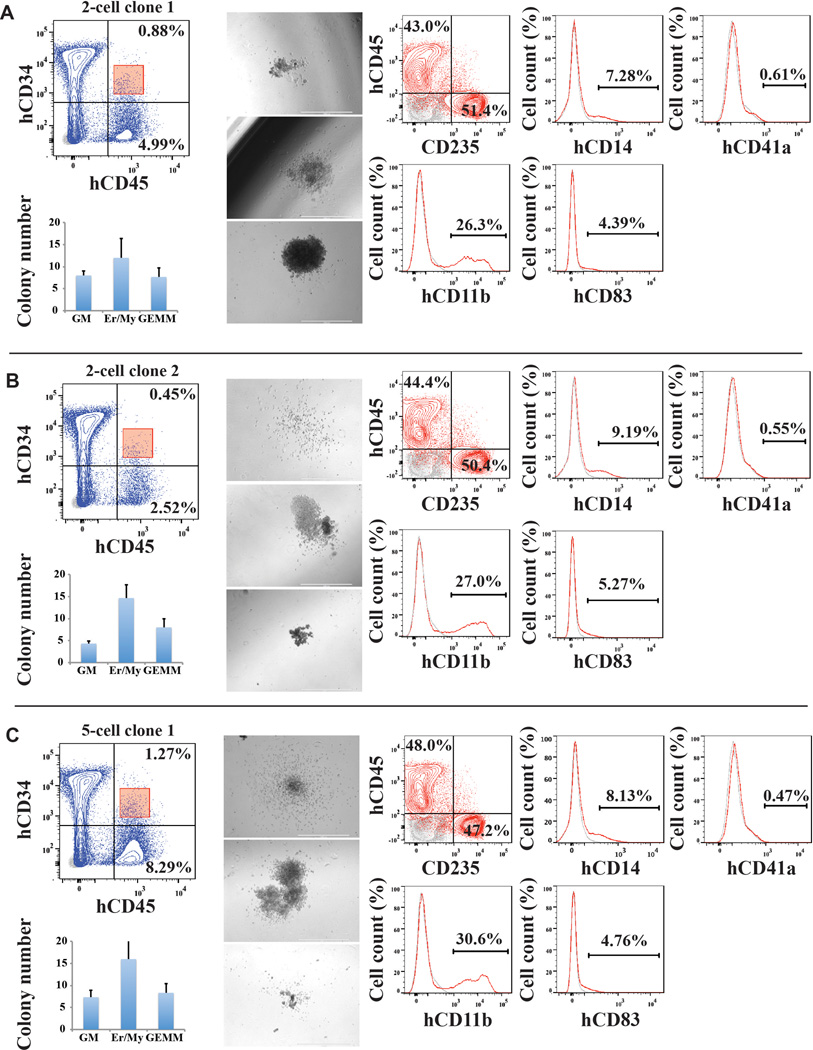
C. Clonal CFC assay of BM hCD45+hCD34+cells(n=3; left plot). Emerging colonies were counted and classified (middle table). CFC colonies derived from single plated hCD45+hCD34+ cell comprise of mixed-lineage erythroid and myeloid progenies(right plots).
D. Reprogrammed cells isolated from the host retained their multi-lineage potential in vitro; secondary CFC assay.
E. Schematic representation of steps of reprogramming of ECs into rEC-hMPP by FGRS.
To examine whether individual rEC-hMPP cells retained clonal multilineage potential, we isolated hCD45+hCD34+cells from BM of secondarily robustly engrafted mouse 15-weeks post-transplantation and then assessed the multilineage CFC activity in clonal (1 cell/well), oligo-clonal (2 and 5 cells/well) and bulk (1000 cells/well) sorted cells (Figure 5C,D). All single-cell derived colonies displayed multilineage differentiation, including CD33+/CD14+/CD11b+myeloid, CD41+ megakaryocytic and CD235+erythroid progenies (Figure 5C,D), indicating that engrafted rEC-hMPPs from secondary transplants retained their MPP potential. Thus, individual cells within the rEC-hMPPs have the immunophenotypic and functional attributes of HSPC-like/self-renewing hMPPs (Figure 5E).
Notably, rEC-hMPPs isolated from primary and secondary engrafted mice manifested no evidence of malignant transformation (Extended Data Figure 8; 9; 10A) or genetic abnormalities or (Extended Data Figure 10B).
Discussion
The availability of engraftable autologous human cells offers the potential to cure a wide spectrum of benign and malignant hematologic disorders. Prior efforts using pluripotent stem cells have been obstructed by low efficiency and poor engraftment2,3,41,42. Here, we have taken advantage of ontological link between endothelial and hematopoietic development to efficiently reprogram mature, fetal and adult ECs into engraftable self-renewing hMPPs without transitioning through a potentially destabilizing pluripotent intermediate. Just as support from non-hemogenic vascular cells important for EHT during development, we found the instructive contribution of the vascular niche was central to reprogramming ECs to hematopoietic cells.
Differentiating pluripotent stem cells or expanding AGM-derived cells to engraftable hematopoietic cells has been inefficient, when stromal cells have been used for niche-like support2,3,41. This could be due to: 1) poor inductive function of stromal cells in serum-free culture; and/or 2) distinguishing features of ECs that resemble the hematogenic niche cells that support EHT11–15. For example, E4ECs produce the proper stoichiometry of inductive angiocrine factors, including Notch, BMP and c-Kit pathways4 that are important for EHT43. Thus, adult organ-specific pro-hematopoietic vascular niches, such as HUVECs11–15,36, BM13, hepatic and splenic sinusoids8 may share functional characteristics with EHT-inductive niche cells. The rEC-hMPPs can engraft primary and secondary recipient mice with individual cells capable of differentiating to multiple hematopoietic lineages. However, the influence of recipient microenvironmental signals and temporal aspects of reprogramming influence the outcome of xenograft studies. NSG mice lack the proper niches for T-cell differentiation and we were not able to determine whether engrafted rEC-hMPPs could give rise to T-cells. We found that temporally restricted expression of SPI1, along with sustained FGR, increased the ability of the rEC-hMPP to differentiate to lymphoid lineages suggesting that sustained SPI1 interfered with lymphogenesis. Notably, even transient expression of FGRS is sufficient to activate endogenous TFs. The age of recipient mice was also important because transplantation of neonatal (2-week old) NSG mice enhanced lymphoid engraftment by rEC-MPPs. Therefore, temporal and chronological expression of FGRS-TFs with proper stoichiometry combined with vascular niche signals appears to increase the yield of rEC-hMPPs with authentic multilineage long-term self-renewing HSPC potential.
Direct reprogramming of ECs into engraftable HSPCs orchestrated by the inductive signals conveyed by tissue-specific vascular niches offers an innovative way to decipher the hierarchy of TFs and micro environmental cues that guide hematopoietic development. Our approach lays a foundation for engineering engraftable autologous rEC-hMPPs and potentially true HSCs for treatment of patients with hematological disorders.
Methods Summary
Endothelial cells (ECs) were reprogrammed into hematopoietic cells by transduction with transcription factors (TFs) and vascular niche induction. To establish vascular niche-platform, ECs were purified and transduced with a lentiviral vector expressing the adenoviral E4ORF1 gene (E4ECs, VeraVecs, Angiocrine Bioscience, NY, NY). Purified CD45−CD133−cKit−CD31+and clonal populations of CD45− CD144+CD31+CD62E+ full term human umbilical vein ECs (HUVECs) and adult primary human dermal microvascular ECs (hDMEC) were cultured in EC-growth medium. Then, HUVECs or hDMECs were transduced with lentiviral vectors expressing GFP and combination of TFs: FOSB, GFI1, RUNX1, and SPI1 (FGRS). After 3 days, GFP+ FGRS-transduced ECs were plated in co-culture with 30–50% subconfluent E4EC-monolayers supplemented with serum-free hematopoietic media composed of StemSpan SFEM, 10% KnockOut Serum Replacement, 5ng/ml FGF-2, 10ng/ml EGF, 20ng/ml SCF, 20ng/ml FLT3, 20ng/ml TPO, 20ng/ml IGF-1, 10ng/ml IGF-2, 10ng/ml IL-3 and 10ng/ml IL-6. After 3 to 4 weeks of co-culture, outgrown GFP+ reprogrammed ECs into human multipotent progenitor cells (rEC-hMPPs) formed typical grape-like hematopoietic colonies. After 4 weeks, human CD45+rEC-MPPs were FACS sorted for: 1) immunophenotypic analyses, 2) methylcellulose-CFC assay, 3) molecular profiling, 4) comparative genomic hybridization and 5) transplanted retroorbitally into primary sublethally-irradiated (275 Rads) 6-week old NSG mice or sublethally-irradiated (100 Rads) 2-week old mice neonates. After three months, human CD45+ cells (hCD45+ cells) derived from bone marrow (BM) or whole BM of the primary engrafted mice was used for transplantation into secondary recipients. After 3 months of primary and 6 months of the secondary transplantation, engrafted hCD45+ cells in BM, spleen and peripheral blood of mice were FACS sorted and processed for: 1) multi-variate immunophenotypic analyses, 2) clonal and oligo-clonal CFC assay, and 3) molecular profiling. Tissues of the engrafted mice were processed for histological examination to rule out any malignant transformation.
Methods
Fetal and adult endothelial cells (ECs) used for reprogramming
Full term human umbilical vein ECs (HUVECs) were obtained as previously described32,33. Multiple purified populations of CD45-CD133-cKit-CD31+HUVECs were isolated from separate umbilical cords (n=8) and were cultured in Endothelial Growth Media (EM): Medium 199 (Thermo Scientific: #FB-01), 20% Fetal Bovine Serum (Omega Scientific), 20 µg/ml endothelial cell supplement (Biomedical Technologies: #BT-203), 1XPen/Strep, and 20 units/ml Heparin (Sigma: # H3149-100KU). Multiple batches (n=3) of adult primary human dermal microvascular endothelial cells (hDMEC) were purchased from ScienCell Research Laboratories (cat #2020). In addition, cultured HUVECs were passaged for 3 to 5 times and then CD45− CD144+ CD31+ CD62E+ HUVECs were sorted for clonal analyses to rule out contamination with pre-existing hemogenic ECs.
For reprogramming experiments, transduced HUVECs or hDMECs were co-cultured with E4ECs in serum-free hematopoietic media (HM) formulated as StemSpan SFEM (Stemcell Technologies), 10% KnockOut Serum Replacement (Invitrogen), 5ng/ml bFGF (FGF-2), 10ng/ml EGF, 20ng/ml SCF (soluble Kit-ligand), 20ng/ml FLT3, 20ng/ml TPO, 20ng/ml IGF-1, 10ng/ml IGF-2, 10ng/ml IL-3, 10ng/ml IL-6 (all from Invitrogen, eBioscience, or Peprotech).
Manufacturing of vascular niche platform
To establish the vascular niche monolayers, HUVECs were purified and transduced with lentiviral vector carrying a cassette of adenoviral E4ORF1 gene (E4ECs) as previously described31 or obtained as VeraVecs from Angiocrine Bioscience, New York, NY. E4ECs proliferate in serum-free and xenobiotic-free conditions only supplemented with minimal angiogenic factors. All naïve ECs that are non-transduced with E4ORF1 are depleted during passaging in serum-free conditions. Confluent monolayers of E4ECs are contact inhibited, non-transformed and propagate as homogenous monolayers providing ideal instructive niche for reprogramming and sustaining FGRS-transduced ECs into rEC-hMPPs.
Reprogramming of ECs into MPPs (rEC-MPPs)
ECs were reprogrammed into hematopoietic cells by transduction with transcription factors (TFs) and vascular niche induction. Purified populations of CD45−CD133−cKit−CD31+and clonal CD45− CD144+CD31+CD62E+ full term HUVECs and adult primary hDMEC were cultured in the EC-growth medium (EM). Then, HUVECs or hDMECs were transduced with lentiviral vectors expressing GFP and combination of transcription factors (TFs), FOSB, GFI1, RUNX1, and SPI1 (FGRS) and maintained in EM. After 3 days, GFP+ FGRS transduced ECs were plated in co-culture with 30 to 50% subconfluent E4ECs monolayers supplemented with serum-free hematopoietic media (HM) composed of StemSpan SFEM, 10% KnockOut Serum Replacement, 5ng/ml FGF-2, 10ng/ml EGF, 20ng/ml SCF, 20ng/ml FLT3, 20ng/ml TPO, 20ng/ml IGF-1, 10ng/ml IGF-2, 10ng/ml IL-3, 10ng/ml IL-6. After 3–4 weeks of co-culture the outgrown GFP+ reprogrammed ECs into human multipotent progenitor cells (rEC-hMPPs) formed typical grape-like hematopoietic colonies. At the end of 4 weeks, human CD45+ rEC-MPPs were FACS sorted for: 1) immunophenotypic analyses, 2) methylcellulose-CFC assay, 3) molecular profiling, 4) comparative genomic hybridization and 5) transplanted retroorbitally into primary sublethally-irradiated (275 Rads) 6-week old NSG mice or sublethally-irradiated (100 Rads) 2-week old mice neonates. After three months, human CD45+ cells (hCD45+ cells) derived from bone marrow (BM) or whole BM of the primary engrafted mice was used for transplantation into secondary recipients. After 3 months of primary and 6 months of the secondary transplantation, engrafted hCD45+ cells in BM, spleen and peripheral blood of mice were FACS sorted and processed for: 1) multi-variate immunophenotypic analyses, 2) multi-cell and clonal methylcellulose-CFC assay, and 3) molecular profiling. Tissues of the engrafted mice were processed for histological examination to rule out any malignant transformation.
Increasing Efficiency of Reprogramming
To increase efficiency of the reprogramming, we developed a strategy to select those subsets of ECs that were most likely transduced with the proper stoichiometry of all four FGRS. We initially focused on generating ECs transduced with GFI1, SPI1 and FOSB TFs because their expression in ECs is negligible (Extended Data Fig. 1). To accomplish this, we transduced 5×106 ECs with FGRS lentiviral “cocktail” marked by puromycin resistance (SPI1) or GFP (FOSB and GFI1). We then applied puromycin selection for 2 days to enrich SPI1-expressing cells and sorted them for GFP expression to enrich for SPI1+GFP+ (FOSB/GFI1) ECs. Subsequently, we transduced these GFP+ puromycin resistant cells with RUNX1, seeded into 12-well plates, and expanded them for two days (105 cells per plate, n=3). We then re-plated 104 of the GFP+ puromycin resistant cells on serum-free E4EC vascular niche layer in hematopoietic media and quantified the number of hematopoietic clusters that emerge after ~20 days of co-culture. We found that GFP+ puromycin resistant cells yielded 156.0±3.6 (n=3) hematopoietic-like colonies per 104 re-plated cells suggesting that the efficiency of reprogramming was at least 1.5% (156/104). This calculation assumes that each colony originates from a single reprogrammed cell. The efficiency is likely much higher in cells expressing the appropriate stoichiometric quantities of each factor.
Identification of viral integration on a single-cell and single-colony level
To identify the presence of viral integration on a single-cell level, we sorted human CD45+ cells from the marrow of rEC-hMPP engrafted mice into a 96-well plate 1 cell per well into a lysis buffer for the Phi29 (multiple displacement amplification – MDA) based whole-genome amplification (WGA). To do single-cell WGA, we used a commercially available kit REPLI-g (Qiagen, Cat.No. 150343). Each WGA reaction product was followed by a set of PCR reaction with primers specific to the CMV promoter and a transgene (FOSB, GFI1, RUNX1, SPI1). All PCR reactions were conducted separately. We used empty wells (no cells sorted) as controls for non-specific amplification. WGA products of the control wells were used for PCR reactions with primers specific to the CMV promoter and a transgene.
To identify the presence of viral integration on a single-cell level, we captured expanding colonies from the plates for CFC assay. Fourteen days after the start of CFC assay 3 distinct cell aggregations/colonies were detected and analyzed (three upper images). Four PCR reactions were performed for each amplified colony using their genomic DNA as template. Cells from the colonies were re-suspended and washed twice in excessive amounts of PBS (10ml) and transferred into the lysis buffer for the WGA. All following procedures were the same as those described for the single-cell viral integration identification.
CMV primer: 5’-cgcaaatgggcggtaggcgtg-3’
FOSB primer: 5’-gctctgctttttcttcctccaact-3’
GFI1 primer: 5’-ccagggccccacacggtcggtagc-3’
RUNX1 primer: 5’ttgcggtgggtttgtgaagac-3’
SPI1 primer: 5’cggatcttcttcttgctgcctgtc-3’
Clonal reprogramming of HUVECs to rEC-hMPPs
HUVECs were isolated from umbilical cord and grown in EC-growth medium. After 2 to 3 passages, CD144+CD31+CD62E(E-selectin)+CD45− HUVECs were FACS sorted into 96-well plates at 1, 2, 5 and 10 cells per well densities for clonal expansion. We used CD62E (E-selectin) surface marker to sort mature activated ECs. Passaging of HUVECs results in upregulation of E-selectin in 40 to 60% of the HUVECs. Expanding clonal populations of selected cells were subsequently transduced with the FGRS TFs followed by replating onto the E4EC monolayers to reprogram them into rEC-hMPPs. Hematopoietic activity of clonally derived CD45+CD34+rEC-hMPPs was assessed using standard methylcellulose-CFC assay.
RNA-Seq processing and analysis
Total RNA was prepared using the Applied Biosystems Arcturus PicoPure RNA isolation kit. The quality of the extracted RNA was checked on an Agilent Technologies 2100 Bioanalyzer. The extracted RNA was used for sequencing using Illumina HiSeq2000. The sequencing output was checked for quality using Illumina pipeline. PE 51×2 and SE 51 reads were mapped to the human genome (hg18) using TopHat (http://tophat.cbcb.umd.edu/) default parameters. RefSeq transcript levels (FPKMs) were then quantified using CuffLinks (http://cufflinks.cbcb.umd.edu/) with upper-quartile normalization and sequence-specific bias correction. For heatmap visualization we determined the maximum FPKM of each transcript across the samples shown. FPKMs were then divided by this number to produce scaled expression values. Heatmaps of gene expression and gene expression clustering were built using GENE-E matrix visualization and analysis platform (http://www.broadinstitute.org/cancer/software/GENE-E/). Clustering of gene expression in the heat maps was conducted using one minus Pearson correlation as dissimilarity measure between transcription profiles. GEO accession number: GSE57662. The GSE57662 study can be reviewed at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57662.
Comparative Genomic Hybridization (CGH)
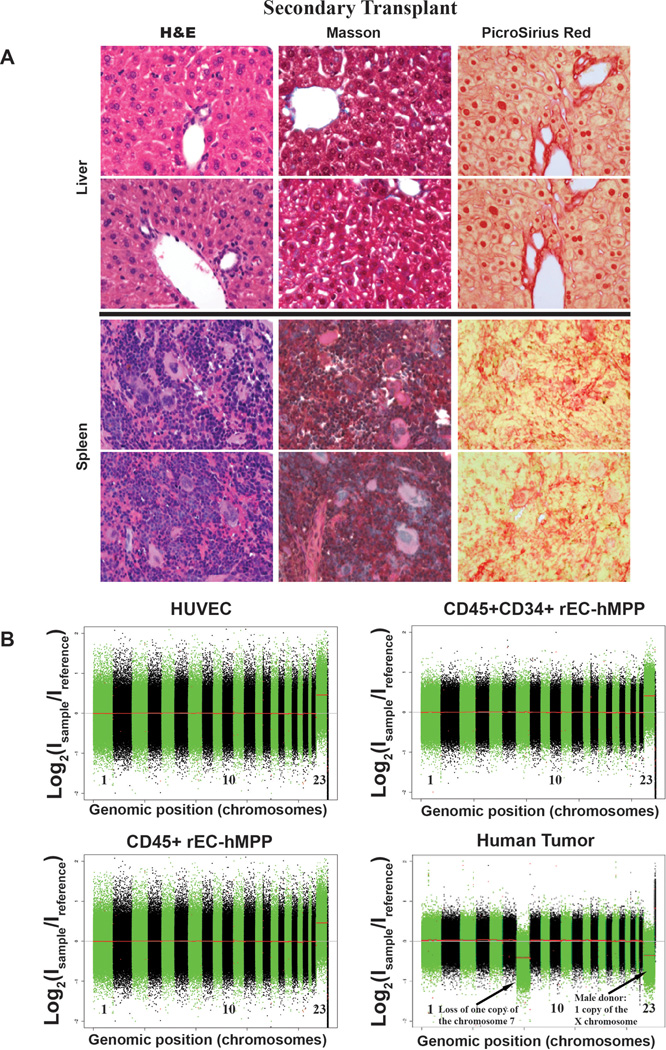
Genomic DNA was extracted from HUVECs, FACS sorted CD45+ rEC-hMPPs, and CD45+CD34+ rEC-hMPPs sorted from the BM of the NSG mice. Before DNA extraction, CD45+CD34+ rEC-hMPPs sorted from the BM were expanded for 72 hours in vitro. As a positive control of chromosomal rearrangements we used a CGH array of a leukemic cell line with a duplication of the chromosome 7 and a deletion of the chromosome 10. Extracted DNA was digested, labeled by random priming and hybridized to the Agilent 1M CGH arrays. The arrays were scanned in an Agilent DNA microarray scanner and obtained data was visualized using Feature Extraction software (version 10.7; Agilent).
Differentiation and reprogramming of human embryonic stem cells (hES)
We used a transgenic hES reporter line that specifically identifies differentiated EC derivatives via a fluorescent reporter driven by a fragment of the human VE-cadherin promoter1,35. To augment endothelial commitment, hES differentiation was initiated in co-culture with E4EC vascular niche cells, described above. One day before plating hES to begin differentiation, MEF conditioned medium was replaced with hES culture medium without FGF-2 and supplemented with 2 ng/ml BMP4. The next day, hES cells were plated directly onto E4EC monolayers in hES culture medium (without FGF-2, plus 2 ng/ml BMP4) and left undisturbed for 48 hours. This point of culture was considered as differentiation day zero. Cells were sequentially stimulated with recombinant cytokines in the following order: day 0 to 7 -supplemented with 10 ng/ml BMP4; day 2 to 14 - supplemented with 10 ng/ml VEGF-A; day 2 to 14 -supplemented with 5 ng/ml FGF-2; day 7 to 14 - supplemented with 10 µM SB-431542. At day 14 of culture, FACS sorting was used to purify the fraction of hES-derived ECs co-expressing the vascular specific CD144 (VE-cadherin) reporter and CD31. These cells were transduced with the FGRS cocktail and 2–3 days later plated on a layer of serum-free E4EC monolayers. The extent of reprogramming was assessed by flow cytometry.
Phagocytosis assay
The rEC-hMPPs generated from 3 to 4 weeks, were cultured in the presence of M-CSF (10ng/ml), SCF (10ng/ml), Flt-3 (10ng/ml), TPO (10ng/ml), and 10% FBS for additional two weeks with E4EC vascular niche layer. We observed an increase in size and granularity of the cultured cells (data not shown). The culture was washed with PBS twice to remove non-adherent cells. Growth media mixed with green fluorescent beads (GFB) at a low concentration of 1 µl/ml was applied to the attached cells for one hour at 37°C.After the incubation, the cells were washed twice with PBS and live cells were stained with the monocytic CD14 antibody. Cells were fixed and stained with DAPI for nuclear visualization. We visualized GFB inside CD14+ cells, but not in CD144 (VE-cadherin)+ endothelial cells (Extended Data Figure 2G).
Purification of human cord blood stem and progenitors cells (HSPCs)
Human umbilical cord blood was obtained under the IRB protocol Stage “Specific Differentiation of Hematopoietic Stem Cells into Functional Hemangiogenic Tissue” (Weill Cornell Medical College IRB # 09060010445). Cord blood mononuclear cells were purified by density gradient using Ficoll-Paque (GE) and enriched for CD34+HSPC using magnetic separation using anti-CD34 microbeads (Miltenyi) or FACS sorting. Further purification was achieved by negative selection of Lin+ cells using Human Progenitor Cell Enrichment Kit (StemCell Technologies) or FACS sorting. RNA from FACS sorted Lin−CD34+CD45+ cells was isolated by using Arcturus PicoPure RNA isolation kit (Applied Biosystems; This kit was used for all RNA extraction procedures).
Lentiviral vectors
Candidate transcription factors (TFs) were subcloned into either pLVX-IRES-ZsGreen1 lentivector (Clontech), pLOC lentivector (OpenBiosystems), or LV105 lentivector (Genecopoeia). For inducible expression of mouse or human FGRS factors we used Tet-On 3G inducible lentiviral vectors (Clontech). Lentiviral particles were packaged as previously described7. In short, human embryonic kidney 293FT (HEK293FT) cells were co-transfected with a lentivector and two helper plasmids psPAX2, and pMD2.G (Trono Lab through Addgene) in the equal molar ratio. Supernatant was collected 48–52 hours post- transfection, filtered and concentrated using Lenti-X concentrator (Clontech). Viral titers were determined in limiting dilution experiments using HUVECs as target cells. We used either the number of GFP+ cells, or the number of formed colonies in the presence of selection antibiotics (puromycin) as a read-out for the number of infectious viral particles per volume. We used an average MOI of 5 to 10 for infection of ECs.
Flow Cytometry
Flow cytometry analysis was performed on a Becton Dickenson LSRII SORP, and fluorescence activated cell sorting (FACS) was performed on an Aria II SORP. Antibodies used were raised against human CD45, CD34, CD14, CD31, CD43, CD90, CD41a, CD33, CD19, CD3, CD4, CD8, CD235, CD45RA, CD83, CD11b, CD38, LIN cocktail, CD117, CD133, CD144 (BD Pharmingen, eBioscience) or mouse CD45 (eBioscience.) Voltage adjustments and compensation was performed with CompBeads (BD Pharmingen), and gating was performed on fluorophore minus one (FMO) controls and unstained controls.
The list of antibodies used in our experiments:
Anti-human antibodies obtained from EBioscience:
CD45 Cat# 47–0459-42; clone HI30, CD34 Cat# 25–0349-42; clone 4H11, CD33 Cat# 48–0337-42; clone p67.6, CD19 Cat# 12–0199-41; clone HIB19, CD3 Cat# 93-0037–42; clone OKT3, CD4 Cat# 17–0048-41; clone OKT4, CD8 Cat# 8048-0087-025; clone SK1, CD43 Cat# 17–0439-73; clone eBio84-3C1, CD83 Cat# 25–0839-41; clone HB15e, CD11b Cat# 12–0118-41; clone ICRF44, LIN Cat# 22–7778-72, CD31 Cat# 11–0319-42; clone WM59, CD31 Cat# 48–0319-42; clone WM59
Anti-human antibodies obtained from BD Pharmigen:
CD90 Cat# 561971; clone 5E10, CD3 Cat# 557851; clone SK7, CD14 Cat# 557742, CD14 Cat# 555399, CD235A Cat# 340947, CD45RA Cat# 347723, CD41a Cat# 555466, CD38 Cat# 646851, CD117 Cat# 333944, CD33 Cat# 333946, CD144 Cat# 560410; clone 55-7H1, FLK1(VEGF-R2) Cat# 560871; clone 89106
Anti-human antibodies obtained from BioLegend:
Lin Cat# 348805
Anti-mouse antibodies obtained from Ebioscience:
CD45 Cat# 25–0451-82; clone 30-f11
Statistics and Animals
All statistics is presented as average ±standard deviation. To identify statistical significance all groups of data were compared using paired student t-test.
Experiments were repeated for at least three times. Number of repeats is demonstrated in all figure legends.
Animal experiments contain at least three animals per group. The number of animals is described in all figure legends and the text of the paper.
We included all tested animals for quantification. Representative images and flow cytometry plots are shown in the figures.
Age and sex-matched animals were allocated in all corresponding experimental groups. All NSG animals for transplantation experiments were female. All ages are specified in the text. Animals were chosen according to their age and their sex (females only). A description of every experiment states the age of the animals used in the experiment.
Transplanted animals were not individually labeled. Hence, subgroups of transplanted animals for organ engraftment were chosen blindly, without prior knowledge of the level of engraftment.
Animal experiments were performed under the guidelines set by the Institutional Animal Care and Use Committee (IACUC).
Extended Data
Extended Data Figure 1. Screening strategy for identification of minimal set of transcriptions factors (TFs) for reprogramming ECs into hematopoietic cells.
A. Candidate genes tested for reprogramming of HUVECs into hematopoietic colonies. To identify TFs that drive EHT transition, we performed RNA-seq of HUVECs and Lin-CD34+ umbilical cord HSPCs to select TFs differentially expressed by HSPCs, but not by HUVECs. We then screened various combinations of differentially expressed TFs to identify a minimal set capable of reprogramming ECs to hematopoietic cells. Levels of expression [log2(RNA-seq value)] in HUVECs and freshly purified CD34+Lin− cord blood cells.
B. One-by-one elimination of TFs experiment revealed a minimal set of TFs; FOSB, GFI1, RUNX1, and SPI1, (FGRS) capable of generating hematopoietic colonies in the HUVEC culture. A pulled set of 26 TFs minus one TF was evaluated for the ability to evoke formation of hematopoietic clusters (day 21 to 25; n=3). Asterisks show statistically significant (p<0.05) reduction of the number of hematopoietic clusters in the transduced HUVECs compared to the full set of TFs. Control represents non-transduced HUVECs. Transduced cells were cultured on a layer of serum-free E4EC monolayers.
C. One-by-one elimination of the FGRS factors shows that all four FGRS-TFs are necessary and sufficient for generation of hematopoietic colonies (day21 to 25; n=3).
“All” in B and C: all TFs are present. “Control” in B and C: all TFs are absent.
D. Schema for reprogramming of ECs into human multipotent progenitor cells (rEC-hMPPs). Clonal or bulk populations of HUVECs or hDMECs were transduced with the FGRS and after 3 days were replated on subconfluent monolayers of E4EC ECs. The emerging colonies of hematopoietic cells were subjected to 1) multi-variate immunophenotypic analyses, 2) clonal and oligo-clonal CFC assay, and 3) molecular profiling (RNA-seq). Tissues of the engrafted mice were processed for histological examination to rule out any malignant transformation.
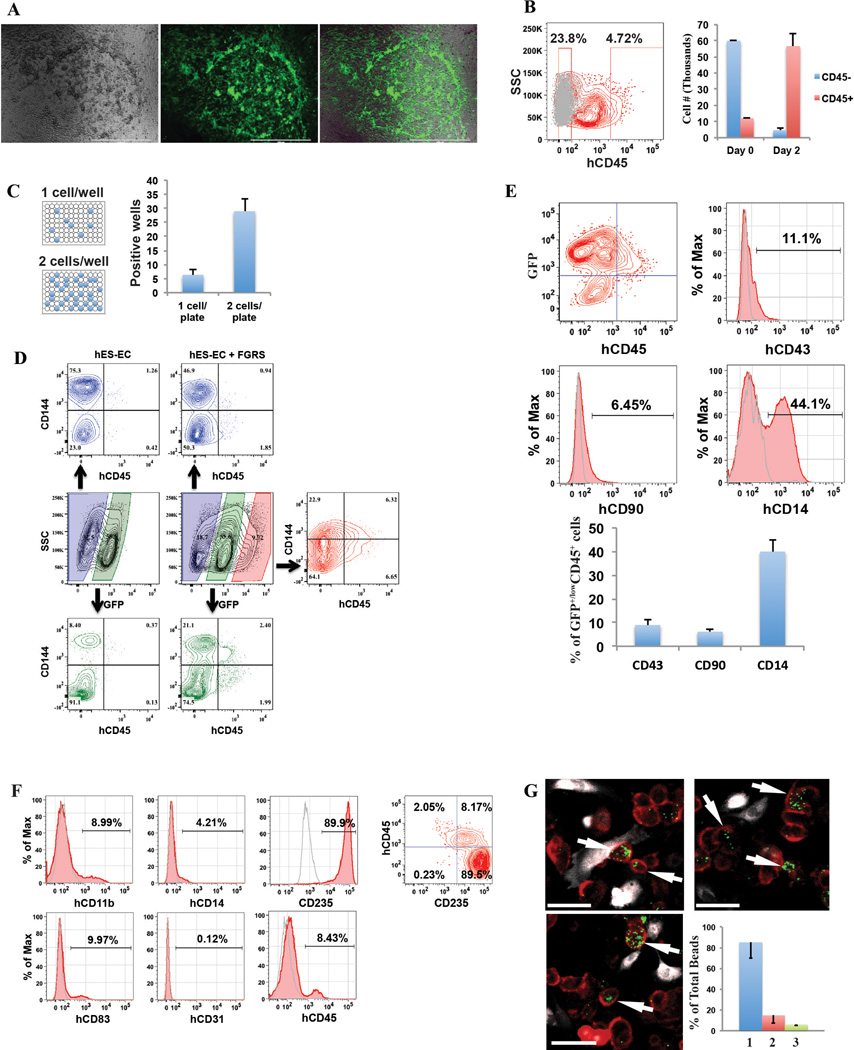
Extended Data Figure 2. FGRS-transduction and vascular-induction reprogram HUVECs, but not hES-ECs, to proliferating functional rEC-MPPs.
A. Multi-colony niche-like structure that physically separates developing hematopoietic colonies from surrounding E4EC vascular niche. The emerging multi-colony sinusoidal-like structures create a unique cellular interface between E4EC monolayers and transduced ECs giving rise to hematopoietic clusters (n=4, scale bar is 1000 µm.)
B. Expansion potential of reprogrammed CD45+hematopoietic cells. CD45+ (12×103) and CD45- (60 ×103) cells were sorted into separate wells and expanded for two days. We observed 5-fold expansion of CD45+ cells (56.6 ×103±7.9 ×103; n=3) and dramatic reduction of CD45− cells number (4.6×103±1.0 ×103; n=3).
C. Clonal expansion of CD45+ cells. CD45+cells were FACS sorted into 96-well plates at the density of 1 and 2 cells/well. After seven days of culture, we observed CD45+ cell expansion in 6.3±2.1 wells (93.1±14.5 cells/well) of 1-cell sort and 29.0±4.3 wells (112.1±21.2 cells/well) of the 2-cell sort (n=3). The difference between cell number in 1 and 2-cell sort was statistically not significant (p=0.78) suggesting that the difference in the number of wells with detected cell expansion was rather due to survival of sorted cells than a reflection of the number of cells sorted into a well.
D. FGRS-induced generation of hematopoietic cells by hES-derived endothelium (hES-EC). Representative experiment demonstrating that transduced hES-EC (Right 4 panels) generated significantly higher number of CD45+CD144- cells compared to control non-transduced hES-EC (left 3 panels).
E. Lineage-specific surface marker analysis of the GFP+CD45+ population of rEC-hMPPs. GFP+CD45+population showed that some of these cells expressed lineage specific surface markers such as CD43+ (8.96%±2.3; n=3), CD90+ (Thy-1+) (6.15%±1.13; n=3) and CD14+ (40.0%±4.95; n=3) (representative flow cytometry measurements; left four panels, statistics for all experiments is in the right-hand bar-graph, n=3).
F. Immunophenotypic analysis of CFU colonies grown in the CFC assay performed in Figure 2C,D.
G. Macrophages differentiated from rEC-hMPPs are functional capable of phagocytosis.
The images (upper row and lower left) show groups of firmly plastic-adherent CD14+ cells (red staining) with clearly visible phagocytosed green fluorescent beads (GFB; green). Endothelial CD144+(VE-cadherin) cells (white staining) were not co-localized with beads. Majority (85.1%±15.1) of GFBs was localized inside CD14+ cells (lower left graph 1). A smaller population of GFBs was distributed outside CD14+ and CD144(VE-cadherin)+ cells (14.8±7.43%; lower left graph 2). The percentage GFBs co-localized with endothelial cells was negligible (4.8±0.83%; lower left graph 3), n=9.
Scale bars are 25 µm.
Extended Data Figure 3. Naïve HUVECs are devoid of hemogenic potential capable of spontaneous differentiation into MPPs.
We performed two sets of experiments to exclude the possibility that rEC-hMPPs were derived from spontaneously differentiating HUVECs with hemogenic or hemangioblastic potential25,34.
A to B: In optimal pro-hematopoietic cultures, naïve non-transduced ECs fail to spontaneously differentiate into rEC-MPPs.
A. We grew non-FGRS-transduced HUVECs in the serum-free media used for reprogramming. Neither serum withdrawal, nor addition of hematopoietic cytokines induced formation of CD45+CD34+ cells and HUVECs sustained their vascular identity. Indeed, serum withdrawal increases number of CD34+ HUVECs; SF-serum free, CK-cytokines cocktail (see methods), SB - TGFβ inhibitor SB431542.
B. Serum withdrawal suppresses HUVEC proliferation. Inhibition of TGFβ signaling (SB) combined with cytokine cocktail (see methods) restores proliferative potential of HUVECs in serum free media. Difference between proliferation of HUVECs in serum free media and all other conditions is statistically significant (asterisk; p<0.005). Statistical significance between pairs of different conditions is shown with blue arrows and p-values, where p<0.005 is statistically significant. Therefore, human rEC-hMPPs originate from reprogrammed ECs, but not cytokine-mediated outgrowth of contaminating pre-existing hemogenic ECs.
C to E: Clonal reprogramming of non-hemogenic HUVECs into rEC-hMPPs using FGRS-transduction and vascular-induction.
We performed EC clonal reprogramming experiments to exclude the possibility that rEC-hMPPs were derived from spontaneously differentiating HUVECs with pre-existing hemogenic or hemangioblastic potential25,34.
C. Since E-selectin is only expressed in ECs, we generated clonal cultures of CD45−CD144+CD31+CD62E(E-selectin)+ ECs32,33. To this end, CD144+CD31+CD62E+CD45−HUVECs were sorted into 96-well plates at 1, 2, 5 and 10 cells per well densities for clonal expansion. Proliferating clones were transduced with FGRS and induced with serum-free E4ECs. D. These clonal cultures yielded rEC-hMPPs comparable to bulk HUVEC cultures. The numbers of hematopoietic-like colonies emerging from 1-cell, 2-cell, 5-cell, and 10-cell clones are not statistically different (p>0.05). E. An example of a hematopoietic-like colony derived from a 1-cell clone #2. Therefore, it is unlikely that rEC-hMPPs are derived through spontaneous differentiation of pre-existing ECs with hemogenic or hemangioblastic potential.
Extended Data Figure 4. Clonal reprogramming of non-hemogenic HUVECs into immunophenotypic and functional rEC-hMPPs using FGRS-transduction and vascular-induction.
A,B,C: CFC assay of clonally reprogrammed rEC-hMPPs. CD45+CD34+rEC-hMPPs were sorted out (red gate in FACS plots; upper left) and plated for CFC assay. Typical hematopoietic colonies arose in the assay (middle column microphotographs; x4 magnification). FACS plots on the right show immunophenotypic analysis of the cells that arose in the CFC assay, demonstrating differentiation into human CD45−CD235+erythroid CD11b+macrophage CD14+monocytic, CD41a+megakaryocytic and CD83+dendritic progenies. The graph in the left lower corner shows quantification of the CFC assay (n=3). Identical panels are shown for two 2-cell clones and one 5-cell clone. A total of 3 independent clones are shown. Thus, given the high efficiency of clonal reprogramming of mature authentic ECs into rEC-hMPPs, it is unlikely that rEC-hMPPs are spontaneously derived from a very rare population of a pre-existing hemogenic or hemangioblastic HUVECs.
Extended Data Figure 5. Single cell analysis of lentiviral integration into engrafted rEC-MPPs.
CD45+ cells were sorted into a 96-well plate (1 cell/well), lysed in corresponding well for whole genome amplification (WGA) using Phi29 enzyme (see methods). Amplified DNA is shown for all 21 cells in the top two gels. Amplified DNA was used as a template for PCR reactions with a forward primer specific for the CMV promoter and reverse primer specific for the coding sequence of a reprogramming factor. T-test PCR with a lentiviral vector. EW – empty well – no template DNA. Red asterisks show failed PCR amplification of viral integration. PCR products are visible as low molecular weight bands labeled as 1 – FOSB, 2 – GFI1, 3 – RUNX1, 4 – SPI1.
Extended Data Figure 6. Conditional expression of FGRS is sufficient for optimal generation of rEC-hMPPs with multilineage potential, including T-Cell lymphoid cells.
A to C: Conditional expression of mouse inducible FGRS factors activates endogenous human FGRS in HUVECs sustaining functional hematopoietic cell fate of rEC-hMPPs.
A. To test whether FGRS-induced reprogramming triggered expression of endogenous FGRS genes24, HUVECs were transduced with lentivirus expressing FGRS-Tet-On and a trans-activator, and grown on E4EC-vascular niche for 18–22 days (n=4) in the presence of doxycycline. Doxycycline was removed from the culture medium after 18–22 days to shut off the expression of mouse FGRS and cells were cultured for additional 7–10 days. Human CD45+CD34+ cells were FACS isolated for CFC assay and whole-transcriptome deep sequencing (RNA-seq). CFC assay revealed emergence of hematopoietic colonies with cells expressing human CD235, CD11b, CD83, and CD14. B. Comparison of transcriptional profiles of the human FGRS expression in human HUVECs, hCD45+ rEC-hMPPs programmed using inducible mouse FGRS, CD45+CD34+ rEC-hMPPs, 22 weeks post-transplantation, hDMEC-derived CD45+CD34+rEC− hMPPs after 15 weeks post-secondary engraftment and naïve CD34+Lin+ cells purified from cord blood.
C. Analysis of whole-transcriptome RNA-Seq of rEC-hMPPs derived using inducible mouse FGRS (n=3). All RNA-Seq reads were aligned against human and mouse FGRS sequences. RNA-Seq reads that align to human FGRS sequences – “Map to Human”, RNA-Seq reads that align to mouse FGRS sequences – “Map to Mouse”, and RNA-Seq reads that align to mouse FGRS sequences without a possibility to align to human sequences – “Map to mouse Only”.
D to E: Optimizing differentiation of rEC-hMPPs into lymphoid progeny
D. The number of T-lymphoid progeny of engrafted rEC-hMPP was negligibly small, raising the possibility that constitutive SPI1 expression prevents rEC-hMPP from differentiating into T-cells39,40. To test this, HUVECs were transduced with lentiviral vectors expressing GFP and that constitutively express FGR-TFs with a Tet-inducible SPI1 (FGR+SPI1-Tet-On construct) for 3 days followed by replating for E4EC-induction. After 27 days of FGR and doxycycline-induced SPI1 expression on E4ECs, GFP+CD45+ hematopoietic-like colonies emerged. Then, doxycycline was withdrawn and the reprogrammed cells were cultured serum-free with Delta-like-4 expressing OP9-stroma (OP9-DLL4) supplemented with IL-7, IL-11, and IL-2. There is an increase of the number of GFP+CD45+ cells emerging during reprogramming of HUVECs by FGR+SPI1-Tet-On construct and E4EC-induction.
E. rEC-hMPPs differentiate into CD3+, CD19+ and CD14+ hematopoietic cells in the absence of exogenous expression of SPI1. After 3 weeks, the numbers of the myeloid and lymphoid cells were quantified by flow cytometry. We were able to reliably detect a small fraction of CD3+ cells (0.16±0.01%; n=3), a larger number of CD19+ (1.17±0.13%; n=3) and CD14+ (16.46±1.02%; n=3) cells. Thus, generation of lymphoid cells from rEC-hMPPs could be optimized by transient expression of TFs.
Extended Data Figure 7. Adult human hDMECs-derived rEC-hMPPs are capable of in vivo primary and secondary multilineage engraftment.
A. Immunophenotypic analysis of cells grown in the CFC assay (from Figure 4B). These panels show quantification of surface marker expression in the cells isolated from the colonies in CFC assay (n=3). hDMECs differentiated into hCD45−CD235+ erythroid, CD11b+CD14+ monocyte/macrophage and CD83+ dendritic cell progenies. Minimal CD144 (VE-cadherin) was detected.
B. Analysis of peripheral blood (PB) of mice at 4, 6, and 12 weeks post-primary transplantation (Figure 5A) revealed circulating hCD45+ and their hCD33+, hCD14+myeloid and hCD45−hCD235+erythroid progenies (n=6). Mouse CD45 (mCD45+) cells were excluded from analyses. Mouse cells – blue. Human cells – red.
C. Analysis of spleen of mice at 14 weeks post-primary transplantation (Figure 5A) revealed presence of hCD45+ (Red gate) and their lymphoid (hCD19+, hCD56+) and myeloid (hCD11b+, hCD41a+) progenies (n=3). Mouse CD45 (mCD45+ cells, blue populations).
D. Analysis of mice BM at 14 weeks post-primary transplantation (Figure 5A). Lin−CD45RA cells (blue gate) was analyzed for CD38 and CD90 expression (green and red gates) and subsequently examined for human CD45 and CD34 expression. This analysis revealed presence of hCD34+cells with small populations of both Lin−CD45RA−CD38−CD90+CD34+ and Lin−CD45RA−CD38−CD90−CD34+ cells satisfying phenotypic definition of human HSCs and MPP, respectively (n=3).
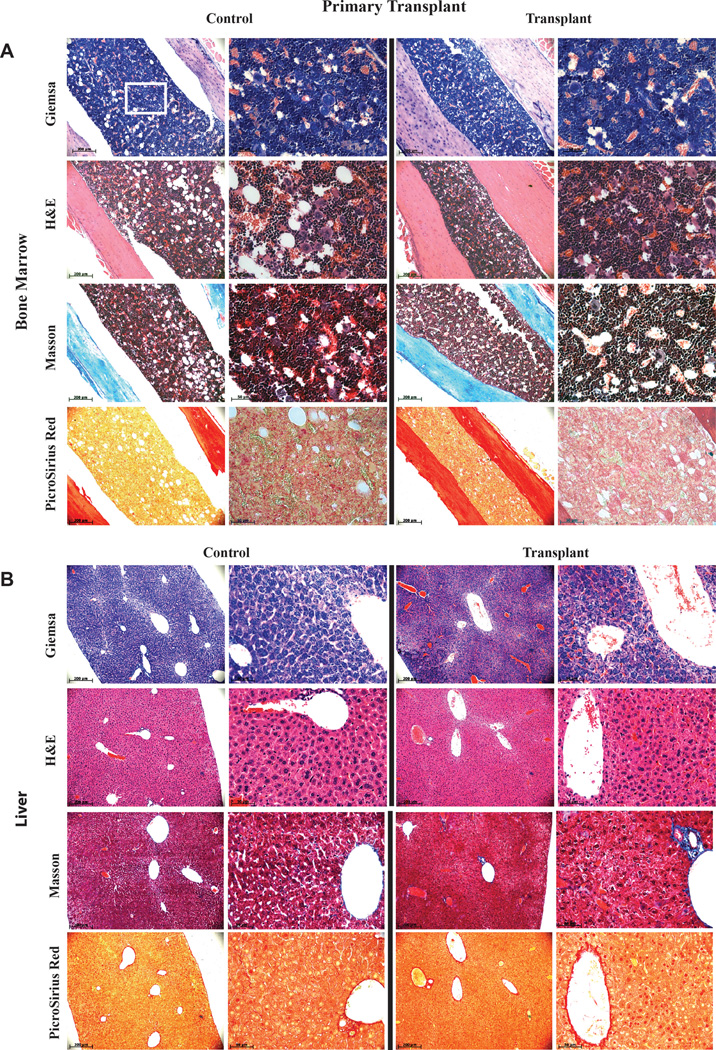
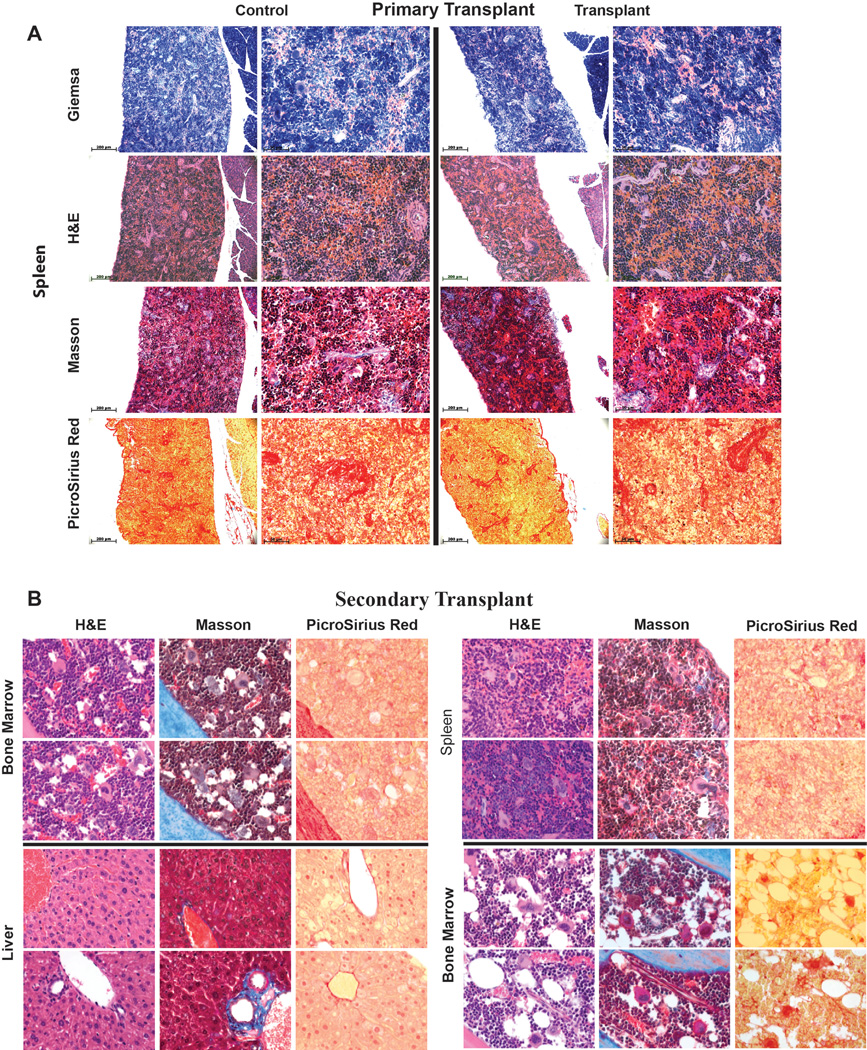
Extended Data Figure 8. Analysis of bone marrow (BM) and liver of primary transplanted mice for signs of malignant transformation.
Analysis of BM (A) and liver (B) of mice 10 months after primary transplantation (from Figure 3B) of HUVEC derived rEC-hMPPs for signs of malignant transformation. The level of fibrosis was determined using Masson and PicroSirus staining. The architectonic geometry of the BM was determined by sequential multi-cross sectional Wright Giemsa and Hematoxylin-Eosin (H&E) staining and compared to age control non-transplanted NSG mice. We did not observe any evidence of fibrosis or alteration of the geometry of the bone marrow or liver of the transplanted mice. Furthermore, no recipient mouse manifested any anatomical or symptomatic evidence of leukemias, lymphomas or myeloproliferative neoplasm (MPN): lymphadenopathy, organomegaly, illness or hemorrhage. Circulating hCD45+ cells in PB displayed no evidence of lympho/myeloproliferation or dysplasia. Furthermore, microscopic architecture of BM and liver was normal and without fibrotic remodeling or aberrant deposition of collagen or desmin. All images were acquired using color CCD camera. The scale bar is 200µm for low-resolution images in the left columns and 50 µm for high-resolution images in the right columns. Upper left image (Giemsa Control) shows a white square in the center that corresponds to the portion of the image shown at high resolution on the right (the same Giemsa Control sample). This rule applies to all shown images.
Extended Data Figure 9. Analysis of spleen of primary transplanted mice and BM, spleen and liver of secondary transplanted mice for signs of malignant transformation.
Analysis of spleen of mice 10 months after primary transplantation (from Figure 3B) of HUVEC derived rEC-hMPPs as well as BM (n=2), spleen- and liver- (n=2, Also Extended Data Figure 10A) of mice that were engrafted with secondary transplanted hDMEC-derived rEC-hMPP cells 15 weeks post-transplantation (from Figure 5B) for signs of malignant transformation. The level of fibrosis was determined using Masson and PicroSirus stainings. The architectonic geometry of the BM was determined by sequential multi-cross sectional Wright Giemsa and Hematoxylin-Eosin (H&E) staining and compared to age control non-transplanted NSG mice. We did not observe any evidence of fibrosis or alteration of the geometry of the bone marrow, spleen or liver of the transplanted mice. Furthermore, no recipient mouse manifested any anatomical or symptomatic evidence of leukemias, lymphomas or myeloproliferative neoplasm (MPN): lymphadenopathy, splenomegaly/organomegaly, illness or hemorrhage. Circulating hCD45+ cells in PB displayed no evidence of lympho/myeloproliferation or dysplasia. Furthermore, microscopic architecture of BM, spleen and liver was normal and without fibrotic remodeling or aberrant deposition of collagen or desmin.
All images were acquired using color CCD camera. In primary transplants the scale bar is 200µm for low-resolution images in the left columns and 50 µm for high-resolution images in the right columns. Upper left image (Giemsa Control) shows a white square in the center that corresponds to the portion of the image shown at high resolution on the right (the same Giemsa Control sample). This rule applies to all shown images (Primary Transplant). All images in Secondary Transplant are acquired at 60x magnification. All images are acquired at 60x magnification. Top rows of images for each organ are secondary transplants; bottom rows of images for each organ are controls.
Extended Data Figure 10. Analysis of liver and spleen of secondary transplanted mice for signs of malignant transformation and analyses of rEC-MPPs for genetic stability.
A. Analysis of liver and spleen of secondary transplanted mice for signs of malignant transformation. Repeat analysis of spleen and liver of mice that were engrafted with secondary transplanted hDMEC-derived rEC-hMPP cells 15 weeks post-transplantation for signs of malignant transformation (from Figure 5B). The level of fibrosis was determined by Masson and PicroSirus stainings. The architectonic geometry of the BM was determined by sequential multi-cross sectional Hematoxylin-Eosin (H&E) staining and compared to age control non-transplanted NSG mice. We did not observe any evidence of fibrosis or alteration of the geometry of the spleen or liver of the transplanted mice. Furthermore, no recipient mouse manifested any anatomical or symptomatic evidence of leukemias, lymphomas or myeloproliferative neoplasm (MPN): lymphadenopathy, splenomegaly/organomegaly, illness or hemorrhage. Circulating hCD45+ cells in PB displayed no evidence of lympho/myeloproliferation or dysplasia. Furthermore, microscopic architecture of BM, spleen and liver was normal and without fibrotic remodeling or aberrant deposition of collagen or desmin.
All images are acquired at 60x magnification. Top rows of images for each organ are secondary transplants; bottom rows of images for each organ are controls.
B. Comparative genomic hybridization analysis (CGH) shows that rEC-hMPPs are genetically stable both in vitro and in vivo. Genomic DNA was extracted from HUVECs, CD45+rEC-hMPPs (35 days post-transduction) or in CD45+CD34+rEC-hMPP sorted from the engrafted NSG BM (24-weeks post-transplantation) and expanded for 72 hours in vitro. A human tumor sample was used as positive control of chromosome rearrangement. Extracted DNA was digested, labeled by random priming and hybridized to the Agilent 1M CGH arrays. The arrays were scanned in an Agilent DNA microarray scanner and obtained data was visualized using Feature Extraction software (version 10.7; Agilent). No genomic abnormalities were identified in CD45+rEC-hMPPs (or in CD45+CD34+rEC-hMPPs engrafted in NSG BM. Hence, rEC-hMPPs remain genetically stable in vitro and in vivo and are not transformed.
Acknowledgements
V.M.S. is supported by Empire State Stem Cell Board (ESSCB) and New York State Department of Health (NYSDH: C026878). S.R. is supported by Ansary Stem Cell Institute (ASCI), HHMI, ESSCB/NYSDH: C024180, C026438, C026878, C028117, NHLBI:R01HL097797, R01HL119872,U01 HL099997NIDDK:R01DK095039,NCI:U54CA163167, Qatar National Priorities Research Foundation grant NPRP08-663-3-140 and the Qatar Foundation BioMedical Research Program. J.M.S. is suppored by grants from NCI:CA159175 and CA163167, NHLBI: HL119872 and HL055748, Starr Foundation and a Leukemia & Lymphoma Society Scholar award. J.M.B. is supported by, American Society of Hematology Scholar Award, NHLBI U01-HL099997 and Angiocrine Bioscience and ASCI. We acknowledge keycontribution of Dr. Jenny Z. Xiang for enabling and executing molecular profiling and Eric Gars for superb technical support. We appreciate Dr. William Schachterle for invaluable recommendations and edits of the manuscript.
Footnotes
Contributions
V.M.S. and S.R. conceived and designed the project, performed experiments, analyzed the data and wrote the manuscript. R.L., Y.L., and J.M.B. performed experiments interpreted, and analyzed data. J.M.S. interpreted, analyzed data and wrote the manuscript. D.J., O.E., A.K., performed the experiments and analyzed the data. All authors commented on the paper.
References
- 1.Rafii S, et al. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. Blood. 2013;121:770–780. doi: 10.1182/blood-2012-07-444208. [DOI] [PubMed] [Google Scholar]
- 2.Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat Biotechnol. 2013;31:416–418. doi: 10.1038/nbt.2571. [DOI] [PubMed] [Google Scholar]
- 3.Choi KD, et al. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell reports. 2012;2:553–567. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira CF, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 6.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 7.Sandler VM, Lailler N, Bouhassira EE. Reprogramming of embryonic human fibroblasts into fetal hematopoietic progenitors by fusion with human fetal liver CD34+ cells. PLoS One. 2011;6:e18265. doi: 10.1371/journal.pone.0018265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolan DJ, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding BS, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding BS, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler JM, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch- dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler JM, et al. Development of a vascular niche platform for expansion of repopulating human cord blood stem and progenitor cells. Blood. 2012;120:1344–1347. doi: 10.1182/blood-2011-12-398115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper AT, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2- mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi H, et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nature cell biology. 2010;12:1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulos MG, et al. Endothelial jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell reports. 2013;4:1022–1034. doi: 10.1016/j.celrep.2013.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avecilla ST, et al. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat Med. 2004;10:64–71. doi: 10.1038/nm973. [DOI] [PubMed] [Google Scholar]
- 17.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doan PL, et al. Tie2(+) bone marrow endothelial cells regulate hematopoietic stem cell regeneration following radiation injury. Stem Cells. 2013;31:327–337. doi: 10.1002/stem.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 21.North TE, et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto M, Porayette P, Yoder MC. Overcoming obstacles in the search for the site of hematopoietic stem cell emergence. Cell Stem Cell. 2008;3:583–586. doi: 10.1016/j.stem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eilken HM, Nishikawa S, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 24.Swiers G, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single- cell level. Nature communications. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes KE, et al. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon-Keylock S, Sobiesiak M, Rybtsov S, Moore K, Medvinsky A. Mouse extraembryonic arterial vessels harbor precursors capable of maturing into definitive HSCs. Blood. 2013;122:2338–2345. doi: 10.1182/blood-2012-12-470971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen MJ, et al. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lancrin C, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314–322. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- 30.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 31.Seandel M, et al. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc Natl Acad Sci U S A. 2008;105:19288–19293. doi: 10.1073/pnas.0805980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rafii S, et al. Isolation and characterization of human bone marrow microvascular endothelial cells: hematopoietic progenitor cell adhesion. Blood. 1994;84:10–19. [PubMed] [Google Scholar]
- 33.Rafii S, et al. Human bone marrow microvascular endothelial cells support long-term proliferation and differentiation of myeloid and megakaryocytic progenitors. Blood. 1995;86:3353–3363. [PubMed] [Google Scholar]
- 34.Wu X, Lensch MW, Wylie-Sears J, Daley GQ, Bischoff J. Hemogenic endothelial progenitor cells isolated from human umbilical cord blood. Stem Cells. 2007;25:2770–2776. doi: 10.1634/stemcells.2006-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James D, et al. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat Biotechnol. 2010;28:161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginsberg M, et al. Efficient Direct Reprogramming of Mature Amniotic Cells into Endothelial Cells by ETS Factors and TGFbeta Suppression. Cell. 2012;151:559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 38.Notta F, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 39.Zou GM, Chen JJ, Yoder MC, Wu W, Rowley JD. Knockdown of Pu.1 by small interfering RNA in CD34+ embryoid body cells derived from mouse ES cells turns cell fate determination to pro-B cells. Proc Natl Acad Sci U S A. 2005;102:13236–13241. doi: 10.1073/pnas.0506218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 41.Ledran MH, et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Doulatov S, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcelo KL, et al. Hemogenic endothelial cell specification requires c-Kit, Notch signaling, and p27-mediated cell-cycle control. Dev Cell. 2013;27:504–515. doi: 10.1016/j.devcel.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]