Abstract
As a persistent organic contaminant, perfluorooctanesulphonic acid (PFOS) has been widely detected in the environment, wildlife, and humans. The present study revealed that zebrafish embryos exposed to 16 µM PFOS during a sensitive window of 48–96 hour post-fertilization (hpf) disrupted larval morphology at 120 hpf. Malformed zebrafish larvae were characterized by uninflated swim bladder, less developed gut, and curved spine. Histological and ultrastructural examination of PFOS-exposed larvae showed structural alterations in swim bladder and gut. Whole genome microarray was used to identify the early transcripts dysregulated following exposure to 16 µM PFOS at 96 hpf. In total, 1,278 transcripts were significantly misexpressed (p < 0.05) and 211 genes were changed at least two-fold upon PFOS exposure in comparison to the vehicle exposed control group. A PFOS-induced network of perturbed transcripts relating to swim bladder and gut development revealed that misexpression of genes were involved in organogenesis. Taken together, early life stage exposure to PFOS perturbs various molecular pathways potentially resulting in observed defects in swim bladder and gut development.
Keywords: Zebrafish embryo, perfluorooctanesulfonic acid, swim bladder, gut, developmental toxicity
1. Introduction
Perfluorinated compounds (PFCs) are a class of persistent contaminants widely used as surfactants, lubricants, adhesives, fire retardants, propellants, and medicines (Renzi et al., 2013). Perfluorooctanesulphonic acid (PFOS), an end product of the breakdown of multiple PFCs, is widely detected in wildlife, humans and the environment (Giesy and Kannan, 2001; Houde et al., 2006; Zhang et al., 2011b). Although PFOS is generally found at low levels in surface water, the chemical is characterized by high bioaccumulation and negligible elimination (Kannan et al., 2005a). As a consequence, higher concentrations of PFOS have been detected in a variety of fish species. For example, PFOS was detected in the liver of wild Gibel carp at levels of up to 9,031 µg/kg wet weight (Hoff et al., 2005) and average PFOS concentrations detected in fish tissue were 8850-fold greater than those measured in surface water (Sinclair et al., 2006). Additionally, high concentrations of PFOS were also detected in fish eggs (145–381ng/g) in lake whitefish from Michigan waters in the United States (Kannan et al., 2005b), which suggests oviparous transfer of this compound (Kannan et al., 2005b).
Zebrafish (Danio rerio) is a freshwater fish that is extensively used as a model organism for various research fields due to their small size, embryonic transparency, and rapid developmental cycle (Hill et al., 2005; Jang et al., 2013). The availability of its complete genome sequence enables the construction of zebrafish microarrays that permit global gene expression analysis (Pichler et al., 2003). Monitoring the expression of thousands of genes simultaneously through microarray analysis allows researchers to identify biological pathways perturbed by chemical exposure (Mathavan et al., 2005). For these exact same reasons, zebrafish have been used for studying PFOS toxicity (Huang et al., 2010; Shi et al., 2008). Zebrafish embryos exposed to 1–5 mg/l PFOS from 4–132 hpf exhibit spinal curvature, uninflated swim bladder, reduced hatching rates, and decreased blood flow and body length (Shi et al., 2008). PFOS-exposed zebrafish embryos are subject to increased cell death, muscle lesions, and abnormal swimming behaviors (Huang et al., 2010). Among various reported phenotypic changes, we previously found alteration of gut and swim bladder from both acute and chronic PFOS exposure (Chen et al., 2013; Huang et al., 2010; Wang et al., 2011), yet the mechanisms that underlie these effects are not well understood. Previous studies have reported widespread proteomic (Shi et al., 2009) and microRNA expression Zhang et al., 2011a) changes associated with acute PFOS exposure in embryonic zebrafish; however, studies on transcriptional changes upon PFOS exposure are still lacking. In the present study, we characterized gene expression changes induced by developmental exposure to PFOS to identify signaling networks that may contribute to adverse morphological outcomes.
2. Materials and methods
2.1. Fish husbandry and embryo collection
Wildtype (AB strain) zebrafish were raised and kept at standard laboratory conditions of 28°C on a 14:10 dark/light photoperiod in a recirculation system according to standard zebrafish breeding protocols (Westerfield, 1993). Water supplied to the system was filtered by reverse osmosis (pH 7.0–7.5), and Instant Ocean® salt was added to the water to raise the conductivity to 450–1000 µS/cm (system water). The fish were fed three times daily with zebrafish diet (Zeigler, Aquatic Habitats, Apopka Florida) and a live artemia (Jiahong Feed Co., Tianjin, China). Zebrafish embryos were obtained from adults in tanks with a sex ratio of 1:1, and spawning was induced in the morning when the light was turned on. Embryos were collected within 0.5 h of spawning and rinsed in an embryo medium (EM: 0.137 M NaCl, 5.4 mM KCl, 0.25 mM Na2HPO4, 0.44 mM KH2PO4, 1.3 mM CaCl2, 1.0 mM MgSO4 and 4.2 mM NaHCO3) (Westerfield, 1993). Fertilized embryos with normal morphology were staged under a dissecting microscope SMZ 1500 (Nikon, Japan) according to the standard methods (Kimmel et al., 1995).
2.2. PFOS stock solutions and exposure protocols
Perfluorooctanesulphonic acid (PFOS; CAS # 1763-23-1, purity >96%) was purchased from Sigma-Aldrich Chemical (St. Louis, MO, USA) and dissolved in 100% dimethyl sulfoxide (DMSO) to prepare PFOS stock solutions of 32 mM. A serial dilution was made in 100% DMSO that was 1,000 times more concentrated to allow for a 1:1,000 dilution with EM to create a serial dilution with a final DMSO concentration of 0.1%. The control also received 0.1% DMSO (v/v in EM).
2.3. Sensitive exposure period screening
To determine which developmental stage is most sensitive to PFOS-induced malformations, embryos/larvae were waterborne exposed to PFOS (8, 16, 32 µM) in 6-well plates (20 embryos per well with 5 mL solution) from 0–48 hpf or 48–96 hpf. The chemical solution was not changed during the exposure window, and there were three biological repeats. At the end of each exposure period, the embryos or larvae were rinsed three times with EM and transferred to 96-well plates (1 embryo per well with 200 µL solution) for continuous development until 120 hpf, where the incidence of various malformation was scored. The embryos in one repeat were from the same well in the 6-well-plates when they were exposed.
2.4. Histological examination of the larval swim bladder and gut
For hematoxylin and eosin (HE) staining, embryos were exposed to 0.1% DMSO or 16 µM PFOS from 48 to 96 hpf, rinsed three times with EM and continuously developed until 120 hpf in EM. These larvae were fixed overnight with 4% paraformaldehyde (PFA) at 4°C, and then dehydrated in graded series of ethanol solutions prior to paraffin embedding. Embedded larvae were sectioned (5µm longitudinal sections) and stained with HE. Fifteen embryos were used for each treatment group. Images were obtained with a confocal microscope FV1000 (Olympus, Japan) and images were captured using a FITC filter.
2.5. Transmission electron microscopic examination of the larval swim bladder and gut
Embryos were exposed to 0.1% DMSO or 16 µM PFOS from 48 to 96 hpf then transferred to EM until 120 hpf. At 120 hpf, larvae were fixed in 2.5% glutaraldehyde at 4°C for 48 h, rinsed in 0.1M PBS, and then set in 1% osmium tetroxide at 37°C for 1 h. Larvae were then stained in 1% uranyl acetate at 37°C for 1 h. Samples were dehydrated through an ethanol series (50%, 75% and 100%), transferred to acetone, and embedded in pure resin prior to sectioning. The plastic blocks were sectioned transversely to obtain 1 µm using a LKB2008 instrument and ultrathin sections of interest were selected using light microscopy. The ultrathin sections of 80 nm made by a POWER TOME XL instrument were collected on 200-mesh copper grids and stained with lead citrate for 10 min. The sections were analyzed with a Hitachi H-7500 transmission electron microscope (TEM).
2.6. NimbleGen microarray
Embryos at 48 hpf were exposed to 0.1% DMSO or 16 µM PFOS for 48 h, and RNAs were then extracted from embryos at 96 hpf. There were six biological replicates per treatment group with each replicate consisted of pooled tissue from 40 larvae. A total of 12 samples were assessed on a single chip that contains 12 individual arrays. Total RNA was isolated with TRIzol Reagent (Life Technologies) according to the manufacturer’s instructions. The quantity and quality of RNA were determined using the Nanodrop-1000 Spectrophotometer and gel electrophoresis. All RNA samples passed the concentration and quality requirements (A260/A280≥1.8 and A260/A230≥1.8). For microarray processing, 10 µg of total RNA was reverse transcribed using SuperScriptIII and oligo primer (Invitrogen), and double stranded cDNA was synthesized and purified using a Qiagen MinElute PCR Purification spin column. Double stranded cDNA was labeled with Cy5 dNTP, and samples were then hybridized to 12×135K zebrafish gene expression arrays (Roche Nimblegen, Madison, WI) and scanned using the Axon GenePix Pro 4200A scanner (Molecular Devices, Sunnyvale, CA) according to the manufacturer’s instruction. The labeling, hybridizing, and scanning steps were finished at the IBEST DNA Sequencing Analysis Core of the University of Idaho, with the details stated in our previous study (Tal et al., 2012).
2.7. Microarray data processing and pathway design
The raw data were extracted, background subtracted, and quantile normalized (Bolstad et al., 2003) using NimbleScan v2.5 software. Gene calls were generated using the Robust Multichip Average (RMA) algorithm as previously described (Irizarry et al., 2003). Principal component analysis of all genes on array was used to evaluate if samples are outliers within each treatment group by correlation. Statistical analysis was performed using an unpaired t-test with 5% FDR in GeneSpring GX v10.0 (Agilent Technologies) to generate significant gene lists. Importing the statistically significant gene list into the Multi-Experiment Viewer (MEV) produced a bi-hierarchical clustering heat map. Individual clusters were further analyzed with the Database for Annotation, Visualization and Integrated Discovery (DAVID (http://david.abcc.ncifcrf.gov/home.jsp) to determine common and unique functional pathways (Dennis Jr et al., 2003). A zebrafish nimblegen background and individual cluster gene lists were uploaded into DAVID using entrez gene identifiers. Functional annotation of clustering using levels 3, 4, and 5 of the gene ontology category of biological processes was applied to each gene list. Only biological processes receiving an enriched score greater than 1 were noted on the bi-hierarchical clustering heat map. Zebrafish mRNA sequences on the microarray were blasted on the NCBI website to find the human orthologs with the highest blast score before subjecting them to Ingenuity Pathways Analysis (IPA, Ingenuity® Systems). The identified genes were mapped to corresponding gene objects in the Ingenuity Pathways Knowledge Base to generate networks, bio-functions, and canonical pathways.
2.8. Quantitative RT-PCR validation
Quantitative real time PCR (qRT-PCR) was used to confirm expression changes resulting from the microarray analysis. A subset of RNAs from the same samples used for the microarray analysis was used for qRT-PCR validation. cDNA was prepared from 5 µg of total RNA per group using a Prime Script® RT reagent Kit (Takara, Japan) following the manufacturer’s instructions. qRT-PCR using gene-specific primers (Table 1, Sunny Biotechnology) was conducted on an Eppendorf Mastercycler® Realplex2. Gradient annealing temperature studies were initially completed to confirm the optimal annealing temperature for each primer set. The reaction mixtures included 10 µl powerTM SYBR Green® supermix, 0.4 µl of each primer, 4.2 µl of ddH2O, and 5 µl of cDNA. The thermal cycle reaction was performed using standard procedures - 95°C for 30s, 40 cycles of 95°C for 5s and 60°C for 30s and the data were collected at the end of each extension step. The gene expression levels were measured in a total of three biological replicates per treatment group (n=3, with 40 embryos per replicate). For each biological replicate, three technicalrepeats were used to reduce sampling error. mRNA levels were calculated and normalized against housekeeping gene β-actin using the equation: fold change = 2−ΔΔCT (Schmittgen and Livak, 2008). Gel electrophoresis and thermal denaturation (melt curve analysis) were used to confirm product specificity. To compare the results from the microarray and qRT-PCR, gene expression profiles were displayed as a fold change relative to the vehicle control group.
Table 1.
Primers used for qPCR expression validation.
| Target | Forward (F) and reverse (R) sequence | PCR (bp) |
|---|---|---|
| β-actin | F: AAGCAGGAGTACGATGAGTC | 238 |
| R: TGGAGTCCTCAGATGCATTG | ||
| ace2 | F: GGCCCTTTCACCTGACAAAGCT | 184 |
| R: GCCTTCCCATACATGCAGACGC | ||
| xdh | F: AGGAGGTTGTGGAGCCTGCACT | 134 |
| R: CCTCCACGGTTGTTACCGCACA | ||
| dhx58 | F: TGCGTTACGGGCTGTTGACCA | 118 |
| R: TCTTTGCGCACTTCCCGTCCA | ||
| nkx2-1b | F: GCTGGTACGGAACGAATCCTGAC | 135 |
| R: TCAGTGGACCCATGCCTTTACCA | ||
| tipin | F: AAACTGGGCCCATCGCCTGT | 121 |
| R: TGGCATGTCCAACCGAATCCGT | ||
| anxa5 | F: GAAGCCTCCAAGAAATAC | 158 |
| R: GTCAAGCAAGTCCACCTC | ||
| acta2 | F: ACCAAGTGGCTAAATACCC | 108 |
| R: CAGTGCTTTCTTCGTCGTC | ||
| cyp19a | F:CTTCAGATTGGACTGGCTGCACAA | 180 |
| R: TTCTCTGCGCTCAGCTCTCCA |
2. 9. Statistical analysis
Sigmoidal regression was used to generate the dose–response curves for EC50 calculation (Origin 8.0, OriginLab). For gene expression comparisons, an unpaired t-test with 5% FDR was performed (SPSS, Chicago, IL, USA). All data are reported as means ± standard error (SEM) unless otherwise stated.
3. Results
3.1. PFOS exposure produces uninflated swim bladder and less developed gut
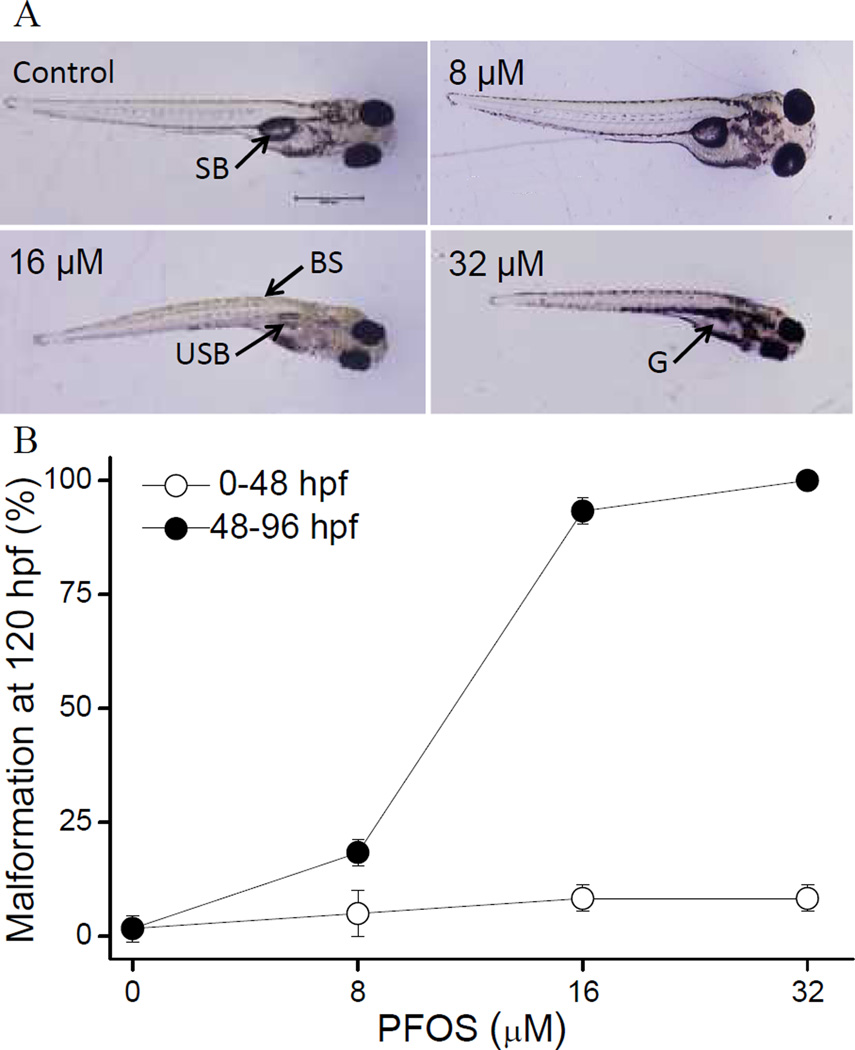
PFOS exposure during 48–96 hpf resulted in several distinct malformations including an uninflated swim bladder, less developed gut, and curved spine at 120 hpf (Fig. 1A–B). Typically, malformed larvae presented with all three types of malformations together. However, larvae developmentally exposed to PFOS from 8–48 hpf did not develop any obvious malformations, even at a concentration of 32 µM (Fig.1A). All the embryos survived during our experiment and no mortality occurred (data not shown). For embryos exposed from 48–96 hpf, all malformations were scored at 96 and 120 hpf. At 96 hpf, 16 µM PFOS treated larvae appeared morphologically normal, while about 25% of 32 µM group larvae showed some malformations at this time point. At 120 hpf, both 16 µM and 32 µM resulted approximately 100% malformation. Thus, 16 µM dose was selected for gene expression analyses.
Fig. 1.
Transient exposure to PFOS results in window-specific morphological effects. (A) PFOS exposure (0–32 µM) induces morphological defects at 120 hpf. SB: swim bladder; BS: bent spine; USB: uninflated swim bladder; G: gut. (B) Embryos were exposed to 0–32 µM PFOS or DMSO control from 0–48 or 48–96 hpf. Graph shows incidence of malformations at 120 hpf. Data represent 3 biological repeats with 20 embryos per treatment group.
3.2. PFOS induced histological alteration in swim bladder and gut section
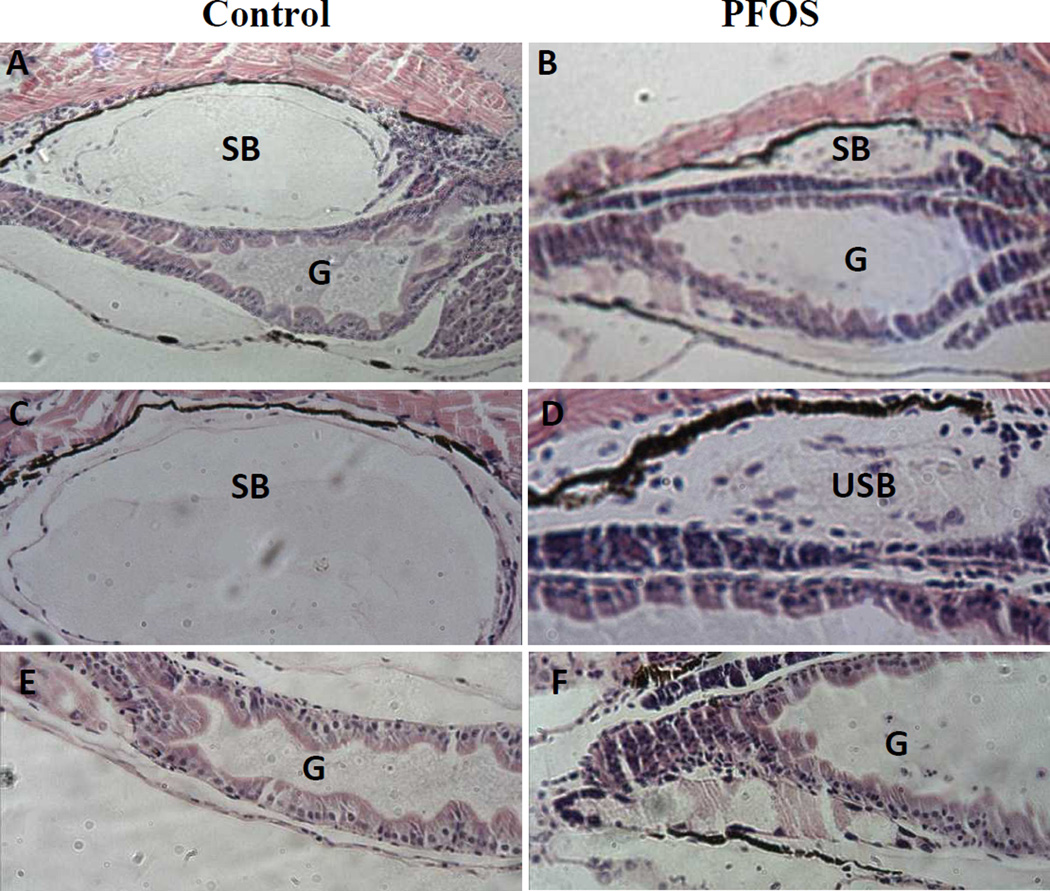
Histologically, PFOS exposure altered the structures of swim bladder and gut relative to vehicle controls (Fig. 2). Compared to vehicle control larvae, the swim bladders in PFOS-exposed larvae were smaller (uninflated), but still showed three distinct layers (Fig. 2C–D). However, the inner wall of the swim bladder cavity was less smooth with some caved shapes (Fig. 2D). On the contrary, PFOS-exposed larvae showed a larger gut tube than the control and displayed a non-uniform inner structure, which was shape uniform in the control larvae (Fig. 2E–F).
Fig. 2.
PFOS induced histological alteration in swim bladder and gut section. Representative histological sections of the gut and swim bladder from DMSO control or 16 µM PFOS exposed larvae at 120 hpf. There are 3 biological repeats with 5 embryos per treatment group. SB: swim bladder; USB: uninflated swim bladder; G: gut.
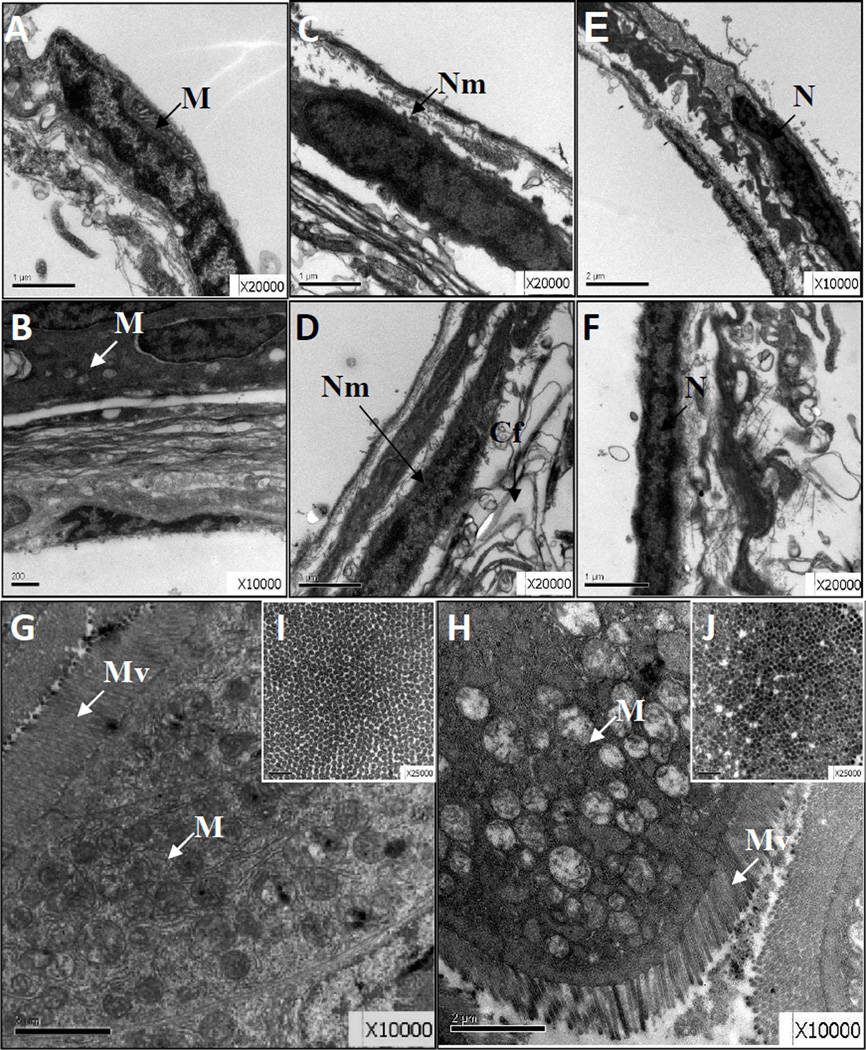
When examined with TEM, the inner cells in PFOS-exposed larvae swim bladder showed pyknosis and mitochondrial vacuole changes when compared with controls (Fig.3A–B). All three layers of cells showed mild apoptosis such as nuclear shrinkage and nuclear envelope gap expansion in PFOS-exposed larvae relative to controls (Fig. 3C vs. Fig. 3D). In the 11 middle yolk layer, the cytoplasm content was decreased, the mosaic-like structure was significantly reduced, the arrangement of collagen fibers was partly disordered, and mild edema was found (Fig. 3C, E vs. Fig. 3D, F). For the guts in the control group, the intestine mucosal epithelial cells were mainly column-shaped and closely connected, with oval nuclei, uniform chromatin, and abundant organelles of mitochondria and endoplasmic reticulum, and a surface arranged with rich, uniform microvilli and an intact basement membrane (Fig. 3G, I). In comparison, the columnar epithelial cells in the PFOS-exposed larvae had partially pyknotic nuclei, increased heterochromatin, partial mitochondrial vacuolation, mild dilated endoplasmic reticulum, and uneven surface microvilli though the intercellular junctions in the PFOS-exposed larvae were closed and the basement membrane was intact (Fig. 3H, J).
Fig. 3.
PFOS induced ultrastructure alteration in swim bladder and gut section. Representative TEM images showing the swim bladder (A–F) and gut (G–J) region for DMSO control (A, C, E, G, I) and PFOS exposed larvae (B, D, F, H, J). The vertical (G, H) and transect (H, J) sections of the gut region are shown. There are 3 biological repeats with 5 embryos per treatment group. Cf: collagen fibers; M: mitochondria; N: nucleus; Nm: nuclear envelope; Mv: microvilli.
3.3. PFOS exposure leads to differential gene expression at 96 hpf
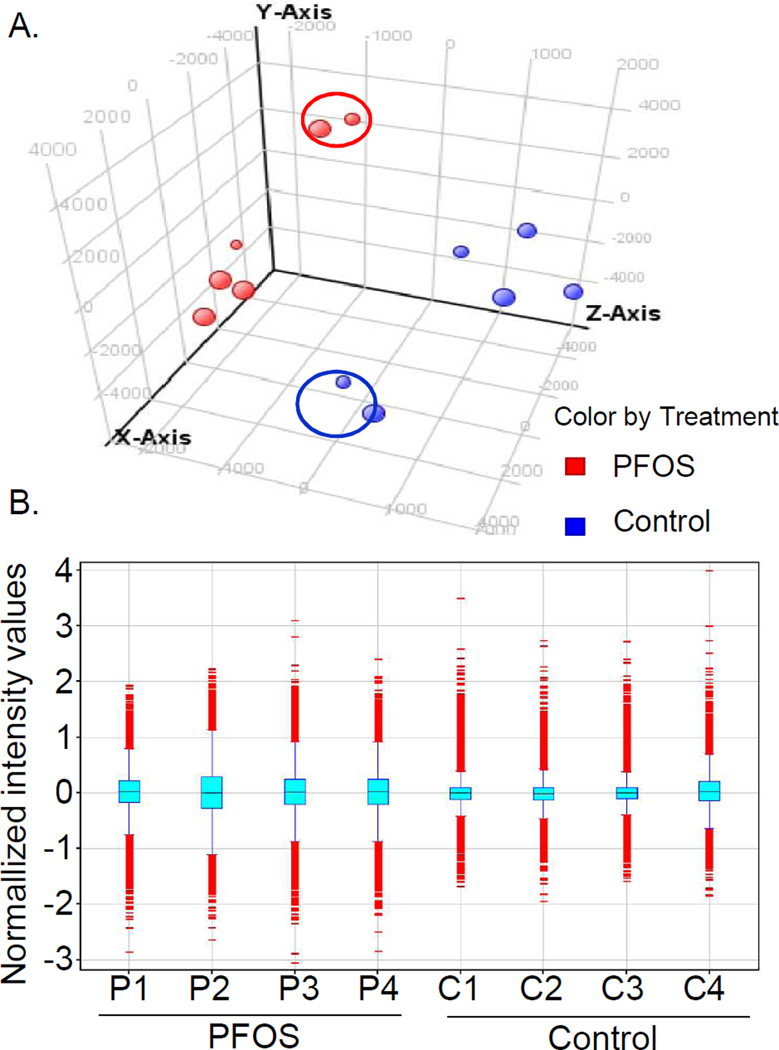
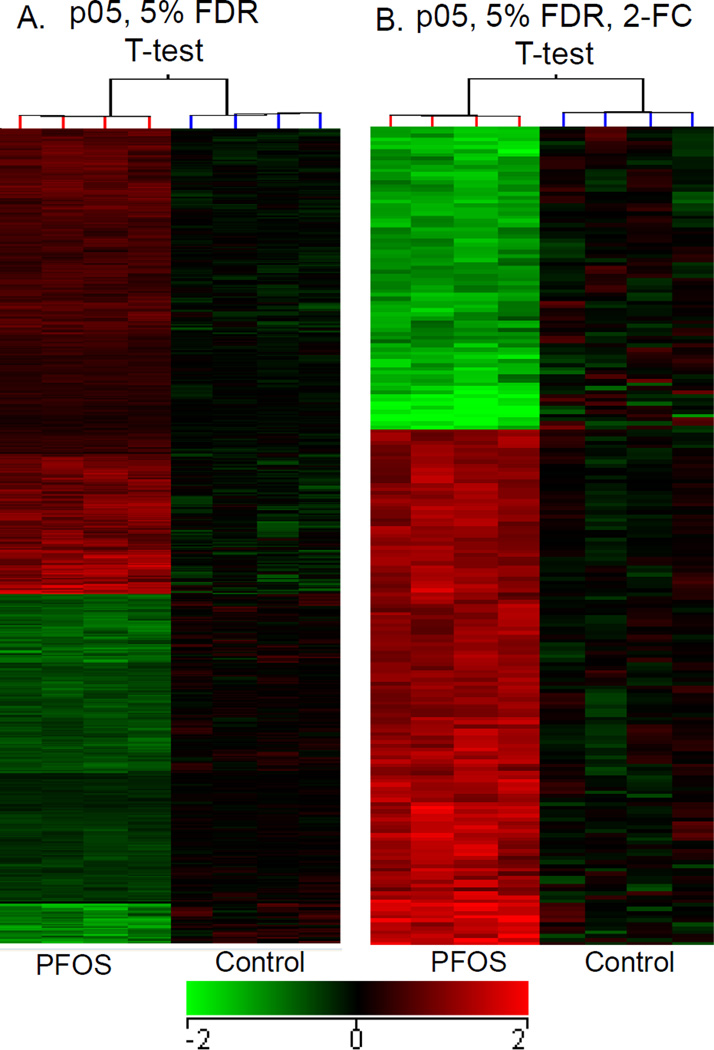
To identify gene expression changes following PFOS exposure during development, global microarray analysis was conducted using RNA isolated at 96 hpf from larvae exposed to PFOS from 48–96 hpf. A principal component analysis of all genes on the array shows separation of the two treatment groups into distinct clusters with four outliers (circles, Fig. 4A). The box plot of normalized data shows consistency in the interquartile range across the biological replicates without outliers (Fig. 4B). Statistical analysis of the differentially expressed transcripts was performed both with and without outliers (Table 2). In the analysis with all six repeats, 162 transcripts were significantly misexpressed (p<0.05) and 5 transcripts were changed more than two-fold by PFOS as compared with the control. When removing the four outliers from the analysis, 1,278 transcripts were significantly misregulated (p<0.05) and 211 genes were changed at least two-fold by PFOS as compared with the control group (Table 2 and Fig. 5).
Fig. 4.
Principal components and normalized plot analyzed PFOS-perturbed genomic mRNA expression in compare with the controls. (A) Principal components analysis of all genes on the array confirms that samples 2 and 5 are outliers within each treatment group. This analysis uses non-transformed data and shows variation among biological replicates. (B) Box plot of normalized data shows consistency in the interquartile range across biological replicates excluding the outliers.
Table 2.
Numbers of differentially expressed genes in PFOS-exposed larvae relative to controls.
| Analysis | All samples (n = 6) | No outliers* (n = 4) | |
|---|---|---|---|
| p05, 5% FDR | T-test | 162 | 1278 |
| p05, 5% FDR, 2-FC | T-test | 5 | 211 |
| 2-FC only | None | 13 | 595 |
Outlier samples (Control 2, 5; PFOS 2, 5) were removed for statistical analysis and calculation of fold-change
Fig. 5.
Hierarchical clustering analyzed PFOS-perturbed genomic mRNA expression in compare with the controls. (A) It showed the changed transcripts at p < 0.05 and (B) those with at least two-fold gene expression changes between the control and PFOS exposed embryos when assessed at 96 hpf. Values represent Log2 fold-changes (p < 0.05 by T-test with 5% FDR).
To validate the array data, nine transcripts involved in organogenesis or metabolic processes were selected for validation by qRT-PCR. In general, the comparison of mRNA abundance determined by the microarray and qRT-PCR revealed similar trends for all examined transcripts (Fig. 6).
Fig. 6.
PFOS-misexpressed the mRNA expression of 8 genes in compare with the controls. qRT-PCR validation of PFOS-regulated transcripts in 96 hpf zebrafish. The mean fold change relative to the controls for the microarray and qPCR are graphed for comparison. The gene name (when known) or the sequence ID was listed for each transcript. Data are representative of 3 biological replicates with 40 embryos per replicate.
3.4. PFOS exposure resulted in the misexpression of organogenesis and developmental network related transcripts
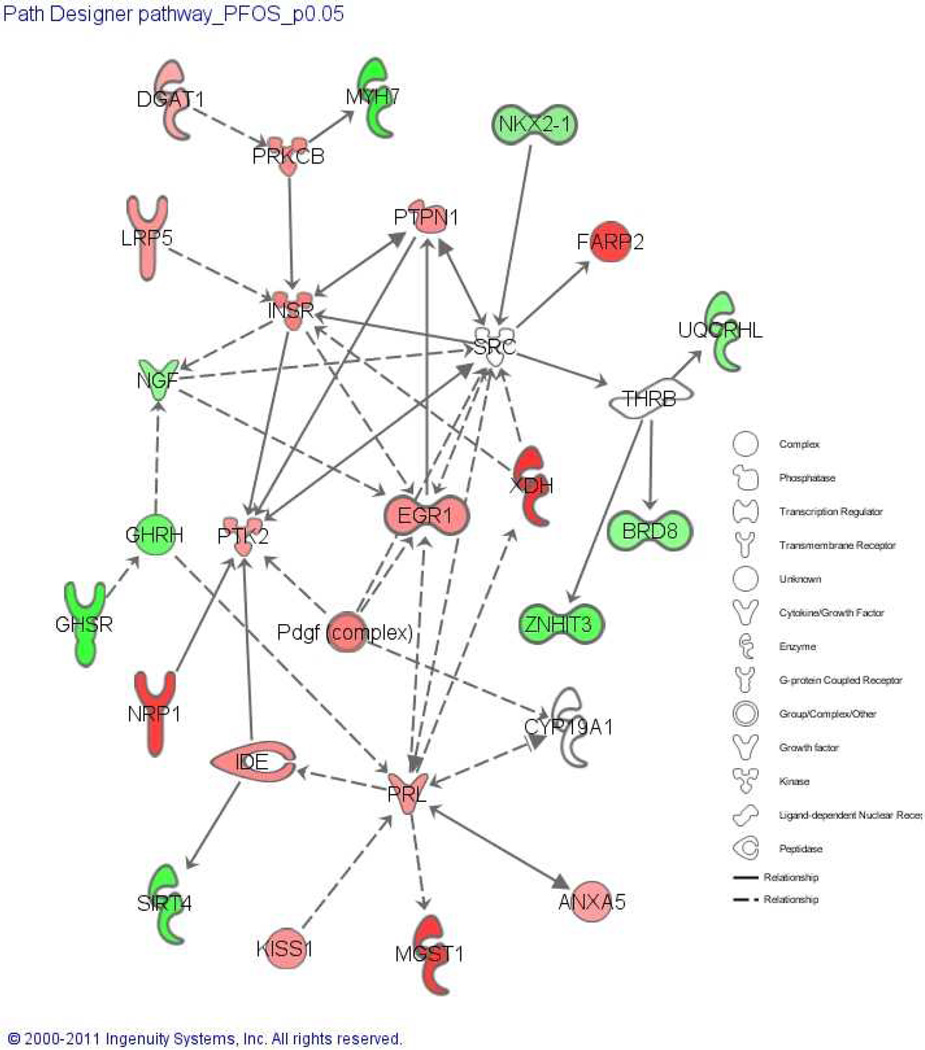
Differentially expressed transcripts were analyzed for enriched biological processes (Table 3). Genes significantly elevated by PFOS exposure were associated with nucleic and macromolecule metabolism, cell differentiation and proliferation, neuron differentiation and development, and voltage-gated channels (Table 3). In contrast, downregulated genes were associated with cellular protein metabolic processes, macromolecular complex assembly, protein-DNA complex assembly, and positive regulation of translation and multicellular organism growth (Table 3). We also used IPA to identify pathways that are significantly altered compared to the control using genes significantly changed in PFOS group. The top toxicity pathways perturbed by PFOS exposure were mechanisms of gene regulation by peroxisome proliferators via PPARα, decreases of transmembrane potential of mitochondria and mitochondrial membrane, and cardiac necrosis/cell death (Supplemental Table 1). Specific analysis of transcripts related to swim bladder and gut development were used to build a PFOS-perturbed network (Fig.7). A total of 16 transcripts were upregulated and are labeled in red (e.g., xdh, ide, lrp, insr, and anxa5). An additional 9 transcripts were downregultated by PFOS exposure and are labeled in green (e.g., cyp19a1, brd8, and nkx2-1a).
Table 3.
PFOS perturbed functional enrichment of biological process GO terms for up- and down-regulated genes in dataset.
| Up-regulated term | Count | P-value | % |
| Regulation of nucleic metabolic process | 79 | 0.0289 | 14.13 |
| Negative regulation of cellular process | 54 | 0.0511 | 9.66 |
| Phosphate metabolic process | 38 | 0.0708 | 6.80 |
| Negative regulation of macromolecule metabolic process | 28 | 0.0339 | 5.01 |
| Macromolecular complex assembly | 26 | 0.0441 | 4.65 |
| Regulation of cell differentiation | 22 | 0.0236 | 3.94 |
| Cellular component morphogenesis | 20 | 0.0208 | 3.58 |
| Positive regulation of cell proliferation | 19 | 0.0315 | 3.40 |
| Neuron differentiation | 18 | 0.0976 | 3.22 |
| Neuron development | 16 | 0.0495 | 2.86 |
| Cell projection morphogenesis | 13 | 0.0607 | 2.33 |
| Blood vessel development | 12 | 0.0824 | 2.15 |
| Positive regulation of transferase activity | 12 | 0.0468 | 2.15 |
| Regulation of cell motion | 10 | 0.0662 | 1.79 |
| Voltage-gated channel | 8 | 0.0987 | 1.43 |
| Cellular response to hormone stimulus | 8 | 0.0634 | 1.43 |
| Nucleosome organization | 6 | 0.0987 | 1.07 |
| Cellular glucan metabolic process | 5 | 0.0195 | 0.89 |
| Insulin receptor signaling pathway | 5 | 0.0195 | 0.89 |
| Regulation of fibroblast proliferation | 4 | 0.0724 | 0.72 |
| Regulation of transporter activity | 4 | 0.0608 | 0.72 |
| Positive regulation of osteoblast differentiation | 4 | 0.0362 | 0.72 |
| Regulation of carbohydrate catabolic process | 4 | 0.0127 | 0.72 |
| Down-regulated term | Count | p Value | % |
| Cellular protein metabolic process | 57 | 0.0824 | 13.41 |
| Macromolecular complex assembly | 24 | 0.0069 | 5.65 |
| Protein-DNA complex assembly | 6 | 0.0317 | 1.41 |
| Response to temperature stimulus | 5 | 0.0886 | 1.18 |
| Positive regulation of translation | 3 | 0.0729 | 0.71 |
| Positive regulation of multicellular organism growth | 3 | 0.0590 | 0.71 |
| Response to vitamin D | 3 | 0.0525 | 0.71 |
| Growth hormone secretion | 3 | 0.0036 | 0.71 |
Fig. 7.
PFOS-perturbed organogenesis and developmental network. It is constructed from differentially regulated transcripts related to swim bladder and gut development. Red and green shading indicate up- and down-regulated transcripts at 96 hpf relative to the baseline, respectively. The intensity of shading indicates the magnitude of regulation.
4. Discussion
In the present study, malformations induced by acute PFOS exposure during a sensitive window of 48–96 hpf included uninflated swim bladder, less developed gut, and bent spine. These observations were consistent with previous findings (Huang et al., 2010; Shi et al., 2008; Wang et al., 2011). Further histology and TEM analysis revealed detailed structural changes in swim bladder and gut associated with PFOS acute exposure. Transcriptional analysis identified several potential pathways and candidate genes involved in the PFOS perturbed organogenesis.
The selection of sensitive window revealed that embryos at the developmental window of 48–96 hpf are more sensitive to PFOS exposure than those at 8 to 48 hpf as a dose of 16 µM led to 100% malformation in embryos exposed to PFOS during 48–96 hpf yet a dose of 32 µM did not cause any malformation for embryos exposed between 8 to 48 hpf. One possible reason for the relative resistance to PFOS of embryos at earlier developmental stage could be due to slower PFOS absorption prior to 48 h and more rapid PFOS accumulation in embryos after 48 h as we have shown previously (Huang et al., 2010). Alternatively, candidate receptors that mediate PFOS-induced toxicity may not become evident till 48 hpf (Bardet et al., 2002). Future studies are necessary to identify the underlying cause for this different window sensitivity to PFOS exposure.
In the present study, whole genomic microarray analysis was used to identify transcripts that are differentially expressed by PFOS exposure. We observed that a total of 1,278 transcripts were significantly affected by PFOS exposure and the biological processes enriched included metabolic processes. These include nucleus, phosphate, macromolecule, cellular glucan and protein metabolism. The digestive system plays a critical role in metabolic processes (DeWitt and Kudsk, 1999) thus the perturbed metabolic process may result from malformed digestive organs (e.g., the zebrafish gut) upon PFOS exposure. The microarray findings we reported here are consistent with the analysis of the proteomic changes identified (Shi et al., 2009) following developmental PFOS exposure (till 192 hpf) as energy metabolism and lipid transport/steroid metabolic process were also implicated in the latter study. Our findings were also in good agreement with an earlier study in PFOS exposed carps where altered genes in the liver were mainly involved in energy metabolism, reproduction, and stress response (Hagenaars et al., 2008). A PFOS-induced network of perturbed transcripts relating to swim bladder and gut development revealed misexpression of insulin-degrading enzyme (ide), cytochrome P450-family 19-subfamily a- polypeptide 1 (cyp19a) and NK2 homeobox 1b (nkx2-1b), all these genes are involved in organogenesis (Donoghue et al., 2000; Lieb et al., 2006; Wendl et al., 2002). Confirmation of alterations at the protein/enzyme level is the next step in the assessment, but is beyond the scope of this study.
Similar to the mammalian lung (Spooner and Wessells, 1970), the zebrafish swim bladder arises from an outgrowth of the foregut endoderm, and is in close temporal and spatial proximity to the liver and pancreas (Field et al., 2003). Prenatal PFOS exposure affects lung development in perinatal rats (Grasty et al., 2005). In the present study, we observed altered structure and gene expression in swim bladder-related transcripts. The observation that swim bladder was one of the main targets for PFOS induced developmental toxicity in zebrafish also corroborates previous findings that liver and lung are two primary target organs of PFOS (Hagenaars et al., 2008; Luebker et al., 2005). Gene expression profiling in the liver and lung of PFOS-exposed mouse fetuses revealed that PFOS-dependent changes are primarily related to activation of PPARα (Rosen et al., 2009). A similar mechanism was proposed for PFOS induced gene expression changes associated with lipid metabolism and cholesterol biosynthesis (Lau et al., 2007) and hepatomegaly changes in lymphoid organs (DeWitt et al., 2009). PFOS has also been shown to affect peroxisomal fatty acid β-oxidation pathway by altering peroxisomal membrane permeability to allow fatty acid influx (Hu et al., 2005). Although we did not observe significant alteration of PPARα in the present study, IPA 15 analysis revealed that mechanism of gene regulation by peroxisome proliferators via PPARα was the top 1 toxicity pathway perturbed by PFOS exposure (Supplemental Table 1). Further analysis of PPAR signaling identified significant expression changes of transcripts related to canonical pathway of PPARα/RXRα activation (Supplemental Fig. 1). Future functional validation is necessary to uncover whether PPARα-dependent signaling plays a functional role in PFOS induced morphological changes in zebrafish larvae.
The zebrafish gut is ventral to the swim bladder, and it forms in early somite stages (10–18 hpf), giving rise to the organs of the digestive tract and its accessory organs such as liver and pancreas at the pharyngula and hatching stages (48–72 hpf) (Wallace et al., 2005; Wallace and Pack, 2003). PFOS exposure induced multiple structural abnormalities in the gut including nuclei pyknosis, heterochromatin, and uneven surface microvilli in the columnar epithelial cells. This is the first study to report abnormal gut morphology upon exposure to PFOS. More studies are needed to delineate mechanisms underlying gut abnormalities upon exposure to PFOS during embryonic development.
It is known that oxidative stress can induce cellular damage and this form of cellular stress involves in many biological and pathological processes (Carnevali et al., 2003; MacNee, 2000). Previous studies indicated that oxidative stress plays an important role in developmental toxicity by PFOS exposure (Liu et al., 2009; Qian et al., 2010; Wei et al., 2008). More recently, prenatal PFOS exposure in rats from gestation day 1 to day 21 induced significant induction of oxidative stress in postnatal pups, representing by increased malondialdehyde level, decreased glutathione content, and declined superoxide dismutase activity (Chen et al., 2012). In fish, reactive oxygen species (ROS)-induced oxidative stress is thought to contribute to abnormal development during embryogenesis (Yamashita, 2003). PFOS exposure to embryonic zebrafish from 4 to 96 hpf caused hypergeneration of ROS, which in turn induced phase II detoxification enzymes and nuclear factor erythroid 2 related factor 2 (nrf2) pathway against oxidative stress to protect oxidative damage (Shi and Zhou, 2010). Findings in our study showed that oxidative stress and nrf2-mediated oxidative stress response signaling are significantly perturbed by PFOS exposure, e.g., mitogen-activated protein kinase 3 (mapk3), janus kinase 2 (jak2), and aldehyde dehydrogenase family 1 member L2 (aldh1l2) were significantly up regulated. Together, these findings indicate that oxidative stress may play an important role in PFOS induced developmental toxicity.
In summary, our study demonstrates that early life stage exposure to PFOS perturbs zebrafish embryonic swim bladder and gut development. Early life stage exposure to PFOS perturbs numerous molecular pathways, collectively leading to the morphological defects observed in the swim bladder and gut of PFOS exposed larvae.
Supplementary Material
Acknowledgements
We thank Zhouxi Fang for technical help with TEM. This work was supported by funding from the National Natural Science Foundation of China (No. 21277104), the Key Project of Zhejiang Provincial Natural Science Foundation (LZ13B070001), and the National Institute of Environmental Health Sciences grants #P30 ES00210 and P42 ES016465.
References
- Bardet P, Horard B, Robinson-Rechavi M, Laudet V, Vanacker J. Characterization of oestrogen receptors in zebrafish (Danio rerio) J Mol Endocrinol. 2002;28:153–163. doi: 10.1677/jme.0.0280153. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Carnevali S, Petruzzelli S, Longoni B, Vanacore R, Barale R, Cipollini M, Scatena F, Paggiaro P, Celi A, Giuntini C. Cigarette smoke extract induces oxidative stress and apoptosis in human lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2003;284:L955–L963. doi: 10.1152/ajplung.00466.2001. [DOI] [PubMed] [Google Scholar]
- Chen J, Das SR, La Du J, Corvi MM, Bai C, Chen Y, Liu X, Zhu G, Tanguay RL, Dong Q. Chronic PFOS exposures induce life stage–specific behavioral deficits in adult zebrafish and produce malformation and behavioral deficits in F1 offspring. Environ Toxicol Chem. 2013;32:201–206. doi: 10.1002/etc.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Zhang L, Yue J-q, Lv Z-q, Xia W, Wan Y-j, Li Y-y, Xu S-q. Prenatal PFOS exposure induces oxidative stress and apoptosis in the lung of rat off-spring. Reprod Toxicol. 2012;33:538–545. doi: 10.1016/j.reprotox.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- DeWitt JC, Shnyra A, Badr MZ, Loveless SE, Hoban D, Frame SR, Cunard R, Anderson SE, Meade BJ, Peden-Adams MM. Immunotoxicity of perfluorooctanoic acid and perfluorooctane sulfonate and the role of peroxisome proliferator-activated receptor alpha. Crit Rev Toxicol. 2009;39:76–94. doi: 10.1080/10408440802209804. [DOI] [PubMed] [Google Scholar]
- DeWitt RC, Kudsk KA. The gut's role in metabolism, mucosal barrier function, and gut immunology. Infect Dis Clin North Am. 1999;13:465–481. doi: 10.1016/s0891-5520(05)70086-6. [DOI] [PubMed] [Google Scholar]
- Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Field HA, Dong PDS, Beis D, Stainier DYR. Formation of the digestive system in zebrafish. ii. pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/s0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- Giesy JP, Kannan K. Global distribution of perfluorooctane sulfonate in wildlife. Environ Sci Technol. 2001;35:1339–1342. doi: 10.1021/es001834k. [DOI] [PubMed] [Google Scholar]
- Grasty RC, Bjork JA, Wallace KB, Wolf DC, Lau CS, Rogers JM. Effects of prenatal perfluorooctane sulfonate (PFOS) exposure on lung maturation in the perinatal rat. Birth Defects Res B Dev Reprod Toxicol. 2005;74:405–416. doi: 10.1002/bdrb.20059. [DOI] [PubMed] [Google Scholar]
- Hagenaars A, Knapen D, Meyer IJ, van der Ven K, Hoff P, De Coen W. Toxicity evaluation of perfluorooctane sulfonate (PFOS) in the liver of common carp (Cyprinus carpio) Aquat Toxicol. 2008;88:155–163. doi: 10.1016/j.aquatox.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Teraoka H, Heideman W, Peterson RE. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol Sci. 2005;86:6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- Hoff PT, Van Campenhout K, Van de Vijver K, Covaci A, Bervoets L, Moens L, Huyskens G, Goemans G, Belpaire C, Blust R, De Coen W. Perfluorooctane sulfonic acid and organohalogen pollutants in liver of three freshwater fish species in Flanders (Belgium): relationships with biochemical and organismal effects. Environ Pollut. 2005;137:324–333. doi: 10.1016/j.envpol.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Houde M, Bujas TA, Small J, Wells RS, Fair PA, Bossart GD, Solomon KR, Muir DC. Biomagnification of perfluoroalkyl compounds in the bottlenose dolphin (Tursiops truncatus) food web. Environ Sci Technol. 2006;40:4138–4144. doi: 10.1021/es060233b. [DOI] [PubMed] [Google Scholar]
- Hu W, Jones PD, Celius T, Giesy JP. Identification of genes responsive to PFOS using gene expression profiling. Environ Toxicol Pharmacol. 2005;19:57–70. doi: 10.1016/j.etap.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Huang H, Huang C, Wang L, Ye X, Bai C, Simonich MT, Tanguay RL, Dong Q. Toxicity, uptake kinetics and behavior assessment in zebrafish embryos following exposure to perfluorooctanesulphonicacid (PFOS) Aquat Toxicol. 2010;98:139–147. doi: 10.1016/j.aquatox.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ooi SL, Wu Z, Boeke JD. Use of mixture models in a microarray-based screening procedure for detecting differentially represented yeast mutants. Stat Appl Genet Mol Biol. 2003;2:1002. doi: 10.2202/1544-6115.1002. [DOI] [PubMed] [Google Scholar]
- Jang GH, Hwang MP, Kim SY, Jang HS, Lee KH. A systematic in-vivo toxicity evaluation of nanophosphor particles via zebrafish models. Biomaterials. 2013 doi: 10.1016/j.biomaterials.2013.09.054. [DOI] [PubMed] [Google Scholar]
- Kannan K, Tao L, Sinclair E, Pastva SD, Jude DJ, Giesy JP. Perfluorinated compounds in aquatic organisms at various trophic levels in a Great Lakes food chain. Arch Environ Contam Toxicol. 2005a;48:559–566. doi: 10.1007/s00244-004-0133-x. [DOI] [PubMed] [Google Scholar]
- Kannan K, Tao L, Sinclair E, Pastva SD, Jude DJ, Giesy JP. Perfluorinated Compounds in Aquatic Organisms at Various Trophic Levels in a Great Lakes Food Chain. Arch Environ Contam Toxicol. 2005b;48:559–566. doi: 10.1007/s00244-004-0133-x. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lau C, Anitole K, Hodes C, Lai D, Pfahles-Hutchens A, Seed J. Perfluoroalkyl acids: a review of monitoring and toxicological findings. Toxicol Sci. 2007;99:366–394. doi: 10.1093/toxsci/kfm128. [DOI] [PubMed] [Google Scholar]
- Lieb W, Graf J, Götz A, König IR, Mayer B, Fischer M, Stritzke J, Hengstenberg C, Holmer SR, Döring A. Association of angiotensin-converting enzyme 2 (ACE2) gene polymorphisms with parameters of left ventricular hypertrophy in men. J Mol Med (Berl) 2006;84:88–96. doi: 10.1007/s00109-005-0718-5. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu W, Song J, Yu H, Jin Y, Oami K, Sato I, Saito N, Tsuda S. A comparative study on oxidative damage and distributions of perfluorooctane sulfonate (PFOS) in mice at different postnatal developmental stages. J Toxicol Sci. 2009;34:245–254. doi: 10.2131/jts.34.245. [DOI] [PubMed] [Google Scholar]
- Luebker DJ, Case MT, York RG, Moore JA, Hansen KJ, Butenhoff JL. Two-generation reproduction and cross-foster studies of perfluorooctanesulfonate (PFOS) in rats. Toxicology. 2005;215:126–148. doi: 10.1016/j.tox.2005.07.018. [DOI] [PubMed] [Google Scholar]
- MacNee W. Oxidants/antioxidants and COPD. J Chest. 2000;117:303S–317S. doi: 10.1378/chest.117.5_suppl_1.303s-a. [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KRK, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS genet. 2005;1:e29. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler FB, Laurenson S, Williams LC, Dodd A, Copp BR, Love DR. Chemical discovery and global gene expression analysis in zebrafish. Nat Biotechnol. 2003;21:879–883. doi: 10.1038/nbt852. [DOI] [PubMed] [Google Scholar]
- Qian Y, Ducatman A, Ward R, Leonard S, Bukowski V, Lan Guo N, Shi X, Vallyathan V, Castranova V. Perfluorooctane sulfonate (PFOS) induces reactive oxygen species (ROS) production in human microvascular endothelial cells: role in endothelial permeability. J Toxicol Environ Health A. 2010;73:819–836. doi: 10.1080/15287391003689317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi M, Guerranti C, Giovani A, Perra G, Focardi SE. Perfluorinated compounds: Levels, trophic web enrichments and human dietary intakes in transitional water ecosystems. Mar Pollut Bull. 2013 doi: 10.1016/j.marpolbul.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Schmid JE, Das KP, Wood CR, Zehr RD, Lau C. Gene expression profiling in the liver and lung of perfluorooctane sulfonate-exposed mouse fetuses: comparison to changes induced by exposure to perfluorooctanoic acid. Reprod Toxicol. 2009;27:278–288. doi: 10.1016/j.reprotox.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shi X, Du Y, Lam PK, Wu RS, Zhou B. Developmental toxicity and alteration of gene expression in zebrafish embryos exposed to PFOS. Toxicol Appl Pharmacol. 2008;230:23–32. doi: 10.1016/j.taap.2008.01.043. [DOI] [PubMed] [Google Scholar]
- Shi X, Yeung LW, Lam PK, Wu RS, Zhou B. Protein profiles in zebrafish (Danio rerio) embryos exposed to perfluorooctane sulfonate. Toxicol Sci. 2009;110:334–340. doi: 10.1093/toxsci/kfp111. [DOI] [PubMed] [Google Scholar]
- Shi X, Zhou B. The role of Nrf2 and MAPK pathways in PFOS-induced oxidative stress in zebrafish embryos. Toxicol Sci. 2010;115:391–400. doi: 10.1093/toxsci/kfq066. [DOI] [PubMed] [Google Scholar]
- Sinclair E, Mayack DT, Roblee K, Yamashita N, Kannan K. Occurrence of perfluoroalkyl surfactants in water, fish, and birds from New York State. Arch Environ Contam Toxicol. 2006;50:398–410. doi: 10.1007/s00244-005-1188-z. [DOI] [PubMed] [Google Scholar]
- Spooner BS, Wessells NK. Mammalian lung development: interactions in primordium formation and bronchial morphogenesis. J Exp Zool. 1970;175:445–454. doi: 10.1002/jez.1401750404. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tilton SC, Philbrick KA, Iwaniec UT, Turner RT, Waters KM, Tanguay RL. MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J. 2012;26:1452–1461. doi: 10.1096/fj.11-194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- Wang M, Chen J, Lin K, Chen Y, Hu W, Tanguay RL, Huang C, Dong Q. Chronic zebrafish PFOS exposure alters sex ratio and maternal related effects in F1 offspring. Environ Toxicol Chem. 2011;30:2073–2080. doi: 10.1002/etc.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chan LL, Wang D, Zhang H, Wang J, Dai J. Proteomic analysis of hepatic protein profiles in rare minnow (Gobiocypris rarus) exposed to perfluorooctanoic acid. J Proteome Res. 2008;7:1729–1739. doi: 10.1021/pr7008059. [DOI] [PubMed] [Google Scholar]
- Wendl T, Lun K, Mione M, Favor J, Brand M, Wilson SW, Rohr KB. Pax2. 1 is required for the development of thyroid follicles in zebrafish. Development. 2002;129:3751–3760. doi: 10.1242/dev.129.15.3751. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio)M. Westerfield Eugene, OR; 1993. [Google Scholar]
- Yamashita M. Apoptosis in zebrafish development. Comp Biochem Physiol B Biochem Mol Biol. 2003;136:731–742. doi: 10.1016/j.cbpc.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li YY, Zeng HC, Wei J, Wan YJ, Chen J, Xu SQ. MicroRNA expression changes during zebrafish development induced by perfluorooctane sulfonate. J Appl Toxicol. 2011a;31:210–222. doi: 10.1002/jat.1583. [DOI] [PubMed] [Google Scholar]
- Zhang W, Lin Z, Hu M, Wang X, Lian Q, Lin K, Dong Q, Huang C. Perfluorinated chemicals in blood of residents in Wenzhou, China. Ecotoxicol Environ Saf. 2011b;74:1787–1793. doi: 10.1016/j.ecoenv.2011.04.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.