Abstract
Oxidative stress plays an important role in the development of obesity and obesity-associated metabolic disorders. As an endogenous antioxidant enzyme, superoxide dismutase 3 (SOD3) has the potential to affect diet-induced obesity and obesity associated complications. In the current work, we overexpressed SOD3 in C57BL/6 mice fed a high fat diet to study its effect on high fat diet-induced obesity, fatty liver and insulin resistance. We demonstrated that the Sod3 gene transfer blocked high fat diet induced obesity, fatty liver and insulin resistance. Real Time PCR analysis of adipose and liver tissues revealed that overexpression of the Sod3 gene suppressed expression of pro-inflammatory genes in adipose tissue including F4/80, Tnfα, Cd11c, Mcp1 and Il6, and increased expression of anti-inflammatory genes such as adiponectin. In the liver, high levels of SOD3 activity in animals enhanced expression of the genes responsible for energy expenditure including Cpt1α, Cpt1β, Pgc1α, Pgc1β and Ucp2. These results suggest that overexpression of the Sod3 gene through gene transfer is an effective approach in preventing diet induced obesity and obesity-associated complications.
Keywords: superoxide dismutase 3, gene therapy, obesity, fatty liver, insulin resistance
Introduction
Obesity is a causative factor in the development of several diseases including diabetes, cardiovascular diseases, metabolic disorders, and cancer.1 Reactive oxygen species (ROS) and increased oxidative stress in adipose tissue play a major role in the development of obesity, fatty liver, glucose intolerance and insulin resistance, the early events in obesity-associated metabolic disorders.2 In the diabetic condition, oxidative stress impairs glucose uptake in muscle and adipose tissue, and decreased insulin secretion from pancreatic beta cells.3 Increased ROS also underlies the pathophysiology of hypertension, atherosclerosis and cardiovascular diseases.4
Superoxide dismutases (SODs) are enzymes primarily responsible for the maintenance of oxidation-reduction homeostasis. SODs utilize the superoxide anion in dismutation reaction and generate hydrogen peroxide; this is further metabolized to oxygen and water by catalase and glutathione peroxidase. Among the three types of SODs, SOD1 and SOD2 are intracellular enzymes which function in the cytoplasm and mitochondria, and SOD3 is an extracellular enzyme exerting extracellular antioxidant effect. A mutation isoform of SOD3, Arg213Gly, with an inactive function was shown to be associated with higher BMI, hypercholesterolemia and hyperlipidemia.5 SOD3 has been shown to be negatively related to insulin resistance in type 2 diabetes6 and metabolic syndrome.7 Overexpression of the Sod3 gene has been found to be effective in protecting islet function in the pancreas against oxidative stress after transplantation.8 Transfer of the Sod3 gene has also been shown to have a therapeutic response in tissue repair9 and anti-inflammation.10
In the present work, we focused on the activity of SOD3 in blocking high fat diet (HFD)-induced obesity, insulin resistance and development of a fatty liver. Our data show that hydrodynamic Sod3 gene transfer blocked HFD-induced weight gain, insulin resistance and alleviated fatty liver. At the biochemical level, overexpression of Sod3 suppressed the expression of pro-inflammation genes and increased expression of the genes responsible for energy metabolism.
Results
Characterization of sod3 gene expression and impacts of hydrodynamic gene delivery
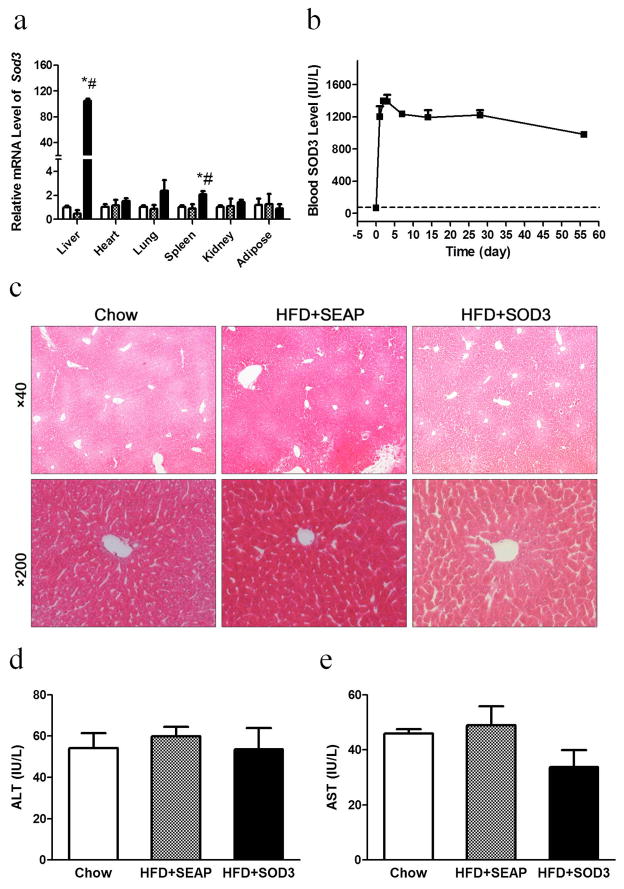
Sod3 gene expression and the impacts of the hydrodynamic procedure on animals were examined 7 days after gene transfer. Figure 1a shows that the liver is the predominant site that expressed the Sod3 gene. Compared to other organs, including the heart, lung, spleen and kidneys, the mRNA levels for the SOD3 are more than 100-fold higher in the liver than those of other organs which show background levels similar to those of control animals injected with pLIVE-SEAP plasmids. Results from a time course (Figure 1b) show that SOD3 activity in the blood reaches the peak level at 1,400 IU/L 3 days after hydrodynamic injection, slightly decreases thereafter, and remains at about 900 IU/L level at the end of the 8-week experiment. Based on H&E staining (Figure 1c) and blood concentration of liver specific enzymes ALT (Figure 1d) and AST (Figure 1e), no liver damage was seen. These results suggest that hydrodynamic gene transfer is safe and effective in introducing and expressing the Sod3 gene in the liver.
Figure 1. Impacts of hydrodynamic gene delivery.
Mice fed regular chow or a HFD were hydrodynamically injected with pLIVE-SOD3 or pLIVE-SEAP plasmids and sacrificed one week later. (a) mRNA levels of sod3 in collected organs. (b) Blood concentrations of SOD3 as function of time. (c) Images of H&E staining of liver sections; (d) Blood concentrations of alanine aminotransferase (ALT); (e) Blood concentrations of aspartate aminotransferase (AST). (□), animals fed chow; (
 ), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow group, # p<0.05 compared with SEAP group.
), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow group, # p<0.05 compared with SEAP group.
Hydrodynamic transfer of Sod3 gene blocks high fat diet-induced weight gain
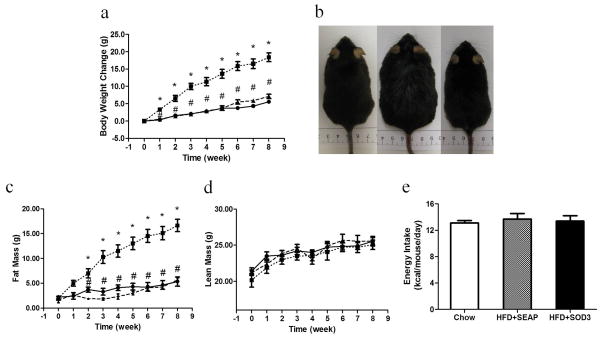
The impact of Sod3 gene transfer on weight gain in animals was examined. Results in Figure 2a show that control animals injected with pLIVE-SEAP plasmids gained about 15 g at the end of an 8-week HFD feeding compared to 5 g in animals injected with pLIVE-SOD3 plasmids. There is no statistical difference in body weights between animals fed a regular chow and those who underwent Sod3 gene transfer. The difference between HFD-fed control and SOD3 treated animals is visually differentiable (Figure 2b). These results suggest that hydrodynamic delivery of pLIVE-SOD3 plasmids completely blocked HFD-induced weight gain. Results from body composition analysis show that the difference between control and SOD3 treated animals is primarily fat mass (Figure 2c) and the lean mass of all animals is not different during the 8-week period (Figure 2d) among the three groups of animals. There is no difference in energy intake among the animals calculated based on the average food intake per animal per day (Figure 2e).
Figure 2. Sod3 gene transfer blocks high fat diet induced obesity in C57BL/6 mice.
(a) Time dependent increase in body weight; (b) Photo-images of animals fed regular chow (left), high fat diet with hydrodynamic transfer of pLIVE-SEAP (middle) and high fat diet with hydrodynamic delivery of pLIVE-SOD3 at the end of 8th week feeding; (c) Time dependent increase of fat mass; (d) Time dependent change of lean mass; (e) Time dependent energy intake by animas. (●) Animals fed regular chow. (■), animals fed a high fat diet and injected with pLIVE-SEAP plasmid; (▲), animals fed a high fat diet and injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
Sod3 gene transfer represses fat and macrophage accumulation in white adipose tissues
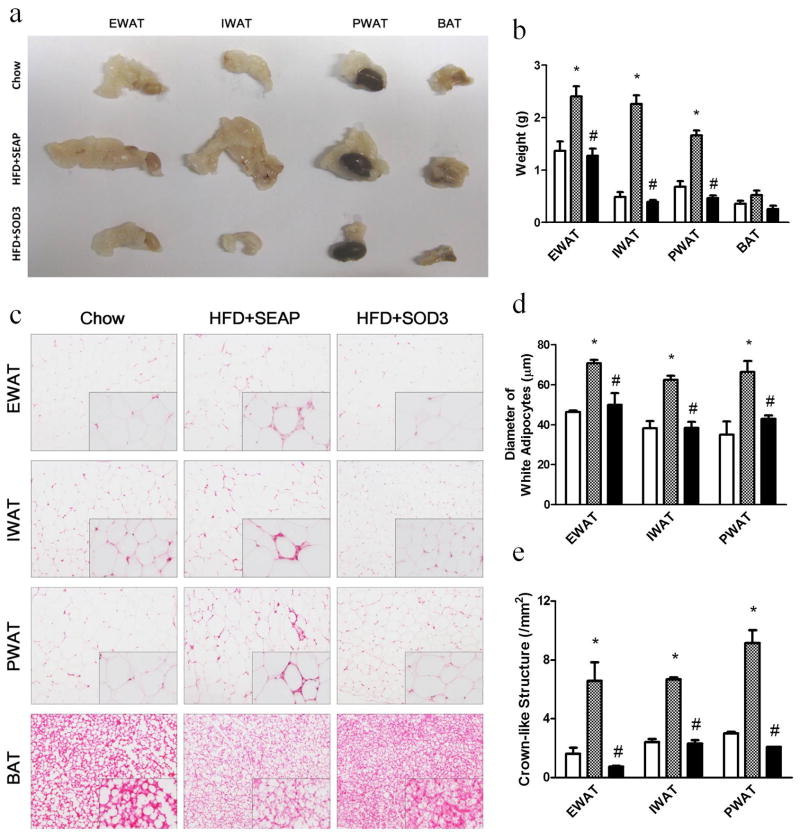
Adipose tissues were collected from the Sod3 gene injected and control animals to study the impact of Sod3 gene transfer. Figure 3a shows the relative amount and size of white and brown adipose tissue in SOD treated and control animals. Compared to animals fed a regular chow, the epididymal (EWAT), inguinal (IWAT), and perirenal (PWAT) white adipose tissues are significantly bigger in size (Figure 3a) and heavier (Figure 3b) in HFD-fed control mice. There is no difference between regular chow fed mice and HFD-fed mice injected with the Sod3 gene. The average weight of combined white adipose tissues in HFD-fed control animals is 2.1 g, 2.4- and 3.0-fold heavier than that of regular mice and animals who underwent Sod3 gene transfer, respectively. No statistical difference is seen in size or the total weight of brown adipose tissue among the three groups of animals. Images in Figure 3c show the shape and size of adipocytes in white and brown adipose tissue. The average diameter of adipocytes in HFD-fed control animals is 66.6 ± 1.9 μm, compared to animals fed a regular chow (42.7 ± 2.2) or in HFD-fed mice with the Sod3 gene transfer (43.9 ± 2.9) (Figure 3d). The crown-like structure, a sign of macrophage infiltration in adipose tissue, is evident in HFD-fed control animals (Figure 3c insert), but scarce in animals who have undergone Sod3 gene transfer or were fed a regular chow. The relative number of crown-like structures identified in HFD-fed control animals is approximately 7.3 ± 0.6 /mm2 compared to 1.7 ± 0.3 in animals injected with the Sod3 gene and 2.3 ± 0.2 in those fed a regular chow (Figure 3e). A whitish color of brown adipose tissue sections in HFD-fed control mice suggests that fat content in brown adipose tissue in HFD-fed control animals is higher than that of animals with Sod3 gene overexpression and those fed a regular chow.
Figure 3. Sod3 gene transfer suppresses fat accumulation and represses crown-like structure in adipose tissue.
(a) Images of different fat pads, one kidney was included when photographed the perirenal white adipose tissue; (b) Weight of adipose tissues; (c) Images of H&E staining of EWAT, IWAT, PWAT and BAT; (d) Average diameter of adipocytes in white adipose tissue calculated from 500 randomly selected cells from 5 tissue slides; (e) Number of crown-like structures in white adipose tissue collected from different location. EWAT: epididymal white adipose tissue, IWAT: inguinal white adipose tissue, PWAT: perirenal white adipose tissue, BAT: brown adipose tissue. (□), animals fed a regular Chow; (
 ), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
Sod3 gene transfer blocks high fat diet-induced fatty liver
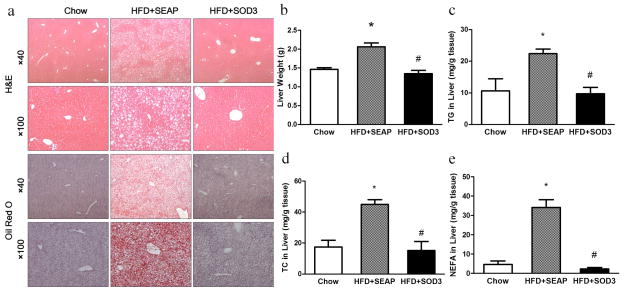
One of the most common physiological changes associated with obesity is an increase in fat content in the liver and the development of fatty liver. Histological images of mouse livers from HFD-fed control mice and animals who received Sod3 gene transfer show significant differences in the number and size of vacuoles in liver sections with H&E staining (Figure 4a). Higher lipid content is indicated in HFD-fed control animals than those of animals injected with the Sod3 gene and regular mice. Results from Oil-red O staining concur with the results of the H&E staining. The average liver weight of HFD-fed control animals is about 2.0 g, compared to approximately 1.5 g in mice with SOD3 overexpression or regular mice (Figure 4b). Similarly, Sod3 gene transfer also results in normal ranges of triglycerides (Figure 4c) and cholesterol (Figure 4d) and non-esterifized fatty acids (Figure 4e) compared to regular mice, and about half that of HFD-fed control mice. These results suggest that sod3 gene transfer blocked HFD-induced hepatic fat accumulation.
Figure 4. Sod3 gene transfer prevents high fat diet induced fatty liver.
(a) Representative images of H&E and oil-red O staining of the liver sections; (b) Liver weight; (c) Relative triglyceride level in the liver; (d) Relative cholesterol level in the liver; (e) Relative nonesterified fatty acids level in the liver. TG: triglyceride, TC: total cholesterol, NEFA: nonesterified fatty acid. (□), animals fed a regular Chow; (
 ), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
Sod3 gene transfer blocks high fat diet-induced glucose intolerance and insulin resistance
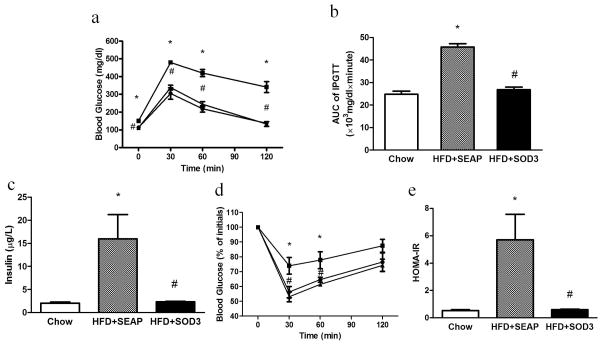
An intraperitoneal glucose tolerance test (IPGTT) and insulin tolerance test (ITT) were performed to assess the effect of Sod3 gene transfer on animals fed a HFD. Results in Figure 5a suggest that 8-week HFD feeding has caused animals injected with the control plasmid to become glucose intolerant, compared to a normal glucose profile in mice who underwent Sod3 gene transfer which is identical to that of regular mice. The AUC of the IPGTT confirms significant glucose intolerance in HFD-fed control animals and a normal glucose sensitivity in animals with Sod3 gene transfer (Figure 5b). A direct measurement of serum insulin levels (Figure 5c) and the results from ITT (Figure 5d) suggest that HFD-fed control mice have developed insulin resistance at the end of the 8-week HFD feeding. In contrast, animals who underwent Sod3 gene transfer have normal insulin levels and exhibit the same insulin sensitivity as regular mice fed a regular chow (Figure 5e).
Figure 5. Sod3 gene transfer improves glucose intolerance and alleviates high fat diet induced insulin resistance.
(a) Glucose profile in IPGTT; (b) AUC of IPGTT; (c) Fasting insulin level in the mice; (d) Glucose profile of ITT; (e) HOMA-IR calculated by fasting insulin level and fasting glucose. IPGTT: intraperitoneal glucose tolerance test, AUC: area under curve, ITT: insulin tolerance test, HOMA-IR: Homeostasis model assessment of insulin resistance. (●) animals fed a regular Chow; (■) animals fed a high fat diet and injected with pLIVE-SEAP plasmid; (▲) animals fed a high fat diet and injected with pLIVE-SOD3. (□), animals fed a regular Chow; (
 ), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with Chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with Chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
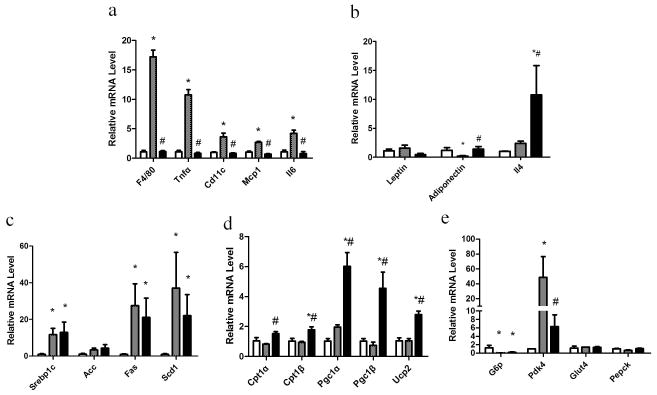
Sod3 gene transfer affects expression of critical genes involved in inflammation and energy metabolism
A decrease in crown-like structures in white adipose tissue (Figure 3e) exposed to sod3 gene transfer in HFD-fed animals suggests that overexpression of the Sod3 gene may suppress HFD-induced inflammation in adipose tissue. To test this possibility, we compared the mRNA levels of genes with known functions in inflammation including F4/80, Tnfα, Cd11c, Mcp1 and Il6. Results in Figure 6a show higher mRNA levels of the selected genes in white adipose tissue in obese mice injected with control plasmid. As expected, mRNA levels of the same group of genes in animals exposed to Sod3 gene transfer are identical to that of regular mice. The mRNA levels of the adiponectin gene in obese mice appear lower than in regular mice and those exposed to Sod3 gene transfer (Figure 6b). The mRNA levels in white adipose tissue of the leptin gene are lower in mice injected with the Sod3 gene, in contrast to a significant increase in Il4 mRNA level. Similar experiments were performed in the mouse liver to assess the effects of Sod3 gene transfer on HFD-fed animals on expression of genes involved in lipid and glucose metabolism. With the exception of Acc, the mRNA levels of genes involved in de novo lipogenesis in the liver including Srebp1c, Fas and Scd1 are higher in the two HFD-fed animal groups (Figure 6c). mRNA levels of genes involved in lipolysis and energy expenditure including Cpt1α, Cpt1β, Pgc1α, Pgc1β and Ucp2 are significantly higher in animals who underwent Sod3 gene transfer than those of HFD-fed control animals (Figure 6d). Expression of genes involved in glucose metabolism such as G6p, a key gene for gluconeogenesis, is decreased in HFD-fed mice, while mRNA levels of Pdk4, a key gene repressing glucose oxidation, is lower in animals who underwent Sod3 gene transfer compared to that of HFD-fed control mice. The mRNA levels of other genes involving in glucose metabolism such as Glut4 and Pepck are not statistically different among the three groups of animals (Figure 6e).
Figure 6. Sod3 gene transfer reduces the inflammation and enhances the energy expenditure.
Animals were sacrificed at the end of 8 week feeding and tissue from white adipose tissue and liver were collected for RNA isolation. mRNA levels of selected gene were quantified by Real Time PCR. (a) Relative mRNA levels of inflammation related genes in adipose tissue; (b) Relative mRNA levels of anti-inflammatory genes in adipose tissue; (c) Relative mRNA levels of genes involved in de novo lipogenesis in the liver; (d) Relative mRNA levels of genes involved in lipolysis and energy metabolism in the liver; (e) Relative mRNA levels of genes involved in glucose metabolism in the liver. (□), animals fed a regular Chow; (
 ), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
), high fat diet fed mice injected with pLIVE-SEAP; (■), high fat diet-fed mice injected with pLIVE-SOD3. Data represent mean ± SEM (n=5). * p<0.05 compared with chow mice; # p<0.05 compared with pLIVE-SEAP injected animals.
Discussion
Chronic inflammation is closely associated with obesity, and an increase in fat intake fuels the ROS generation in the adipose tissue and boosts macrophage infiltration.2 We have previously shown that intraperitoneal injections of clodronate liposomes to eliminate macrophages blocks HFD-induced weight gain and insulin resistance,11 although the exact molecular mechanism responsible for the response is not fully understood. In the current work, we investigate the impacts of overexpression of the Sod3 gene focusing the influence of SOD3 on HFD-induced obesity and development of insulin resistance and fatty liver. We used a hydrodynamics-based procedure to obtain a high level of SOD3 gene expression and clear responses in adipose tissue and the liver. Real Time-PCR analysis showed strong Sod3 gene expression in the liver (Figure 1a), resulting in sustained levels of SOD3 in the blood, over 900 IU/ml for the 8-week period of HFD feeding (Figure 1b). H&E staining of liver sections and examination of blood concentrations of ALT and AST confirm that the hydrodynamics-based procedure for gene transfer is safe (Figures 1c–e).
Sustained high levels of SODs resulted in suppression of HFD-induced weight gain in animals compared to control mice with transfer of pLIVE-SEAP plasmid (Figure 2a). Results from an MRI analysis show that lack of HFD-induced weight gain is associated with lower fat mass with Sod3 gene transfer (Figure 2c), in agreement with the observation of smaller size of white adipose tissue and smaller diameter of adipocytes compared to that of control animals injected with the control plasmid (Figures 3a–3d). These results suggest that the function of SOD3 is specific in blocking adipogenesis.
It has been previously shown that pro-inflammatory cytokines such as TNFα, MCP1 and IL6 are associated with the development of obesity, and anti-inflammatory cytokines, such as adiponectin, inhibit the process.12 At the pro-inflammatory state, macrophages are converted from the M2 (anti-inflammatory state) to M1 type (inflammatory state). M1 macrophages, which express F4/80+/CD11c+, will surround the hypertrophic or necrotic adipocytes and form crown-like structures, causing the adipose tissue remodeling and pathological fat pad expansion.13 In full agreement with these previous studies, our data show that in HFD-fed control mice, there were more inflammatory macrophages in the adipose tissue (Figures 3c, 3e) with an increase in transcription of pro-inflammation marker genes such as Tnfα, Mcp1, Il6, F4/80 and Cd11c (Figure 6a). In contrast, mice transferred with Sod3 gene showed a lower mRNA level of the same set of genes and an elevation of adiponectin and Il4 expression (Figure 6b). Adiponectin has the anti-inflammatory effect and is capable of suppressing NF-κB signal transduction,14 consequently up-regulating the anti-inflammatory cytokine IL10 expression15 and down-regulating the pro-inflammatory cytokine TNFα expression.16 It can also maintain the macrophages with M2 type characteristics,17 a function similar to that of IL4.18 Together, this set of data suggests that overexpression of SOD3 elevated the level of transcription of anti-inflammatory genes and blocked chronic inflammation in adipose tissue.
Adipose tissue inflammation is a critical determinant leading to the development of insulin resistance and type 2 diabetes.19 Insulin resistance results in the decrease of glucose oxidation, which leads to glucose intolerance and hyperinsulinemia. Previous studies have shown that the plasma SOD3 level was inversely related to fasting glucose level, body mass index (BMI) and insulin resistance.6 A study from Tamai et al. has shown that the genetic variant of the Sod3 gene was associated with insulin resistance and the susceptibility to type 2 diabetes; a missense mutation (Ala40Thr) showed diagnosis of type 2 diabetes at an earlier age and lower insulin sensitivity.20 Overexpression of SOD3 and other two endogeneous antioxidant enzymes (SOD1 and glutathione peroxidase) protected pancreatic islets from oxidative stress.21 Factors such as the C/EBP enhancer, which regulate the expression of genes involved in insulin resistance could also elevate gene expression and protein levels of SOD3,22 which was thought a compensatory effect for improving insulin sensitivity. Our data are in agreement with the conclusions derived from these previous studies and are in support that overexpression of the Sod3 gene could prevent the HFD-induced insulin resistance (Figure 5).
Fatty liver is a common complication of obesity. Previous studies have shown that the SOD3 level is associated with fatty liver and related diseases. Madan et al. showed SOD3 levels in serum decreased in the patients with fatty liver disease compared to the health control.23 Similar results were obtained in obese rats.24 Inversely, injection of high doses of adenoviral SOD3 vectors (3 × 1010 PFU) protected the liver from injury induced by oxidative stress in rats.25 Some antioxidants such as resveratrols decreased the severity of non-alcoholic fatty liver disease in rats, which was accompanied with elevation of SOD3.26 High SOD3 levels also provided beneficial effects against non-alcoholic steatohepatitis27 and liver cirrhosis.28 In accordance with these data, our results demonstrate that overexpression of the Sod3 gene suppresses the ectopic fat accumulation in the liver and protects the liver from HFD-induced development of fatty liver (Figure 4).
Previous studies have shown that adiponectin gene expression was significantly reduced in ob/ob mice,29 monkeys30 and humans.31 A decrease in serum levels of adiponectin correlates with insulin resistance and glucose intolerance.32 Adiponectin, in addition to the anti-inflammation effect, has beneficial effects on the metabolic disorders. It can modulate the expression of Cpt1,33 Pgc1,34 and Ucp2,35 a set of genes involved in energy expenditure. Our results in Figure 6d demonstrate that SOD3 overexpression significantly increased expression of Cpt1α, Cpt1β, Pgc1α, Pgc1β and Ucp2, genes responsible for energy expenditure. No statistical difference was seen in Sod3 injected and HFD-fed control animals with respect to the mRNA levels of genes involved in cholesterol synthesis (Srebp1c), fatty acid biosynthesis (Acc, Fas, Scd1), and glucose metabolism (G6p, Glut4, Pepck). Significant elevation in mRNA levels of the Pdk4 gene was seen in HFD-fed control mice and its level was reduced by Sod3 gene transfer. Further studies are needed to identify genes and metabolic pathways by which the SOD applies its effect on the prevention of HFD-induced weight gain, insulin resistance and the development of a fatty liver.
In summary, in this study, we demonstrate that overexpression of the Sod3 gene produces a variety of beneficial effects against diet-induced obesity and its complications, including fatty liver and insulin resistance. The protective effect is achieved by suppressing expression of genes responsible for inflammation in adipose tissue and increasing expression of genes for energy expenditure. Our findings also demonstrate that overexpression of SOD3 via gene transfer could result in a protective effect against the development of obesity and obesity-associated insulin resistance and fatty liver and be considered an effective approach in preventing HFD-induced obesity. If same level of SOD3 gene expression can be achieved in human, the strategy employed in this study could lead to effective prevention of obesity resulted from a high fat diet.
Materials and Methods
Plasmid vectors
Plasmids carrying either mouse Sod3 or secreted embryonic alkaline phosphotase (SEAP) gene were cloned into a pLIVE plasmid vector from Mirus Bio (Madison, WI). The mouse Sod3 gene was purchased from Thermo Fisher Scientific Biosciences (Middletown, VA), digested with restriction enzymes Sac I and Xho I, and the Sod3 containing fragment inserted into multiple cloning sites of the pLIVE vector. The insertion in the new plasmid was confirmed by DNA sequencing (University of Georgia Genomics Facility). The plasmids were isolated using the method of cesium chloride-ethidium bromide gradient centrifugation and kept in saline at 80 °C until use.
Animals and animal experiments
C57BL/6 mice (male, 9 weeks old, 24~26 g) were purchased from Charles River Laboratories (Wilmington, MA) and housed under standard conditions with a 12 h–12 h light-dark cycle. The high fat diet (60% calories from fats, 20% from carbohydrates and 20% from proteins) was purchased from Bio-Serv (Frenchtown, NJ; #F3282). Three groups of mice were employed (n=5 for each), one group was fed normal chow, and the remaining two groups were fed a HFD. HFD-fed animals received hydrodynamic injections of either pLIVE-SEAP plasmid (control) or pLIVE-SOD3 plasmids. Mice were fed continuously for 8 weeks and the body weight and composition, and food intake were measured weekly. One week before being sacrificed, animals were fasted for 6 h followed by a glucose tolerance test. The same animals were allowed to rest for two days before being fasted for 4 h prior to an insulin sensitivity test. Animals were sacrificed at end of the 8th week and blood and various tissue samples were collected for biochemical and histological examinations.
Assessment of sod3 gene expression
Animals were hydrodynamically injected via the tail vein with 10 μg/mouse of pLIVE-SOD3 or pLIVE-SEAP plasmid DNA per mouse on day one according to the standard procedure.36 Approximately 50 μl of blood was collected from the tail using Microvette (Newton, NC) at different times and serum was prepared. SOD3 activity in the blood was determined using a previously established procedure by McCord et al.37 Reagents for the SOD3 enzyme assay including xanthine, xanthine oxidase, cytochrome C3+ and SOD standard were purchased from Sigma (St. Louis, MO).
Real Time-PCR for mRNA analysis
Animals were sacrificed at desirable times and the heart, lung, liver, spleen, kidneys and adipose tissues were collected. TRIzol reagents from Invitrogen (Carlsbad, CA) were used for extraction of total RNA from all tissues with the exception of the adipose tissue where the specialized kits from QIAGEN (Valencia, CA) for RNA isolation were used. cDNA was synthesized using a First Strand cDNA Synthesis System for Quantitative RT-PCR, the commercial kits from OriGene (Rockville, MD). RT-PCR was performed with SYBR Green as detection reagent and the ΔΔCt method for data analysis.38 GAPDH mRNA was used as an internal control. All primers (Table 1) were ordered from Sigma (St. Louis, MO). Melting curve analysis of all quantitative PCR products was performed and showed a single DNA duplex.
Table 1.
Primer sequences for Real Time PCR analysis of gene expression
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Acc | ATGGGCGGAATGGTCTCTTTC | TGGGGACCTTGTCTTCATCAT |
| Cd11c | CTGGATAGCCTTTCTTCTGCTG | GCACACTGTGTCCGAACTCA |
| Cpt1α | CTCCGCCTGAGCCATGAAG | CACCAGTGATGATGCCATTCT |
| Cpt1β | GCACACCAGGCAGTAGCTTT | CAGGAGTTGATTCCAGACAGGT |
| F4/80 | TGACTCACCTTGTGGTCCTAA | CTTCCCAGAATCCAGTCTTTCC |
| Fas | GGAGGTGGTG ATAGCCGGTAT | TGGGTAATCCATAGAGCCCAG |
| G6p | CGACTCGCTATCTCCAAGTGA | GTTGAACCAGTCTCCGACCA |
| Glut4 | GTGACTGGAACACTGGTCCTA | CCAGCCACGTTGCATTGTAG |
| Ifnγ | ATGAACGCTACACACTGCATC | CCATCCTTTTGCCAGTTCCTC |
| Il1β | GCAACTGTTCCTGAACTCAACT | ATCTTTTGGGGTCCGTCAACT |
| Il6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| Mcp1 | TTAAAAACCTGGATCGGAACCAA | GCATTAGCTTCAGATTTACGGGT |
| Pdk4 | AGGGAGGTCGAGCTGTTCTC | GGAGTGTTCACTAAGCGGTCA |
| Pepck | CTGCATAACGGTCTGGACTTC | CAGCAACTGCCCGTACTCC |
| Pgc1α | TATGGAGTGACATAGAGTGTGCT | CCACTTCA ATCCACCCAGAAAG |
| Pgc1β | TCCTGTAAAAGCCCGGAGTAT | GCTCTGGTAGGGGCAGTGA |
| Scd1 | TTCTTGCGATACACTCTGGTGC | CGGGATTGAATGTTCTTGTCGT |
| Sod3 | AGGTGGATGCTGCCGAGAT | TCCAGACTGAAATAGGCCTCAAG |
| Srebp-1c | GCAGCCACCATCTA GCCTG | CAGCAGTGAGTCTGCCTTGAT |
| Tnfα | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| Ucp-1 | AGGCTTCCAGTACCATTAGGT | CTGAGTGAGGCAAAGCTGATTT |
| Ucp-2 | ATGGTTGGTTTCAAGGCCACA | CGGTATCCAGAGGGAAAGTGAT |
| Ucp-3 | CTGCACCGCCAGATGAGTTT | ATCATGGCTTGAAATCGGACC |
Determination of plasma concentrations of alanine aminotransferase (ALT), aspartate aminotransferase (AST) and insulin
Blood samples were collected from heart cavities immediately after euthanizing mice. Sera were isolated by centrifugation at 5,000 rpm for 5 min. ALT and AST concentrations were determined following the manufacture’s instruction (Thermo Scientific, Middletown, VA). Insulin levels in the blood were measured using a commercial ELISA kit from Mercodia Inc. (Winston Salem, NC).
Measurement of triglycerides (TG), total cholesterol (TC) and non-esterified fatty acid (NEFA) in the liver
The measurement was performed according to the Folch method.39 In brief, approximately 100 mg of liver sample were freshly collected and homogenized in a tube containing 20x of liver volume of solution consisting of chloroform and methanol (2:1). Tissue homogenates were incubated at 4°C overnight and centrifuged at 12,000 rpm for 20 min. Supernatants were collected, air dried and dissolved in 1% Triton-isopropanol. The amounts of TG, TC and NEFA were determined using commercial kits from Thermo-Scientific (Middletown, VA), Genzyme (Boston, MA), and Wako (Tokyo, Japan), respectively.
Intra-peritoneal glucose tolerance test (IPGTT) and insulin tolerance test (ITT)
Mice were fasted for 6 h before IPGTT. Glucose in 0.9% saline was injected (i.p.) at 2.0 g/kg, and the time-point was set as 0 min. Blood glucose levels at 0, 30, 60 and 120 min were measured using glucose test strips and glucose meters. For ITT, mice were fasted for 4 h prior to injection of insulin (Humulin, 0.8 U/kg) from Eli Lilly (Indianapolis, IN), and blood glucose was measured at 0, 30, 60 and 120 min after insulin injection. A homeostasis model for assessing insulin resistance (HOMA-IR) was employed to define the degree of insulin resistance. [HOMA-IR=fasting glucose (mg/dl) * fasting insulin (μg/L)/405].
H&E staining
Freshly collected tissues from animals were fixed overnight in 10% neutral formalin solution. After dehydration and embedding, tissue sections were made at 5~7 μm in thickness and dried at 37°C for 1 h. H&E staining was performed according to the instruction of a commercial kit from BBC Biochemical (Atlanta, GA). Tissue slides were examined using an optical microscope and diameters of adipocytes in white adipose tissue were measured using the NIS-Elements imaging platform from Nikon Instruments Inc. (Melville, NY).
Oil-red O staining
Freshly collected liver samples were frozen in liquid nitrogen. Frozen sections were made using a Cryostat at 8 μm in thickness and fixed in 10% neutral buffered formalin for 30 min. Slides were washed with phosphate-buffered saline and rinsed with 60% isopropanol for 5 min before being stained with freshly prepared Oil-red O working solution from Electron Microscopy Sciences (Hatfield, PA) for 30 min. Isopropanol (60%) was used to rinse the sections for 5 min followed by counterstain with hemotoxylin for 1 min. An optical microscope (ECLIPSE Ti; Nikon, Melville, NY) was employed to examine the slides and selected structures were photographed.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance, and a p value below 0.05 (p<0.05) is considered statistically different. Data were expressed as mean ± SEM.
Acknowledgments
The study was supported in part by grants from NIH (RO1EB007357 and RO1HL098295). Ran Cui is a recipient of Scholarship (No. 201306260063) from the China Scholarship Council. The authors would like to thank Ms. Ryan Fugett for proofreading and English editing of the manuscript.
Footnotes
Conflict Interest
The authors declare no conflict interest.
References
- 1.Matsuda M, Shimomura I. Increased oxidative stress in obesity: implications for metabolic syndrome, diabetes, hypertension, dyslipidemia, atherosclerosis, and cancer. Obes Res Clin Pract. 2013;7:e330–e341. doi: 10.1016/j.orcp.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114:1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H, Jin X, Lam K, Yan SK. Oxidative stress and diabetes mellitus. Clin Chem Lab Med. 2011;49:1773–1782. doi: 10.1515/CCLM.2011.250. [DOI] [PubMed] [Google Scholar]
- 4.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10:453–471. doi: 10.1038/nrd3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marklund SL, Nilsson P, Israelsson K, Schampi I, Peltonen M, Asplund K. Two variants of extracellular superoxide dismutase: relationship to cardiovascular risk factors in an unselected middle_aged population. J Intern Med. 1997;242:5–14. doi: 10.1046/j.1365-2796.1997.00160.x. [DOI] [PubMed] [Google Scholar]
- 6.Adachi T, Inoue M, Hara H, Maehata E, Suzuki S. Relationship of plasma extracellular-superoxide dismutase level with insulin resistance in type 2 diabetic patients. J Endocrinol. 2004;181:413–417. doi: 10.1677/joe.0.1810413. [DOI] [PubMed] [Google Scholar]
- 7.Isogawa A, Yamakado M, Yano M, Shiba T. Serum superoxide dismutase activity correlates with the components of metabolic syndrome or carotid artery intima-media thickness. Diabetes Res Clin Pract. 2009;86:213–218. doi: 10.1016/j.diabres.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann TG, Wheeler MD, Froh M, Schwabe RF, Bunzendahl H, Samulski RJ, et al. Effects of three superoxide dismutase genes delivered with an adenovirus on graft function after transplantation of fatty livers in the Rats. Transplantation. 2003;76:28–37. doi: 10.1097/01.TP.0000065299.29900.17. [DOI] [PubMed] [Google Scholar]
- 9.Laurila JP, Castellone MD, Curcio A, Laatikainen LE, Haaparanta-Solin M, Gronroos TJ, et al. Extracellular superoxide dismutase is a growth regulatory mediator of tissue injury recovery. Mol Ther. 2009;17:448–454. doi: 10.1038/mt.2008.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottfredsen RH, Goldstrohm DA, Hartney JM, Larsen UG, Bowler RP, Petersen SV. The cellular distribution of extracellular superoxide dismutase in macrophages is altered by cellular activation but unaffected by the naturally occurring R213G substitution. Free Radic Biol Med. 2014;69:348–356. doi: 10.1016/j.freeradbiomed.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bu L, Gao M, Qu S, Liu D. Intraperitoneal injection of clodronate liposomes eliminates visceral adipose macrophages and blocks high-fat diet-induced weight gain and development of insulin resistance. AAPS J. 2013;15:1001–1011. doi: 10.1208/s12248-013-9501-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteiro R, Azevedo I. Chronic inflammation in obesity and the metabolic syndrome. Mediators Inflamm. 2010 doi: 10.1155/2010/289645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi N, Argueta JGM, Masuhiro Y, Kagishita M, Nonaka K, Saito T, et al. Adiponectin inhibits Toll-like receptor family-induced signaling. FEBS lett. 2005;579:6821–6826. doi: 10.1016/j.febslet.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Kumada M, Kihara S, Ouchi N, Kobayashi H, Okamoto Y, Ohashi K, et al. Adiponectin specifically increased tissue inhibitor of metalloproteinase-1 through interleukin-10 expression in human macrophages. Circulation. 2004;109:2046–2049. doi: 10.1161/01.CIR.0000127953.98131.ED. [DOI] [PubMed] [Google Scholar]
- 16.Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 17.Ohashi K, Parker JL, Ouchi N, Higuchi A, Vita JA, Gokce N, et al. Adiponectin promotes macrophage polarization toward an anti-inflammatory phenotype. J Biol Chem. 2010;285:6153–6160. doi: 10.1074/jbc.M109.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, et al. Fat Treg cells: a liaison between the immune and metabolic systems. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamai M, Furuta H, Kawashima H, Doi A, Hamanishi T, Shimomura H, et al. Extracellular superoxide dismutase gene polymorphism is associated with insulin resistance and the susceptibility to type 2 diabetes. Diabetes Res Clin Pract. 2006;71:140–145. doi: 10.1016/j.diabres.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Mysore TB, Shinkel TA, Collins J, Salvaris EJ, Fisicaro N, Murray-Segal LJ, et al. Overexpression of glutathione peroxidase with two isoforms of superoxide dismutase protects mouse islets from oxidative injury and improves islet graft function. Diabetes. 2005;54:2109–2116. doi: 10.2337/diabetes.54.7.2109. [DOI] [PubMed] [Google Scholar]
- 22.Adachi T, Inoue M, Hara H, Suzuki S. Effects of PPARgamma ligands and C/EBPbeta enhancer on expression of extracellular-superoxide dismutase. Redox Rep. 2004;9:207–12. doi: 10.1179/135100004225005985. [DOI] [PubMed] [Google Scholar]
- 23.Madan K, Bhardwaj P, Thareja S, Gupta SD, Saraya A. Oxidant stress and antioxidant status among patients with nonalcoholic fatty liver disease (NAFLD) J Clin Gastroenterol. 2006;40:930–935. doi: 10.1097/01.mcg.0000212608.59090.08. [DOI] [PubMed] [Google Scholar]
- 24.Carmiel-Haggai M, Cederbaum AI, Nieto N. A high-fat diet leads to the progression of non-alcoholic fatty liver disease in obese rats. FASEB J. 2005;19:136–138. doi: 10.1096/fj.04-2291fje. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler MD, Katuna M, Smutney OM, Froh M, Dikalova A, Mason RP, et al. Comparison of the effect of adenoviral delivery of three superoxide dismutase genes against hepatic ischemia-reperfusion injury. Hum Gene Ther. 2001;12:2167–2177. doi: 10.1089/10430340152710513. [DOI] [PubMed] [Google Scholar]
- 26.Bujanda L, Hijona E, Larzabal M, Beraza M, Aldazabal P, García-Urkia N, et al. Resveratrol inhibits nonalcoholic fatty liver disease in rats. BMC Gastroenterol. 2008 doi: 10.1186/1471-230x8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurent A, Nicco C, Tran Van Nhieu J, Borderie D, Chéreau C, Conti F, et al. Pivotal role of superoxide anion and beneficial effect of antioxidant molecules in murine steatohepatitis. Hepatology. 2004;39:1277–1285. doi: 10.1002/hep.20177. [DOI] [PubMed] [Google Scholar]
- 28.Laviña B, Gracia-Sancho J, Rodríguez-Vilarrupla A, Chu Y, Heistad DD, Bosch J, et al. Superoxide dismutase gene transfer reduces portal pressure in CCl4 cirrhotic rats with portal hypertension. Gut. 2009;58:118–125. doi: 10.1136/gut.2008.149880. [DOI] [PubMed] [Google Scholar]
- 29.Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 30.Hotta K, Funahashi T, Bodkin NL, Ortmeyer HK, Arita Y, Hansen BC, et al. Circulating concentrations of the adipocyte protein adiponectin are decreased in parallel with reduced insulin sensitivity during the progression to type 2 diabetes in rhesus monkeys. Diabetes. 2001;50:1126–1133. doi: 10.2337/diabetes.50.5.1126. [DOI] [PubMed] [Google Scholar]
- 31.Diez JJ, Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol. 2003;148:293–300. doi: 10.1530/eje.0.1480293. [DOI] [PubMed] [Google Scholar]
- 32.Menzaghi C, Trischitta V, Doria A. Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56:1198–1209. doi: 10.2337/db06-0506. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Wu L, Wang C, Liu L, Zhao Y. Adiponectin modulates carnitine palmitoyltransferase-1 through AMPK signaling cascade in rat cardiomyocytes. Regul Pept. 2007;139:72–79. doi: 10.1016/j.regpep.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, et al. Adiponectin and AdipoR1 regulate PGC-1 [agr] and mitochondria by Ca2+ and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 35.Zhou M, Xu A, Tam PK, Lam KS, Huang B, Liang Y. Upregulation of UCP2 by adiponectin: the involvement of mitochondrial superoxide and hnRNP K. PloS One. 2012;7:e32349. doi: 10.1371/journal.pone.0032349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Song YK, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 37.McCord JM, Fridovich I. Superoxide dismutase an enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]