Abstract
Objective
Statins stimulate transcription of proprotein convertase subtilisin/kexin type 9 (PCSK9), a negative regulator of the low-density lipoprotein receptor (LDLR), thus blunting the cholesterol-lowering effects of statin treatment. Though there is inter-individual variation in PCSK9 statin response, little is known about ancestral and other genetic factors that could contribute to this variation.
Methods
We measured plasma PCSK9 before and after six weeks of 40 mg/day simvastatin in 901 participants of the Cholesterol and Pharmacogenetics (CAP) clinical trial and tested phenotypic and genetic factors for correlation with PCSK9 statin response.
Results
Statin-induced changes in plasma LDL-cholesterol, total cholesterol, and apolipoprotein B were all significantly correlated with statin-induced changes in PCSK9. A detailed examination of associations of genetic ancestry with PCSK9 statin response revealed that Ashkenazi Jews had smaller statin-induced increases in PCSK9 levels than other self-reported Caucasians (p=0.016). Using genome-wide association analysis, we found that the “G” minor allele of rs13064411 in the WD repeat domain 52 (WDR52) gene was significantly associated with greater statin-induced increases in plasma PCSK9 in CAP Caucasians (p=8.2×10−8).
Conclusions
Overall, these results suggest that genetic ancestry and rs13064411 genotype contribute to inter-individual variation in PCSK9 statin response in Caucasians.
Keywords: PCSK9, simvastatin, low-density lipoprotein cholesterol (LDL-C), ancestry, rs13064411, WDR52, Ashkenazi Jews
Introduction
Statins lower intracellular cholesterol levels by inhibiting 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), the rate-limiting cholesterol biosynthesis enzyme, thus stimulating transcription of sterol regulatory element binding transcription factor 2 (SREBF2; aka SREBP2) target genes, including the low-density lipoprotein receptor (LDLR) and proprotein convertase subtilisin/kexin type 9 (PCSK9) [1]. Since PCSK9 plays a role in the degradation of LDLR [2], upregulation of PCSK9 partially interferes with the increased clearance of LDL particles from the blood by LDLR following statin treatment. This has made PCSK9 a therapeutic target for lowering LDL-cholesterol (LDL-C) levels and cardiovascular disease risk in individuals treated with statins further than is possible with statin treatment alone [3–5] or in individuals who are not treated with statins [6,7].
In the endogenous state, plasma PCSK9 protein levels are positively correlated with LDL-C and total cholesterol (TC) levels [8], and these correlations have been reported to be stronger in men than women [9]. However, this intrinsic correlation between plasma PCSK9 and LDL-C levels is abolished by statin treatment [10,11]. The statin-induced changes in plasma PCSK9 have been reported to be inversely correlated with baseline PCSK9 concentrations [12] and statin-induced change of plasma LDL-C levels [11] and to depend on the dose and duration of statin treatment [13]. We have shown that LDL-C response to statins differs between populations of Caucasian and African American ancestry [14], but the potential effects of ancestral background on PCSK9 statin response have not been evaluated. Furthermore, though a modest number of genetic loci have been associated with LDL-C statin response to date [15–18], no genetic factors have been identified that contribute to the substantial inter-individual variation in plasma PCSK9 statin response [10–12,19].
In the present study, we measured plasma PCSK9 protein levels in 901 participants of the Cholesterol and Pharmacogenetics (CAP) clinical trial [14] at baseline and after six weeks of treatment with 40 mg/day simvastatin. Using genome-wide SNP genotypes and other clinical measurements, we investigated genetic predictors of plasma PCSK9 response to statin as well as the correlation of PCSK9 statin response with lipid phenotypes to better understand the basis for inter-individual variation in statin response of this protein.
Methods
Study population and serum lipid and lipoprotein measurements
The CAP trial (NCT00451828) of 40 mg/day simvastatin for 6 weeks and the characteristics of the study population (N=944) have been described previously [14]. Importantly, participants were classified as African American if they self-reported at least three African American grandparents and were classified as Caucasian if they self-reported at least three Caucasian (white) grandparents. Serum total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), triglycerides (Tg), high-density lipoprotein cholesterol (HDL-C), and apolipoprotein B (APOB) were measured using standard techniques [14].
PCSK9 protein measurements in serum
Levels of PCSK9 protein were measured in plasma samples isolated from 901 CAP participants before and after six weeks of 40mg/day simvastatin treatment by BG Medicine Inc. in collaboration with Merck using a colorimetric ELISA assay modified from Merck’s DELFIA assay [20]. Plates were coated overnight at 4°C and then blocked for one hour at room temperature. Duplicate samples, controls, and calibrators were diluted 1:1000 prior to being incubated with Merck’s AX213 anti-PCSK9 antibody for one hour at 37°C with shaking. After four washes, the AX1-Biotin secondary antibody was added at a final concentration of 1 μg/ml for one hour at room temperature with shaking. After washing, Streptavidin-HRP conjugate was added for 30 minutes without shaking at room temperature and then washed again four times. TMB substrate was added for 20 minutes at room temperature without shaking, and the reaction was terminated with stop solution before plates were read at 450 nm. To minimize experimental variation between samples taken at different time points from the same participant, all samples from each participant were located next to each other on the same plate. Two controls were included on each of the 52 plates that were run over 13 days, and the precision of these measurements was 10.06% at 1.34 nM and 10.53% at 4.67 nM.
Covariate and correlation statistical analyses
Covariates were individually tested for effects on phenotypes using Wilcoxan rank-sum test for binary covariates (sex, smoking status, and self-reported ethnicity) or Spearman’s rank-order correlation (ρ) for continuous covariates (age and BMI). Axes of ancestry variation were tested for correlation with phenotypes while correcting for other significant covariates using multiple linear regression or using Spearman’s rank-order correlation (ρ) when no other covariates were detected to have an effect on the phenotype. Correlations between lipid and PCSK9 phenotypes were calculated using Spearman’s rank-order correlation (ρ) and tested for interaction with sex using multiple linear regression with a sex by PCSK9 interaction term. Analyses were performed in JMP version 9.0.0.
Genotyping of CAP participants
The majority (N=587) of CAP self-reported Caucasians were genotyped using the Illumina HumanHap300 (N=305) or HumanHap610-Quad (N=282) beadchips as previously described [16]. Most self-reported African Americans (N=290) and self-reported Caucasians (N=549) were also genotyped at 196,726 SNPs using the Cardio-MetaboChip [21] in separate batches.
Ancestry analyses
Of the 196,475 autosomal Metabochip loci, 75,820 were not genotyped in all 839 individuals and 46,044 had a minor allele frequency less than 1%, leaving 112,218 completely genotyped common SNPs. Subsequently, SNPs that were correlated (r2>0.3) with at least one other SNP in a 150 SNP window (step size 15 SNPs) were eliminated using Plink [22], leaving 50,745 SNPs. Pairwise identity-by-state (IBS) distances between all individuals were computed and the resulting matrix of IBS distances was used to perform multidimensional scaling (MDS) analysis in Plink [22].
We generated a second MDS plot using 300K+ genome-wide genotypes from 587 CAP self-reported Caucasians along with publicly available genotype data of human genome diversity panel (HGDP) [23] (Yoruba, Maya, Han, and all European and Middle Eastern populations except the Mozabite) and Ashkenazi Jew [24] reference populations genotyped with the Illumina HumanHap650Y (N=367) or Illumina HumanHap610-Quad (N=18) platforms, respectively. Following the same SNP filtering criteria as above, 169,995 SNPs survived the filters and were used to generate the pairwise IBS matrix.
CAP self-reported Caucasians with genome-wide SNP data were classified based on the nearest reference population if they were sufficiently close to at least one reference population (<3 SD from the mean of the population along each of the first three ancestry axes). For this purpose, one outlying individual from the Bedouin population was excluded when calculating that population’s means and standard deviations. The closest reference population was selected for each relevant individual by minimizing the distance along the first three genome-wide ancestry axes. One-way analysis of variance (ANOVA) was used to compare the delta log PCSK9 values among population groups, and a two-tailed t-test was used to compare the Ashkenazi Jewish subset to the rest of the self-reported Caucasians.
Genome-wide association study and imputation
Starting with the 630,247 markers present on one or both genome-wide genotyping platforms (HumanHap300 or HumanHap610-Quad), we first filtered based on missingness and Hardy-Weinberg equilibrium at each marker on each platform separately. On the 610K-Quad platform, 31,018 markers were missing genotypes in over 5% of individuals, and 186 of the remaining markers were substantially out of Hardy-Weinberg equilibrium (p<1×10−6). On the 300K platform, 1,036 markers had greater than 5% missingness and 40 of the remaining markers were out of Hardy-Weinberg equilibrium (p<1×10−6). Removing markers that had greater than 5% missingness and/or were out of Hardy-Weinberg equilibrium on at least one platform left 600,688 markers. The genotype datasets from both platforms were then combined, all markers were tested for deviations from Hardy-Weinberg equilibrium again using the combined sample set (N=60 removed with p<1×10−6), and markers with minor allele frequencies under 1% (N=30,206) were removed before conducting a genome-wide association study on the remaining markers (N=570,422).
We used linear regression with an additive model to conduct the association analysis of the delta log PCSK9 phenotype from 562 self-reported Caucasians with the 570,422 remaining SNPs using the third genome-wide ancestry axis as a covariate in Plinkv1.07 [22]. Quantile-quantile plots and Manhattan plots were created based on standard methods. The Bonferroni threshold for genome-wide significance after correcting for 570,422 tests was 8.77×10−8 in our study.
Genotypes of 39,164 chromosome 3 SNPs that were not monomorphic in the CAP Caucasians or triallelic in dbSNP and were not A/T or C/G SNPs were used for imputation using the 1000G PhaseIv3 EUR haplotypes [25] as a reference. Phased chromosome 3 haplotypes of the 587 CAP Caucasians with genome-wide genotype data were estimated in MaCH using 20 rounds and 200 states [26]. Chromosome 3 genotypes were then imputed using minimac with 5 rounds and 200 states [27]. An association study of imputed SNP allelic dosages in the region surrounding rs13064411 on chromosome 3 with delta log PCSK9 was conducted using mach2qtl [26] with the third genome-wide ancestry axis as a covariate, and association results were filtered to exclude markers with an imputation R2<0.3.
Results
Influence of ancestry and other covariates on PCSK9 levels and statin response
Using measurements of serum PCSK9 before and after statin treatment in 901 participants of the CAP clinical trial, we first tested for covariate associations with statin-induced changes in plasma PCSK9. Though age, smoking status, and ancestry have been associated with statin-induced changes in plasma LDL-C in the CAP population [14], age, BMI, gender, smoking status, and self-reported ancestry (Caucasian vs. African American) did not significantly influence statin-induced changes in PCSK9. However, at baseline and during statin treatment, women, smokers, and self-reported African Americans had higher PCSK9 protein levels than men, non-smokers, and self-reported Caucasians, respectively (N=901; Supplementary Figure S1, Supplemental Digital Content 1, http://links.lww.com/FPC/A756 ).
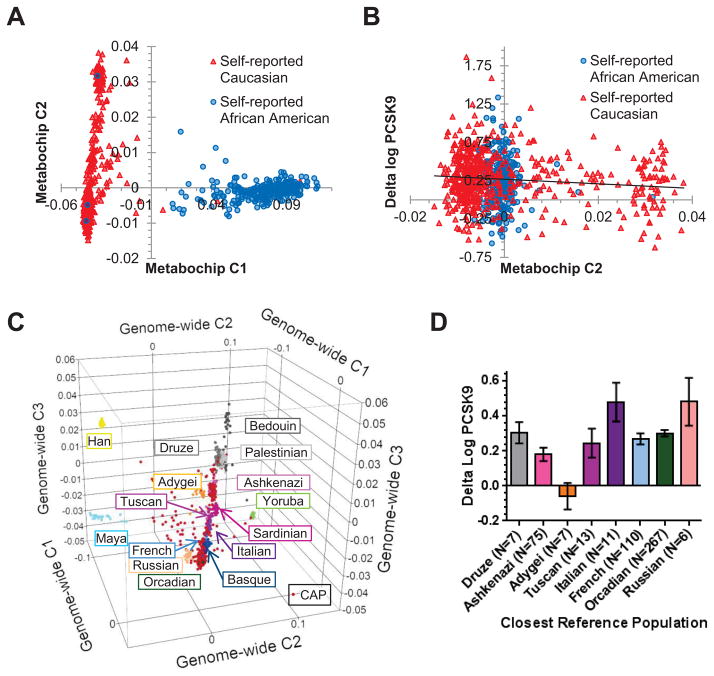
To more precisely evaluate the effect of ancestry, we used genetic data from the majority of our participants to determine if any major axes of ancestry variation were associated with statin-induced changes in plasma PCSK9 levels. We first used genotype data from the metabochip [21] to create a multidimensional scaling plot using PLINK [22] for 839 CAP participants (Figure 1A). The metabochip was chosen for this purpose because we did not have genotype data from a more comprehensive genome-wide platform available for the self-reported African American participants. We then tested whether the first two axes of ancestry variation from the multi-dimensional scaling plot were associated with statin-induced changes in plasma PCSK9 levels. The first dimension, C1, which separated the majority of the self-reported Caucasians from the self-reported African Americans, was not correlated with statin-induced changes in PCSK9 but was significantly correlated with baseline PCSK9 after correction for gender and smoking status (N=798, p=3.3×10−5) as expected from the self-reported ancestry findings. Notably, the second dimension, C2, which mainly separated the CAP self-reported Caucasians from one another, was significantly correlated with statin-induced changes (delta log) in plasma PCSK9 (N=798, Spearman’s ρ=−0.07, p=0.049, Figure 1B). For comparison, delta log LDL-C was correlated with the first ancestry dimension after correction for age and smoking status (N=798, p=0.013 but not with the second ancestry dimension (p>0.2), consistent with previous findings in CAP using the self-reported ancestry data [14].
Figure 1. Statin-induced changes in PCSK9 vary with ancestry in the CAP clinical trial population.
A) First two dimensions (C1,C2) of ancestry multi-dimensional scaling (MDS) plot for N=798 CAP participants using metabochip genotypes. B) Correlation of delta log PCSK9 with metabochip ancestry dimension two in CAP (Spearman’s ρ=−0.07, p=0.049, N=798). C) First three dimensions (C1–C3) of ancestry MDS plot for N=587 CAP self-reported Caucasians with selected Human Genome Diversity Panel and Ashkenazi Jewish reference populations. D) Delta log PCSK9 of CAP participants who were categorized as closest to different reference populations (ANOVA p=0.0016). Bars are mean ± SE.
To clarify distribution along the second dimension of the metabochip multi-dimensional scaling plot, we created a second multi-dimensional scaling plot with additional genome-wide SNP genotype data from the self-reported Caucasian participants (N=587) [16] compared to publicly available data from reference populations in the Human Genome Diversity Panel (N=367 individuals from 14 populations) [23] and from Ashkenazi Jews (N=18) [24]. On this plot, the first dimension, C1, separated the European and Middle Eastern populations from the African, Asian, and Native American populations and the second dimension, C2, separated the African from the Asian and Native American populations (Figure 1C, Supplementary Figure S2A, Supplemental Digital Content 2, http://links.lww.com/FPC/A757 ). The third dimension, C3, mainly served to separate European and Middle Eastern populations from one another along a north-south axis (Figure 1C; Supplementary Figure S2B, Supplemental Digital Content 2, http://links.lww.com/FPC/A757) and corresponded almost perfectly to the second dimension from the previous metabochip-based plot (r2=0.90, Supplementary Figure S2C, Supplemental Digital Content 2, http://links.lww.com/FPC/A757).
To assess the relative contributions of individual populations to the correlation of statin-induced changes in PCSK9 with ancestral variation along the European to Middle Eastern axis, we categorized all of the CAP Caucasians based on the reference populations that they were closest to, if any (Supplementary Figure S2D, Supplemental Digital Content 2, http://links.lww.com/FPC/A757). Only eight population categories contained more than five CAP self-reported Caucasians, and, as expected, average statin-induced changes in plasma PCSK9 differed significantly among them (ANOVA p=0.0016, Figure 2D). Furthermore, the 75 CAP individuals predicted to have Ashkenazi Jewish ancestry had significantly smaller increases in plasma PCSK9 levels with statin treatment than the rest of the CAP self-reported Caucasians on average (p=0.016, Supplementary Figure S2E, Supplemental Digital Content 2, http://links.lww.com/FPC/A757). For comparison, variation in statin-induced changes in plasma LDL-C did not differ among the major CAP Caucasian ancestry classification groups (ANOVA p>0.75).
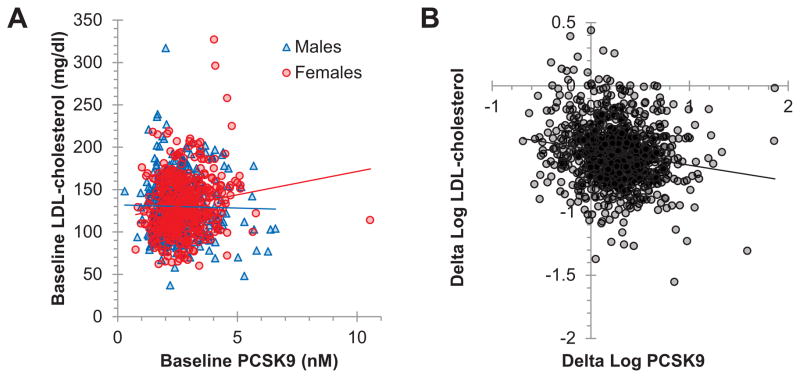
Figure 2. Correlations of baseline and statin-induced changes in LDL-cholesterol with PCSK9.
A) Endogenous levels of LDL-C and PCSK9 were measured in CAP serum samples (N=901) before simvastatin treatment and their correlations were calculated for the complete dataset (Spearman’s ρ=0.089, p=0.0073) and for males (Spearman’s ρ=0.032, p=0.50) and females (Spearman’s ρ=0.15, p=0.0021) separately. Trendlines from linear regression are shown for each gender. B) Statin-induced changes (log(Statin)-log(Baseline)) in LDL-C were compared to corresponding changes in PCSK9 using Spearman’s rank-order correlation (Spearman’s ρ= −0.16, p=8.7×10−7). For illustrative purposes, one point (coordinates 0.236, −2.773) was not shown, but this point was included in statistical analyses. Linear regression trendline is shown.
Correlation of PCSK9 and lipid levels before and after statin treatment
A modest correlation was observed between plasma PCSK9 and LDL-C levels at baseline in CAP (Spearman’s ρ=0.089, p=0.0073), but, in contrast to a previous report [9], this correlation was stronger in women (sex by PCSK9 interaction p=0.011, Figure 2A). The correlation between PCSK9 and LDL-C levels disappeared with statin treatment (p>0.9), consistent with previous observations [10,11]. As expected, statin-induced changes in PCSK9 levels were strongly inversely correlated with baseline PCSK9 levels (Spearman’s ρ= −0.45, p=8.8×10−46). Statin-induced changes in plasma PCSK9 levels were also significantly inversely correlated with statin-induced changes in LDL-C (Spearman’s ρ= −0.16, p=8.7×10−7) (Figure 2B, Table 1). Consistent with the results for LDL-C, we found that statin-induced changes in plasma total cholesterol and APOB, but not triglycerides or HDL-cholesterol, were correlated with statin-induced changes in plasma PCSK9 levels (Table 1). Average levels and statin-induced changes of these phenotypes in the CAP population are shown in Supplementary Table S1 (Supplemental Digital Content 3, http://links.lww.com/FPC/A758). We did not identify a statistically significant gender interaction for the correlation of statin-induced changes in LDL-C and PCSK9 (sex by PCSK9 interaction p=0.08).
Table 1.
Spearman’s correlations of statin-induced changes in plasma lipid phenotypes with statin-induced changes (delta log) in plasma PCSK9 in CAP (N=901).
| Lipid Phenotype | Spearman’s ρ | p |
|---|---|---|
| Delta log LDL-cholesterol | −0.1630 | 8.7×10−7 |
| Delta log APOB | −0.1443 | 1.4×10−5 |
| Delta log total cholesterol | −0.1357 | 4.4×10−5 |
| Delta log HDL-cholesterol | −0.0152 | 0.65 |
| Delta log triglycerides | −0.0007 | 0.98 |
Genome-wide association study of plasma PCSK9 statin response
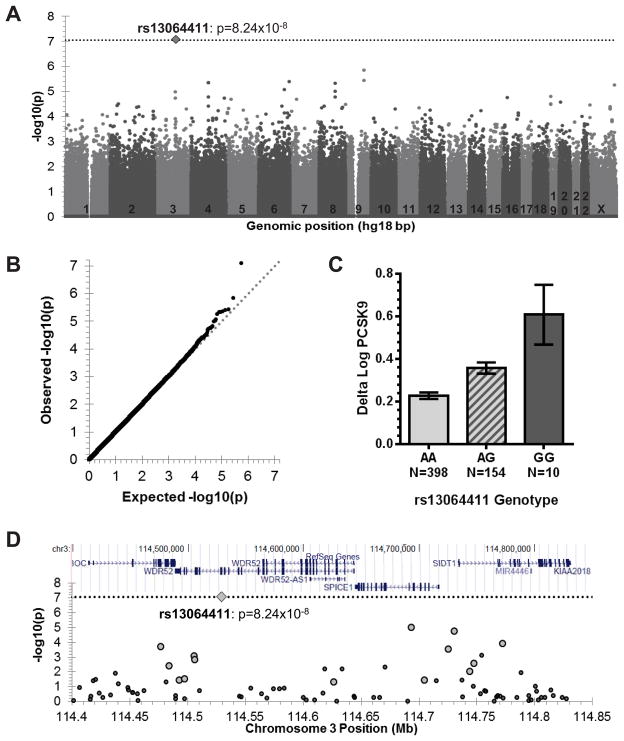
Given the inter-individual variation in plasma PCSK9 statin response and the association of ancestry with this response, we sought to identify specific genetic factors that influence the response by conducting a genome-wide association study in CAP self-reported Caucasian participants (N=562). Using the third genome-wide ancestry axis (C3) as a covariate, we detected evidence for association of rs13064411 on chromosome 3 with delta log PCSK9 (p=8.2×10−8, Bonferroni-adjusted p=0.047, Figures 3A–B, Tables 2 and S2, Supplemental Digital Content 4, http://links.lww.com/FPC/A759 ). The “G” minor allele of rs13064411 was associated with greater statin-induced increases in plasma PCSK9 levels in CAP self-reported Caucasians (Figure 3C, Supplementary Figures S3A–B, Supplemental Digital Content 5, http://links.lww.com/FPC/A760 ). Each rs13064411 “G” allele was associated with a mean statin-induced increase in plasma PCSK9 of approximately 0.4 nM or 23% after adjusting for the effects of the third genome-wide ancestry axis. The same allele was weakly associated with greater statin-induced decreases in plasma LDL-C levels (N=562, p=0.08, Supplementary Figures S3C-D, Supplemental Digital Content 5, http://links.lww.com/FPC/A760 ). In an attempt to replicate our results, we genotyped rs13064411 in the African American subset (N=287) of the CAP population. While there was a trend for greater statin-induced increases in PCSK9 levels in the 17 rs13064411 G/A heterozygotes compared to the remaining 270 A/A homozygotes, this difference was not statistically significant (p=0.22). However, our power to detect such an association was only 41%, assuming that the effect size was as large in African Americans as it was in the Caucasians. rs13064411 is a synonymous substitution in the WD repeat domain 52 (WDR52) gene (Figure 3D) and is in strong linkage disequilibrium with several SNPs in WDR52 and neighboring genes (Supplementary Figure S4, Supplemental Digital Content 6, http://links.lww.com/FPC/A761 ). Additional SNPs in this region were imputed using a 1000 Genomes Project European reference panel. The SNP with the strongest association with statin-induced increase in PCSK9 (rs66866534, p= 7.6×10−8) was in intron 2 of WDR52 (Supplementary Figure S5, Supplemental Digital Content 7 http://links.lww.com/FPC/A762 ).
Figure 3. GWAS of statin-induced changes in PCSK9.
A) Manhattan plot of delta log PCSK9 GWAS p- values. Dotted line indicates Bonferroni threshold for statistical significance of p=8.76×10−8. B) Quantile- quantile plot of observed versus expected GWAS p-values, with dotted line indicating y=x. Genomic control estimate was 1 for these data. C) Mean ± standard error of statin-induced changes in PCSK9 by rs13064411 genotype. D) RefSeq genes in genomic region surrounding rs13064411 taken from the UCSC genome browser (above) and the association of SNPs in this region with delta log PCSK9 (below). Larger symbols correspond to markers that are correlated with rs13064411 (r2>0.3).
Table 2.
Delta log PCSK9 GWAS results (p<1×10−5) using an additive model. Genome-wide ancestry dimension 3 was used as a covariate, and markers that were poorly genotyped, were substantially out of Hardy-Weinberg equilibrium, or had minor allele frequencies less than 1% were excluded. The Bonferroni-adjusted p-value for the rs13064411 association was 0.047.
| SNP | Chr | hg18 bp | Closest Gene | Effect Allele | Freq | N | Beta | P |
|---|---|---|---|---|---|---|---|---|
| rs13064411 | 3 | 114529330 | WDR52 | G | 15.3% | 562 | 0.142 | 8.24E-08 |
| rs16914931 | 9 | 100132137 | GABBR2 | A | 12.1% | 273 | 0.198 | 1.45E-06 |
| rs16914943 | 9 | 100134174 | GABBR2 | C | 14.4% | 272 | 0.180 | 3.75E-06 |
| rs4710191 | 6 | 167492532 | GPR31 | G | 43.3% | 560 | −0.0854 | 4.12E-06 |
| rs17336353 | 4 | 95735539 | PDLIM5 | G | 19.0% | 272 | 0.151 | 4.62E-06 |
| rs11097432 | 4 | 95798728 | PDLIM5 | C | 18.7% | 272 | 0.158 | 4.63E-06 |
| rs7814749 | 8 | 87790465 | CNGB3 | A | 37.1% | 562 | −0.0913 | 4.74E-06 |
| rs566103 | X | 144017211 | SPANXN1 | G | 23.5% | 560 | −0.12 | 5.73E-06 |
| rs6903961 | 6 | 140195104 | LOC100132735 | A | 16.7% | 562 | −0.111 | 8.69E-06 |
| rs1372170 | 8 | 87815035 | CNGB3 | A | 43.6% | 560 | −0.0853 | 9.73E-06 |
Discussion
We report here that Ashkenazi Jewish ancestry is associated with attenuation of statin-induced increases in plasma PCSK9 levels. We also find a genome-wide significant association with genetic variation in the WD repeat domain 52 (WDR52) locus, with the minor allele correlating with enhanced statin-induced increases in PCSK9. In addition, we confirm that simvastatin-induced changes in plasma PCSK9 are inversely correlated with changes in plasma LDL-cholesterol, total cholesterol, and APOB in participants of the CAP clinical trial.
To our knowledge, this is the first study that reveals an influence of ancestral background on the magnitude of statin-induced changes in plasma PCSK9 levels. This highlights the importance of using genetic data to generate a detailed picture of ancestral substructure within a study population to determine the effects, if any, of ancestral background on phenotypes of interest. Though ancestry self-report information can be useful, especially in studies for which genetic data is limited or unavailable, our findings illustrate some of the limitations of ancestry self-report data, both in terms of accuracy and precision. For instance, we find that PCSK9 statin response varies among self-reported Caucasian populations (Figure 1D), illustrating that even subtle differences in ancestry can affect drug response phenotypes. Different drug response phenotypes can be affected differently by ancestry, as illustrated by the fact that, unlike PCSK9 statin response, LDL-cholesterol statin response does not differ among Caucasians but does differ between Caucasians and African Americans [14]. The difference in PCSK9 statin response that we observe between self-reported Caucasians with and without Ashkenazi Jewish ancestry likely arises from multiple underlying genetic differences that have accumulated during the period of genetic isolation of the Ashkenazi Jewish population [28]..
Though rs13064411 is located in an exon of WDR52, it is in substantial linkage disequilibrium with SNPs in at least two other nearby genes, spindle and centriole associated protein 1 (SPICE1) and SID1 transmembrane family, member 1 (SIDT1), and may have longer range effects on other genes if it tags genetic variation that influences a regulatory element. Further studies such as replication in an independent population and functional follow-up are necessary to validate the relationship between the WDR52 locus and PCSK9 statin response, as well as to identify the gene(s) underlying this association. Though the association did not replicate in our African American cohort, the low minor allele frequency and small sample size of this population makes it difficult to determine if this was due to true lack of a relationship or to a lack of power. In addition, there could be genetic modifiers that differ between populations of African and European ancestry that could influence the strength of the association.Since plasma LDL-C is a well-established marker of cardiovascular disease risk, in this study we determine its correlation with PCSK9 levels at baseline and after statin treatment, as well as the correlation of statin-induced changes in both measures. Consistent with previous observations, there is a modest correlation between PCSK9 and LDL-C at baseline that is abolished by statin treatment [10,11]. In addition, we find a significant interaction of gender with this correlation, with the two measurements more tightly related in women than men. Interestingly, while others have also observed a gender interaction, the correlation was reported to be stronger in men than in women [9]. It is possible that different proportions of pre- and post-menopausal women or other unknown covariates are affecting the correlations differently in each study and thus analysis of additional independent populations will be necessary to ascertain a true gender discordance in the PCSK9:LDL-C relationship. As has been observed previously, we also see significant correlations of statin-induced changes in PCSK9 with changes in LDL-C and related measures in CAP, but these do not differ significantly by gender as was previously observed [29].
Overall, this study shows that simvastatin-induced changes in PCSK9 are inversely correlated with simvastatin-induced changes in LDL-C and are influenced by underlying genetic variation, both globally in terms of ancestral background and locally in terms of genetic variation at the WDR52 locus.
Supplementary Material
Acknowledgments
We would like to thank Joshua D. Smith, Mark Rieder, and Deborah Nickerson for performing the 300K and 610K-Quad genotyping, Kent Taylor and Mark Goodarzi for performing the Metabochip genotyping, and Merck and BG Medicine for quantification of plasma PCSK9. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This work was funded by an investigator-initiated grant from Merck, NHLBI U19 HL069757, NHLBI R01 HL104133, NHLBI T32 HL098057, and AHA post-doctoral fellowship 12POST10430005.
Footnotes
Conflicts of interest
Dr. Krauss has received research funding from Sanofi-Regeneron.
References
- 1.Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, et al. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 2.Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, et al. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 3.Raal F, Scott R, Somaratne R, Bridges I, Li G, Wasserman SM, et al. Low-density lipoprotein cholesterol-lowering effects of AMG 145, a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 serine protease in patients with heterozygous familial hypercholesterolemia: the Reduction of LDL-C with PCSK9 Inhibition in Heterozygous Familial Hypercholesterolemia Disorder (RUTHERFORD) randomized trial. Circulation. 2012;126:2408–2417. doi: 10.1161/CIRCULATIONAHA.112.144055. [DOI] [PubMed] [Google Scholar]
- 4.Giugliano RP, Desai NR, Kohli P, Rogers WJ, Somaratne R, Huang F, et al. Efficacy, safety, and tolerability of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 in combination with a statin in patients with hypercholesterolaemia (LAPLACE-TIMI 57): a randomised, placebo-controlled, dose-ranging, phase 2 study. The Lancet. 380:2007–2017. doi: 10.1016/S0140-6736(12)61770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth EM, McKenney JM, Hanotin C, Asset G, Stein EA. Atorvastatin with or without an Antibody to PCSK9 in Primary Hypercholesterolemia. N Engl J Med. 2012;367:1891–1900. doi: 10.1056/NEJMoa1201832. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan DOAGSR, et al. Effect of a monoclonal antibody to pcsk9 on low-density lipoprotein cholesterol levels in statin-intolerant patients: The gauss randomized trial. JAMA. 2012;308:2497–2506. doi: 10.1001/jama.2012.25790. [DOI] [PubMed] [Google Scholar]
- 7.Koren MJ, Scott R, Kim JB, Knusel B, Liu T, Lei L, et al. Efficacy, safety, and tolerability of a monoclonal antibody to proprotein convertase subtilisin/kexin type 9 as monotherapy in patients with hypercholesterolaemia (MENDEL): a randomised, double-blind, placebo-controlled, phase 2 study. Lancet. 2012;380:1995–2006. doi: 10.1016/S0140-6736(12)61771-1. [DOI] [PubMed] [Google Scholar]
- 8.Alborn WE, Cao G, Careskey HE, Qian YW, Subramaniam DR, Davies J, et al. Serum proprotein convertase subtilisin kexin type 9 is correlated directly with serum LDL cholesterol. Clin Chem. 2007;53:1814–1819. doi: 10.1373/clinchem.2007.091280. [DOI] [PubMed] [Google Scholar]
- 9.Mayne J, Raymond A, Chaplin A, Cousins M, Kaefer N, Gyamera-Acheampong C, et al. Plasma PCSK9 levels correlate with cholesterol in men but not in women. Biochem Biophys Res Commun. 2007;361:451–456. doi: 10.1016/j.bbrc.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 10.Welder G, Zineh I, Pacanowski MA, Troutt JS, Cao G, Konrad RJ. High-dose atorvastatin causes a rapid sustained increase in human serum PCSK9 and disrupts its correlation with LDL cholesterol. J Lipid Res. 2010;51:2714–2721. doi: 10.1194/jlr.M008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Awan Z, Seidah NG, MacFadyen JG, Benjannet S, Chasman DI, Ridker PM, et al. Rosuvastatin, proprotein convertase subtilisin/kexin type 9 concentrations, and LDL cholesterol response: the JUPITER trial. Clin Chem. 2012;58:183–189. doi: 10.1373/clinchem.2011.172932. [DOI] [PubMed] [Google Scholar]
- 12.Berthold HK, Seidah NG, Benjannet S, Gouni-Berthold I. Evidence from a Randomized Trial That Simvastatin, but Not Ezetimibe, Upregulates Circulating PCSK9 Levels. PLoS One. 2013;8:e60095. doi: 10.1371/journal.pone.0060095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo YL, Liu J, Xu RX, Zhu CG, Wu NQ, Jiang LX, et al. Short-Term Impact of Low-Dose Atorvastatin on Serum Proprotein Convertase Subtilisin/Kexin Type 9. Clinical drug investigation. 2013 doi: 10.1007/s40261-013-0129-2. [DOI] [PubMed] [Google Scholar]
- 14.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, et al. Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ Cardiovasc Genet. 2009;2:173–181. doi: 10.1161/CIRCGENETICS.108.818062. [DOI] [PubMed] [Google Scholar]
- 16.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chasman DI, Giulianini F, Macfadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic Determinants of Statin Induced LDL-C Reduction: The JUPITER Trial. Circ Cardiovasc Genet. 2012 doi: 10.1161/CIRCGENETICS.111.961144. [DOI] [PubMed] [Google Scholar]
- 18.Deshmukh HA, Colhoun HM, Johnson T, McKeigue PM, Betteridge DJ, Durrington PN, et al. Genome-wide association study of genetic determinants of LDL-c response to atorvastatin therapy: importance of Lp(a) J Lipid Res. 2012;53:1000–1011. doi: 10.1194/jlr.P021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Careskey HE, Davis RA, Alborn WE, Troutt JS, Cao G, Konrad RJ. Atorvastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J Lipid Res. 2008;49:394–398. doi: 10.1194/jlr.M700437-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Ni YG, Di Marco S, Condra JH, Peterson LB, Wang W, Wang F, et al. A PCSK9-binding antibody that structurally mimics the EGF(A) domain of LDL-receptor reduces LDL cholesterol in vivo. J Lipid Res. 2011;52:78–86. doi: 10.1194/jlr.M011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 24.Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 25.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci U S A. 2010;107:16222–16227. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayne J, Dewpura T, Raymond A, Cousins M, Chaplin A, Lahey KA, et al. Plasma PCSK9 levels are significantly modified by statins and fibrates in humans. Lipids Health Dis. 2008;7:22. doi: 10.1186/1476-511X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.