Summary
Developmental alterations of excitatory synapses are implicated in autism spectrum disorders (ASDs). Here, we report increased dendritic spine density with reduced developmental spine pruning in layer V pyramidal neurons in postmortem ASD temporal lobe. These spine deficits correlate with hyperactivated mTOR and impaired autophagy. In Tsc2+/- ASD mice where mTOR is constitutively overactive, we observed postnatal spine pruning defects, blockade of autophagy, and ASD-like social behaviors. The mTOR inhibitor rapamycin corrected ASD-like behaviors and spine pruning defects in Tsc2+/ mice, but not in Atg7CKO neuronal autophagy deficient mice or Tsc2+/-:Atg7CKO double mutants. Neuronal autophagy furthermore enabled spine elimination with no effects on spine formation. Our findings suggest that mTOR regulated autophagy is required for developmental spine pruning, and activation of neuronal autophagy corrects synaptic pathology and social behavior deficits in ASD models with hyperactivated mTOR.
Introduction
Autism Spectrum Disorders (ASDs) are characterized by impaired social interactions, communication deficits, and repetitive behaviors. Multiple ASD susceptibility genes converge on cellular pathways that intersect at the postsynaptic site of glutamatergic synapses (Bourgeron, 2009; Peca et al., 2012), implicating abnormalities in dendritic spines in ASD pathogenesis. Consistently, increased spine density is observed in frontal, temporal, and parietal lobes in ASD brains (Hutsler and Zhang, 2010) and changes in synaptic structure are detected in multiple ASD model mice (Zoghbi and Bear, 2012). It remains however unclear why spine pathology occurs and how it is associated with the onset and progression of ASD related symptoms.
Postnatal synaptic development in mammalian cerebral cortex is a dynamic process involving concurrent formation and elimination/pruning (Purves and Lichtman, 1980; Rakic et al., 1986). Synapse formation exceeds pruning at early ages, yielding excessive excitatory synapses essential for the assembly of neural circuits. Synaptic elimination subsequently outpaces formation, resulting in net spine pruning from childhood through adolescence. Consistently, the density of dendritic spines peaks in early childhood and is followed by a steep decline during late childhood and adolescence to adult levels (Penzes et al., 2011), a process that provides selection and maturation of synapses and neural circuits.
While ASDs exhibit striking genetic and clinical heterogeneity, multiple ASD syndromes are caused by mutations in genes that act to inhibit mammalian target of rapamycin (mTOR) kinase, including Tsc1/Tsc2, NF1, and Pten (Bourgeron, 2009). Synaptic mTOR integrates signaling from various ASD synaptic and regulatory proteins, including SHANK3, FMRP and the glutamate receptors mGluR1/5 (Peca et al., 2012; Bourgeron, 2009). Overactive mTOR signaling may produce an excess of synaptic protein synthesis, which could indicate a common mechanism underlying ASD. Synapses, however, must balance protein synthesis and degradation to maintain homeostasis and support plasticity (Bingol and Sheng, 2011). An important means for removing damaged organelles and degrading long-lived or aggregate-prone proteins is macroautophagy (autophagy hereafter), a process downstream of mTOR signaling that involves the formation of autophagosomes to capture and transport cytoplasmic components to lysosomes. The activation of mTOR inhibits autophagy at an early step in autophagosome formation (Kim et al., 2011). In support of a role for autophagy dysregulation in ASD etiology, a recent study identified ASD-associated exonic copy number variation mutations in genes coding for proteins involved in autophagic pathways (Poultney et al., 2013).
Autophagy has been implicated in synaptic remodeling in C. elegans (Rowland et al., 2006) and Drosophila (Shen and Ganetzky, 2009), but a role in mammalian synaptic development is unexplored. We hypothesized that autophagy remodels synapse maturation downstream of mTOR, and autophagy deficiency downstream of overactivated mTOR contributes to ASD synaptic pathology. We found a higher spine density in basal dendrites of layer V pyramidal neurons in ASD patients than in controls. The increased spine density was associated with a defect in net postnatal spine pruning that was correlated with hyperactivated mTOR and impaired autophagy. Using Tsc1/2 mutant ASD mice and Atg7CKO neuronal autophagy deficient mice, we find that aberrant autophagy and mTOR hyperactivation underlies ASD-like synaptic pathology and correcting autophagy signaling could normalize developmental dendritic spine pruning defects and social behaviors.
Results
Dendritic spine pruning deficits in ASD human brain
We measured dendritic spines of basal dendrites of layer V pyramidal neurons in the superior middle temporal lobe, Brodmann Area 21 (BA21), a region implicated in ASD due to its participation in brain networks involved in social and communicative processes, including language, social and speech perception, auditory and visual processing, and comprehension of intentions (Redcay, 2008; Zahn et al., 2007). Abnormalities in ASD temporal lobe have been confirmed by functional imaging and pathological studies, including disturbed gene transcription profiles (Garbett et al., 2008; Voineagu et al., 2011), increased dendritic spine densities in pyramidal neurons (Hutsler and Zhang, 2010), and reduced functional specialization (Shih et al., 2011).
We compared dendritic spine morphology in ASD patients and controls (Demographic data in Table S1) using the Golgi-Kopsch technique. In the adolescent group, only males were examined to exclude effects of hormone status. No correlation was revealed between spine density and potential confounding factors, including postmortem interval (PMI), seizure history, cause of death, brain pH, or tissue storage (Table S2). As in previous studies (Harris et al., 1992), dendritic protrusions with the ratio of head/neck diameter > 1 were classified as spines. The spines were characterized by a neck 0.9–3.0 μm long and a spine head diameter of 0.5-2.0 μm (Figs S1A and S1B). The average spine head diameter (p=0.519) and spine length (p=0.819) from individual neurons were similar in ASD patients and controls at all ages examined (Figs S1C and S1D). The mean net spine density per individual was significantly higher in ASD patients than in controls (Figs 1A and 1B: mean ± SD: 8.81 ± spines/10 μm vs. 11.32 ± spines/10 μm, p=0.017, two-tailed t-test).
Fig 1. Dendritic spine pruning in temporal lobe of ASD patients and controls.
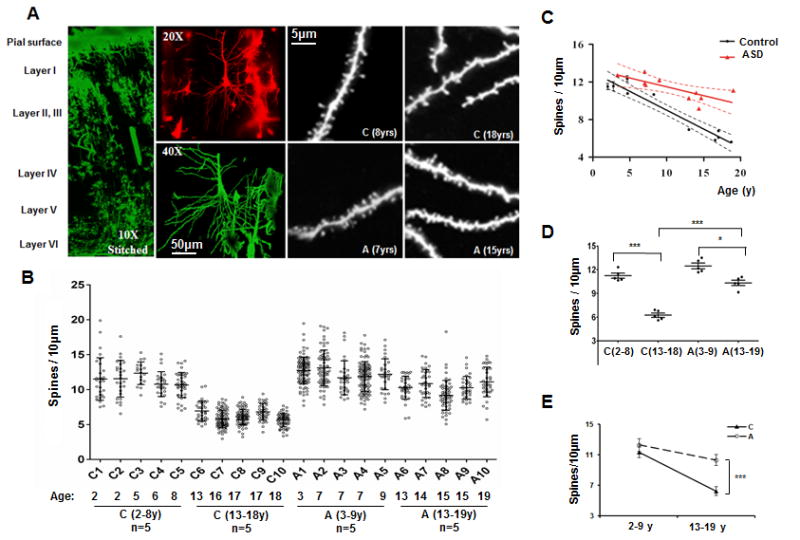
(A)Representative Golgi images for postmortem human temporal lobe (left, 10X, stitched from 9 separate image stacks), Layer V pyramidal neurons with basal dendritic tree (upper middle, 20X, pseudocolored in red; lower middle, 40X, pseudocolored in green; scale bar: 50 μm). The right four panels (100X, scale bar: 5 μm) are representative images of proximal basal dendritic segments from 2 control subjects (C, aged 8 yrs and 18 yrs) and 2 ASD cases (A, aged 7 yrs and 15 yrs); (B) Distribution of spine density (mean ± SD) in basal dendrites after the 1st bifurcation. Age and diagnosis are indicated for each sample. Controls aged 2-8yrs [C(2-8yrs)]: n=5; controls aged 13-18yrs [C(13-18yrs)]: n=5; ASD cases aged 2-8yrs [A(2-8yrs)]: n=5; ASD cases aged 13-18yrs [A(13-18yrs)]: n=5. Each point represents the average spine density for each individual neuron measured from each individual; (C) A linear regression of spine density with age in the control subjects (n = 10) and ASD patients (n = 10). The number of spines per 10 μm was plotted against the age of each individual. Broken lines indicate 95% confidence intervals; (D) Spine density (Mean ± SD) for the controls and ASD patients in childhood and adolescence. Each point represents the mean spine density for an individual. 2-way ANOVA, Bonferroni post hoc test. ***, p<0.001, *, p<0.05; (E) The decrease of spine density with age was greater in the controls than the ASD patients (Mean ±SD). ***, p<0.001.
Linear regression of spine density with age indicated a substantially greater level of net spine pruning in controls (slope = -0.40 spines / 10 μm /year, R2 = 0.93) than in ASD patients (slope = -0.19 / 10 μm / year, R2 = 0.55; difference from linear regression of controls, F = 9.4, p = 0.007) (Fig 1C). Due to the limited number of brain samples available, we grouped patients and controls into two age categories: childhood (2-9y) and adolescence (13-20y) (Figs 1D and 1E). Analysis revealed profound effects of both disease and age on spine density (p<0.001, 2-way ANOVA, effect of disease: F (1, 16) =73.11, p<0.001; effect of age: F (1, 16) =145.7, p<0.001; disease X age interaction: F (1, 16) =28.35, p<0.001). The spine density was slightly higher in childhood ASD patients than controls (mean ± SD: 12.32 ± 0.60 spines /10 μm in ASD cases vs. 11.37 ± 0.68 spines /10 μm in controls) but markedly higher in adolescent ASD patients than controls (10.33 ± 0.74 spines/10 μm in ASD cases vs. 6.24 ± 0.59 spines/10 μm in controls). From childhood through adolescence, dendritic spines decreased by ∼45% in control subjects but only by ∼16% in ASD patients (Fig 1E), demonstrating a developmental defect in net spine pruning in ASD.
Disturbed mTOR-autophagy signaling and spine pruning in ASD
To test the hypothesis that mTOR-autophagy signaling is disturbed in ASD and associated with ASD spine pathology, we performed western blot analysis of phospho-mTOR (p-mTOR), total mTOR (t-mTOR), phospho-S6 (p-S6), total S6 (t-S6), and the autophagosome marker LC3 and p62 (Figs 2A and 2F) in frozen BA21 brain samples from age-, gender-, PMI- matched ASD patients and controls (Demographic data in Table S3). To determine the relationship between mTOR activity and density of dendritic spines, we examined the protein levels of postsynaptic marker PSD95 and the presynaptic protein synapsin I (Fig 2A).
Fig 2. Dysregulated mTOR-autophagy signaling and spine pruning in ASD temporal lobe.
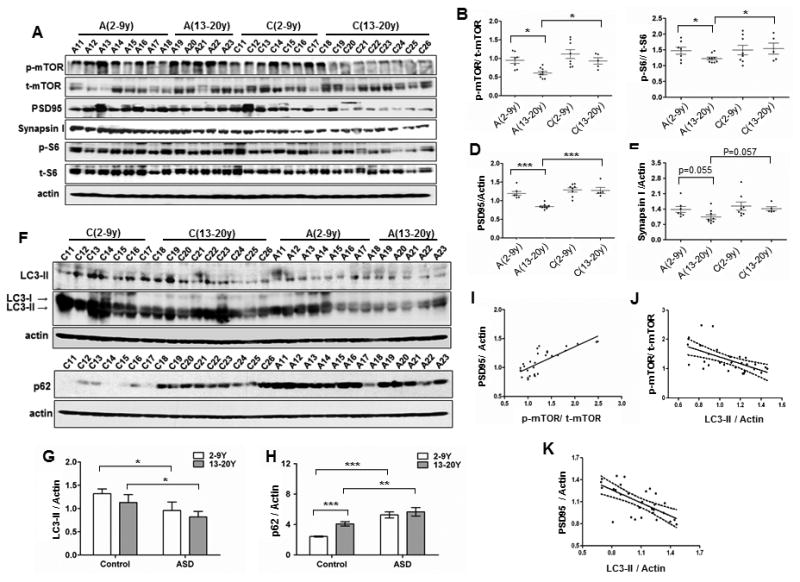
(A) Representative western blots of p-mTOR, t-mTOR, p-S6, t-S6, PSD95 and synapsin I in temporal lobe of ASD patients and control subjects aged 2-9y (ASD, n = 8; Controls, n=7) and 13-19y (ASD, n = 5; Controls, n=9). A, ASD patients; C, controls; (B-E) The relative density (Mean±SD) for p-mTOR (B) and p-S6 (C) were normalized to t-mTOR and t-S6, respectively. PSD95 (D) and synapsin I (E) levels were normalized to actin and are presented as scatter plots for ASD patients and controls in two age groups. Each point represents each individual subject. ** p<0.01; *** p<0.001 (2-way ANOVA, Bonferroni's post hoc test); (F) Western blot of autophagy markers, LC3-II and p62, in temporal lobe of ASD patients and control subjects aged 2-9 yrs and 13-20 yrs; (G) LC3-II levels normalized to actin in controls and patients. ** p<0.01; *** p<0.001 (2-way ANOVA, Bonferroni post hoc test); (H) p62 levels normalized to actin in controls and patients. ** p<0.01; *** p<0.001 (2-way ANOVA, Bonferroni post hoc test); (I) Correlation between p-mTOR and PSD95 (r2 =0.598, P<0.001); (J) Correlation between p-mTOR and LC3-II in individuals younger than 10 yrs (R2 =0.347, p<0.0001), indicating that LC3-II is regulated by mTOR in both ASD patients and controls; (K) Correlation between LC3-II and PSD95 in individuals younger than 10 years (R2 =0.422, p<0.0001), suggesting a relationship between synaptic structure protein levels and autophagy.
No effects of PMI, cause of death, brain pH or length of storage on protein levels were detected (Table S2). We observed a decrease in p-mTOR level with age in controls, and higher p-mTOR level in ASD patients than in controls at 13-20 y (Fig 2B). We observed similar changes in p-S6, a reporter for mTOR activity (Fig 2C). PSD95 protein level was higher in controls aged 2-10 y than controls aged 13-20 y, consistent with normal developmental spine pruning. This decrease in PSD95 level with age was absent in ASD patients (Fig 2D), consistent with the lower spine pruning in ASD patients. The presynaptic marker synapsin I exhibited a near-significant decrease with age in controls but not in patients (Fig 2E). Levels of p-mTOR were correlated with PSD95 (Fig 2 I), indicating that lower mTOR activity is associated with a higher dendritic spine density in children and adolescents.
To determine if impaired autophagy underlies the spine pruning deficit in human ASD, we characterized basal autophagy in postmortem tissue of the temporal lobe in patients with ASD. The level of LC3-II, a biomarker that indicates the abundance of autophagosomes, was significantly lower in ASD patients than controls throughout childhood and adolescence (Figs 2F and 2G), while the level of p62, a protein substrate for autophagy, was higher in both childhood and adolescent ASD patients that controls (Figs 2F and 2H). These data suggest a low level of basal autophagy in ASD temporal cortex throughout development. LC3-II and p62 protein levels were not correlated with any confounding factor for tissue preservation but were correlated with seizure activity (Table S2), a common feature of ASD.
The impairment in autophagy in ASD patients was confirmed by immunolabel of LC3-positive puncta in BA21 layer V: the cellular area occupied by LC3 puncta and the integrated intensity of LC3 puncta were lower in pyramidal neurons from ASD patients than age-matched controls during both early childhood and adolescence (Figs S1E, S1F, S1G). Decreased autophagy in cortical neurons was confirmed by the accumulation of autophagy substrates p62 and ubiquitin (Ub) (Figs S1H, S1I, S1J), and is thus an early feature of ASD. Higher levels of p-mTOR in both control and patients with ASD were strongly associated with lower levels of LC3-II (Fig 2J), suggesting that the lower level of autophagy in ASD patients was attributable to high mTOR activity. Higher LC3-II levels were strongly associated with lower levels of PSD95 (Fig 2K). These data indicate that mTOR-dependent autophagy is negatively correlated with spine density in human brain during childhood and adolescence.
mTOR dysregulation causes spine pruning defects in TSC-deficient mouse models of ASD
We then investigated whether mTOR hyperactivation and resulting inhibition of autophagy causes ASD-like dendritic spine pathology in ASD animal models. We focused on mutations in genes encoding tuberous sclerosis complexes TSC1 (hamartin) and TSC2 (tuberin), proteins that form a heterodimer that constitutively inhibits Rheb to inactivate mTOR (Ehninger and Silva, 2011). Mutations in both Tsc1 and Tsc2 cause mTOR hyperactivation and ASD-like behaviors in mice (Chevere-Torres et al., 2012; Tsai et al., 2012; Goorden et al., 2007; Sato et al., 2012).
Tsc2+/- mice however exhibit normal social preference in a 3-chamber test (Ehninger et al., 2008; Ehninger et al., 2012) but deficient social interaction in a dyadic reciprocal social test (Sato et al., 2012). This discrepancy in social behaviors could be due to differences in testing protocols, the gender, the age or genetic background of the testing mice. We thus characterized ASD-like social behaviors In P30-35 adolescent male Tsc2+/- mice maintained in a B6/C57 background. We observed no motor defects or anxiety-like behaviors in open field (Figs S2A-S2F). In the novel object recognition test, Tsc2+/- mice spent less time exploring the novel object than their wild type (wt) littermates, with no difference in time spent exploring the familiar object (Fig 3A). Tsc2+/- mice, however, did not exhibit ASD-like repetitive behaviors (Fig 3B). Sociability was assessed during a dyadic social interaction with a novel (non-cagemate) mouse matched for sex and genotype (see Supplementary Information). Tsc2+/- mice spent less time sniffing the stimulus mouse (Fig 3C), indicating impaired social interactions. Social deficits were confirmed using a 3-chamber social test. While Tsc2+/- mice showed a preference for interacting with a social target compared with non-social target (Fig 3D, left), the preference index (the ratio of time sniffing mouse vs. empty cage) was decreased (Fig 3D, right). In the social novelty test, Tsc2+/- mice spent a similar amount of time sniffing both novel and familiar social targets (Fig 3E, left), with decreased preference index (the ratio of time sniffing a stranger mouse vs. a familiar mouse, Fig 3E, right), indicating a reduced preference for social novelty.
Fig 3. Spine pruning defects in Tsc1/2 deficient cortical projection neurons.
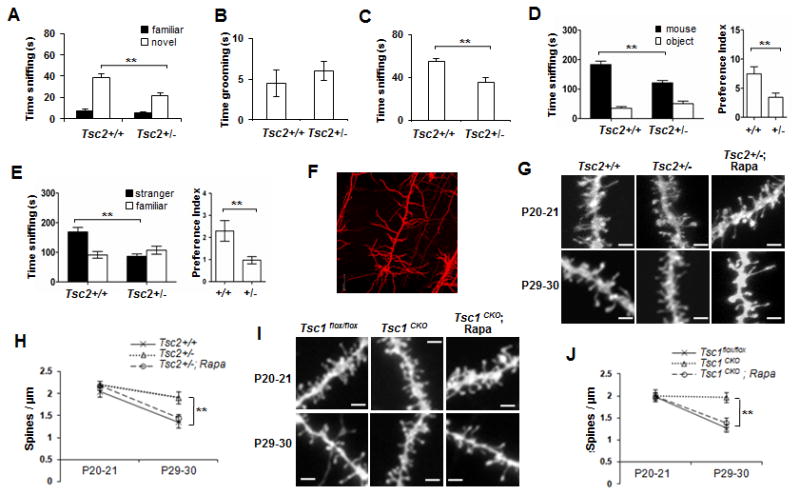
(A-D) Social behaviors in P30-33 male adolescent TSC2+/- mice. Tsc2+/+wt: n=15, Tsc2+/-: n=14. (A) Novel object recognition test showing time spent investigating a familiar vs. novel object; (B) ASD-like repetitive behavior; (C) Dyadic reciprocal social interaction test showing time spent sniffing a stimulus mouse; (D) Sociability in the 3-chamber test showing time spent (left) and preference (right) for sniffing a stimulus mouse or an object; (E) Social novelty in the 3-chamber test showing time spent (left) and preference (right) for sniffing a stranger mouse vs. a familiar mouse. Compared to wt, ** p<0.01; * p<0.05 (unpaired t test); (F) A confocal image of a DiI-labeled layer V cortical pyramidal neuron. Scale: 20 μm; (G) Typical confocal images of DiI labeled dendrites in wt, Tsc2+/- and rapamycin treated Tsc2+/- mice at P19-20 and P29-30. Rapamycin was administered at 3mg/kg/day intraperitoneally from P21 to P28, and the mice were labeled for spine analysis on P29-30. Scale bar: 2 μm. (H) Spine pruning in Tsc2+/- mice. ** compared with wt at P29-30, p<0.01 (2-way ANOVA, Bonferroni post hoc test). n=7-10 mice per group; (I) Representative images of DiI labeled dendrites from Tsc1CKO mutants and Tsc1flox/flox controls. Scale bar: 2 μm; (J) Spine density in Tsc1flox/flox and Tsc1CKO mice at P19-20 and P29-30. ** compared to P29-30 Tsc1flox/flox controls, p<0.01 (2-way ANOVA, Bonferroni post hoc test).
The density of dendritic spines in pyramidal neuron basal dendrites of layer V A1/S2 in temporal cortex, which is thought to be analogous to the primate primary auditory cortex (A1) and secondary somatosensory cortex (S2) (Benavides-Piccione et al., 2002), was examined by DiOlistic labeling (Fig 3F). A higher spine density was found in adolescent Tsc2+/- mice than in wt (Figs 3G and 3H). The greater spine density in Tsc2+/- layer V cortex at P30 was confirmed by increased immunolabel for the presynaptic marker, synaptophysin, and the postsynaptic marker, PSD95 (Figs S2G and S2H). We also observed increased PSD95 and F actin-labeled puncta along the dendrites of mature Tsc2+/- primary neuronal cultures (Figs S2I and S2J).
Net spine pruning normally occurs in mice after the 3rd postnatal week (Zuo et al., 2005), and so we compared spine densities between P19-20 and P29-30. If the lack of TSC and hyperactivation of mTOR led to spine overgrowth, an increase in spine density would be expected prior to spine pruning, in our case, at P19-20. DiOlistic labeling revealed similar numbers of spines at P19-20 in Tsc2+/- and wt mice, but far more spines in P29-30 Tsc2+/- mice than in wt (Figs 3G and 3H). The soma size and basal dendritic tree complexity were similar in wt and Tsc2+/- mutants (Fig S3A). These results suggest that there is a period of massive spine pruning between the ages of P19-20 and P29-30 in wt, but a lack of normal pruning in Tsc2+/- mice. Inhibition of mTOR by i.p. injection of rapamycin showed no effects in wt controls (Fig S3B) but corrected the pruning defect in Tsc2+/- mutants to the control level.
To confirm the effect of the Tsc deficiency on dendritic spine pruning, we used a Tsc1 conditional knockout mouse line (Tsc1CKO), in which the Tsc1 gene was depleted from pyramidal neurons in the forebrain by crossing Tsc1flox/flox mice to CamKII-Cre mice. As CamKII promoter driven Cre recombination begins in layer II-III cortical pyramidal neurons at P19-20 but is substantial in all cortical layers at P23-30 (Fig S4A), the level of TSC1 is normal in deep cortical layers at the start of this time window for spine pruning and depleted thereafter (Figs S4B &S4C). This mouse line allows us to model an appropriate developmental period for evaluating roles for cell autonomous effects of neuronal mTOR in the regulation of developmental spine pruning. At P19-20, the density of spines in basal dendrites of A1/S2 layer V pyramidal neurons was equivalent for both Tsc1flox/flox control and Tsc1CKO mice (p>0.05), but spines in control mice were significantly less dense than in Tsc1CKO mice at P29-30 (p<0.01, Figs 3I and 3J). Although soma size was slightly greater (15%) in P29-30 Tsc1CKO pyramidal neurons, the number of primary basal dendrites was similar for control and Tsc1CKO mice (Figs S4D, S4E and S4F). Rapamycin treatment did not affect spine density in Tsc1flox/flox control mice (Fig S5B), but corrected spine pruning in Tsc1CKO mice to control levels (Figs 3I and 3J), with no effect on basal dendritic branching. Thus, both Tsc1CKO and Tsc2+/- mutants showed a lack of efficient postnatal spine pruning, indicating that TSC inhibition of mTOR is required for postnatal spine pruning. The effects of TSC1 deletion on spine density in pyramidal neurons at P30 is consistent with those reported in vivo in Purkinje cells (Tsai et al., 2012).
Autophagy deficiency in Tsc2+/- mutant neurons
We then addressed whether autophagy remodels dendritic spines downstream of mTOR. We confirmed suppression of basal autophagy due to mTOR disinhibition in the Tsc mutant mouse brain by 1) a decrease in protein levels of LC3-II in Tsc2+/- cortex, which was normalized by rapamycin (Fig 4A); 2) an increase in the level of phospho-S6 (pS6), indicating mTOR hyperactivation, and a reduction in GFP-LC3 puncta, a fluorescent marker for autophagosomes, in cortical pyramidal neurons from Tsc2+/-: GFP-LC3 mice (Fig S5A), indicating impaired autophagy. Rapamycin normalized pS6 levels and numbers of GFP-LC3 puncta; 3) decreased immunolabel for endogenous LC3 in Tsc2+/- mutant primary neuronal cultures (Fig S5B); 4) an accumulation of autophagy substrates, including p62 (Komatsu et al., 2007a), lipid droplets and damaged mitochondria (Martinez-Vicente et al., 2010), in Tsc2+/- primary neuronal cultures (Figs S5 B, C, D, E); 5) accumulation of p62- and Ub- positive inclusions in pyramidal neurons in Tsc1CKO mouse brain (Fig S5F).
Fig 4. TSC ablation downregulates autophagic activity and rapamycin reconstitutes normal autophagy.
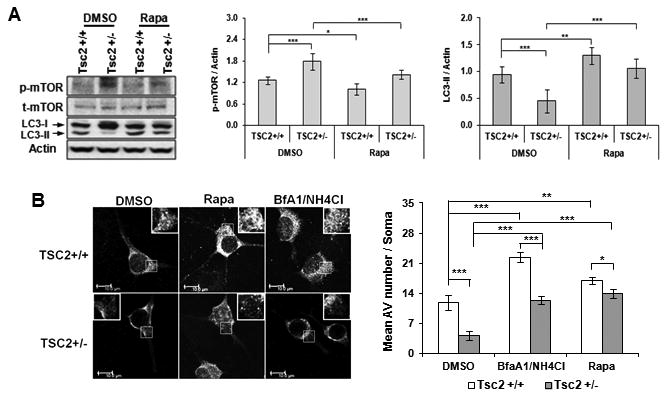
(A)Western blot analysis of p-mTOR, t-mTOR and LC3-II in P29 Tsc2+/- mouse brain. Tsc2 wt and Tsc2+/-mice were i.p. injected with DMSO vehicle or rapamycin from p20 to p28. Right: quantification of p-mTOR and LC3-II levels. Mean ± SD. * p<0.05; ** p<0.01; *** p<0.001 (2-way ANOVA, Bonferroni post hoc test). n=5-6 animals per group. Tsc2+/- mutant cortex on P29-30 showed increased p-mTOR levels and decreased levels of LC3-II. Inhibiting mTOR with rapamycin decreased p-mTOR and increased LC3II in both wild type and mutant lines; (B) Impaired autophagic flux in Tsc2+/-; GFP-LC3 cortical neurons. Right: mean number of GFP-LC3 puncta per soma of cortical neurons. 8-10 neurons per group in triplicates were analyzed. Scale bar: 10 μm. * p<0.05; ** p<0.01; *** p<0.001 (2-way ANOVA, Bonferroni post hoc test).
Fig 5. Dendritic spine pruning defects and ASD like behaviors in Atg7CKO mice.
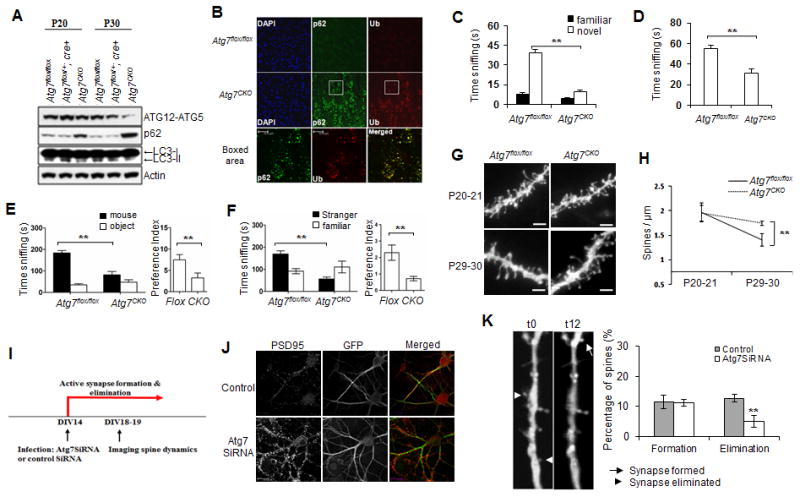
(A)Western blot analysis of autophagy markers in the Atg7CKO cortex. Brain homogenates from P19-20 and P29-30 mice were immunoblotted with antibodies against Atg12-Atg5, LC3 and autophagy substrate p62. Data shown are representative of three separate experiments. Loss of autophagy was indicated by a decrease in levels of Atg5-12 conjugation and LC3-II protein, and an increase in p62 protein; (B) Immunofluorescent labeling of p62 and ubiquitin (Ub) in P30 Atg7CKO mouse cortex. Scale bar: 10 μm. (C-F) ASD like social behaviors in Atg7CKO mice. Atg7flox/flox males, n=15; Atg7CKO males, n=13; (C) Novel object recognition test showing time spent sniffing a familiar object vs. a novel object; (D) Dyadic social interaction test showing the time testing mice spent sniffing a stimulus mouse. ** Compared to Atg7flox/flox; p<0.01; unpaired t-test; (E) Sociability in the 3-chamber test showing time spent (left) and preference (right) for a stimulus mouse or an object; (F) Social novelty in the 3-chamber test showing time spent (left) and preference (right) for sniffing a stranger mouse vs. a familiar mouse. Compared to wt, ** p<0.01 (unpaired t test); (G) Dendritic segments from Atg7flox/flox and Atg7CKO pyramidal neurons at P19-20 and P29-30. n=7-10 animals per group. Scale bar: 2 μm; (H) Fewer spines were pruned in Atg7CKO mice. ** Compared to P29-30 Atg7flox/flox, p<0.01 (2-way ANOVA, Bonferroni post hoc test). (I) Timeline of infection and spine analysis; (J) Cultured control and Atg7 SiRNA lentiviral infected mouse hippocampal neurons at DIV20, visualized by GFP and PSD95 fluorescence. Atg7SiRNA expressing neurons exhibited more PSD95 puncta than controls; (K) Spine formation and elimination in control and Atg7SiRNA infected neurons during a 12-hour time window at DIV19-20.
We analyzed autophagy flux in Tsc2+/-:GFP-LC3 neurons in comparison to Tsc2 wt : GFP-LC3 neurons (Fig 4B). Cultured primary neurons were treated with bafilomycin (BafA1) and NH4Cl to inhibit lysosomal hydrolase and block autophagosome-lysosome fusion. We reasoned that if autophagy flux were impaired by mTOR hyperactivation, BafA1/NH4CL blockade of lysosomal degradation would produce a lower accumulation of autophagosomes in Tsc2+/- neurons than in wt. As expected, there was more accumulation of GFP-LC3 puncta in BafA1/NH4CL treated wt neurons than Tsc2+/- neurons, confirming a failure of autophagosome induction in Tsc2+/- neurons (Fig 4B). Rapamycin normalized autophagosome formation in Tsc2+/- neurons. These findings support the hypothesis that basal neuronal autophagy is depressed due to mTOR hyperactivation in Tsc mutant ASD mouse models.
Autophagy deficiency results in ASD like social behaviors and spine pruning defects
To investigate if neuronal autophagy deficiency produces ASD-like behaviors and dendritic spine pathology, we generated forebrain excitatory neuronal specific autophagy-deficient mice by crossing Atg7flox/flox mice to CamKIIa-cre mice. Atg7 is an E1-like activating enzyme required for autophagosome formation (Komatsu et al., 2006). A deficit in autophagy was confirmed by western blot analysis of conjugated ATG5-12, p62 and LC3-II proteins (Fig 5A) and by immunolabel of p62 and ubiquitin (Ub) (Figs 5B and 6A), proteins that form aggregates after autophagy inhibition (Komatsu et al., 2006; Komatsu et al., 2007). At P20 there were no differences in LC3-II and conjugated ATG5-12 levels between genotypes, although p62 levels were higher in Atg7flox/flox:CamKII-Cre (Atg7CKO) autophagy-deficient mice (Fig 5A). By P30, however, Atg7CKO mice exhibited less conjugated ATG5-12 and LC3-II protein and more p62 protein than the Atg7flox/flox controls. Atg7CKO mice displayed occasional p62/Ub positive inclusions in pyramidal neurons at P20 and prominent p62/Ub positive aggregates in layer II-III and layer V-VI pyramidal neurons at P30 (Fig 5B). The results confirm a loss of autophagy between P20 and P30 in cortical pyramidal neurons in Atg7CKO mice.
Fig 6. Autophagy deficiency underlies spine pruning defects and ASD-like social deficits in Tsc2+/- mice.
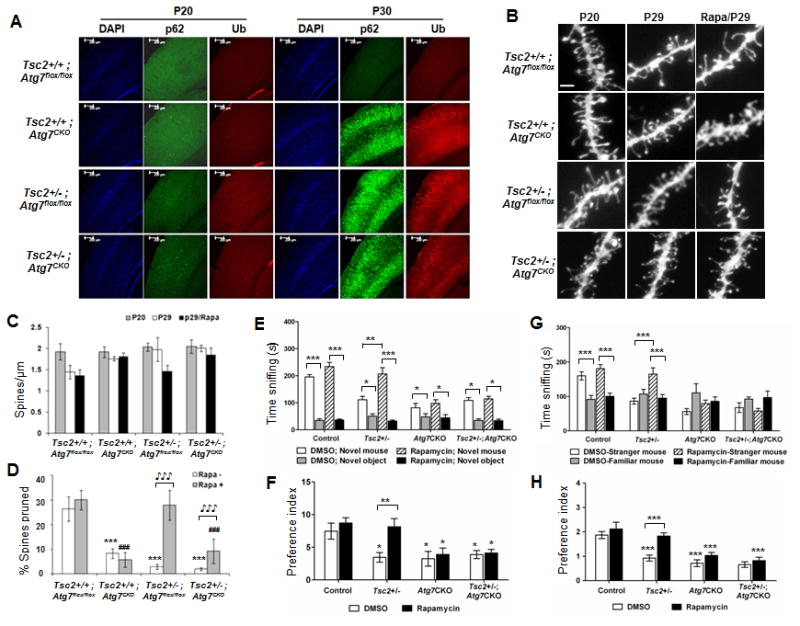
(A) p62 and Ub positive immunolabeled inclusions in autophagy deficient neurons. Note that P62+ and Ub+ inclusions were occasionally present in Tsc2+/-, Atg7CKO and Tsc2+/-:Atg7CKO cortex at p20, but appear in a majority of cortical neurons in these three lines at P30. No p62+ and Ub+ inclusions were seen in P30 cortical neurons from Atg7flox/flox control mice; (B-D) Rapamycin normalized dendritic spine pruning in an autophagy-dependent manner; (B) Representative images of dendrites from different mouse lines treated with DMSO vehicle or rapamycin. Scale: 2 μm; (C) Graphic representation of dendritic spine densities in each condition; (D) Percentage of spines pruned in control and mutant mouse lines. Percentage change in mean spine density (MSD) between P20 and P29 was calculated as: (MSD (P20) - MSD (P29)) / MSD (P20) *100%. Rapamycin rescued spine pruning deficits in Tsc2+/- mice, but not in Atg7CKO and had relatively little effect in Tsc2+/-:Atg7CKO double mutants. n=7-10 animals per group. *** Compared to DMSO treated Atg7flox/flox controls, p<0.001; ### Compared to rapamycin treated Atg7flox/flox controls, p<0.001; (IMG) compared to DMSO vehicle controls, p<0.001 (2-way ANOVA, Bonferroni post hoc test); (E-H) Autophagy deficiency blocks the rescue by rapamycin on ASD like social behaviors in Tsc2+/- mice. n=10-14 animals per group; (E, F) Sociability: DMSO treated Tsc2+/-, Atg7CKO and Tsc2+/-:Atg7CKO mice each spent less time sniffing social target versus non-social target (E) and exhibited decreased preference (F) for the social target vs. non-social target. Rapamycin treatment ameliorated impaired sociability in Tsc2+/- mice, but not in Atg7CKO and Tsc2+/-:Atg7CKO mice; (G, H) Social novelty: DMSO treated Tsc2+/-, Atg7CKO and Tsc2+/-:Atg7CKO mice all spent less time sniffing novel mice during social novelty test (G) and displayed a decrease in preference for social novelty (H). Rapamyin treatment prevented the loss of preference for social novelty in Tsc2+/- mice, but not in Atg7CKO and Tsc2+/-:Atg7CKO mice. *, p<0.05; **, p<0.01; ***, p<0.001 (2-way ANOVA, Bonferroni post hoc test).
Atg7CKO mice exhibited ASD-like social recognition (Fig 5C) and social interaction deficits similar to those seen in Tsc2+/- mutants. During dyadic encounters, they spent less time sniffing stimulus mice than their control littermates (Fig 5D). In the 3-chamber test, Atg7CKO mice displayed impaired preference for sniffing the social target (Fig 5E) and for social novelty (Fig 5F). However, these mice did not show stereotyped repetitive behaviors, motor defects, or anxiety-like behaviors (Figs S6A, B, C, D, E, F,and G).
We did not observe significant changes in the size of neuronal soma and the number of primary basal dendrites during this developmental period (Figs S6H,S6I and S6J). At P19-20, Atg7CKO basal dendrites from layer V A1/S2 pyramidal neurons exhibited a similar number of spines as those in the Atg7flox/flox control mice (Figs 5G and 5H). In contrast, by P29-30 the Atg7CKO basal dendrites exhibited more spines than controls. Thus, basal neuronal autophagy is required for normal spine pruning during postnatal development and for the development of normal social behaviors. The pruning defect is unlikely to result from abnormalities in microglia and astrocytes, as we observed no activation of microglia or astrocytes at any age in Atg7CKO mice (Figs S6K and S6L).
Autophagy mediates spine elimination in primary cultures of hippocampal neurons
The increased spine density and reduced spine pruning in Atg7CKO mice may result from an increase in synapse formation or a decrease in synapse elimination. Primary cultures of hippocampal neurons have been used as an in vitro system to investigate the formation, maturation and pruning of dendritic spines, in which dendritic spines are formed and pruned during a developmental period similar to that in vivo in mouse brain (Orefice et al., 2013; Ko et al., 2011; Papa et al., 1995), with spine density increases between 6-10 days in vitro (DIV6-10), peaks at DIV14-21 and decreases after weeks 3 or 4 in vitro in neuronal cultures. We thus infected CA1 hippocampal neurons with a lentivirus expressing EGFP-Atg7SiRNA or an EGFP control virus at DIV14-15, a period of active synapse formation and stabilization in cultures. Three to four days after infection, the cultures were fixed and stained for postsynaptic marker PSD95 (Figs 5I and 5J). Neurons expressing Atg7SiRNA exhibited a higher level of PSD95 puncta at DIV19-20 than neurons infected with control virus (Fig 5J), suggesting an increased spine density. We calculated the rate of spine genesis and spine pruning at DIV19-20 during a 12-hour time window. In control neurons infected with viral vector controls, ∼12% of spines were formed and ∼13% eliminated, indicating equivalent rates of spine formation and elimination that reflect stabilized spine densities (Fig 5K). In contrast, ∼12% of spines were formed, but only ∼5% of pre-existing spines were pruned in Atg7siRNA infected neurons (p<0.05, t-test). Therefore, Atg7 knockdown produces excessive dendritic spines by inhibiting elimination, but exerted no effect on formation.
Autophagy deficiency underlies spine pruning defects in Tsc2+/- mice
We then asked whether autophagy deficiency underlies spine pruning defects in Tsc2+/- mice. mTOR regulates a number of downstream biological processes including protein synthesis, autophagy, ribosome biogenesis, and activation of transcription leading to lysosome biogenesis or mitochondrial metabolism. To disentangle autophagy from other downstream effectors of mTOR, we crossed Tsc2+/- mice to the Atg7CKO mice to produce a Tsc2+/-:Atg7CKO double mutant line. We hypothesized that if neuronal autophagy were responsible for spine pruning, rapamycin treatment during the fourth week would reconstitute normal autophagy and pruning in the Tsc2+/- mice, but would not do so in Atg7CKO and Tsc2+/-:Atg7CKO double mutant mice.
We observed high levels of p62 and ubiquitin in Tsc2+/-, Atg7CKO and Tsc2+/-:Atg7CKO double mutant cortices at P30, consistent with a deficit of autophagy between P20 and P30 (Fig 6A). We imaged basal dendrites of layer V A1/S2 pyramidal neurons in all lines at P20 to provide a baseline. We then treated mice from all lines with DMSO vehicle or rapamycin from P21 to P28. On P29, basal dendrites of layer V A1/S2 pyramidal neurons were labeled and analyzed (Figs 6B and 6C). In the DMSO vehicle treated mice, the percentage of spines pruned between P21 and P29 was 26%, 8%, 3% and 2% in the four mouse lines respectively (Fig 6D, 2-way ANOVA, Bonferroni's post hoc test; genotype × treatment interaction: F(3,16)=15.38, p<0.001; effect of treatment: F(1,16)=32.16, p<0.001; effect of genotype: F(3,16)=56.17, p<0.001). Therefore, basal levels of autophagy appeared responsible for ∼70% [(26-8)/26] of postnatal spine pruning in control mice. No significant effect of rapamycin on spine pruning in Atg7flox/flox control mice was observed. Rapamycin reversed spine pruning defects in Tsc2+/- mice to the level of control mice, but did not rescue spine pruning in Atg7CKO mice or in Tsc2+/-:Atg7CKO double mutants, demonstrating that neuronal autophagy is required for spine elimination in Tsc2+/- mice. Note that a relatively small fraction (8%/26% = 30%) of spine pruning was preserved in Atg7CKO mice, indicating that additional mechanisms independent from neuronal autophagy are responsible for the remainder of spine elimination during postnatal development. Consistently, we found that rapamycin rescued spine pruning in Tsc2+/-: Atg7CKO mice to this relatively low level of neuronal autophagy-independent pruning.
Rapamycin normalizes social deficits in Tsc2+/- mice but not in Atg7 conditional knockouts
We next examined whether rapamycin rescued social deficits in Tsc2+/- mice, Atg7CKO mice and the double mutants. Sociability and social novelty were tested with the 3-chamber testing paradigm as above. DMSO vehicle produced no effect on any mouse line (Figs 3, 5, and 6). Atg7flox/flox control mice treated with vehicle preferred to sniff the novel mouse more than the non-social object in the sociability test (Figs 6E and 6F), and preferred to sniff the stranger mouse more than the familiar mouse in the social novelty test (Figs 6G and 6H). In contrast, each mutant mouse line displayed impaired preferences for sociability and social novelty.
The preferences of the Atg7flox/flox control mice were unaffected by rapamycin. In contrast, rapamycin normalized the sociability and social novelty preferences of the Tsc2+/- mice. Rapamycin, however, did not normalize preferences of either the Atg7CKO mice or the double mutants (Sociability test, Fig 6F, p<0.01, 2-way ANOVA, Bonferroni's post hoc test. genotype × treatment interaction: p>0.05, effect of treatment: F(1,85)=4.156, p<0.05; effect of genotype: F(3,85)=6.775, p<0.001); Social novelty test, Fig 6H, p<0.01, 2-way ANOVA: genotype X treatment interaction: F(3,85)=3.085, p<0.05; effect of treatment: F(1,85)=13.02, p<0.001; effect of genotype: F(3,85)=25.64, p<0.001). Thus, rapamycin rescued impaired sociability and social novelty preferences of Tsc2+/- mice, but did not rescue behaviors in Atg7CKO mice in which neuronal autophagy cannot be activated. Note that rapamycin failed to reverse the impaired social behaviors in Tsc2+/-:Atg7CKO double mutants, although spine pruning deficits were partially reversed.
Discussion
Dendritic spine pruning defect in the ASD brain
We assessed spine density across development and confirm an increase in basal dendrite spine density in layer V pyramidal neurons in ASD temporal lobe. Layer V pyramidal neurons are the major excitatory neurons that form cortical-cortical and cortical-subcortical projections. Basal dendrites receive excitatory and inhibitory inputs from local sources, and excitatory cell types target this compartment almost exclusively (Spruston, 2008). The increase in basal dendrite spine density suggests an enhanced local excitatory connectivity, a feature of ASD (Belmonte et al., 2004) proposed to cause failure in differentiating signals from noise, prevent development of normal long-range cortical-cortical and cortical-subcortical communications, and underlie neocortical excitation/inhibition imbalance (Sporns et al., 2001; Gogolla et al., 2009).
Note that while signs of ASD can often be detected at 12-18 months, ∼82% of ASD diagnoses occur at 4 years or older (http://www.cdc.gov/ncbddd/autism/data.html), and CNS tissues from very young ASD patients are extremely rare. As human brain samples from ASD patients cannot be identified prior to diagnosis, pathological analysis cannot determine if increased spine density precedes symptoms. We therefore relied on correlations among an age range of available pathological specimens, synaptic density, and biochemical markers for analysis. It is remarkable that the only available brain sample suitable for morphological study of a very young diagnosed ASD patient (age = 3Y) displayed a synaptic density higher than any control subject. We find that a defect in net spine pruning was responsible for the abnormally high synaptic densities in childhood and adolescent ASD, an observation confirmed in animal models. A variety of results indicate that this deficit is due in large part to a loss of mTOR-dependent autophagy in neurons. While synapse formation outpaces synapse elimination at young ages, yielding the highest synaptic density in early life, significantly reduced cortical autophagy was also apparent in the youngest diagnosed ASD patient (age = 2 Y, frozen tissue), as indicated by low levels of the autophagic vacuole marker LC3-II and increased level of autophagy substrates p62. An ongoing deficiency in autophagy and impaired spine elimination at younger ages would be expected to increase net spine density and interfere with the dynamic turnover of synapses that organizes neural circuits. Interruption of this maturational organization of the brain would lead to a persistence of immature or formation of aberrant circuits in ASD.
The near-linear decrease in spine number from all cases between the ages of 2 to 19 years indicates that spine pruning in temporal lobe occurs over the first two decades, and that net loss of synapses is substantially greater in controls than ASD patients. The spine densities declined during the first and second decade by 41% in normal controls but only by 16% in ASD patients, a level independently confirmed by analysis of pre- and post-synaptic markers. This deficit may contribute to abnormalities in cognitive functions that humans acquire in their late childhood, teenage or early adult years, such as the acquisition of executive skills such as reasoning, motivation, judgment, language, and abstract thought (Goda and Davis, 2003; Sternberg and Powell, 1983). Many children diagnosed with ASD reach adolescence and adulthood with functional disability in these skills, in addition to social and communication deficits (Seltzer et al., 2004). The extended duration for normal spine pruning in human brain may provide an opportunity for therapeutic intervention of multiple functional domains associated with ASD after the disease is diagnosed.
While our study examined a single brain region, spine pruning during early postnatal development occurs in cerebral cortex, cerebellum, olfactory bulb, and hippocampus (Purves and Lichtman, 1980; Shinoda et al., 2010). As ASD-related neuropathology involves disruptions in connectivity across the brain, it is likely that additional ASD brain regions may feature spine pruning defects during different periods of synaptic development. Nevertheless, the disorganization of synaptic connectivity in the temporal lobe, a central node in the social brain network (Gotts et al., 2012), may compromise function of a network of anatomically distinct brain regions that underlie global brain dysfunction and ASD-like social deficits (Normand et al., 2013; Tsai et al., 2012).
mTOR regulated autophagy and ASD synaptic pathology
The genetic heterogeneity of ASD encourages the identification of steps that converge on common pathways to produce the clinical syndrome. Dysregulated mTOR signaling has been identified in autism, Fragile X syndrome, tuberous sclerosis, neurofibromatosis, and PTEN mediated macroencephaly (Peca et al., 2012; Bourgeron, 2009)., each of which features altered dendritic spine densities. mTOR inhibitors, including rapamycin and its analogues, have been examined in clinical trials for treating ASD and neuropsychological deficits in children with TSC (Sahin, 2012).
We find that ASD brains exhibit both disrupted mTOR signaling and synaptic defects. It is highly unlikely that these patients possessed TSC mutations, and so our findings suggest that mTOR signaling provides a common convergent mechanism in ASD. mTOR signaling, however, contributes to protein synthesis required for neuronal survival, development, synaptic plasticity, learning, and memory (Hoeffer and Klann, 2010), and prolonged use of mTOR inhibitors may cause adverse effects (Rodrik-Outmezguine et al., 2011). An important goal is to identify specific signaling pathways downstream of mTOR that may provide more precise targets. For example, a link has been established between eIF4E dependent translational control downstream of mTOR and ASD-like phenotypes in mouse models (Santini et al., 2012). We provide evidence from postmortem brain that autophagy deficiency, which is a consequence of mTOR overactivation, strongly correlates with ASD dendritic spine pathology. The reduction of mTOR-regulated neuronal autophagy is further consistent with our recent findings of a lack of autophagic mitochondrial turnover in ASD brains (Tang et al., 2013).
We have confirmed in mouse models that inhibition of neuronal autophagy produced ASD-like inhibition of normal developmental spine pruning and ASD-like behaviors. Pharmacological inhibition of mTOR activity normalized ASD-like spine pruning deficits and ASD-like behaviors in mice largely by activating neuronal autophagy. As these data suggest a direct link between mTOR-regulated autophagy and pruning of synaptic connections during postnatal development, developing targeted means to enhance autophagy downstream of mTOR during development may provide the basis for novel ASD therapeutics.
Autophagy and spine pruning
The precise control of synapse pruning could be achieved by multiple signaling systems that converge to eliminate synaptic connections. This could involve the targeted degradation of synaptic components. Recent evidence suggests that neuronal activity decreases dendritic spine number in part through activation of the myocyte enhancer factor 2 (MEF2) transcription factor (Pfeiffer et al., 2010), which promotes ubiquitin-proteasome system- (UPS-) dependent degradation of the synaptic scaffolding protein PSD95 (Tsai et al., 2012). In addition to the UPS, which is primarily responsible for the degradation of short-lived cytosolic proteins, neurons rely on lysosomal-dependent degradation mechanisms for the turnover of long-lived synaptic proteins and damaged organelles. Ablation of autophagy genes ATG7 or ATG5 causes neurodegeneration associated with aberrant organelles and ubiquitin-rich inclusions in neuronal cell bodies (Hara et al., 2006; Komatsu et al., 2006), as well as disrupted membrane homeostasis in axon terminals (Komatsu et al., 2007; Hernandez et al., 2012).
Using an in vitro primary neuronal culture system, we find that autophagy regulates spine elimination but not spine formation during developmental pruning of dendritic spines. Autophagy may remodel dendritic spines by directing internalized postsynaptic membrane neurotransmitter receptors, including GABA-A (Rowland et al., 2006) and AMPAR (Shehata et al., 2012), toward lysosomal degradation. Although autophagy was classically considered an ‘in-bulk’ process, evidence now supports selectivity mediated via recognition of post-translational modifications by molecules that bind cargo and components of the autophagic machinery. p62 is the most extensively characterized cargo-recognizing molecule, and binds preferentially to an ubiquitin linkage (Lys63) on the surface of ubiquitinated protein aggregates, poly-ubiquitinated proteins and organelles. In addition, autophagy may degrade proteins that suppress spine elimination, and the loss of autophagy could accumulate proteins that block spine pruning, for example by releasing translationally suppressed synaptic mRNA for local protein synthesis (Banerjee et al., 2009).
As neuronal autophagy is responsible for ∼70% of postnatal net spine elimination, it is likely that basal autophagy regulates spine elimination in cooperation with additional regulatory mechanisms downstream of mTOR, including eIF4E dependent translational control and neuronal outgrowth (Santini et al., 2012), and other non-neuronal intrinsic regulatory mechanisms including neuroimmune disturbances and astrocyte activation (Garbett et al., 2008; Voineagu et al., 2011; Paolicelli et al., 2011; Schafer et al., 2012; Chung et al., 2013). Defective neuronal autophagy can be induced by infected microglia (Alirezaei et al., 2008), pointing to the possibility of glial non-cell autonomous autophagic regulation of spine morphogenesis. In addition, changes in mTOR-autophagy signaling and spine pruning defects may represent a secondary mechanism in response to an imbalance between excitatory and inhibitory neurotransmission, identified in both Mecp2 mutant mice and Tsc1 deficient mice and implicated in ASD-associated stereotypies and social behavioral deficits (Chao et al., 2010; Fu et al., 2011; Yizhar et al., 2011). Altered synaptic function is consistent with our recent finding that chronic lack of neuronal autophagy enhances evoked neurotransmitter release and rate of synaptic recovery (Hernandez et al., 2012).
In summary, we find that many ASD brains exhibit both disrupted mTOR signaling and synaptic defects during childhood and adolescence, suggesting that mTOR signaling may provide a common mechanism involved in ASD synaptic pathology (Sawicka and Zukin, 2012). We further demonstrated that ASD behaviors and synaptic deficits are elicited by altered mTOR signaling via an inhibition of autophagy required for normal developmental spine pruning. The results indicate a direct link between mTOR-autophagy and pruning of synaptic connections during postnatal development, and suggest that targeting neuronal autophagy could provide therapeutic benefit.
Experimental Procedures
ASD like social behavioral tests
Mice were tested for novel object recognition and social interactions, anxiety-like behaviors, exploratory locomotion behaviors, and self grooming repetitive behavior. Sociability and social novelty were tested in a 3-chamber testing paradigm. Procedures were approved by Columbia University IACUC.
Biochemistry, DiOlistic labeling, Golgi staining and immunohistochemistry
Mouse and human brain tissue were lysed with 1X RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors, and subjected to western blot analysis. Neurons in mouse brain were labeled with DiI using a Helios gene gun system at 120 psi. Fluorescent image stacks were acquired with a Leica multiphoton system. Neuronal morphology in postmortem human brain was analyzed by Golgi-Kopsch technique. Images were reconstructed with Imaris FilamentTracer Module (Bitplane).
Full Methods and associated references are in the Supplemental Information.
Supplementary Material
Acknowledgments
This study was supported by the Simons Foundation. Additional support for D.S. is from DOD TSCRP (TS110056) and the Parkinson's Disease and JPB Foundations, for G.T. from NIMH (K01MH096956), for M.L.C. from AHA, for A.J.D. from NIMH (MH64168), for F.C. from NIH (DP2OD001674-01), for O.A. from NIH (NS049442). We thank the Autism Tissue Portal, Harvard Brain Bank and Maryland NICHD Brain & Tissue Bank for kindly providing us brain tissues for the present study.
Footnotes
Author contributions G.T. and D.S. conceived and designed the study. G.T. and M.L.C. performed and analyzed DiOlistic labeling experiments; K.G. and F.C. designed, performed and analyzed all behavioral experiments; G.T., C.B. performed mouse breeding, and E.K. made neuronal cultures. G.T. and A.S, performed biochemistry, Golgi staining, immunolabeling of mouse and human brains and establishing neuronal cultures. S.H.K and M.S performed data analysis. G.R., A.J.D and J.E.G supervised brain sample selection, Golgi staining and data interpretation in human subjects. Z.Y., A.Y. and O.A. assisted with the design of autophagy and behavioral study. G.T., J.E.G., B.S.P. and D.S. wrote the manuscript. All authors read and approved with the final version.
Author Information The authors declare no competing financial interests
Supplemental information includes extended experimental procedures, ten Figures, and three tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alirezaei M, Kiosses WB, Flynn CT, Brady NR, Fox HS. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PloS One. 2008;3:e2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER. Structural changes accompanying memory storage. Annu Rev Physiol. 1993;55:397–426. doi: 10.1146/annurev.ph.55.030193.002145. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Sheng M. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chévere-Torres I, Maki JM, Santini E, Klann E. Impaired social interactions and motor learning skills in tuberous sclerosis complex model mice expressing a dominant/negative form of tuberin. Neurobiol Dis. 2012;45:156–164. doi: 10.1016/j.nbd.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, Smith SJ, Barres BA. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/-mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Silva AJ. Rapamycin for treating Tuberous sclerosis and Autism spectrum disorders. Trends Mol Med. 2011;17:78–87. doi: 10.1016/j.molmed.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Sano Y, de Vries PJ, Dies K, Franz D, Geschwind DH, Kaur M, Lee YS, Li W, Lowe JK, et al. Gestational immune activation and Tsc2 haploinsufficiency cooperate to disrupt fetal survival and may perturb social behavior in adult mice. Mol Psychiatry. 2012;17:62–70. doi: 10.1038/mp.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Cawthon B, Clinkscales W, Bruce A, Winzenburger P, Ess KC. GABAergic Interneuron Development and Function Is Modulated by the Tsc1 Gene. Cerebral cortex. 2011;22:2111–2119. doi: 10.1093/cercor/bhr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, Mirnics K, Persico AM. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda Y, Davis GW. Mechanisms of synapse assembly and disassembly. Neuron. 2003;40:243–264. doi: 10.1016/s0896-6273(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Gogolla N, Leblanc JJ, Quast KB, Südhof TC, Fagiolini M, Hensch TK. Common circuit defect of excitatory-inhibitory balance in mouse models of autism. J Neurodev Disord. 2009;1:172–181. doi: 10.1007/s11689-009-9023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/- mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62:648–655. doi: 10.1002/ana.21317. [DOI] [PubMed] [Google Scholar]
- Gotts SJ, Simmons WK, Milbury LA, Wallace GL, Cox RW, Martin A. Fractionation of social brain circuits in autism spectrum disorders. Brain. 2012;135:2711–2725. doi: 10.1093/brain/aws160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, Torres CA, Setlik W, Cebrián C, Mosharov EV, Tang G, Cheng HC, Kholodilov N, Yarygina O, Burke RE, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 2012;74:277–284. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Soler-Llavina GJ, Fuccillo MV, Malenka RC, Südhof TC. Neuroligins /LRRTMs prevent activity- and Ca2+/calmodulin-dependent synapse elimination in cultured neurons. J Cell Biol. 2011;194:323–334. doi: 10.1083/jcb.201101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007a;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci. 2007b;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: An evaluation of the hypothesis. Annu Rev Neurosci 2000. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand EA, Crandall SR, Thorn CA, Murphy EM, Voelcker B, Browning C, Machan JT, Moore CI, Connors BW, Zervas M. Temporal and mosaic Tsc1 deletion in the developing thalamus disrupts thalamocortical circuitry, neural function, and behavior. Neuron. 2013;78:895–909. doi: 10.1016/j.neuron.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orefice LL, Waterhouse EG, Partridge JG, Lalchandani RR, Vicini S, Xu B. Distinct roles for somatically and dendritically synthesized brain-derived neurotrophic factor in morphogenesis of dendritic spines. J Neurosci. 2013;33:11618–11632. doi: 10.1523/JNEUROSCI.0012-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Spine pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Papa M, Bundman MC, Greenberger V, Segal M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peça J, Feng G. Cellular and synaptic network defects in autism. Curr Opin Neurobiol. 2012;22:866–872. doi: 10.1016/j.conb.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Šimic G, Rasin MR, Uylings HB, Rakic P, Kostovic I. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci. 2011;108:13281–13286. doi: 10.1073/pnas.1105108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y, De Rubeis S, Durand S, Stevens C, Rehnström K, et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet. 2013;93:607–619. doi: 10.1016/j.ajhg.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Lichtman JW. Elimination of synapses in the developing nervous system. Science. 1980;210:153–157. doi: 10.1126/science.7414326. [DOI] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science. 1986;232:232–235. doi: 10.1126/science.3952506. [DOI] [PubMed] [Google Scholar]
- Redcay E. The superior temporal sulcus performs a common function for social and speech perception:implications for the emergence of autism. Neurosci Biobehav Rev. 2008;32:123–142. doi: 10.1016/j.neubiorev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–59. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006;26:1711–1720. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M. Targeted treatment trials for tuberous sclerosis and autism: no longer a dream. Curr Opin Neurobiol. 2012;22:895–901. doi: 10.1016/j.conb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Kasai S, Kobayashi T, Takamatsu Y, Hino O, Ikeda K, Mizuguchi M. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat Commun. 2012;3:1292. doi: 10.1038/ncomms2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Zukin RS. Dysregulation of mTOR signaling in neuropsychiatric disorders: therapeutic implications. Neuropsychopharmacology. 2012;37:305–306. doi: 10.1038/npp.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer MM, Shattuck P, Abbeduto L, Greenberg JS. Trajectory of development in adolescents and adults with autism. Ment Retard Dev Disabil Res Rev. 2004;10:234–247. doi: 10.1002/mrdd.20038. [DOI] [PubMed] [Google Scholar]
- Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci. 2012;32:10413–10422. doi: 10.1523/JNEUROSCI.4533-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P, Keehn B, Oram JK, Leyden KM, Keown CL, Müller RA. Functional differentiation of posterior superior temporal sulcus in autism: a functional connectivity magnetic resonance imaging study. Biol Psychiatry. 2011;70:270–277. doi: 10.1016/j.biopsych.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda Y, Tanaka T, Tominaga-Yoshino K, Ogura A. Persistent synapse loss induced by repetitive LTD in developing rat hippocampal neurons. PLoS One. 2010;5:e10390. doi: 10.1371/journal.pone.0010390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman GM. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Sternberg RJ, Powell JS. The development of intelligence. In: Mussen PH, editor. Handbook of Child Psychology. 4th. JohnWiley and Sons; 1983. pp. 341–419. [Google Scholar]
- Tang G, Gutierrez Rios P, Kuo SH, Akman HO, Rosoklija G, Tanji K, Dwork A, Schon EA, Dimauro S, Goldman J, Sulzer D. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 2013;54:349–361. doi: 10.1016/j.nbd.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, Huber KM. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, Stehfest K, Fudim R, Ramakrishnan C, Huguenard JR, Hegemann P, Deisseroth K. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R, Moll J, Krueger F, Huey ED, Garrido G, Grafman J. Social concepts are represented in the superior anterior temporal cortex. Proc Natl Acad Sci USA. 2007;104:6430–6435. doi: 10.1073/pnas.0607061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4:a009886. doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan WB. Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature. 2005;436:261–265. doi: 10.1038/nature03715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


