Abstract
Epstein-Barr Virus (EBV) persists as a latent infection in many lymphoid and epithelial malignancies, including Burkitt's lymphomas, nasopharyngeal carcinomas, and gastric carcinomas. Current chemotherapeutic treatments of EBV-positive cancers include broad- spectrum cytotoxic drugs that ignore the EBV-positive status of tumors. An alternative strategy, referred to as oncolytic therapy, utilizes drugs that stimulate reactivation of latent EBV to enhance the selective killing of EBV positive tumors, especially in combination with existing inhibitors of herpesvirus lytic replication, like Ganciclovir (GCV). At present, no small molecule, including histone deacetylase (HDAC) inhibitors, have proven safe or effective in clinical trials for treatment of EBV positive cancers. Aiming to identify new chemical entities that induce EBV lytic cycle, we have developed a robust high throughput cell-based assay to screen 66,840 small molecule compounds. Five structurally related tetrahydrocarboline derivatives were identified, two of which had EC50 measurements in the range of 150-170 nM. We show that these compounds reactivate EBV lytic markers ZTA and EA-D in all EBV-positive cell lines we have tested independent of the type of latency. The compounds reactivate a higher percentage of latently infected cells than HDAC inhibitors or phorbol esters in many cell types. The most active compounds showed low toxicity to EBV-negative cells, but were highly effective at selective cell killing of EBV-positive cells when combined with GCV. We conclude that we have identified a class of small molecule compounds that are highly effective at reactivating latent EBV infection in a variety of cell types, and show promise for lytic therapy in combination with GCV.
Keywords: EBV, lytic therapy, HTS, reactivation, small molecule screen, tetrahydrocarbolines
Introduction
Epstein-Barr virus (EBV) is a human herpesvirus that infects over 90% of the world's population (reviewed in (1, 2)). The primary infection is the major cause of infectious mononucleosis, and latent infection can drive the formation of Burkitt's lymphoma (BL), Hodgkin lymphomas, nasopharyngeal carcinoma (NPC), and gastric carcinoma (GC) (reviewed in (3-6)). Latent infection with EBV is a major cause of post-transplant lymphoproliferative disease in immunosuppressed patients (7, 8) and greatly enhances risk of developing non-Hodgkin and primary CNS lymphomas in the HIV-positive population (9, 10). Most EBV associated cancers contain viral DNA that exists predominantly as a latent infection in which only a limited set of viral genes are expressed (11). These latency-associated genes are implicated in host-cell proliferation and survival, and latent EBV can directly promote tumor progression.
A recently proposed approach to treat EBV-positive cancers involves the induction of EBV lytic cycle followed by administration of antiviral drugs (12-14). This targeted “oncolytic therapy” requires the initiation of EBV lytic cycle and expression of viral kinases, which phosphorylate nucleoside analogues (e.g. Ganciclovir, GSV) by converting the pro-drug to an active, selective suicide substrate for the viral and cellular DNA polymerases. This strategy aims to lower side effects associated with standard chemotherapy presently used to treat lymphomas and provides a molecular targeted therapeutic strategy by exploiting the biology of a key etiologic disease factor.
Limitations of viral “oncolytic therapy” is that most existing methods activate the lytic life-cycle in only a small percentage of latently infected cells, are toxic and are cell-type restricted. In addition, out of many known chemical activators of the EBV lytic cycle, only the histone deacetylase inhibitors derived from butyrate analogues have been tested in clinical trials (15, 16). In one clinical trial, arginine butyrate was found to be efficacious but was not tolerated due to toxicity, while sodium butyrate (NaB) was found to have unsuitable pharmacokinetics (15, 16). More recent studies have screened clinically approved drugs for potential activators for latent EBV and identified bortezomib as an activator of latent EBV in a limited number of EBV positive BL cells (17). Whether bortezomib will activate EBV in a broad range of cell and tumor types, and whether it will be tolerated in clinical applications, remains to be determined.
Due to the need for improved efficacious therapeutics with lower toxicity and a decreased potential for recurrence of EBV-positive lymphoid malignancies, we have developed a cell-based system for the high throughput screening of thousands of compounds. This system utilizes an EBV positive BL cell line (MutuI,) carrying a stable episome-based reporter construct that responds with high-sensitivity and dynamic range to the EBV-encoded lytic cycle activator protein ZTA. We have screened 66,840 compounds in total and found a number of structurally related molecules that robustly activate EBV lytic cycle gene expression in multiple cell types, including BLs, LCLs, and NPC derived cell lines. These newly identified reactivators synergize with GCV to eliminate EBV-positive cells while maintaining low toxicity in the absence of the antiviral drug or with treatment of EBV negative cells.
Results
Development, optimization, and validation of a cell-based high-throughput screening assay
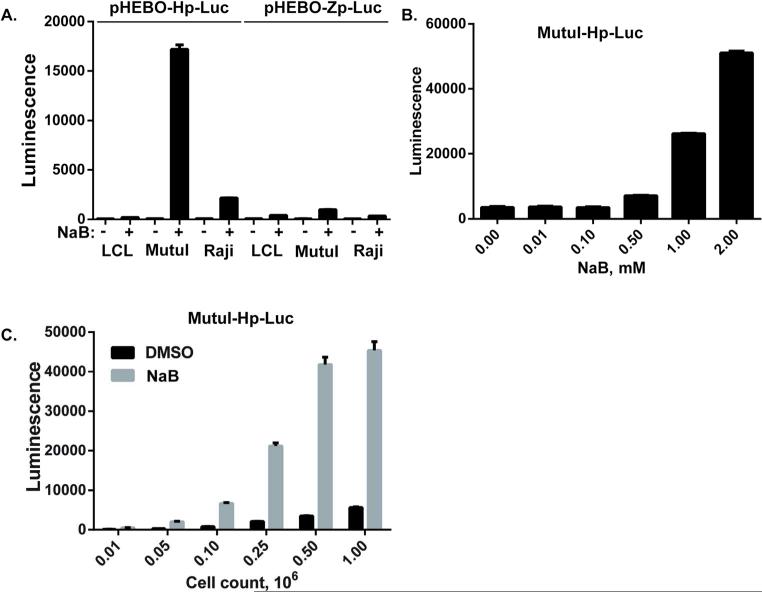
To develop a high-throughput, cell-based assay for identification of small molecule activators of EBV lytic cycle gene expression, we generated stable cell lines containing reporter genes under the control of EBV lytic cycle promoters for BZLF1 (Zp) or BHLF1 (Hp). BZLF1 is the immediate early gene encoding the ZTA lytic activator. BHLF1 is an early lytic gene highly responsive to ZTA transcriptional activation. To create reporter constructs that can be maintained as a chromatinized episomes similar to latent EBV genome, we utilized the OriP-containing, hygromycin resistant vector pHEBO (18). EBV promoters Hp and Zp were cloned upstream of the luciferase reporter gene in pHEBO to generate pHEBO-Hp-Luc and pHEBO-Zp-Luc. These plasmids were then used to generate stable episomal reporters in EBV positive cell lines LCLs, MutuI, and Raji. To test the responsiveness of the Hp- and Zp-luciferase reporters to EBV lytic cycle activation in each cell type, we assayed luciferase expression in cells treated with or without sodium butyrate (NaB). We found that pHEBO-Hp-Luc in MutuI cells provided the most robust response (>20 fold) to NaB relative to other reporters and cell lines (Fig. 1A). The pHEBO-Hp-Luc MutuI stable cell line (referred to as MutuI-Hp-Luc) was therefore selected for further development of a high-throughput assay. For these experiments, we used NaB as a positive control compound for lytic activation and DMSO as a negative control. We independently assessed the assays tolerance to DMSO and optimized response to NaB concentration, treatment time, and cell density. We found the assay to tolerate as much as 2% DMSO (data not shown). We tested a range of NaB concentrations to determine the maximum induction of luciferase and dose-dependency of the response. Treating MutuI-Hp-Luc cells with up to 2mM NaB consistently produced high luciferase signals and a signal-to-background of 20-40-fold, with relatively low cell toxicity (Figure 1B). We observed maximum luciferase stimulation achieved by 2 mM NaB after 48 hours of treatment, with longer periods of exposure to 2mM NaB led to a loss of cell viability (data not shown). We therefore chose 48 hour treatment for compound screening. We also determined that 25,000 cells per well for a 384-well plate was sufficient to achieve maximum signal with 2mM NaB (Figure 1C). To quantitatively assess the accumulative effect of these optimizations on assay's precision and accuracy to control conditions and validation for high-throughput screening, we tested the cellular response to DMSO and NaB in replicate 384-well assay plates prepared on 3 different days. For these experiments, one-half of each assay plate (n=192) was treated with DMSO and the other half with 2 mM NaB. The assay consistently yielded a Z-factor >0.7, a signal to noise (S/N) ratio >70, and a signal to background (S/B) ratio >7 (19) (Figure 2A). Based on these data, we concluded that the assay was sufficiently robust for high-throughput screening.
Figure 1. Development of a high-throughput assay for EBV lytic reactivation.
(A) Episomal plasmids pHEBO-Hp-Luc or pHEBO-Zp-Luc were used to generate stable cell lines in EBV positive LCL281, MutuI, or Raji. Stable cell lines were then assayed for luciferase activity (Luminescence) after treatment with DMSO (-) or with 2 mM NaB (+). (B) The MutuI-Hp-Luc stable cell line containing pHEBO-Hp-Luc in MutuI cells was assayed for luciferase-dependent luminescence at 0, 0.01, 0.1, 0.5, 1.0 or 2.0 mM NaB. (C) MutuI-Hp-Luc cells were assayed for optimal cell density in 384 well plates upon treatment with either 0.1% DMSO or 2 mM NaB for 48 hrs. Cells were plated at 0.01, 0.05, 0.1, 0.25, 0.5, or 1.0 x106 cells per ml. Error bars represent standard deviation (SD) from the mean.
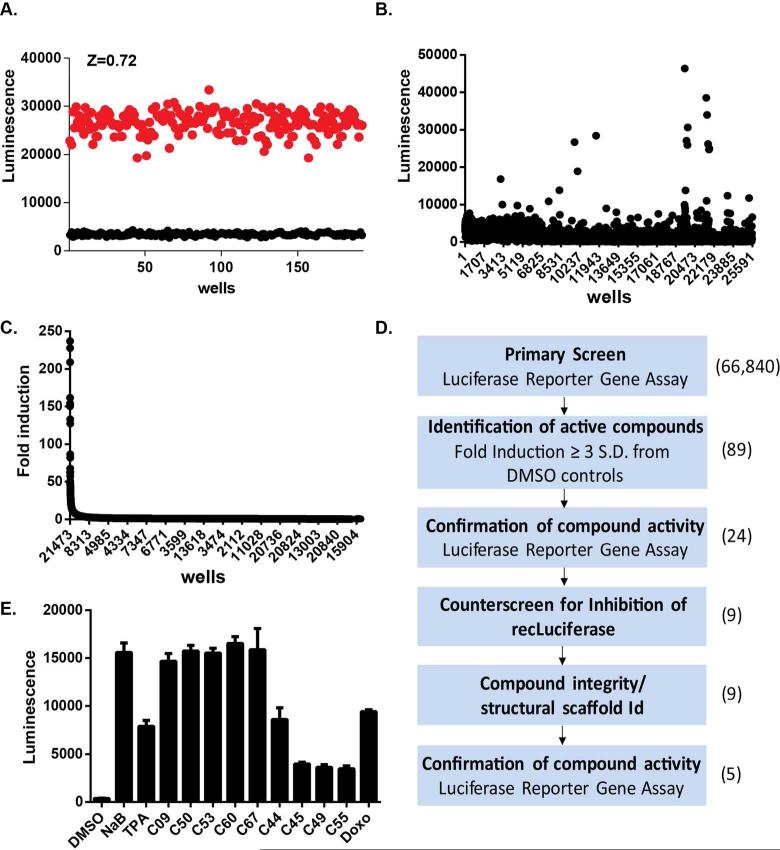
Figure 2. The high-throughput screen (HTS) validation and results.
(A) MutuI-Hp-Luc cells were analyzed in 384 well formats for statistical robustness. Scatter plot of the luciferase-dependent luminescence values for MutuI-Hp-Luc cells treated with DMSO (black) or with 2 mM NaB (red) for 48 hrs. Z-factor was determined to be 0.72. (B) Scatter plot of luminescence values for HTS campaign of ChemDiv library. The well number is indicated on the X-axis. (C) Data from HTS of ChemDiv screen (panel B) re-sorted according to the fold-induction of luminescence relative to DMSO. The x-axis is labeled as Log[comp mM] which indicates a base ten logarithm of compound concentration, in mM (D) Flow chart of the screening campaign and validation studies. (E) Confirmation luciferase assay for 9 positive hits delivered individually in each well at 10 NM. Positive controls include NaB, TPA, and Doxorubicin (Doxo).
High-throughput screening results
To identify small molecules that activate the EBV lytic life cycle, we screened a total of 66,840 compounds, representing known bioactive compounds, natural products, focused libraries aimed at specific target classes (e.g. kinases, receptors, proteases), and 3) diversity sets representing areas of chemical space. The average Z-factor and signal window for DMSO and 2 mM NaB treated wells of control plates throughout the screen was 0.698 and 9.17, respectively. The mean luciferase activity of all test wells in the screen was equal to the average luciferase activity of DMSO treated control wells on each assay plate. Using an initial cutoff of greater than 3 standard deviations from the mean DMSO control signal, we identified 89 candidate activators of the EBV lytic life cycle through this screen (Figures 2B and 2C). To confirm the activity of these candidate hits, we performed a secondary confirmation assay with compounds picked from the library source plates. Each compound was tested in triplicate at a single concentration of 10 uM in MutuI-Hp-Luc cells. We were able to confirm the activity of 24 compounds that induced luciferase expression greater than two fold in this assay. To eliminate the possibility that these compounds acted directly on the luciferase enzyme, and not on EBV, we tested them for their ability to inhibit purified luciferase in vitro. Many small molecules have been shown to inhibit the luciferase enzyme in vitro and stimulate luciferase activity in cell-based screens due to luciferase protein stabilization (20). Fifteen of these 24 compounds with confirmed activity in our cell-based reporter gene assay inhibited recombinant luciferase, and thus were eliminated from further consideration (data not shown). This screen/counterscreen scheme yielded 9 candidate activators of the EBV lytic life cycle for an overall hit rate of 0.013% (summarized in Figure 1D).
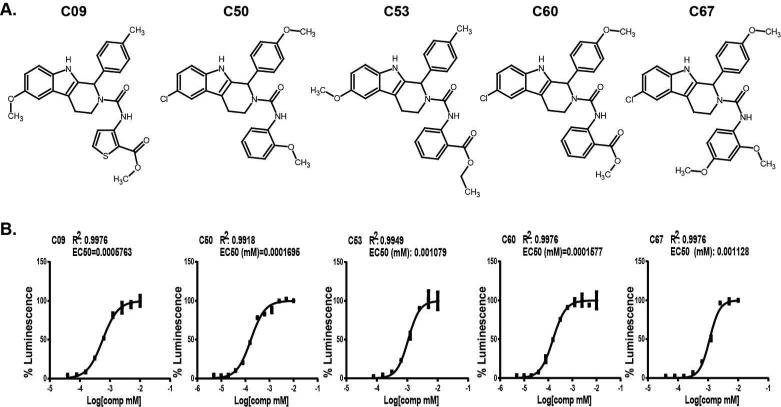
To further investigate the activity of these compounds and the potential mechanism of action, we purchased fresh powder supplies of each compound, confirmed their mass and purity by LC/MS, and retested their activity in our cell-based reporter gene assay. Five out of nine compounds confirmed activity comparable to 2mM NaB (Figure 2E). None of the 5 confirmed candidates showed significant inhibition of recombinant luciferase in vitro (data not shown). Remarkably, all five EBV activators shared similar structure belonging to the same chemical family (Figure 3A). To further characterize the activity of these small molecules we assessed the concentration-dependent response of each compound's activity. As shown in figure 3B, each compound displayed concentration-dependent responses with EC50 values that range between160 nM to 1 uM. C50 and C60 were the most potent activators, with EC50 values at 160-170 nM. In contrast, NaB and arginine butyrate typically required millimolar concentrations to trigger the latent to lytic switch (16, 21, 22) (Figure 1B).
Figure 3. Structure and EC50 analysis of five candidate small molecule activators of EBV.
(A) Chemical structures of five candidate small molecule EBV activators C09, C50, C53, C60, C67. (B) Dose response curves for each compound titrated 2-fold starting from 10NM concentration. EC50 and R-values are indicated in each panel.
Newly identified compounds shown broad tropism for activation of EBV lytic cycle gene expression
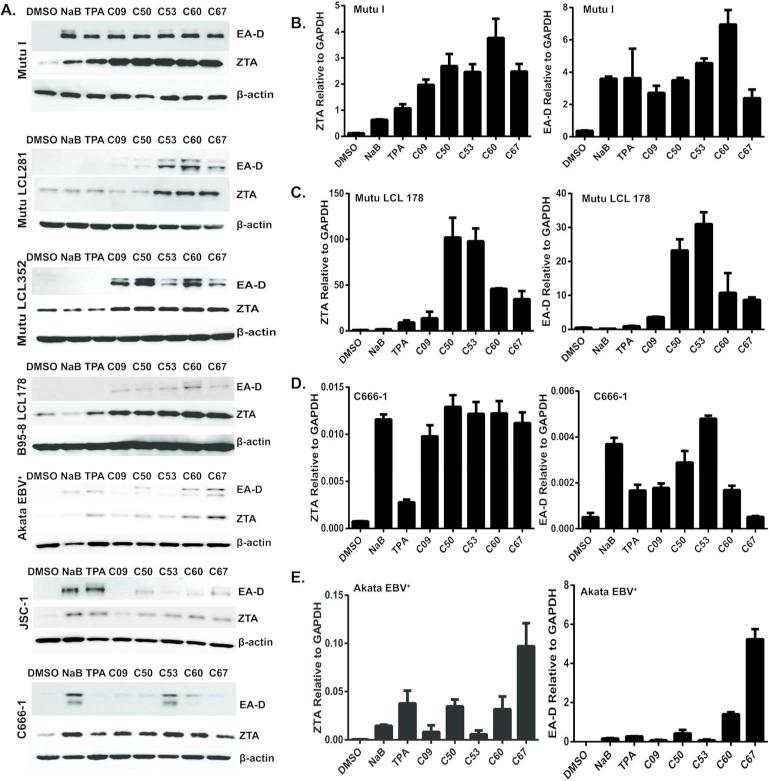
To date, no single EBV activator consistently reactivates EBV in all EBV3 positive cell lines (23, 24). We have observed that some BL cell lines (such as MutuI) can be reactivated with NaB, while LCLs that have been cultured in vitro for several weeks lose their sensitivity to NaB or TPA treatment. We compared a variety of cell lines with different latency types to determine whether the newly identified compounds are only active in MutuI or can be used to initiate lytic expression in other cells (Fig. 4). Compounds C09, C50, C53, C60, C67 were compared with positive controls NaB or TPA, relative to DMSO negative control. We assayed EBV lytic antigens EA-D and ZTA expression by Western blot for MutuI (Type I BL), various LCLs (Type III LCL), Akata (Type I BL cell), JSC1 (KSHV co-infected PEL cell) and C666-1 (Type II NPC cells). We also assayed EA-D (BMRF1 gene) and Zta (BZLF1 gene) expression by RT-PCR for MutuI, Mutu-LCL, C666-1, and Akata cells (Fig. 4B-E). For all cell lines tested, the new compounds were able to upregulate expression of EA-D and ZTA. In several cases, the compounds stimulated EA-D and ZTA to levels equal to or greater than 2 mM NaB treatment. This indicates that these compounds have a broad tropism for activation of EBV lytic cycle gene expression.
Figure 4. Various latency types are switched to lytic cycle by the new compounds.
(A) Western blot analysis of EA-D and ZTA expression in EBV-positive cell lines treated with DMSO, NaB, TPA and the compounds C09, C50, C53, C60, C67 for 72 hours. β–Actin is loading control. Cell lines include MutuI, Mutu-LCL281, Mutu-LCL352, B95.8-LCL178, Akata, JSC1, and C666-1. (B-E). Real-time RT-PCR analysis of EA-D (left panels) and ZTA expression (right panels) in EBV-positive cell lines, treated with DMSO, NaB, TPA or the indicated compounds for 48 hours. Cell lines include MutuI (B), Mutu-LCL178 (C), C666-1 (D), and Akata (E).
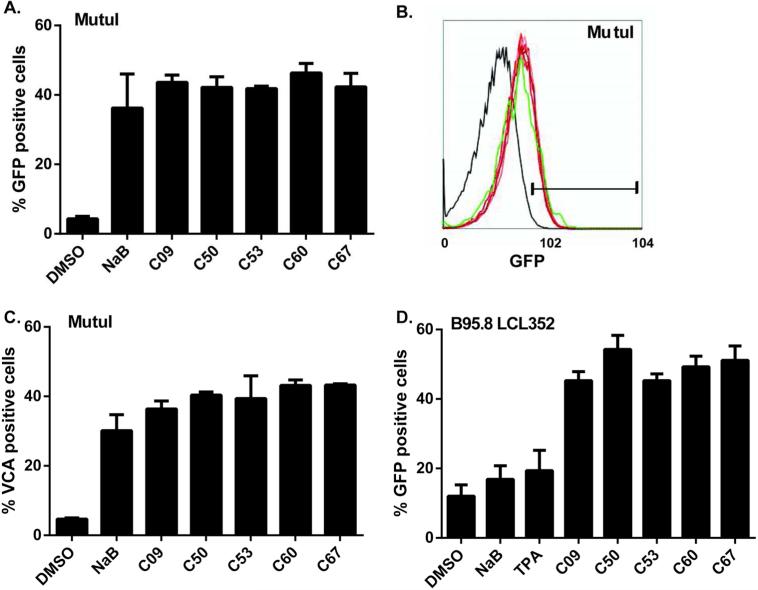
The newly identified compounds boost the percentage of lytic cells in culture
Most chemical activators of EBV lytic gene expression trigger reactivation in only a small proportion (up to 30%) of the cell population (23-25). Triggering lytic reactivation in a higher percentage of refractory cells is an important goal for EBV lytic therapy. To determine the percentage of MutuI and LCL cells reactivated with the newly identified compounds, we have generated a MutuI-Hp-GFP stable cell line to monitor lytic reactivation using fluorescence activated flow cytometry. Immunostaining for viral capsid antigen (VCA) was also used to confirm the findings. We found that 40-50% of MutuI–Hp-GFP cells expressed GFP after 72 hours of treatment with these compound reactivators (Figure 5A and 5B), and a comparable percentage of MutuI cells expressed VCA on their surface after a 96-hour treatment (Figure 5C). Treatment of B95-8-infected LCLs that carry a GFP-EBV genome with these activators induced greater than 50% of the cells to become lytic after a 72-hour treatment (Figure 5D). In all cases, the newly identified compounds induced lytic activation in an equal or greater percentage of cells compared to treatment with 2 mM NaB
Figure 5. Percentage of lytic cells induced by different compounds.
(A) FACS analysis of the percentage of GFP-positive (lytic) cells in MutuI-Hp-GFP cells after 72 hours of treatment with either 2% DMSO, 2 mM NaB, 20 ng/ml TPA or 1 M compounds.
(B) FACS scan of GFP-positive (lytic) MutuI-H-GFP cells after 72 hours of treatment with either DMSO (black), NaB (green) or compounds (different shades of red). Gate separates the GFP-positive population. (C) Percentage of VCA-positive (lytic) MutuI cells determined by FACS analysis after 96 hours of treatment with DMSO, NaB or compounds. (D) Same as in panel A, except for B95.8 LCL352 containing pHEBO-Hp-GFP.
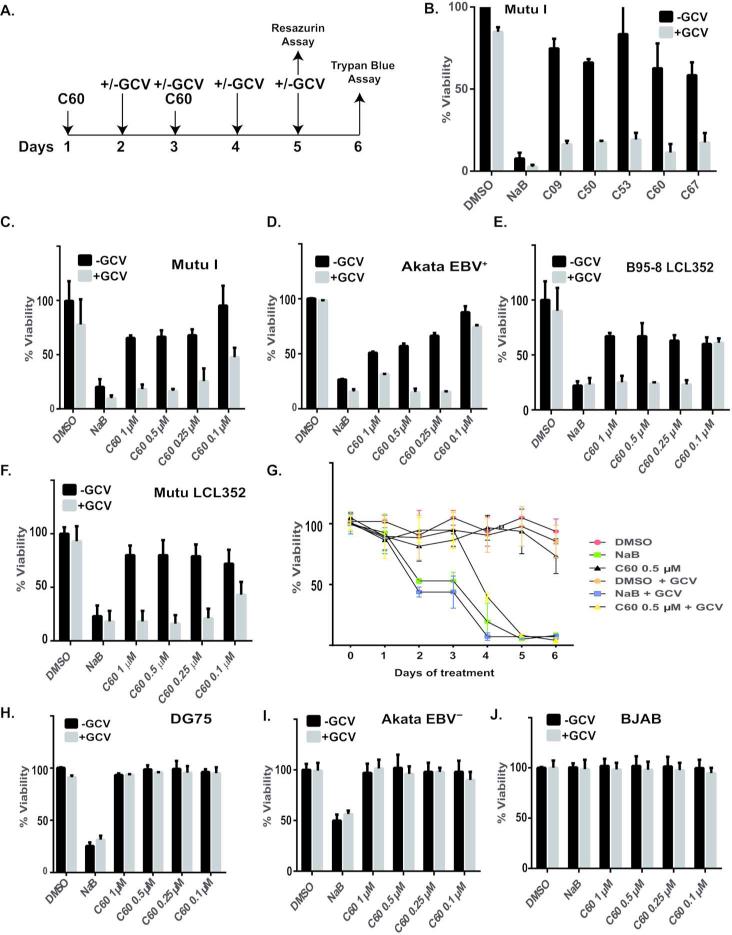
EBV activator C60 and GCV synergize in inducing apoptosis in EBV-positive cells
Combining agents that induce the lytic cycle of EBV with inhibitors of viral lytic replication has been shown to be a promising strategy to eliminate EBV-positive cancers (15, 16). We therefore tested whether combination of the newly identified activators and GCV would increase selective cell killing of EBV infected cells. The experimental design is described in Fig. 6A. We first tested whether combining GCV with a sublethal dose of compounds (1 M) could induce cell death in MutuI cells after six days of treatment. Cell viability was measured by trypan blue dye exclusion. As a single agent GCV reduced MutuI cell viability by ~10% over the course of 6 days. Treatment of MutuI cells with 1 M of the newly identified activators as single agents reduced the viability of cultures by 15 to 30%, possibly due to the fact that EBV reactivation itself reduces host cell viability. The combination of compounds with GCV produced a synergistic effect killing nearly 80% of MutuI cells within 6 days. (Figure 6B)
Figure 6. EBV activator C60 and GCV synergize in inducing apoptosis in EBV-positive cells.
(A) The scheme of experiment. MutuI-Hp-Luc were seeded at 0.2 million per ml and treated with C60. C60 was added again 48 hours later. Freshly made GCV was added on Day 2 for 4 days twice a day, in the morning and 8 hours later. Cell viability was measured either with Resazurin 5 days after the first C60 treatment, or the cells were stained with Trypan blue on Day 6. (B) Percentage of live cells treated with compounds or controls with (gray bars) or without (black bars) GCV, for 6 days, as noted in (A.) Live cells were counted by Trypan Blue stain exclusion method. (C-F) Resazurin cytotoxicity assay on cells treated with compounds and controls for 5 Days, according to experimental scheme in (A). Different concentrations of C60 combined with GCV (gray bars) or without GCV (black bars) were used. Cell viability was measured as percentage of DMSO treated control cells. Cells tested include EBV positive MutuI (C), Akata (D), DG75 (E), Mutu-LCL352 (F). Daily viability assay for MutuI cells treated with either DMSO, NaB, or C60 (0.5 M) with or without GCV. (G-I) Rezazurin cytotoxicity assay on EBV negative cells DG75 (H), Akata EBV- (I), or BJAB (J) treated identically as described for panels C-F.
Compound C60 was selected for dilution studies since it showed the greatest potency in MutuI cell based assays (Fig. 3B), and consistently stimulated EBV lytic reactivation in multiple cell types (Fig. 4). We treated EBV-positive MutuI, Akata, LCLs and EBV-negative Akata, DG75 and BJAB cells with a range of C60 concentrations either as a single agent or in combination with 10ug/ml of GCV according to the scheme (Figure 6A). For these experiments we used a fluorescence-based readout (resazurin/Alomar blue) assay as the indicator of cell viabillty. Synergistic cell killing was observed for EBV positive MutuI (Fig. 6C), EBV-positiveAkata (Fig. 6D) and LCL cells (Fig. 6E and F). In contrast, C60 produced no observable effect on the viability of a EBV negative BL cell line DG75 (Fig. 6H), EBV-negative Akata (Fig. 6I), or BJAB (Fig. 6J) either as a single agent or in combination with GCV. We followed the scheme in Fig.6A to address whether the addition of GCV to either C60 or NaB kills MutuI cells in a time-dependent manner. We took daily measurements of viability of MutuI cells treated with DMSO, NaB or 0.5uM of C60 with or without GCV (Fig. 6G) using resazurin. The synergistic effects of C60 with GCV could be observed by day 4. In contrast, NaB did not synergize with GCV, and it was toxic to cells regardless of EBV positive status, possibly due to its broad function as HDAC inhibitor, and stimulation of many genes unrelated to EBV function (Fig. 6G). These findings indicate that our newly identified activators of the EBV lytic life cycle can function synergistically with GCV for selective killing of EBV positive cells.
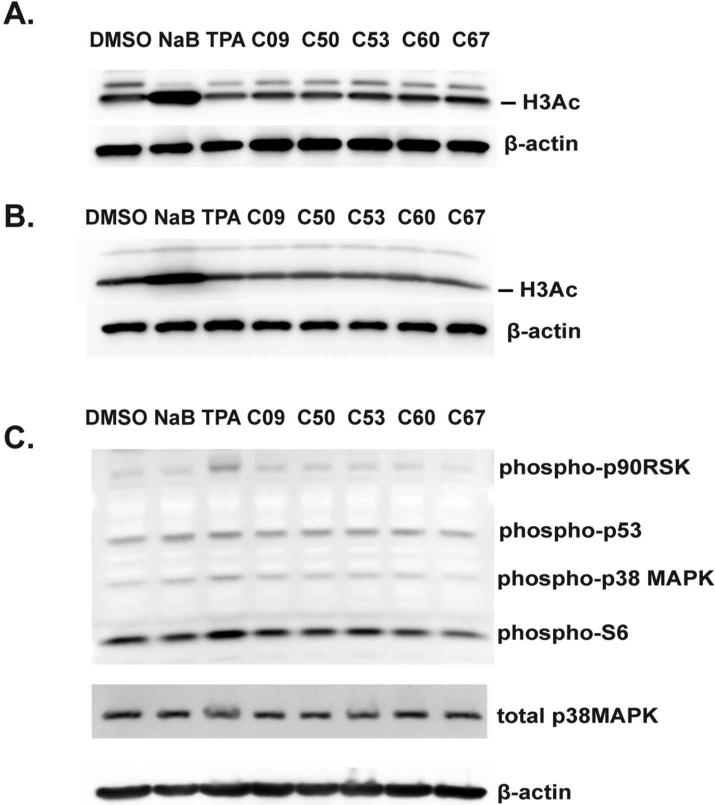
Mechanism of EBV reactivation of the new compounds is distinct from HDAC inhibitors and TPA
Butyrates are known inhibitors of HDACs (26). The mechanism of TPA reactivation of EBV has also been studied in depth (24, 27, 28). TPA first activates PKC and within 30 minutes triggers p38MAPK kinase phosphorylation (29), resulting in upregulation of MAPK pathway. Through activation of Erk1/2, TPA is also known to phosphorylate p53 and ribosomal kinases, such as p90RSK and S6 (29). We have used two cell lines MutuI and Mutu-LCL352 to test whether the newly identified EBV activators function as HDACs by increasing acetylation on histone H3. While NaB caused histone acetylation in both cell lines as expected, none of our EBV activators stimulated H3 acetylation, indicating they do not function as HDACs, like NaB (Figures 7A and 7B). To test whether the newly identified compounds share the mechanism of EBV reactivation with TPA, we used an antibody cocktail to detect p38 MAPK, S6, p90RSK and p53 phosphorylation. As expected, TPA showed an increase in phosphorylation of these kinases. In contrast, the compounds failed to induce detectable levels of phosphorylation of these tragets, suggesting that the mechanisms of EBV activation by the newly identified compounds have a mechanism distinct from TPA (Figure 7C).
Figure 7. Mechanism of EBV reactivation of the new compounds in distinct from HDAC inhibitors and TPA.
(A) MutuI cells were treated with 2% DMSO, 2 mM NaB, 20ng/ml TPA, or 1 μM compounds for 48 hours, immunoblotted, and probed for acetylation of histone H3 and β-actin. (B) Mutu-LCL352 cells were treated and probed as in (A). (C) MutuI cells were treated with controls or compounds for 0.5 hours, immunoblotted, and probed for phosphorylated forms of p90RSK, p53, p38MAPK, S6, total MAPK, and β-actin.
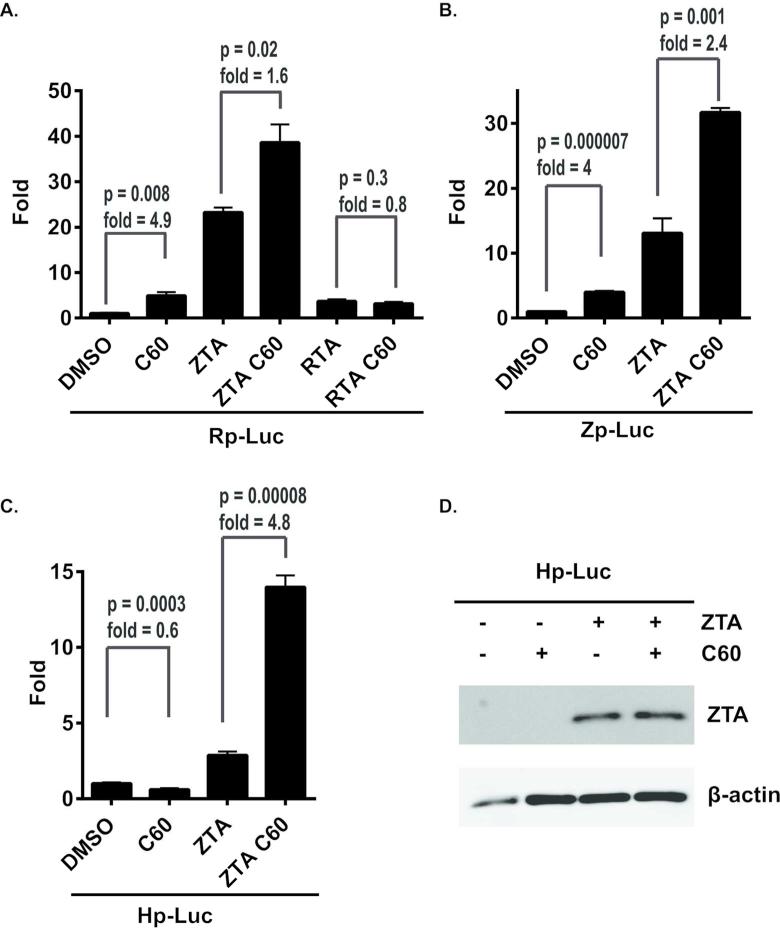
New compounds augment ZTA-dependent transcription activation of viral early promoters
To test whether the class of new compounds act by transcription activation of ZTA(Zp), RTA(Rp) or BHLF1(Hp) promoter, we transfected 293T cells with plasmids containing Zp, Rp, or Hp promoter regions controlling Luciferase reporter and treated them with either DMSO or 1 uM of C60 (Figure 8A-C). C60 alone produced an ~4-fold activation of Zp and Rp (Figure 8A-B), but had slight inhibitory effect on Hp (Figure 8C). To test whether C60 enhances ZTA or RTA-dependent activation, we contransfected cells with ZTA or RTA, and treated them with either DMSO or 1 uM of C60. We found that C60 augmented ZTA activation on all three promoters, increasing luciferase signal 4.8 fold in Hp-Luc cells, 2.4 fold in Zp-Luc and 1.6 fold in Rp-Luc cells (Figure 8A-C). In contrast, C60 had no effect on RTA activation of Rp (Figure 8A). C60 did not increase ZTA protein levels or mobility as indicated by Western blot analysis of transfected cells (Figure 8D). We conclude that C60 can stimulate the core promoter activities of Zp and Rp, as well as further enhance the transcription activation function of ZTA on these viral promoters.
Figure 8. C60 can activate Zp, Rp, and ZTA transcriptional activation function.
(A-C) Luciferase reporter assays with the Rp-Luc (A), Zp-Luc (B), or Hp-Luc (C) treated with either 1 M DMSO or C60 with the addition of ZTA or RTA expression vector as indicated. Error bars represent sdm, and p-values for the C60 response is indicated. (D) Western blot of Zta and β-actin from 293 transfected cells used for luciferase assays.
Discussion
We have developed and validated a high-throughput, 384-well cell-based assay that measures the ability of a small molecule compound to switch latent EBV to lytic cycle gene expression. The assay was robust and sensitive, featuring Z-factors consistently exceeding 0.7, signal-to-noise ratio of 70.2 and signal-to-background ratio of 7.7. We have used luciferase as a reporter placed under the control of the EBV BHLF1 promoter (Hp) to indirectly measure the ZTA expressed by reactivated EBV. Using this assay, we were able to screen 66,840 compounds that resulted in the discovery of 5 compounds with activities consistently comparable to NaB. All five “candidate” compounds possess structural similarity. Two of these compounds have EC50 activity measured between 150-170 nM, making them possible candidates for further drug development.
Identification of new activators of EBV is important for the improvement of EBV-lytic therapies. Most of the existing methods for activating EBV are frequently cell line specific or are limited by the low percentage of cells that can be reactivated. The new compounds we have identified have improved cell tropism and increase frequency of activation relative to standard reactivating reagents, NaB and TPA. We found that C60 and its analogues can induce lytic activation in a wide variety of EBV positive cells, including BLs, LCLs, PELs, and NPC derived cell lines. We also found that these newly identified compounds can induce nearly 50% of MutuI and over 50% of LCL cell - the highest percentage that has been achieved to date for chemical reactivation methods. The mechanisms that restrict the cell type and population percentage are not well understood. Cell-type specific signaling pathways, like B-cell receptor signaling and unfolded protein response pathways, have been implicated in control of EBV reactivation in some cell types (30, 31). Epigenetic controls, including DNA methylation and histone deacetylation have been implicated in other cell types (22, 24). In addition, host cell factors, like STAT3 and EBF1 have also been implicated in the control of lytic cycle reactivation frequency (25, 32). While we do not yet know the target of the newly identified compounds, we found that C60 was capable of stimulating transcription of the EBV immediate early gene promoters Zp and Rp, as well as augment ZTA transcription activation (Fig. 8). These findings suggest that C60 functions through a pathway that regulates transcription control of viral immediate early promoters and the ZTA transcriptional activator. Future studies will be required to further characterize the molecular targets of these compounds and their mechanisms of activation of EBV lytic cycle gene expression.
Remarkably, all five novel EBV activators share similar structure belonging to the tetrahydrocarboline structural class of compounds. (Figure 3A). Tetrahydrocarbolines are an interesting class of molecules that are known to possess diverse biological activity such as kinesin spindle protein, topoisomerase II, protein tyrosine phosphatase, phosphodiesterase inhibition, and sst3 receptor antagonists (33, 34). The five active molecules provide some initial structure activity relationships (SAR) required for EBV reactivation. All five have substitution on the 6 position of the indole ring with either a chloro or a methoxy group. Also all five are linked through the piperidine nitrogen with a urea linkage terminating with a substituted aromatic ring. The initial lead chemical series is chemically tractable and readily amenable to analog synthesis to flesh out the SAR to improve potency and drug-like properties of this novel class of EBV reactivators.
Out of five novel small molecule EBV activators, C60 is consistently a well-performing EBV lytic cycle inducer in all cell lines tested. Titration of C60 (Fig.5) with GCV showed that C60 synergizes with GCV on killing EBV-positive cells only at concentrations that cause EBV reactivation (Fig.2). Furthermore, this cytotoxic synergy is not observed in EBV-negative cells. In contrast to NaB, all five novel compounds did not produce cell toxicity in EBV negative cells. This selectivity offers possibility for further investigation of in vivo efficacy and toxicity of these compounds.
We have made preliminary attempts to compare the molecular mechanisms of EBV reactivation by the novel compounds to that of NaB. Our data suggests that the identified compounds’ mechanisms of action are distinct from already known came from our observation that the new compounds are able to reactivate EBV even in MutuI and B95.8 LCL cell lines resistant to NaB or TPA. Checking histone H3 acetylation and the activity of p38 MAPK signaling cascade indicated that the new EBV activators are unlikely to be HDAC inhibitors, like NaB, nor stimulate the cellular signaling pathways commonly activated by TPA. The discovery of the molecular target and the signaling affected by the novel EBV activators is ongoing. In conclusion, we have identified a chemical series that can activate EBV from a wide range of latently infected lymphoid and epithelial derived tumor cells. These newly identified reactivators have the potential to improve therapeutic approaches for EBV specific oncolytic therapies.
Methods
Cell culture
Akata (EBV+ BL), BJAB, Akata EBV negative (EBV- BL), JSC-1(EBV+/KSHV+ PEL), DG75 (EBV- BL), MutuI (EBV+ BL), C666-1 (EBV+ NPC), 293T cells were obtained from ATCC. LCL cell lines were made by in vitro immortalization with EBV by infecting B-lymphocytes isolated from blood obtained from different EBV-negative donors with either B85-8 or MutuI EBV strains. LCLs derived from MutuI virus are referred to as Mutu-LCLs. All cell lines had low passage and were cultured for no more than 1 month in RPMI supplemented with 13% heat inactivated FBS, 50Ng/ml Penicillin, and 1% Glutamax (Invitrogen) at 37°C and 10% CO2. Cell concentration was maintained at 0.2-0.8 million per ml, and cell viability was over 90% for each cell line at the time of compound screening or before each treatment. For the assay development, confirmation assays, and dose-response curves, 104 cells were plated in 96 well white plates. Drug treatments of were carried out in RPMI media supplemented with 5% FBS.
Immunoblotting
Cells were incubated with either DMSO, 2mM sodium butyrate (NaB), 20 Ng/ml 12-O-tetradecanoyl phorbol-13- acetate (TPA) or 1 NM of compounds for 72 hours, washed in PBS once and lysed in 1xLaemmli buffer. Protein extract equivalent to 105 cells were loaded on 8-16% SDS-PAGE gels (Invitrogen) and transferred to nitrocellulose membranes (Millipore). Lytic reactivation was detected with a rabbit polyclonal anti-ZTA and a mouse monoclonal anti-EA-D (Abcam) antibodies diluted in 5% evaporated milk and incubated with the membranes for 2 hours at RT, or overnight at 4°C, followed by secondary anti-mouse or rabbit HRP-conjugated (Roche). Rabbit antibodies against phospho-p38 MAPK, -S6, -p90RSK and -p53 West Pico are mixed into a PathScan cocktail (Cell Signaling). West Femto (Thermo) and ECL Prime (GE) were used to detect the proteins with Fujifilm LAS-3000 camera and software.
FACS analysis and immunofluorescence.
For the immunofluorescence staining of viral capsid antigen, cells were fixed with 4% paraformaldehyde in PBS, washed and blocked with PBS containing 1%BSA. Anti-VCA antibody (Millipore) was used at 1:100 followed by anti-mouse R-phycoerythrin (PE)-conjugated antibody (Sigma). FACS analysis was used to analyze the percentage of PE-positive cells. LSR-II instrument () and FACSDiva software (BD Biosciences) were used to analyze the samples. FloJo software was used to create graphs (Tree Star, Inc).
Real-time PCR analysis
Cells were incubated with DMSO, 2mM NaB, 20 Ng/ml phorbol ester (TPA) or 1 NM of compounds for 48 hours. Total RNA was isolated with Trisol reagent and reverse-transcribed with random hexamers and Superscript II RT polymerase (maker). Primers used for amplification of EBV and cellular gene have been published previously (35), and are available upon request.
Luciferase and GFP Reporter Construction
BHLF1 promoter (Hp) or the BZLF1 promoter (Zp) was cloned into pHEBO plasmid into NheI and HindIII restriction sites of pHEBO-Luc plasmid (18, 36). To create pHEBO-GFP, most of Luciferase gene was excised by BsrGI and HindIII sites and replaced with EGFP excised from pEGFP-N1 (Clontech). MutuI cells were transfected with the new pHEBO-Zp-Luc, pHEBO-Hp-Luc or pHEBO-Hp-GFP constructs and transfected cells were selected with 400 Ng/ml of Hygromycin. After 2 weeks of selection, stable hygromycin-resistant cells were seeded at 5000 per well in 384 well white plates and assayed as described below.
For the Rp, Hp and Zp stimulation assays, we used pHEBO-Hp-Luc, while Zp and Rp were inserted into pGL3-Basic plasmids (Promega). ZTA was cloned in pcDNA3 (Invitrogen) and RTA was expressed from pRTS15-RTA (37). All 293T cells were co-transfected with Renilla Luciferase control plasmid expressed from pGL4.74[hRluc/TK] vector (Promega)
High-throughput screening library
The Wistar Molecular Screening Facility small molecule library is a collection of both synthetic and natural products from commercial sources that can be grouped into three categories: 1) known bioactive compounds, including drugs; 2) focused libraries aimed at specific target classes (e.g. kinases, ion channels, GPCRs); and 3) diversity sets representing areas of chemical space. This library is comprised of Spectrum 2000, the NIH clinical collection, a 14,400 compound library from Maybridge, and a 50,000 compound library from ChemDiv. The Maybridge “Hit Finder” library represents lead-like diversity from the larger Maybridge screening collection (56,000). The ChemDiv library consists of a 30,000 compound “Discovery Chemistry” set of diverse lead-like small molecules, and 20,000 compounds from a “Target Specific” set, with subsets of compounds designed to target various enzymes and receptors. The Maybridge and ChemDiv libraries were formatted in self-deconvoluting mixtures (5XY) for high-throughput screening where each compound is represented twice in the screen, and mixed with 4 unrelated compounds in separate wells. This approach enables screens to be done 2.5-fold more efficient with respect to reagent and consumable consumption without sacrificing biological replicates or ability to rapidly identify bioactive compounds (38).
High-throughput screening
Assay plates were prepared by seeding 25,000 MutuI-Hp-Luc cells in 50 ul of RPMI supplemented with 15% FBS in white opaque 384 well tissue culture plates (Corning), using a Biotek Microflo and a 5ul cassette. Fifty nanoliters of test compounds or DMSO was transferred to assay plates using a Janus nanohead MDT (Perkin Elmer). Columns 1, 2, 23, and 24 received DMSO (0.2% final concentration). The remaining 320 wells in columns 3-22 received test compounds at a final concentration of 10 uM cumulative concentration for the 5 compounds/2uM individual compound concentration/0.2% final DMSO concentration. Cells were incubated with compounds for 48 hours at 37°C/5% CO2. Twenty microliters of Steady-Glo Luciferase Reagent (Promega) was added to assay plates and incubated for 15 minutes at room temperature. Plates were centrifuged and luminescence was measured on an Envision Excite multilabel microplate reader, using the ultrasensitive luminescence measurement technology (Perkin Elmer).
Data Analysis and quality control
Data for test compounds was normalized to DMSO plate controls to calculate fold stimulation (i.e. fold stimulation = Test(compound)/Average(DMSO)) and z-score (Z-score =(Test(Compound)-Average(DMSO))/StdDev(DMSO) using OpenHTS (CeuticalSoft). The average fold stimulation and three times the standard deviation of all compounds screened were calculated. The sum of these two aggregate values was used as an initial cutoff to identify active compound wells, i.e. any compound well that exhibited stimulation of luciferase greater than the cutoff was considered to contain an active molecule and used for subsequent deconvolution of the screening mixtures. Orthogonally screening mixtures were deconvoluted at a well level and stratified into 4 categories: 1) actives (i.e. the bioactive compound cleanly maps to a unique well in both the horizontal and vertical dimensions); 2) ambiguous (i.e. the bioactive compound maps to 2 or more wells in either dimension); 3) orphan (i.e. an orthogonal match could not be identified in the second dimension); or 4) inactive (i.e. fold-stimulation < cutoff parameter). Z factor was used to measure the statistical robustness of the high-throughput screening (19). Z factor equals (1-3(σp-σn)/|μp-μn|) where σ is variance, μ is mean, p is positive, and n is negative.
Compound confirmation, counterscreens and quantitative assessment of compound activity
To confirm the activity of candidate compounds identified after deconvolution that exceeded the cutoff parameter, liquid stock from the screening library was retested as single compounds at a final concentration of 10 uM, using assay conditions identical to the primary high-throughput screen. Data quality and compound activity was assessed as described above. To eliminate confirmed actives that may interfere with the assay technology by inhibiting luciferase, we tested the activity of all compounds in a biochemical luciferase assay counterscreen (39). To quantitatively assess the activity and prioritize compounds for further follow-up, fresh liquid stocks prepared from powders were reformatted as a 10 point dilution series spanning at least 3-logs, starting at 10 uM, the initial screening concentration. Data for test compounds was normalized to DMSO and NaB (1 mM) plate controls to calculate fold stimulation (i.e. fold stimulation= test_well/ Avg_DMSO). The relative EC50 for each compound (i.e. the concentration of compound that produced 50% of the maximum effect) was calculated using a 4 point non-linear regression analysis (GraphPad Prism 6). Error bars represent SD for each concentration of a compound. To assist with decision making points, compound activities were stratified into 4 categories: class 1- well defined top and bottom asymptotes; class 2- incomplete stimulation (i.e., no top asymptote-100% induction); class 3-compounds with activity at a single concentration or steep Hill slopes; class 4-inactive. Compounds with activities that fall into class 1 and 2 were prioritized for further analysis.
Cell viability Assays
Assay plates were prepared by seeding 25,000 MutuI cells in 50 ul of RPMI supplemented with 15% FBS in 384 well tissue culture plates (Nunc). One hundred nanoliters of test compounds or DMSO was transferred to assay plates using a Janus nanohead MDT (Perkin Elmer). Cells were incubated for 5 days at 37°C/5% CO2 and viability was monitored by the addition of 20 ul of 500 uM Resazurin (Sigma) and incubation for 4 hours at 37oC. Plates were centrifuged and fluoresence intensity was measured at 590 nm on an Envision Excite multilabel microplate reader (Perkin Elmer)
Acknowledgments
We thank the members of the Lieberman lab and the Wistar Institute Molecular Screening Facility for their advice and technical support. This work was supported by grants from NIH P30 CA 010815-42 (D. Altieri) to the Wistar Institute Cancer Center Core Support, NIH RO1 CA085678 to PML, and the Wistar Institute Cancer Biology Training Grant from the NIH (1T32 CA09171)
References
- 1.Rickinson AB, Kieff E. Epstein-Barr Virus. 5th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 2.Kieff E. Epstein-Barr Virus and its replication. 5th ed. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 3.Thorley-Lawson DA. EBV the prototypical human tumor virus--just how bad is it? The Journal of allergy and clinical immunology. 2005;116:251–261. doi: 10.1016/j.jaci.2005.05.038. quiz 262. [DOI] [PubMed] [Google Scholar]
- 4.Klein E, Kis LL, Klein G. Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions. Oncogene. 2007;26:1297–1305. doi: 10.1038/sj.onc.1210240. [DOI] [PubMed] [Google Scholar]
- 5.Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Seminars in cancer biology. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 6.Fukayama M, Ushiku T. Epstein-Barr virus-associated gastric carcinoma. Pathology, research and practice. 2011;207:529–537. doi: 10.1016/j.prp.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Loren AW, Porter DL, Stadtmauer EA, Tsai DE. Post-transplant lymphoproliferative disorder: a review. Bone marrow transplantation. 2003;31:145–155. doi: 10.1038/sj.bmt.1703806. [DOI] [PubMed] [Google Scholar]
- 8.Gottschalk S, Rooney CM, Heslop HE. Post-Transplant Lymphoproliferative Disorders. Annu Rev Med. 2004 doi: 10.1146/annurev.med.56.082103.104727. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton-Dutoit SJ, Rea D, Raphael M, Sandvej K, Delecluse HJ, Gisselbrecht C, Marelle L, van Krieken HJ, Pallesen G. Epstein-Barr virus-latent gene expression and tumor cell phenotype in acquired immunodeficiency syndrome-related non-Hodgkin's lymphoma. Correlation of lymphoma phenotype with three distinct patterns of viral latency. The American journal of pathology. 1993;143:1072–1085. [PMC free article] [PubMed] [Google Scholar]
- 10.MacMahon EM, Glass JD, Hayward SD, Mann RB, Becker PS, Charache P, McArthur JC, Ambinder RF. Epstein-Barr virus in AIDS-related primary central nervous system lymphoma. Lancet. 1991;338:969–973. doi: 10.1016/0140-6736(91)91837-k. [DOI] [PubMed] [Google Scholar]
- 11.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nature reviews. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez MI, Judde JG, Magrath IT, Bhatia KG. Switching viral latency to viral lysis: a novel therapeutic approach for Epstein-Barr virus-associated neoplasia. Cancer research. 1996;56:969–972. [PubMed] [Google Scholar]
- 13.Kenney S, Ge JQ, Westphal EM, Olsen J. Gene therapy strategies for treating Epstein-Barr virus-associated lymphomas: comparison of two different Epstein-Barr virus-based vectors. Human gene therapy. 1998;9:1131–1141. doi: 10.1089/hum.1998.9.8-1131. [DOI] [PubMed] [Google Scholar]
- 14.Moore SM, Cannon JS, Tanhehco YC, Hamzeh FM, Ambinder RF. Induction of Epstein-Barr virus kinases to sensitize tumor cells to nucleoside analogues. Antimicrobial agents and chemotherapy. 2001;45:2082–2091. doi: 10.1128/AAC.45.7.2082-2091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrine SP, Hermine O, Small T, Suarez F, O'Reilly R, Boulad F, Fingeroth J, Askin M, Levy A, Mentzer SJ, Di Nicola M, Gianni AM, Klein C, Horwitz S, Faller DV. A phase 1/2 trial of arginine butyrate and ganciclovir in patients with Epstein-Barr virus-associated lymphoid malignancies. Blood. 2007;109:2571–2578. doi: 10.1182/blood-2006-01-024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh SK, Perrine SP, Williams RM, Faller DV. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood. 2012;119:1008–1017. doi: 10.1182/blood-2011-06-362434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirley CM, Chen J, Shamay M, Li H, Zahnow CA, Hayward SD, Ambinder RF. Bortezomib induction of C/EBPbeta mediates Epstein-Barr virus lytic activation in Burkitt lymphoma. Blood. 2011;117:6297–6303. doi: 10.1182/blood-2011-01-332379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of biomolecular screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.Thorne N, Shen M, Lea WA, Simeonov A, Lovell S, Auld DS, Inglese J. Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chemistry & biology. 2012;19:1060–1072. doi: 10.1016/j.chembiol.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Countryman J, Gradoville L, Bhaduri-McIntosh S, Ye J, Heston L, Himmelfarb S, Shedd D, Miller G. Stimulus duration and response time independently influence the kinetics of lytic cycle reactivation of Epstein-Barr virus. Journal of virology. 2009;83:10694–10709. doi: 10.1128/JVI.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. Lytic cycle switches of oncogenic human gammaherpesviruses(1) Advances in cancer research. 2007;97:81–109. doi: 10.1016/S0065-230X(06)97004-3. [DOI] [PubMed] [Google Scholar]
- 23.Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. Lytic cycle switches of oncogenic human gammaherpesviruses. Advances in cancer research. 2007;97:81–109. doi: 10.1016/S0065-230X(06)97004-3. [DOI] [PubMed] [Google Scholar]
- 24.Daigle D, Gradoville L, Tuck D, Schulz V, Wang'ondu R, Ye J, Gorres K, Miller G. Valproic acid antagonizes the capacity of other histone deacetylase inhibitors to activate the Epstein-barr virus lytic cycle. Journal of virology. 2011;85:5628–5643. doi: 10.1128/JVI.02659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. Journal of virology. 2010;84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marks PA, Richon VM, Kelly WK, Chiao JH, Miller T. Histone deacetylase inhibitors: development as cancer therapy. Novartis Foundation symposium. 2004;259:269–281. discussion 281-268. [PubMed] [Google Scholar]
- 27.Nakagawa Y. Artificial analogs of naturally occurring tumor promoters as biochemical tools and therapeutic leads. Bioscience, biotechnology, and biochemistry. 2012;76:1262–1274. doi: 10.1271/bbb.120162. [DOI] [PubMed] [Google Scholar]
- 28.Gao X, Ikuta K, Tajima M, Sairenji T. 12-O-tetradecanoylphorbol-13-acetate induces Epstein-Barr virus reactivation via NF-kappaB and AP-1 as regulated by protein kinase C and mitogen-activated protein kinase. Virology. 2001;286:91–99. doi: 10.1006/viro.2001.0965. [DOI] [PubMed] [Google Scholar]
- 29.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nature reviews. Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 30.Amon W, Farrell PJ. Reactivation of Epstein-Barr virus from latency. Reviews in medical virology. 2005;15:149–156. doi: 10.1002/rmv.456. [DOI] [PubMed] [Google Scholar]
- 31.Lai IY, Farrell PJ, Kellam P. X-box binding protein 1 induces the expression of the lytic cycle transactivator of Kaposi's sarcoma-associated herpesvirus but not Epstein-Barr virus in co-infected primary effusion lymphoma. The Journal of general virology. 2011;92:421–431. doi: 10.1099/vir.0.025494-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies ML, Xu S, Lyons-Weiler J, Rosendorff A, Webber SA, Wasil LR, Metes D, Rowe DT. Cellular factors associated with latency and spontaneous Epstein-Barr virus reactivation in B-lymphoblastoid cell lines. Virology. 2010;400:53–67. doi: 10.1016/j.virol.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Pasternak A, Feng Zhe, de Jesus Reynalda, Ye Zhixiong, He Shuwen, Dobbelaar Peter, Bradley Scott A., Chicchi Gary G., Tsao Kwei-Lan, D. T., Eiermann George J., Li Cai, Feng Yue, Wu Margaret, Shao Qing, Zhang Bei B., Nargund Ravi, Mills Sander G., Howard Andrew D., Yang Lihu, Zhou Yun-Ping. Stimulation of Glucose-Dependent Insulin Secretion by a Potent, Selective sst3 Antagonist. ACS Med Chem Lett. 2012;3:289–293. doi: 10.1021/ml200272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daugan A, Grondin P, Ruault C, Le Monnier de Gouville AC, Coste H, Kirilovsky J, Hyafil F, Labaudiniere R. The discovery of tadalafil: a novel and highly selective PDE5 inhibitor. 1: 5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indole-1,3(2H)-dione analogues. Journal of medicinal chemistry. 2003;46:4525–4532. doi: 10.1021/jm030056e. [DOI] [PubMed] [Google Scholar]
- 35.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS pathogens. 2010;6 doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng Z, Chen C-J, Zerby D, Delecluse H-J, Lieberman PM. Identification of acidic and aromatic residues in the Zta activation omain essential for Epstein-Barr virus reactivation. J. Virol. 2001;75:10334–10347. doi: 10.1128/JVI.75.21.10334-10347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragoczy T, Miller G. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. Journal of virology. 2001;75:5240–5251. doi: 10.1128/JVI.75.11.5240-5251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson S, Messick T, Schultz DC, Reichman M, Lieberman PM. Development of a high-throughput screen for inhibitors of Epstein-Barr virus EBNA1. Journal of biomolecular screening. 2010;15:1107–1115. doi: 10.1177/1087057110379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. Journal of medicinal chemistry. 2008;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]