Abstract
Precise regulation of gene expression programs during embryo development requires cooperation between transcriptional factors and histone-modifying enzymes, such as the Gcn5 histone acetyltransferase. Gcn5 functions within a multi-subunit complex, called SAGA, that is recruited to specific genes through interactions with sequence-specific DNA-binding proteins to aid in gene activation. Although the transcriptional programs regulated by SAGA in embryos are not well defined, deletion of either Gcn5 or USP22, the catalytic subunit of a deubiquitinase module in SAGA, leads to early embryonic lethality. Here, we review the known functions of Gcn5, USP22 and associated proteins during development and discuss how these functions might be related to human disease states, including cancer and neurodegenerative diseases.
Keywords: ATAC complex, cancer, development, differentiation, Gcn5, histone acetylation, neurodegenerative disease, PCAF, SAGA, stem cells, Usp22
Histone acetylation has long been linked to active gene transcription. This modification is actively regulated by the opposing actions of histone acetyltransferases (HATs) and deacetylases, which target a number of highly conserved residues in the amino-terminal tail regions of the histones. Gcn5 was identified as the first transcription-related HAT in 1996 [1], and since that time, we have learned much about its biochemical partners and its biological functions.
In yeast, Gcn5 is integrated into SAGA and ADA complexes [2,3], and the major sub-units and structures of the Gcn5-containing complexes are highly conserved across evolution [4,5]. In metazoans, Gcn5 is also part of a second, distinct complex, termed ATAC [6]. This article will focus on recent advances made towards defining Gcn5 functions during mammalian development, as well as the role of particular SAGA and ATAC components in human diseases.
Structure & composition of SAGA
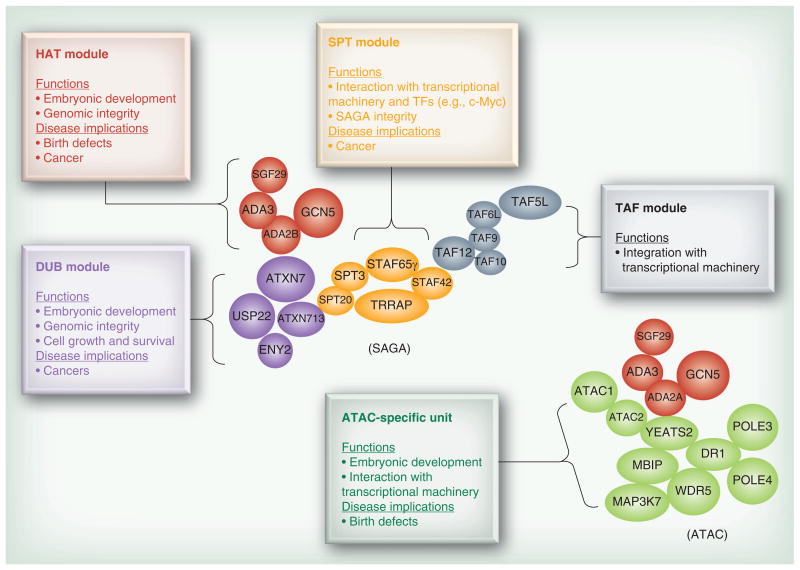
Several excellent reviews of SAGA composition and structure have been written recently, therefore we will provide a brief description here [7–9]. SAGA is organized into several functional submodules (Figure 1), including a histone acetylation center that contains Gcn5 (HAT) together with the Ada proteins, and a deubiquitination (DUB) module that contains Ubp8, in addition to Sgf73, Sgf11 and Sus1 or their mammalian orthologs (Table 1) [10–12]. The best-characterized substrates for SAGA include several acetylation sites in H3 and a ubiquitylation site in H2B. SAGA also contains two modules essential for its architectural integrity and its interactions with transcriptional machinery: the TAF module and the SPT module (Table 1). The composition of SAGA (also referred to as STAGA or TFTC) is largely conserved in mammalian cells (Table 1) [4,13–19]. In addition, Gcn5 is incorporated into a distinct but related complex, called ATAC, which is distinguished by its Ada2a subunit [20,21]. The functions of ATAC are less well defined than those of SAGA, but studies in flies and mice indicate ATAC is important for acetylation on H4K12, 16 [22,23], mitotic progression [24] and normal embryo development [21]. PCAF, which is highly related to Gcn5 [25], is a component of another multi-subunit complex similar to SAGA (Table 1) [26]. The increased complexity of mammalian SAGA and related complexes reflect their involvement in several diverse processes, including embryonic development, stress responses, cell growth, genome integrity, signaling pathways and metabolic control.
Figure 1. Schematics of mammalian Gcn5-containing complexes and their functions and implications in diseases.
The modular structure and composition is based on a model of yeast SAGA [12]. In contrast to SAGA, the physical relationships between subunits in ATAC are less well defined. This figure only summarizes the presence of subunits, not their arrangement within ATAC. POLE3, POLE4 and MAP3K7 are included per [20]. The modules are indicated by the following color codes: red, HAT module; blue, DUB module; orange, SPT module; grey, TAF module; and green, ATAC-specific unit. The disease implications are based on studies using mammalian cells or mouse model systems, as well as from human patient samples, as described in the text. DUB: Deubiquitination; HAT: Histone acetyltransferase; TF: Transcription factor.
Table 1.
Components of SAGA and related complexes in yeast and human.
| Component | Yeast | Human | |||
|---|---|---|---|---|---|
|
| |||||
| ySAGA [2–3,5,7,12,27–30] | yADA [2,3] | hSAGA [7,13,14,16–18,20,31–33] | hPCAF [26,34] | hATAC [20,21] | |
| HAT module | Gcn5 | Gcn5 | GCN5 | GCN5/hPCAF | |
| PCAF | |||||
| Ada2 | Ada2 | ADA2A/B | |||
| ADA2B | |||||
| ADA2A | |||||
| Ada3 | Ada3 | ADA3 | ADA3 | ADA3 | |
| Sgf29 | Sgf29 | SGF29 | SGF29 | SGF29 | |
| Ahc1 | ATAC1 (ZZZ3) | ||||
| Ahc2 | |||||
|
| |||||
| 2nd HAT center | ATAC2 | ||||
|
| |||||
| DUB module | Ubp8 | USP22 | ? | ||
| Sgf73 | ATXN7 | ? | |||
| Sgf11 | ATXN7l3 | ? | |||
| Sus1 | ENY2 | ? | |||
|
| |||||
| TAF module | Taf5 | TAF5L | TAF5L | WDR5 | |
| Taf6 | TAF6L | TAF6L | MAP3K7† | ||
| Taf9 | TAF9 | TAF9 | |||
| Taf10 | TAF10 | TAF10 | DR1 (NC2β) | ||
| Taf12 | TAF12 | TAF12 | POLE3† | ||
|
| |||||
| SPT module | Tra1 | TRRAP | TRRAP | POLE4† | |
| Spt8‡ | |||||
| Ada5/Spt20 | p38IP/SPT20 | MBIP | |||
| Spt7 | STAF65g | YEATS2 | |||
| Spt3 | SPT3 | SPT3 | |||
| SAP130 | SAP130 | ||||
| Ada1 | STAF42 | ||||
Developmental functions of Gcn5 & PCAF
Gcn5 and PCAF are highly similar (75% identical at the protein level) [25] and they are incorporated into similar complexes (Table 1) [26]. However, deletion of Gcn5 or PCAF has very different consequences in mice, indicating these proteins may have different functions in vivo.
Deletion of Gcn5 (Gcn5l2, renamed as Kat2a) results in early embryonic lethality in mice. Gcn5-null embryos show severe growth retardation by 8.5 days post coitum, as well as defects in maintenance of mesodermal lineages due to increased cell death. By contrast, deletion of PCAF (Kat2b) causes no obvious abnormal phenotypes [36,37]. This difference in phenotype may be partially due to differential expression levels and patterns of Gcn5 and PCAF during mouse development [25,36]. However, deletion of both Gcn5 and PCAF results in a more severe phenotype than loss of Gcn5 alone, indicating that PCAF does have some functions redundant to those of Gcn5 early in embryogenesis [36]. Gcn5 and PCAF also have redundant functions in mouse embryonic fibroblasts, which are distinct from those of the CBP/p300 HATs [38]. Although mice heterozygous for the Gcn5-null allele exhibit no defects, mice heterozygous for both Gcn5 and p300 have a more severe phenotype than p300-single hetero zygotes, indicating these HAT family proteins also have both shared and distinct functions during development [39].
Normal Gcn5 expression levels and activity are both required for proper development. Combination of a hypomorphic allele of Gcn5 with a deletion allele in mice to reduce expression below 50% causes homeotic transformations in the skeleton, as well as exencephaly [40,41]. Mice homozygous for a catalytically inactive allele of Gcn5 die in mid-gestation with severe neural tube closure defects [42]. The longer survival time of the Gcn5-hat mutants relative to the null mutants suggests that Gcn5 has functions early in development that are independent of its HAT activity. At least part of the increase in severity of phenotype in the Gcn5-null mice may be due to loss of USP22 and other DUB module components from SAGA upon loss of Gcn5, which gives rise to telomere defects, genome instability and increased cell death [33].
Use of a conditional allele of Gcn5 that is deleted upon exposure to Cre recombinase allows definition of Gcn5 functions at particular stages of development or in particular tissues and lineages. For example, Nestin-Cre-mediated deletion of Gcn5, specifically in neural progenitor cell populations, results in a 25% reduction in brain mass with a microcephaly phenotype similar to that observed in Nestin-Cre-driven knockouts of c-Myc or N-myc [43–46]. Gene expression analysis indicates that about a sixth of the genes whose expression is affected by the loss of Gcn5 are N-myc targets, and Gcn5 is required for maintenance of histone acetylation at these target genes [46]. These findings indicate that Gcn5 is a key transcriptional cofactor for N-myc in neural progenitor cells in the developing brain.
The capability of embryonic stem (ES) cells to self-renew and differentiate makes them an excellent model for studying development in vitro. Gcn5-null ES cells survive and appear to grow normally, except for a delay in progressing through the G2/M phase of the cell cycle [47]. When stimulated to differentiate, Gcn5-null cells form endoderm, ectoderm and mesoderm within embryoid bodies, indicating they are capable of differentiation. However, expression of transcription factors essential for ES cell identity such as Oct4 and Nodal is prematurely lost at early stages of differentiation when Gcn5 is absent, suggesting that Gcn5 is required for maintaining ES cells in a pluripotent, self-renewing state [47]. Gcn5 may also be important in other stem cell-like states. For example, Gcn5 acetylates H1.4K34 in induced pluripotent stem cells, and this modification strongly marks both carcinoma in situ and invasive seminoma nuclei, both of which are precursors of type II testicular germ cell tumors [48]. H1.4K34ac is also dynamically regulated during spermatogenesis with the highest expression levels detected in immature germ cells (spermato gonia) and meiotic spermatocytes [48]. Gcn5, therefore, may also be important for normal sperm development.
Usp22 functions in differentiation & development
Usp22 is also essential for mouse embryonic development [49]. Usp22 and its yeast ortholog, Ubp8, are best characterized in terms for their activity towards H2B, but they also have other substrates [33,50,51]. The lethality of Usp22-null mice was linked to deubiquitylation and stabilization of the Sirt1 deacetylase, which in turn suppressed p53-mediated apoptosis. Although another group did not see any effects on Sirt1 levels upon depletion of Usp22 [49,52]. The full range of Usp22 functions during mouse development are not yet clear.
In mouse ES cells, Usp22 represses the transcription of the pluripotency factor Sox2, thereby promoting differentiation [53]. Loss of Usp22 occupancy at the Sox2 locus is associated with elevated levels of H2Bub and increased transcription of Sox2. Atxn7l3, another component of the DUB module, and Sirt1, also associate with the Sox2 locus, providing a novel example of a HAT complex component that recruits a deacetylase to repress transcription of a target gene.
Developmental functions of ATAC
ATAC was first identified in flies [22], and a highly similar mammalian complex was subsequently defined [20]. ATAC contains a second HAT protein, Atac2, which is specific for lysines in H4, in contrast to Gcn5, which targets H3 and H2B. Atac2-null mice die early in gestation with a phenotype that resembles, but is less severe than, that of Gcn5-null embryos [21], as might be expected since Gcn5 loss affects both ATAC and SAGA. ATAC localizes to different regulatory elements and regulates target genes that are distinct from those of SAGA [54,55]. ATAC also acetylates nonhistone proteins, and in human cells, ATAC controls mitotic progression by acetylating cyclin A [24]. Additional studies are needed to understand how Gcn5 functions are proportioned between the SAGA and ATAC complexes and to define the full range of ATAC functions.
Disease connections
The genetic studies above clearly indicate that Gcn5 and USP22 have important roles during development, which may presage important functions for these proteins in human diseases. So far, findings from a number of laboratories indicate that Gcn5 and SAGA might contribute to both neurodegenerative diseases and to cancers.
Neurodegenerative diseases
The neural tube defects observed in the Gcn5-hat mutant mice and in mice bearing hypomorphic alleles of Gcn5 suggest that Gcn5 or SAGA may be particularly important in neural functions. Indeed, a component of the SAGA DUB module, Atxn7, is implicated in a human neural degenerative disease, spinal cerebellar ataxia type 7 (SCA7). Polyglutamine expansions in ataxin7 (polyQ-Atxn7) are associated with SCA7, which is characterized by both cerebellar and retinal degeneration. Mouse models of SCA7 bearing polyQ-Atxn7 alleles confirm that the polyQ expansions contribute to the pathogenesis of the disease [56]. Reduction of polyQ-Atxn7 expression restores motor function and prevents cerebellar synaptic reorganization in a conditional mouse model [57], suggesting a causative role for polyQ-Atxn7 in the development of SCA7. PolyQ-Atxn7 incorporates into SAGA [19,58,59] and has been reported to inhibit Gcn5 HAT activity, resulting in a dominant-negative effect on SAGA transcriptional activity as a coactivator of photoreceptor genes regulated by the cone-rod homeobox transactivator in SCA7 transgenic mice [19]. Another group reported that polyQ-Atxn7 altered the recruitment of SAGA to photoreceptor genes, leading to changes in chromatin structure and deregulation of these genes, contributing to a subsequent progressive loss of rod photoreceptor function [60].
Gcn5 depletion in SCA7 mice accelerates cerebellar and retinal degeneration, even though cerebellar deletion of Gcn5 in the absence of polyQ-Atxn7 caused only mild ataxia [61]. These findings indicate that loss of Gcn5 may contribute to the time of onset and severity of human SCA7, but is not sufficient to drive disease formation. Loss of Gcn5 impairs the deubiquitinating activity of Usp22 [33], which partners with Atxn7 in the DUB module. Gcn5 loss, then, may exacerbate the SCA7 phenotype by compromising Usp22 functions. PolyQ-Atxn7 may also inhibit USP22 activity in vivo. Additional USP22 mouse models are needed to determine if loss of DUB activity leads to neurodegeneration as seen in SCA7.
PCAF appears to have important neural functions as well. Even though PCAF-null mice are viable without any overt physical phenotypes [36,37], subsequent studies have revealed behavioral alterations in these mice. At a young age, PCAF-null mice exhibit impaired short-term memory, with deficits in learning abilities, spatial memory and recognition memory. As the animals age, contextual long-term memory becomes affected as well [62]. These memory deficits are likely due to morphological changes observed in the hippocampus, a region in the brain that is crucial for multiple forms of memory [63,64]. PCAF-null mice also develop an exaggerated response to acute stress, indicating that PCAF may have a role in controlling emotional states. Interestingly, PCAF involvement in stress response seems to vary among different mouse genetic backgrounds [65], suggesting PCAF-null mice could be potentially valuable for research into genetic variables that affect emotional behaviors and disorders [66,67]. Moreover, mice lacking PCAF develop a resistance to amyloid toxicity [65], such as is associated with human dementias. Although a role for PCAF has not yet been confirmed in human cognitive diseases, these observations indicate that modulating acetyltransferase activity may offer a new way to develop therapies for dementias.
Cancer implications
Several studies using different cell model systems have demonstrated the involvement of Gcn5, USP22 and several other SAGA components in processes that are closely linked to the hallmarks of cancer, including DNA damage repair, cell cycle regulation and post-translational regulation of both oncoproteins and tumor suppressors.
DNA damage response & maintenance of genome integrity
Chromatin compaction can be a barrier to DNA repair [68]. Various factors that modify histones or remodel nucleosomes facilitate repair [69], including Gcn5.
A large-scale screen for DNA damage-responsive histone modifications in human cells revealed that H3K9ac and H3K56ac are rapidly and reversibly reduced in response to DNA damage [70]. The reduction of these two modifications might be caused by enhanced activities of histone deacetylases, or by histone eviction coupled with degradation in response to DNA damage. Gcn5, known to deposit the acetyl mark on H3K9, was reported to also acetylate H3K56 in human cells [70]. Another group, however, found that H3K56Ac is mediated by p300/CBP [71], and that H3K56Ac forms foci at sites of DNA damage in human cells. These different observations might reflect different dosages of DNA-damaging reagents used by the two groups. In any case, these data indicate that these HATs are important to the DNA damage response, whose failure leads to genomic instability that is central to carcinogenesis [72].
Other studies also indicate that Gcn5 is involved in multiple types of DNA repair. After UV-induced DNA damage, Gcn5 is recruited to damage sites by direct association with E2F1, and acetylation of H3K9 by Gcn5 facilitates nucleotide excision repair [73]. In response to γ-irradiation-induced double-strand breaks, cross-talk between serine-139 phosphorylation in H2AX (forming γ-H2AX) and Gcn5-mediated H3 acetylation aids the recruitment of SWI/SNF complexes to facilitate double-strand break repair [74].
Gcn5, Usp22 and SAGA are also important to maintaining the integrity of telomeres. Shelterin proteins protect telomeres from inappropriate recombination. Two shelterin components, TRF1 and Pot1a are regulated by Usp22, which limits their ubiquitination and subsequent turnover by the proteasome. Loss of Gcn5 leads to depletion of Usp22 and the DUB module from SAGA, compromising Usp22 activity, leading to telomere fusions [33].
Regulation of cell growth, proliferation & survival
Several connections have been made between Gcn5, SAGA and the regulation of cell growth and proliferation. For example, SAGA has been defined as a co-activator for the c-Myc oncoprotein in a number of in vitro cell systems [75–77]. TRRAP, STAF65g and GCN5 itself have been reported to interact directly with Myc, thereby recruiting SAGA to Myc target genes [75,77]. The catalytic activity of Gcn5 is required for activation of Myc target genes [77,78]. Gcn5 and other HATs acetylate the Myc protein, increasing its stability [79]. Mouse model systems indicate that Myc induces widespread changes in chromatin structure, including increased acetylation [80], and that Gcn5 and N-Myc share a number of transcriptional targets [46]. Myc often cooperates with other transcription factors, including E2F1, for full activation of its target genes. A recent study of non-small-cell lung cancers revealed that GCN5 is highly expressed in these tumors, compared with matched normal tissues, and that it specifically potentiates lung cancer growth by directly promoting the expression of E2F1, cyclin D1 and cyclin E1 in an E2F1-dependent manner [81].
The Sgf29 component of SAGA contains tandem Tudor domains that serve as ‘readers’ for H3K4 di- or tri-methylation (H3K4me2/3). These interactions are required for recruitment of the SAGA complex and H3 acetylation at target gene promoters [82]. Interestingly, studies in rat hepatoma cell lines indicate that Sgf29 acts downstream of Sry and promotes c-Myc-mediated gene transcription and malignant transformation [31,83], providing yet another link between SAGA functions and Myc.
Gcn5 and PCAF also acetylate the p53 tumor suppressor at Lys320, and this acetylation promotes activation of p53 target genes such as p21 [84]. The involvement of Gcn5 in the functions of both the Myc oncogene and the p53 tumor suppressor indicate that it may both promote and inhibit cancer growth.
Usp22 has strong links to oncogenesis. Usp22 was first identified in microarray screens as part of an 11-gene ‘death from cancer’ signature for highly aggressive, therapy-resistant tumors [85]. This signature includes the Bmi1 polycomb group protein, and several Bmi1 target genes, suggesting that this molecular signature is related to stem cell-like characteristics [86]. Usp22 was later shown to act as an oncogene [87], regulating cell cycle progression, proliferation and apoptosis [50,81]. Increased expression of Usp22 has now been associated with poor prognosis in several additional cancers, including liver cancers, colorectal cancers, breast cancers, esophageal squamous cell carcinoma and oral squamous cell carcinoma [88–90]. If an increase in Usp22 DUB activity can be linked to these cancers, then DUB inhibitors may provide attractive prospects for new therapy development.
Developmental signaling pathways linked to malignancies
Aberrations in signaling pathways that are essential to embryonic development or to stem cell properties are often observed in cancer. These altered pathways may help to sustain populations of cancer-initiating cells (i.e., cancer stem cells), which share many characteristics with ES cells [91,92]. Components of Gcn5 complexes and PCAF may be involved in these signaling pathways. PCAF, for example, is required for Hedgehog-Gli-dependent transcription that drives cell proliferation in medulloblastomas and glioblastomas, and silencing of PCAF reduces the tumor-forming potential of neural stem cells [93]. ADA2a and ADA3 are required for acetylation of β-catenin, which promotes transcriptional functions of β-catenin in lung and colon cancer cell lines [94].
Involvement in metabolic energy pathways in cancer
Normal cells depend primarily on mitochondrial oxidative phosphorylation to generate the energy needed for cellular processes. By contrast, most cancer cells switch to aerobic glycolysis, even in the presence of oxygen, to meet the soaring need for energy to support increased proliferation and growth [95]. Such reprogramming of energy metabolism is termed the ‘Warburg effect’ [72]. Under hypoxia, HIF-1α upregulates glycolytic enzymes to promote glycolysis, whereas p53 inhibits glycolysis and increases oxidative phosphorylation by activating TIGAR and SCO2 gene expression. PCAF mediates differential recruitment of HIF-1α and p53 to the promoters of TIGAR and/or SCO2 genes, thereby tuning the energy needs of the cells to different environmental conditions [96]. Although it is not yet clear whether PCAF is a rate-limiting factor for the glycolysis switch during tumor growth, these findings indicate that PCAF may provide an attractive target for controlling tumor growth and survival.
Conclusion & future perspective
Gcn5, Usp22 and a few other subunits of SAGA have been linked directly or indirectly to neurodegenerative diseases and to cancer. The essential developmental functions of Gcn5 and Usp22 in mouse indicate these factors may also be important for preventing human birth defects, such as exencephaly. PCAF has also been linked to cancer, as well as to energy metabolism, cognitive capacities and psychological behaviors. Clearly, understanding the full range of functions of these factors, and development of ways to modulate those functions, is important to human health.
One area that needs further investigation is definition of the relative distribution of Gcn5, in time and in space, between SAGA and ATAC during mammalian development, and the exact roles of these two complexes in embryogenesis. If Gcn5 levels are limiting in vivo, then changing the balance of its association between these complexes might have significant consequences for developmental programs. Loss or gain of individual subunits of the SWI/SNF ATP-dependent chromatin remodeler complex, for example, leads to a switch in lineage commitment during development [97]. Would the same be true for SAGA and ATAC? If so, might that also impact the functions of Gcn5 in cancer or other diseases?
Another open question is whether alterations in levels or activity of Gcn5, PCAF or Usp22 are sufficient to impact cancer development. Would inhibition of Gcn5, for example, reduce the oncogenic properties of c-Myc overexpression? Is overexpression of USP22, such as is seen multiple human cancers, enough to initiate or drive cancer formation? It will also be important to determine how Gcn5 and USP22 functions are linked in cancer and in other diseases, such as SCA7.
Finally, the connections observed between PCAF and SAGA components and neural processes are fascinating, and may foreshadow discovery of a greater role for these proteins in both neural development and neural functions in adults. It is not clear, for example, how polyQ-Atxn7 affects the activity of USP22, or whether such effects contribute to SCA7. Another question is whether Gcn5 might affect cognitive abilities or emotional state as does PCAF, and if so, whether these two HATs are involved in the same or separate pathways.
Although we have learned a lot about the biochemical activities of these HATs over the last 18 years, we obviously still have a lot to learn about their biological functions. This knowledge will not only advance our understanding of chromatin regulation, but will also advance our understanding of human disease pathways.
Supplementary Material
Executive summary.
SAGA in transcription
Gcn5 is the first transcription-related histone acetyltransferase (HAT) to be described, linking SAGA to gene transcription.
SAGA is organized into functional modules and harbors both HAT (Gcn5) and deubiquitination (Usp22) activities, and the modularity of SAGA is conserved from yeast to humans.
Identification of the second enzymatic activity of SAGA, the deubiquitinase Usp22, broadens the substrates of SAGA and revealed new roles of SAGA in gene regulation, as well as other processes.
Functions of SAGA in mammals & implications in human diseases
Both Gcn5 and Usp22 are essential for mouse embryogenesis, indicating these factors may also be important for preventing birth defects in humans.
Several subunits of SAGA are involved in pathogenesis of neurodegenerative diseases, for instance, polyQ expansion in Atxn7 causes spinal cerebellar ataxia type 7, and Gcn5 deletion accelerates the disease progression in spinal cerebellar ataxia type 7 mice. This provides potential new targets for the development of therapeutic strategies for such diseases.
SAGA interacts with transcription factors such as Myc to regulate gene transcription, cell growth and survival. Several key subunits are indicated in such processes, including TRRAP, Usp22 and Gcn5. The close interactions with Myc link SAGA to an array of human cancers that are driven by this oncoprotein.
Acknowledgments
The authors would like to thank E Koutelou and B Atanassov for helpful comments on the manuscript.
This work was supported by the NIH GM067718 to SYR Dent.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1••.Brownell JE, Zhou J, Ranalli T, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84(6):843–851. doi: 10.1016/s0092-8674(00)81063-6. Identified Gcn5 as the first histone acetyltransferase (HAT) related to transcriptional regulation. [DOI] [PubMed] [Google Scholar]
- 2.Grant PA, Duggan L, Cote J, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11(13):1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 3.Eberharter A, Sterner DE, Schieltz D, et al. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(10):6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand M, Leurent C, Mallouh V, Tora L, Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286(5447):2151–2153. doi: 10.1126/science.286.5447.2151. [DOI] [PubMed] [Google Scholar]
- 5.Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15(2):199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Guelman S, Suganuma T, Florens L, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26(3):871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22(3):374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samara NL, Wolberger C. A new chapter in the transcription SAGA. Curr Opin Struct Biol. 2011;21(6):767–774. doi: 10.1016/j.sbi.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26(6):527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingvarsdottir K, Krogan NJ, Emre NC, et al. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25(3):1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25(3):1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. Both studies demonstrated the second enzymatic activity of SAGA complex (deubiquitylation module) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Lee KK, Sardiu ME, Swanson SK, et al. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. doi: 10.1038/msb.2011.40. The study applied a combination of biochemical approaches, quantitative proteomics and computational methods using wild-type and deletion strains to demonstrate the functional and structural organization of SAGA and ADA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candau R, Moore PA, Wang L, et al. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16(2):593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273(37):23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 15.Smith ER, Belote JM, Schiltz RL, et al. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 1998;26(12):2948–2954. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez E, Palhan VB, Tjernberg A, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21(20):6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barlev NA, Emelyanov AV, Castagnino P, et al. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol. 2003;23(19):6944–6957. doi: 10.1128/MCB.23.19.6944-6957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmlinger D, Hardy S, Sasorith S, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13(12):1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 19.Palhan VB, Chen S, Peng GH, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci USA. 2005;102(24):8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YL, Faiola F, Xu MY, Pan SQ, Martinez E. Human ATAC is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283(49):33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guelman S, Kozuka K, Mao Y, et al. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol. 2009;29(5):1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suganuma T, Gutierrez JL, Li B, et al. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15(4):364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 23.Ciurciu A, Komonyi O, Boros IM. Loss of ATAC-specific acetylation of histone H4 at Lys12 reduces binding of JIL-1 to chromatin and phosphorylation of histone H3 at Ser10. J Cell Sci. 2008;121(Pt 20):3366–3372. doi: 10.1242/jcs.028555. [DOI] [PubMed] [Google Scholar]
- 24.Orpinell M, Fournier M, Riss A, et al. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010;29(14):2381–2394. doi: 10.1038/emboj.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Edmondson DG, Roth SY. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18(10):5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogryzko VV, Kotani T, Zhang X, et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94(1):35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 27.Grant PA, Schieltz D, Pray-Grant MG, Yates JR, 3rd, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2(6):863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 28.Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22(13):4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell DW, Weaver CM, Jennings JL, et al. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24(16):7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Navarro S, Fischer T, Luo MJ, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116(1):75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 31.Kurabe N, Katagiri K, Komiya Y, et al. Deregulated expression of a novel component of TFTC/STAGA histone acetyltransferase complexes, rat SGF29, in hepatocellular carcinoma: possible implication for the oncogenic potential of c-Myc. Oncogene. 2007;26(38):5626–5634. doi: 10.1038/sj.onc.1210349. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Lang G, Ito S, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29(1):92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 33••.Atanassov BS, Evrard YA, Multani AS, et al. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35(3):352–364. doi: 10.1016/j.molcel.2009.06.015. Gcn5 is implicated in maintaining genome integrity by modulating Usp22 activity toward components of the shelterin complex, thus regulating their turnover. It is also the first to reveal that Usp22 has nonhistone substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassilev A, Yamauchi J, Kotani T, et al. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2(6):869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 35.Pray-Grant MG, Schieltz D, McMahon SJ, et al. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22(24):8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26(2):229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 37••.Yamauchi T, Yamauchi J, Kuwata T, et al. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA. 2000;97(21):11303–11306. doi: 10.1073/pnas.97.21.11303. Both studies demonstrated that Gcn5 and PCAF have distinct yet overlapping functions in embryogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin QH, Yu LR, Wang LF, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan HM, Xu AW, Coco C, et al. GCN5 and p300 share essential functions during early embryogenesis. Dev Dyn. 2005;233(4):1337–1347. doi: 10.1002/dvdy.20445. [DOI] [PubMed] [Google Scholar]
- 40.Lin W, Zhang Z, Chen CH, Behringer RR, Dent SY. Proper Gcn5 histone acetyltransferase expression is required for normal anteroposterior patterning of the mouse skeleton. Dev Growth Differ. 2008;50(5):321–330. doi: 10.1111/j.1440-169X.2008.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin W, Zhang Z, Srajer G, et al. Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev Dyn. 2008;237(4):928–940. doi: 10.1002/dvdy.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27(9):3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Bokhoven H, Celli J, Van Reeuwijk J, et al. MYCN haploinsufficiency is associated with reduced brain size and intestinal atresias in Feingold syndrome. Nat Genet. 2005;37(5):465–467. doi: 10.1038/ng1546. [DOI] [PubMed] [Google Scholar]
- 44.Wey A, Knoepfler PS. c-myc and N-myc promote active stem cell metabolism and cycling as architects of the developing brain. Oncotarget. 2010;1(2):120–130. doi: 10.18632/oncotarget.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum. 2010;9(4):537–547. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Cerdeno V, Lemen JM, Chan V, et al. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS ONE. 2012;7(6):e39456. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SYR. Developmental potential of Gcn5(-/-) embryonic stem cells in vivo and in vitro. Dev Dyn. 2007;236(6):1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- 48.Kamieniarz K, Izzo A, Dundr M, et al. A dual role of linker histone H1. 4 Lys 34 acetylation in transcriptional activation. Genes Dev. 2012;26(8):797–802. doi: 10.1101/gad.182014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Lin Z, Yang H, Kong Q, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46(4):484–494. doi: 10.1016/j.molcel.2012.03.024. Demonstrated for the first time that Usp22 is required for mammalian embryogenesis. [DOI] [PubMed] [Google Scholar]
- 50.Atanassov BS, Dent SYR. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011;12(9):924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson MA, Koutelou E, Hirsch C, et al. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol Cell Biol. 2011;31(15):3126–3135. doi: 10.1128/MCB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armour SM, Bennett EJ, Braun CR, et al. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol. 2013;33(8):1487–1502. doi: 10.1128/MCB.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sussman RT, Stanek TJ, Esteso P, Gearhart JD, Knudsen KE, McMahon SB. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2) J Biol Chem. 2013;288(33):24234–24246. doi: 10.1074/jbc.M113.469783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagy Z, Riss A, Fujiyama S, et al. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci. 2010;67(4):611–628. doi: 10.1007/s00018-009-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44(3):410–423. doi: 10.1016/j.molcel.2011.08.037. Demonstrated that mammalian SAGA and ATAC regulate distinct sets of genes, which is one possible explanation for differences in phenotypes observed in Gcn5 and Atac2 null embryos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoo SY, Pennesi ME, Weeber EJ, et al. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron. 2003;37(3):383–401. doi: 10.1016/s0896-6273(02)01190-x. [DOI] [PubMed] [Google Scholar]
- 57.Furrer SA, Waldherr SM, Mohanachandran MS, et al. Reduction of mutant ataxin-7 expression restores motor function and prevents cerebellar synaptic reorganization in a conditional mouse model of SCA7. Hum Mol Genet. 2013;22(5):890–903. doi: 10.1093/hmg/dds495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helmlinger D, Hardy S, Eberlin A, Devys D, Tora L. Both normal and polyglutamine- expanded ataxin-7 are components of TFTC-type GCN5 histone acetyltransferase- containing complexes. Biochem Soc Symp. 2006;(73):155–163. doi: 10.1042/bss0730155. [DOI] [PubMed] [Google Scholar]
- 59.McCullough SD, Xu X, Dent SY, Bekiranov S, Roeder RG, Grant PA. Reelin is a target of polyglutamine expanded ataxin-7 in human spinocerebellar ataxia type 7 (SCA7) astrocytes. Proc Natl Acad Sci USA. 2012;109(52):21319–21324. doi: 10.1073/pnas.1218331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helmlinger D, Hardy S, Abou-Sleymane G, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4(3):e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen YC, Gatchel JR, Lewis RW, et al. Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Hum Mol Genet. 2012;21(2):394–405. doi: 10.1093/hmg/ddr474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maurice T, Duclot F, Meunier J, et al. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology. 2008;33(7):1584–1602. doi: 10.1038/sj.npp.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bird CM, Burgess N. The hippocampus and memory: insights from spatial processing. Nat Rev Neurosci. 2008;9(3):182–194. doi: 10.1038/nrn2335. [DOI] [PubMed] [Google Scholar]
- 64.Van Strien NM, Cappaert NL, Witter MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 2009;10(4):272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- 65.Duclot F, Meffre J, Jacquet C, Gongora C, Maurice T. Mice knock out for the histone acetyltransferase P300/Creb binding protein-associated factor develop a resistance to amyloid toxicity. Neuroscience. 2010;167(3):850–863. doi: 10.1016/j.neuroscience.2010.02.055. [DOI] [PubMed] [Google Scholar]
- 66.Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60(2):84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 67.Jacobson LH, Cryan JF. Feeling strained? Influence of genetic background on depression-related behavior in mice: a review. Behav Genet. 2007;37(1):171–213. doi: 10.1007/s10519-006-9106-3. [DOI] [PubMed] [Google Scholar]
- 68.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128(4):721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 69.Smeenk G, Van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- 70.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28(13):1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 73.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39(4):1390–1397. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29(8):1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20(2):556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flinn EM, Wallberg AE, Hermann S, Grant PA, Workman JL, Wright AP. Recruitment of Gcn5-containing complexes during c-Myc-dependent gene activation. Structure and function aspects. J Biol Chem. 2002;277(26):23399–23406. doi: 10.1074/jbc.M201704200. [DOI] [PubMed] [Google Scholar]
- 77.Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278(22):20405–20412. doi: 10.1074/jbc.M211795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu XH, Vorontchikhina M, Wang YL, Faiola F, Martinez E. STAGA recruits mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28(1):108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patel JH, Du Y, Ard PG, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24(24):10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25(12):2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L, Wei TY, Si XX, et al. Lysine acetyltransferase GCN5 potentiates the growth of non-small cell lung cancer via promotion of E2F1, cyclin D1, and cyclin E1 expression. J Biol Chem. 2013;288(20):14510–14521. doi: 10.1074/jbc.M113.458737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bian CB, Xu C, Ruan JB, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30(14):2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murakami S, Chishima S, Uemoto H, et al. The male-specific factor Sry harbors an oncogenic function. Oncogene. 2013 doi: 10.1038/onc.2013.262. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 84.Barlev NA, Liu L, Chehab NH, et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8(6):1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 85••.Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle. 2005;4(9):1171–1175. doi: 10.4161/cc.4.9.2001. Highlights the correlation of aberrant Usp22 expression with aggressive cancers. [DOI] [PubMed] [Google Scholar]
- 86.Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5(16):1886–1901. doi: 10.4161/cc.5.16.3222. [DOI] [PubMed] [Google Scholar]
- 87.Liu YL, Jiang SX, Yang YM, Xu H, Liu JL, Wang XS. USP22 acts as an oncogene by the activation of BMI-1-mediated INK4a/ARF pathway and Akt pathway. Cell Biochem Biophys. 2012;62(1):229–235. doi: 10.1007/s12013-011-9287-0. [DOI] [PubMed] [Google Scholar]
- 88.Liu YL, Yang YM, Xu H, Dong XS. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J Surg Oncol. 2011;103(3):283–289. doi: 10.1002/jso.21802. [DOI] [PubMed] [Google Scholar]
- 89.Zhang YX, Yao L, Zhang XY, et al. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J Cancer Res Clin Oncol. 2011;137(8):1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Piao SL, Liu YL, Hu J, et al. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS ONE. 2012;7(8):e42540. doi: 10.1371/journal.pone.0042540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karamboulas C, Ailles L. Developmental signaling pathways in cancer stem cells of solid tumors. Biochim Biophys Acta. 2013;1830(2):2481–2495. doi: 10.1016/j.bbagen.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 93.Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squatrito M, Helin K. Histone acetyltransferase PCAF is required for hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013;73(20):6323–6333. doi: 10.1158/0008-5472.CAN-12-4660. [DOI] [PubMed] [Google Scholar]
- 94.Yang M, Waterman ML, Brachmann RK. HADA2a and hADA3 are required for acetylation, transcriptional activity and proliferative effects of beta-catenin. Cancer Biol Ther. 2008;7(1):122–130. doi: 10.4161/cbt.7.1.5197. [DOI] [PubMed] [Google Scholar]
- 95.Heiden MGV, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rajendran R, Garva R, Ashour H, et al. Acetylation mediated by the p300/CBP-associated factor determines cellular energy metabolic pathways in cancer. Inter J Oncol. 2013;42(6):1961–1972. doi: 10.3892/ijo.2013.1907. [DOI] [PubMed] [Google Scholar]
- 97.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.