Abstract
NRAS is the second most frequently mutated gene in melanoma. Previous reports have demonstrated the sensitivity of cancer cell lines carrying KRAS mutations to apoptosis initiated by inhibition of protein kinase C delta (PKCδ). Here, we report that PKCδ inhibition is cytotoxic in melanomas with primary NRAS mutations. Novel small-molecule inhibitors of PKCδ were designed as chimeric hybrids of two naturally-occurring PKCδ inhibitors, staurosporine and rottlerin. The specific hypothesis interrogated and validated is that combining two domains of two naturally-occurring PKCδ inhibitors into a chimeric or hybrid structure retains biochemical and biological activity, and improves PKCδ isozyme selectivity. We have devised a potentially general synthetic protocol to make these chimeric species using Molander trifluorborate coupling chemistry. Inhibition of PKCδ, by siRNA or small molecule inhibitors, suppressed the growth of multiple melanoma cell lines carrying NRAS mutations, mediated via caspase-dependent apoptosis. Following PKCδ inhibition, the stress-responsive JNK pathway was activated, leading to the activation of H2AX. Consistent with recent reports on the apoptotic role of phospho-H2AX, knockdown of H2AX prior to PKCδ inhibition mitigated the induction of caspase-dependent apoptosis. Furthermore, PKCδ inhibition effectively induced cytotoxicity in BRAF-mutant melanoma cell lines that had evolved resistance to a BRAF inhibitor, suggesting the potential clinical application of targeting PKCδ in patients who have relapsed following treatment with BRAF inhibitors. Taken together, the present work demonstrates that inhibition of PKCδ by novel small molecule inhibitors causes caspase-dependent apoptosis mediated via the JNK-H2AX pathway in melanomas with NRAS mutations or BRAF inhibitor-resistance.
INTRODUCTION
Although melanomas account for less than 5% of skin cancer cases, they were responsible for more than 75% of estimated skin cancer deaths in 2012, and the incidence rate has been increasing for the last 30 years.1 While chemotherapeutic treatments have improved response rates in metastatic melanoma, there has been no significant impact on survival for decades.1
Melanoma is highly dependent upon the RAS/RAF/MEK/ERK pathway, one of the three major mitogen-activated protein kinase (MAPK) pathways. The components of this pathway, therefore, can serve as the targets of drugs for late-stage melanomas. BRAF (one of the three RAF isoforms) is the most commonly mutated gene in melanoma (45–55% of melanoma cases), while mutations in NRAS (one of the three RAS isoforms) are observed in 15–30% of melanoma cases.2, 3 The BRAF inhibitor PLX4032 (vemurafenib) shows high activity in patients with BRAF-V600E mutation; however, responders eventually and inevitably became resistant to this drug and relapsed.4 One of the proposed mechanisms of acquired resistance to vemurafenib is reactivation of MEK/ERK signaling independently of BRAF, the suppression of which had been the goal of PLX4032 action, by a variety of compensatory alterations.5, 6 In contrast to BRAF, the oncogenic RAS/GAP switch is an exceedingly difficult target for rational drug discovery, and is now widely considered “un-drugable”.3, 7, 8 An “indirect” approach, targeting a survival pathway required by tumor cells bearing an activated RAS allele, may represent an alternative strategy for NRAS-mutant melanomas.
We previously demonstrated that cancer cells carrying oncogenic KRAS mutations undergo apoptosis when protein kinase C delta (PKCδ) activity is inhibited by means of a chemical inhibitor, RNA interference, or a dominant-negative variant.9–12 Other groups also subsequently validated PKCδ as a target in cancer cells of multiple types with aberrant activation of KRAS signaling.13, 14
PKCδ belongs to the PKC family of serine/threonine protein kinases which are involved in diverse cellular functions, such as proliferation, tumor promotion, differentiation and apoptotic cell death.15 The PKC family is categorized into three subfamilies based on structural, functional and biochemical differences, and activators: the classical/conventional PKCs (α, βI, βII, γ), the novel PKCs (δ, ε, θ, µ), and the atypical PKCs (ζ, λ). The novel PKCs, including PKCδ, are characteristically activated by diacylglycerol (DAG) and are independent of the need for the secondary messenger Ca2+. PKCδ functions as either a pro-apoptotic or an anti-apoptotic/pro-survival regulator depending upon cellular context, such as the specific stimulus or its subcellular localization.15 PKCδ is implicated as an early regulator in certain anti-apoptotic/pro-survival signaling cascades through induction or suppression of downstream substrates, including ERK, AKT and NF-κB. Other context-dependent effectors of PKCδ include JNK, glycogen synthase kinase-3 (GSK3), FLICE-like inhibitory protein (FLIP), cIAP2 and p21Cip1/WAF1. A role for PKCδ as an anti-apoptotic/pro-survival regulator has been reported in various types of cancer cells, including non-small cell lung cancer, pancreatic and colon cancers.16–20 Interestingly, these types of cancers are correlated with high rates of activating mutations in KRAS genes.7, 8 Importantly, unlike many other PKC isozymes, PKCδ is not required for the survival of normal cells and tissues, and PKCδ-null mice are viable, fertile and develop normally.21
Our previous studies demonstrating the synthetic lethal activity of PKCδ inhibition in pancreatic, lung, neuroendocrine and breast cancers, and cancer stem-like cells (CSCs) with KRAS mutations 9–12 suggested the potential of targeting PKCδ in melanomas with an activating NRAS mutation. In this study, we demonstrate that inhibition of PKCδ by siRNA or novel chemical compounds suppresses the growth of melanoma lines with NRAS mutations through induction of caspase-dependent apoptosis. A novel PKCδ inhibitor developed through pharmacophore modeling exerted cytotoxic activity on NRAS-mutant tumors at concentrations one log lower than commercially-available PKCδ inhibitors. This cytotoxicity was mediated by activation of stress-responsive JNK-H2AX pathway, which involves a novel function of phospho-H2AX in mediating the apoptotic response. Furthermore, this study also showed that PKCδ inhibition can effectively inhibit the growth of PLX4032-resistant melanoma cells with BRAF mutations, demonstrating the potential of an approach targeting PKCδ in the substantial fraction of patients with melanoma who currently have only limited treatment options.
RESULTS AND DISCUSSION
PKCδ is a potential therapeutic target in melanoma with NRAS mutation
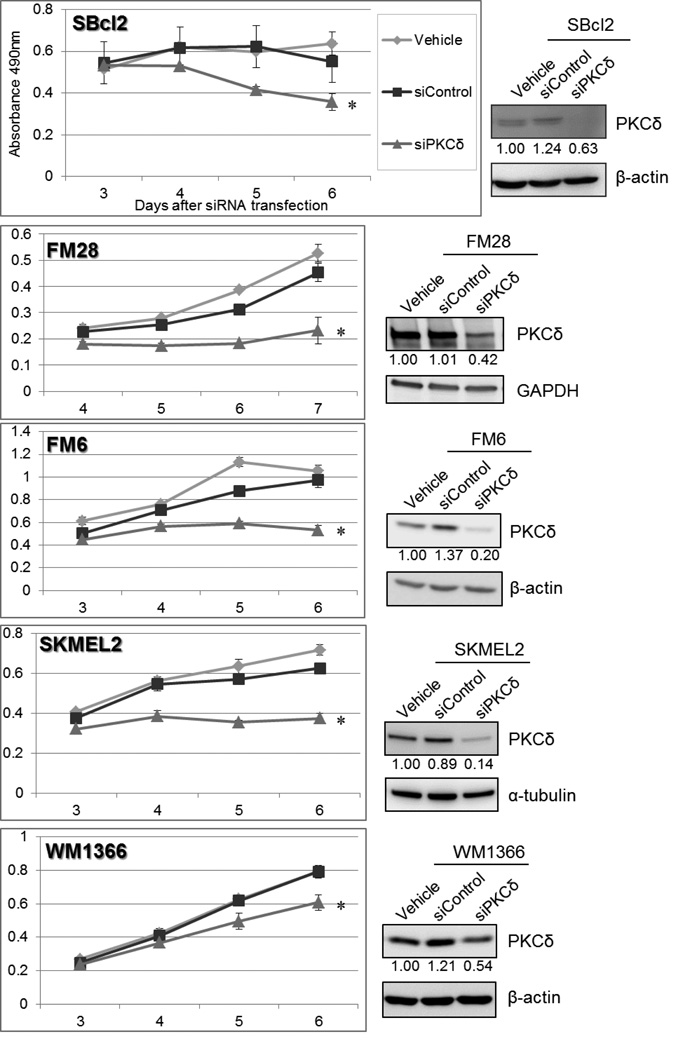
To validate the potential of this approach targeting PKCδ in melanomas with NRAS mutations, we first examined the effect of PKCδ-selective inhibition on cell growth by specifically and selectively knocking down PKCδ protein expression in multiple melanoma cell lines harboring NRAS mutations, using siRNA. The specificity of the PKCδ-specific siRNAs employed herein for PKCδ among all the other PKC isoforms has been previously demonstrated.9–11 Even partial knockdown of PKCδ protein significantly inhibited the proliferation of multiple melanoma cell types with NRAS mutations, including SBcl2, FM28, FM6 and SKMEL2 cells (Figure 1). Interestingly, the degree of protein knockdown did not appear to be the sole factor in determining the degree of growth inhibitory effect by siRNA transfection; some cell lines were more susceptible than others to cell growth inhibition resulting from PKCδ downregulation. No viable cells with chronic suppression of PKCδ could ever be isolated, consistent with our previous demonstration of a requirement for PKCδ activity for the viability in cells bearing mutationally-activated RAS.
Figure 1. Downregulation of PKCδ suppresses cell survival in melanoma cell lines with NRAS mutation.
siRNA targeting PKCδ (“siPKCδ”) or non-targeting siRNA (“siControl”) were transfected into SBcl2 and FM28 (50 nM), SKMEL2 (10 nM), and FM6 and WM1366 (5 nM), after establishing cell line-specific optimal transfection conditions. As a vehicle control, cells were treated in parallel with transfection reagent alone (“vehicle”). MTS assays were performed at 3 or 4 days after siRNA transfection. Each point represents the average of triplicates, and error bars indicate the standard deviations. P values (*) were calculated between vehicle control and siPKCδ on the last assay day (p < 0.006). Downregulation of PKCδ protein on the first assay day was assessed by immunoblot analysis. The relative band intensity of PKCδ is indicated below the image (normalized to loading controls, β-actin, α-tubulin or GAPDH).
These cell survival assays verified that PKCδ is essential for survival of NRAS-mutant melanoma cells.
Development of novel PKCδ inhibitor BJE6-106 (B106)
Potent small molecule inhibitors of PKCδ have not previously been available. Broad (pan) inhibitors of PKC isozymes are generally toxic, as certain PKC isozymes are required for normal physiological functions, and inhibition of such isozymes by a non-selective PKCδ inhibitor can damage normal cells.22, 23 We therefore pursued development of a more potent PKCδ inhibitor with higher PKCδ selectivity in order to explore the therapeutic potential of this approach of targeting PKCδ.
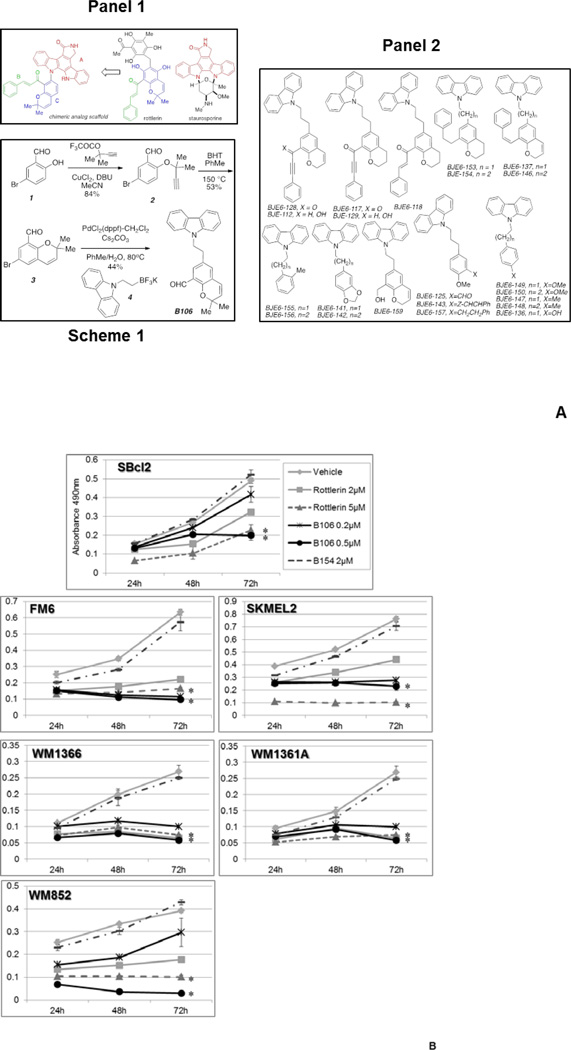
We initially generated a pharmacophore model based on molecular interactions of small molecules with “novel” class PKC isozymes. In the initial pharmacophore model for PKCδ inhibitors, mallotoxin/rottlerin, a naturally-occurring product, with moderate aqueous solubility, and oral bioavailability,24 was used as a prototype structure for a molecule with PKCδ-inhibitory activity (IC50=5µM). Protein structural data for PKCθ, another “novel” PKC isozyme which is also inhibited by mallotoxin/rottlerin, was incorporated (Supplemental Information). Mallotoxin/rottlerin is relatively selective for PKCδ over PKCα (PKCδ IC50:PKCα IC50 is approximately 30:1). We and others have also shown that mallotoxin/rottlerin, at the concentrations employed herein, is not cytostatic or cytotoxic to normal primary cells or cell lines, and is well-tolerated when administered orally or intraperitoneally to mice.9–12, 24 This favorable toxicity profile, combined with its in vivo efficacy, made mallotoxin/rottlerin attractive as a starting point for modification and drug development. We further developed the pharmacophore model using a prototype chimeric structure based on mallotoxin/rottlerin and a more general class of protein kinase C inhibitors (the natural product staurosporine), and incorporating protein structural data for “novel” class PKCs. The strategy was to retain most of the “bottom” part of mallotoxin/rottlerin (Figure 2A, panel 1) which is assumed to give mallotoxin/rottlerin its PKCδ specificity but to vary the “head group” which is assumed to bind to the hinge region of the kinase active site. Numerous “head groups” from known potent kinase inhibitors were tested in the PKCδ model.11 The criteria for selection was that the resulting molecule should form favorable interactions with the hinge region while the “bottom part” retained interactions with the binding site similar to that of staurosporine (from the x-ray crystallographic studies) and mallotoxin/rottlerin (from docking studies into PKCδ). In these 2nd generation of PKCδ inhibitors, the “head” group was made to resemble that of staurosporine, a potent general PKC inhibitor, and other bisindoyl maleimide kinase inhibitors, with domains B (cinnamate side chain) and C (benzopyran) conserved from the mallotoxin/rottlerin scaffold to preserve isozyme specificity. The chromene portion of mallotoxin was combined with the carbazole portion of staurosporine to produce chimeric molecule including KAM1.11 KAM1 was indeed active and more PKCδ-specific than rottlerin/mallotoxin, and showed activity against cancer cells with activation of RAS or RAS signaling, including human neuroendocrine tumors, pancreatic cancers and H460 lung cancer cells.11 KAM1 had an IC50 of 3 µM for PKCδ (similar to mallotoxin/rottlerin) and better isozyme selectivity (IC50 of >150 µM for PKCα) (Table 1).11
Figure 2. PKCδ inhibitors suppress survival in melanoma cell lines with NRAS mutations.
(A) Structure and synthesis of PKCδ inhibitors. Panel 1: Design of mallotoxin/rottlerin-staurosporine hybrids; Scheme 1: Synthesis of B106; Panel 2: 3rd Generation Compounds
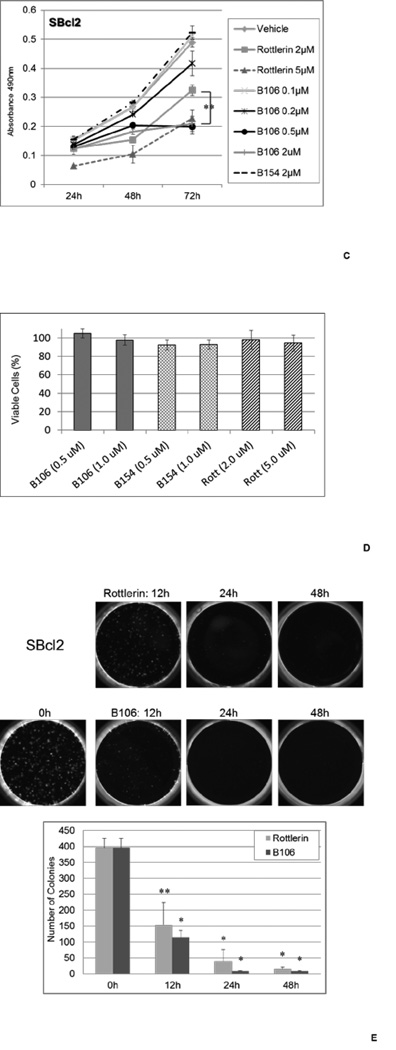
(B) PKCδ inhibitors suppress cell survival in melanoma cell lines with NRAS mutations. SBcl2, FM6, SKMEL2, WM1366, WM1361A and WM852 cells were exposed to rottlerin (2 or 5 µM) or B106 (0.2 or 0.5 µM) for 24, 48 or 72 hr and MTS assays were performed at each time point. DMSO and B154 (2 µM) served as a vehicle control and a negative compound control, respectively. Each point represents the average of triplicates and error bars indicate the standard deviations. P values (*) were calculated between DMSO (vehicle control) and rottlerin 5 µM, or DMSO and B106 0.5 µM in each cell line at 72 hr (p < 0.0002).
(C) Titration of PKCδ inhibitor treatment. The expanded doses of B106 (0.1 µM and 2 µM) in the MTS assay in SBcl2 in Figure 2A are shown. ** indicates a p value < 0.5 between treatment of 2 µM of rottlerin and B106.
(D) Effects of PKCδ inhibitors on primary human melanocytes. Cell survival of human primary melanocytes exposed to the indicated concentrations of the compounds for 72 h (relative to DMSO-treated controls; mean ± SD, n = 3).
(E) PKCδ inhibitors induce irreversible effects on cell growth. SBcl2 cells were treated with rottlerin or B106 at 1 µM for 0, 12, 24 or 48 hr. After these exposure times, the same number of viable cells from each treatment condition was replated at low cell density and cells were cultured in medium without inhibitors for 8 days. Cell colonies were counted. Each point represents the average of triplicates and error bars indicate the standard deviations. P values: ** p<0.01, * p<0.001 compared to time 0 hr.
Table 1. Comparison of properties of PKCδ inhibitors.
In vitro kinase assays demonstrated that 3rd generation PKCδ inhibitor B106 is more potent and more selective for PKCδ over PKCα than rottlerin/mallotoxin or the 2nd generation PKCδ inhibitor KAM1. B154 is used as an inactive (negative control) compound.
| Compounds | Generation | PKCδ IC50 | PKCα IC50 | PKCδ/PKCα Selectivity |
“Ras-specific” Cytotoxicity |
|---|---|---|---|---|---|
| Rottlerin | 1st | 3–5 µM | 75 µM | 28-fold | 3–5 µM |
| KAM1 | 2nd | 3 µM | 157 µM | 56-fold | 3 µM |
| B106 | 3rd | 0.05 µM | 50 µM | 1000-fold | 0.5 µM |
| B154 | 3rd | >40 µM | >100 µM | - | None |
On the basis of structure-activity relationship (SAR) analysis of KAM1 and other 2nd generation compounds, we then generated thirty-six new 3rd generation compounds (Figure 2A, panel 2). These derivatives showed a broad range of PKCδ-inhibitory activity, ranging from IC50 of >40 µM to <0.05 µM (Supplemental Table 1). BJE6-106 (B106) (Figure 2A, Scheme 1), our current lead 3rd generation compound, has an IC50 for PKCδ of <0.05 µM and targeted selectivity over classical PKC isozymes (a 1000-fold PKCδ selectivity over PKCα) (Table 1). BJE6-154 (B154) was among the least potent of the thirty-six compounds studied (PKCδ IC50 of >40 µM) and was used as a negative-control compound with minimal inhibitory activity against PKCδ.
Inhibition of PKCδ activity induces cell growth inhibition in melanoma cell lines with NRAS mutations
To investigate the effect of PKCδ inhibition by small molecule compounds on tumor cell growth, tumor cell survival was assessed in the presence of mallotoxin/rottlerin or B106 using a panel of melanoma cell lines with Q61 NRAS mutations, including SBcl2, FM6, SKMEL2, WM1366, WM1361A and WM852 (Figure 2B, Table 2). Cells were exposed to rottlerin (2 or 5 µM) or B106 (0.2 or 0.5 µM) and viable cells were quantitated at 24, 48 and 72 hr after treatment. Rottlerin consistently inhibited proliferation of all cell lines at 5 µM, and intermediate inhibitory effects were observed at 2 µM. The 3rd generation PKCδ inhibitor B106 effectively inhibited growth of all cell lines tested at 0.5 µM, and at 0.2 µM in some cell lines, which is at least ten times lower than the concentration of rottlerin required to exert the same magnitude of cytotoxic effect. Both inhibitors demonstrated dose-dependent cytotoxic effects, and B106 at 0.5 µM was significantly more active than rottlerin at 2 µM (Figure 2C). Exposure to B154 at 2 µM produced a proliferation curve similar to vehicle (DMSO) treatment in all cell lines, consistent with our hypothesis that the cell growth inhibition induced by B106 resulted from the inhibition of PKCδ activity. Furthermore, B106 produced no statistically-significant effects on the proliferation of primary human melanocytes at concentrations of 0.5 and 1.0 µM, indicating the tumor-specific effect of B106 (Figure 2D).
Table 2.
Confirmed NRAS Q61 mutations of the cell lines.
| Cell Line | Allele | Amino acid | Type |
|---|---|---|---|
| SBcl2 | C181A | Q61K | Homozygous |
| FM6 | C181A | Q61K | Heterozygous |
| FM28 | C181A | Q61K | Homozygous |
| SKMEL2 | A182G | Q61R | Heterozygous |
| WM-1361A | A182G | Q61R | Heterozygous |
| WM-1366 | A182T | Q61L | Heterozygous |
| WM852 | A182G | Q61R | Homozygous |
To assess the irreversible damage done to the cells by PKCδ inhibition in a different manner, clonogenic colony assays were performed using SBcl2 melanoma cells to determine the kinetics of the action of PKCδ inhibitors on the growth and proliferative characteristics of the cells. In contrast to a proliferation assay, which examines potentially temporary and reversible effects on proliferation and survival, clonogenic assays assess irreversible effects of a compound on cell viability and proliferative capacity. Cells were exposed to mallotoxin/rottlerin or B106 for 12, 24 or 48 hr and then re-plated in medium without inhibitors, and the difference in colony-forming ability of cultures was assessed. Both mallotoxin/rottlerin and B106 treatment significantly decreased the number of colonies formed in SBcl2 cells after as little as 12 hr of treatment, and approximately 40-fold reduction in the number of colonies was observed with 48 hr of drug exposure (Figure 2E). These results demonstrate an irreversible cytotoxic effect of these PKCδ inhibitors on tumor cell growth, even after limited and transient exposure to the compounds.
Collectively, these results supported PKCδ as a potential therapeutic target in melanomas with NRAS mutation. The new PKCδ inhibitor B106 demonstrated activity at nanomolar concentrations, and may serve as a lead compound for future modifications.
Inhibition of PKCδ activity triggers caspase-dependent apoptosis
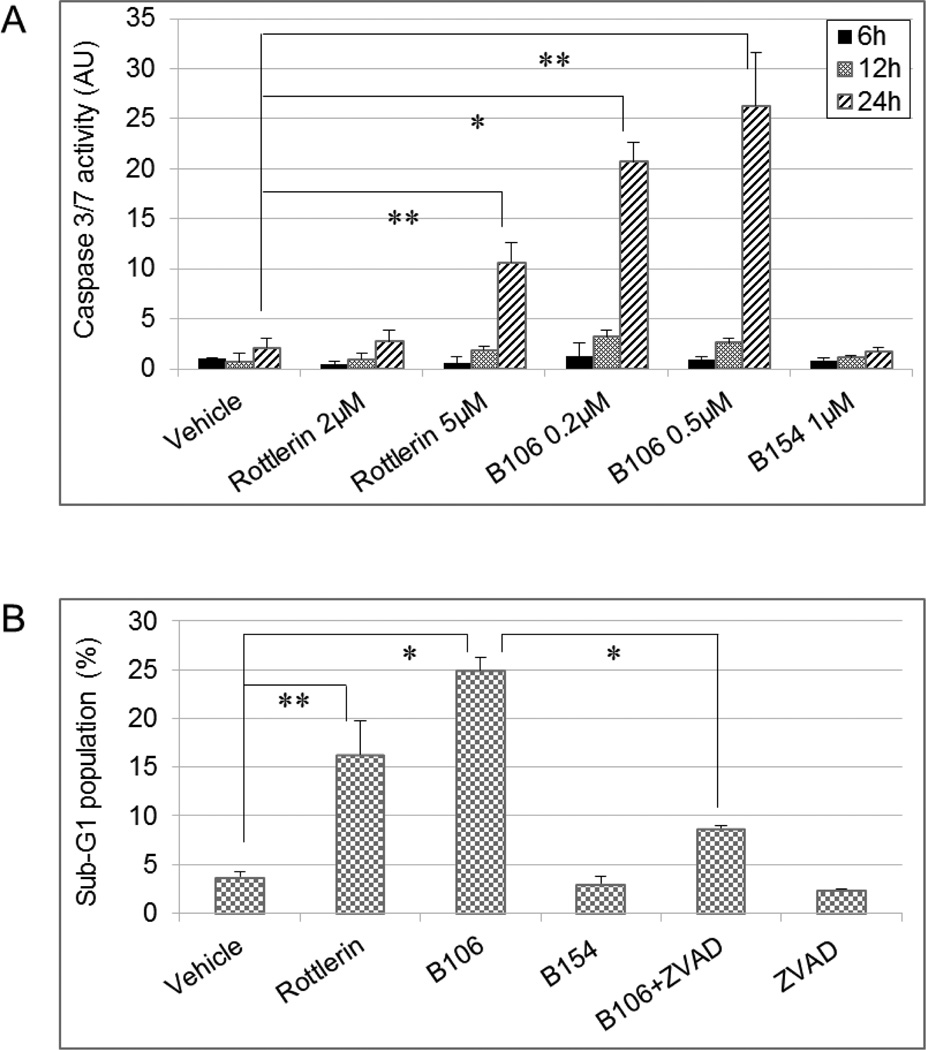
We next determined how PKCδ inhibition results in suppression of tumor cell growth in melanoma. Activated caspase 3 and caspase 7, the ultimate executioners of apoptosis, trigger proteolytic cleavage of crucial key apoptotic proteins, which in turn leads to late apoptotic events, including DNA fragmentation. The activity of effector caspases 3 and 7 was assessed in cells treated with PKCδ inhibitors. Twenty-four hours of exposure to rottlerin (5 µM) or B106 (0.2 and 0.5 µM) significantly increased the activity of caspase 3/7 in SBcl2 cells compared to vehicle (DMSO) (Figure 3A). The effect of B106 on caspase 3/7 activation was greater than that of rottlerin: a 10-fold increase at 0.2 µM and a 12.5-fold increase at 0.5 µM of B106, in contrast to a 5-fold increase by rottlerin at 5 µM. These findings indicated the potential involvement of caspase 3/7-mediated apoptosis in response to PKCδ inhibition.
Figure 3. Inhibition of PKCδ induces caspase-dependent apoptosis.
(A) Effector caspase 3/7 activation by PKCδ inhibition. SBcl2 cells were exposed to rottlerin (2 or 5 µM) or B106 (0.2 or 0.5 µM) for 6, 12 or 24 hr and caspase 3/7 activity was measured. DMSO and B154 (1 µM) served as a vehicle control and a negative compound control, respectively. The average values of triplicates were normalized to those of vehicle-treated sample at 6 hr. Error bars indicate the standard deviations. P values: ** p < 0.003, * p < 0.0002.
(B) DNA fragmentation induced by PKCδ inhibition. SBcl2 cells were treated with rottlerin (5 µM), B106 (0.5 µM) alone, or B106 (0.5 µM) plus the pan-caspase inhibitor Z-VAD-FMK (100 µM) together for 24 hr. The proportion of sub-G1 population was measured by flow cytometry. Values represent the average of duplicates and error bars indicate the standard deviations. p values: ** p < 0.04, * p < 0.004.
As evidence of apoptosis, induction of DNA fragmentation, a hallmark of late events in the sequence of the apoptotic process, in the presence or absence of PKCδ inhibitors was assessed by flow cytometric analysis. The proportion of cells containing a DNA content of less than 2n (fragmented DNA), categorized as the “sub-G1” population and considered in the late apoptotic phase, was significantly higher after treatment with rottlerin at 5 µM and even higher after treatment with B106 at 0.5 µM, whereas B154, a negative-control compound for B106, lacking PKCδ-inhibitory activity, produced no more fragmented DNA than did vehicle control (DMSO), suggesting the effect of B106 on DNA fragmentation was related to inhibition of PKCδ activity (Figure 3B). To determine whether activation of caspases by PKCδ inhibitors was necessary for the observed apoptosis, the pan-caspase inhibitor Z-VAD-FMK (carbobenzoxy-valyl-alanyl-aspartyl-[Omethyl]-fluoromethylketone) was employed. Pre-treatment of cells with Z-VAD-FMK (50 µM) prevented B106-induced caspase 3 cleavage in immunoblot analysis (data not shown). B106-induced DNA fragmentation was significantly abrogated when SBcl2 cells were pretreated with Z-VAD-FMK (100 µM) (Figure 3B). Taken together, these data suggest that PKCδ inhibition attenuates tumor cell growth by inducing caspase-dependent apoptosis in NRAS-mutant melanoma cells.
PKCδ inhibition triggers apoptotic response via the stress-responsive JNK pathway
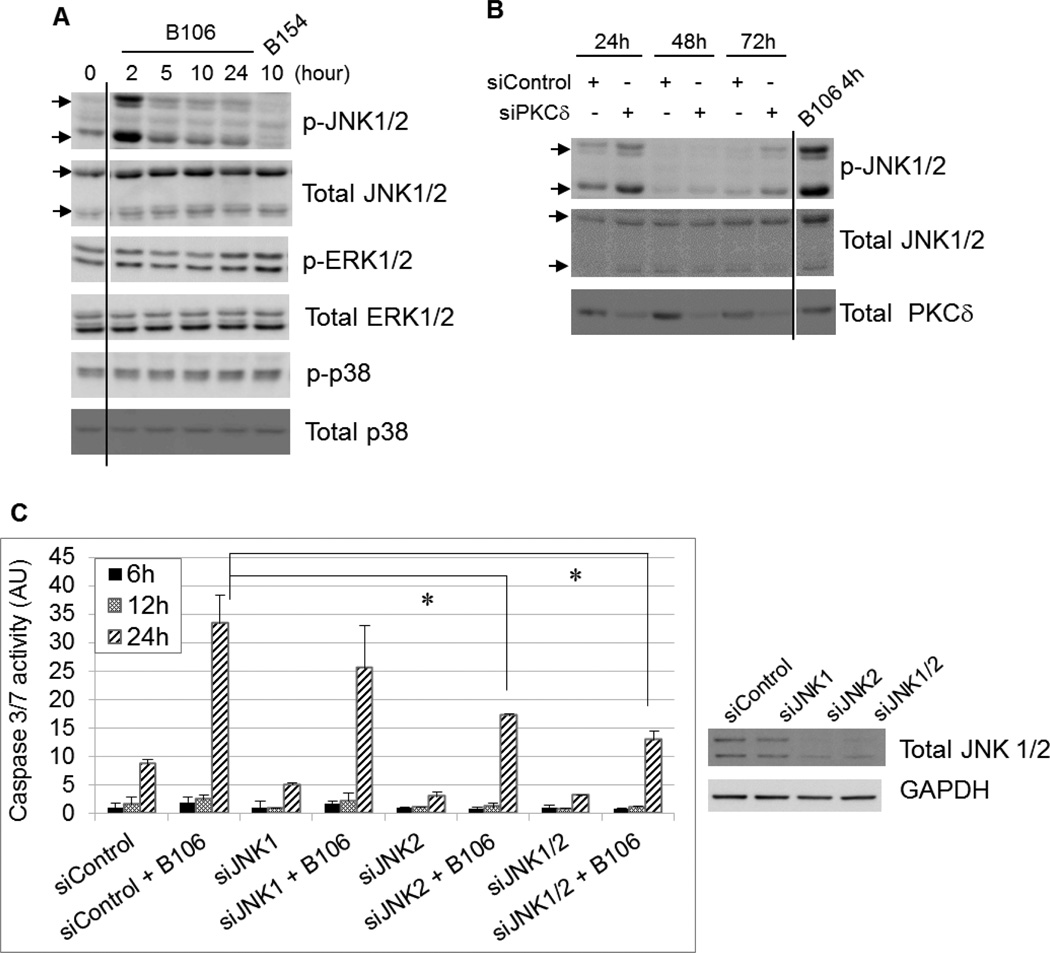
To identify which intracellular signaling pathway PKCδ inhibition employs to induce cytotoxicity, the activation status of known downstream targets of PKCδ was examined after PKCδ inhibition, including MAPKs (ERK, p38 and JNK), AKT, NFκB pathway, cyclin-dependent kinase inhibitors, p53, IAPs, GSK3β or c-Abl. Inhibition of PKCδ activity in SBcl2 cells by B106 induced phosphorylation (activation) of JNK1/2 (T183/Y185) most strongly after two hr of exposure (Figure 4A). In contrast, phosphorylation of the closely-related MAPKs p38 and ERK was not affected by PKCδ inhibitors (Figure 4A). Consistent with these observations generated using chemical inhibitors, selective downregulation of PKCδ by transfection of PKCδ-specific siRNA induced phosphorylation of JNK1/2 at 24 hr, (when effects of siRNA on PKCδ levels were first observed) (Figure 4B). Transfection of PKCδ-specific or negative control siRNA did not affect phosphorylation levels of ERK or p38.
Figure 4. PKCδ inhibition triggers an apoptotic response through activation of JNK.
PKCδ inhibition activates JNK. (A, B) SBcl2 cells were exposed to B106 (1 µM) or the negative control compound B154 (1 µM) for indicated times (A) or transfected with siRNA targeting PKCδ (“siPKCδ”) or non-targeting siRNA (“siControl”) at 5 nM for the indicated times (B). Protein lysates were subjected to immunoblot analysis for levels of phosphorylated or total MAPK proteins. (C) Activation of caspase 3/7 is mitigated by knockdown of JNK prior to B106 treatment. SBcl2 cells were transfected with siRNA targeting JNK1 or JNK2 alone (5 nM), or the combination of JNK1 and JNK2 siRNA (5 nM each), or non-targeting siRNA (10 nM) for 72 hours, and subsequently exposed to B106 (0.5 µM) or vehicle (DMSO) for 6, 12 and 24 hours. Caspase 3/7 activity was measured. The average values of triplicates were normalized to those of the vehicle-treated sample at 6 hours between the pairs exposed to the same siRNA. Error bars indicate the standard deviations. P values: * p < 0.005. Downregulation of JNK1/2 proteins were confirmed by immunoblot analysis at 72 hr. In panels A & B, certain lanes not relevant to this discussion were excised, as indicated by the vertical lines.
Among its pleiotropic cellular activities, JNK is an effector in certain apoptotic responses, and some chemotherapeutic agents, including paclitaxel, cisplatin and doxorubicin, employ the JNK pathway for their cytotoxic activity.25, 26 Because of the data demonstrating that PKCδ inhibition causes caspase-dependent apoptosis (Figure 3) and JNK activation (Figures 4A &B), the effect of inhibition of the JNK pathway during B106 treatment was explored to determine if there is a functional relationship. SBcl2 cells were transfected with non-specific siRNA or siRNA specific for JNK1 or JNK2 alone, or co-transfected with JNK1-plus JNK2-specific siRNA for 72 hr, and then exposed to B106 or DMSO (vehicle) for 6, 12 or 24 hr, followed by measurement of caspase activity (Figure 4C). Analysis at 24 hr after B106 treatment showed that knockdown of JNK2 alone, and co-knockdown of JNK1 and 2, mitigated B106-induced caspase 3/7 activation in rough proportion to the knockdown efficiency of JNK1/2 proteins. These data indicated that JNK is a necessary mediator of the apoptotic response induced by PKCδ inhibition.
PKCδ inhibition activates the MKK4-JNK-H2AX pathway
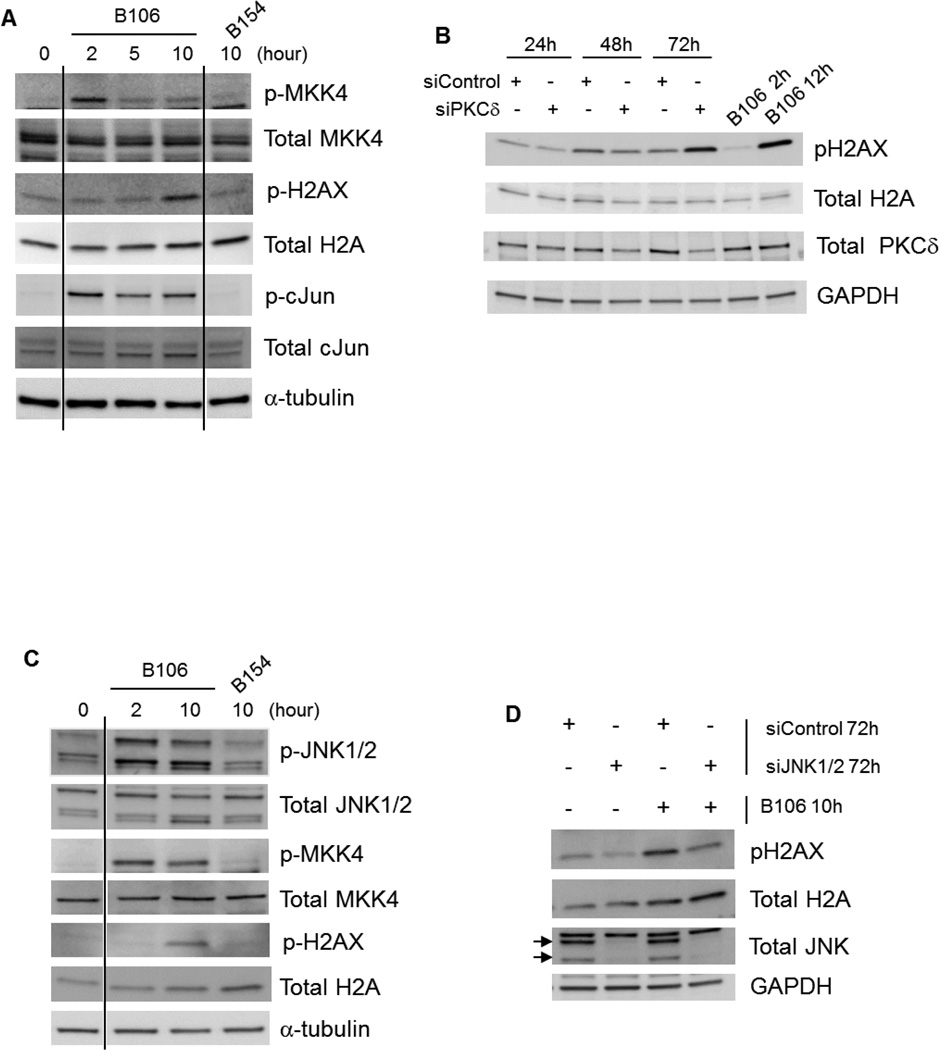
We tested for involvement of known upstream and downstream effectors of the JNK pathway following PKCδ inhibition. The MAPKK kinases MKK4 and MKK7 lie one tier above JNK. MKK4 was activated by B106 (Figure 5A), whereas MKK7 was not phosphorylated in response to B106 (data not shown). Activation of the canonical JNK substrate, c-Jun, was also observed in response to B106 exposure, confirming the activation of the JNK pathway by PKCδ inhibitors (Figure 5A). Furthermore, activation of H2AX (histone H2A variant X), another downstream effector of JNK associated with its apoptotic actions,27 was noted at later time points in response to B106 treatment (Figure 5A). B106 consistently induced H2AX phosphorylation as early as after 10 hr of exposure. The effect of PKCδ inhibition on H2AX activation was further confirmed by selective downregulation of PKCδ with siRNA. Phosphorylation of H2AX was observed at 72 hr after PKCδ siRNA transfection (Figure 5B). PKCδ inhibition by B106 treatment similarly induced phosphorylation of MKK4, JNK and H2AX in NRAS mutant melanoma WM1366 cells (Figure 5C).
Figure 5. PKCδ inhibition activates the MKK4-JNK-H2AX pathway.
(A) Activation of upstream and downstream components of the JNK pathway by B106. SBcl2 cells were exposed to B106 or the negative control compound B154 at 1 µM for the indicated times. Protein lysates were subjected to immunoblot analysis. (B) Selective downregulation of PKCδ results in phosphorylation of H2AX. SBcl2 cells were transfected with siRNA targeting PKCδ (“siPKCδ”) or non-targeting (“siControl”) at 50 nM for the indicated times. Protein lysates were subjected to immunoblot analysis. In panels A & B, certain lanes not relevant to this discussion were excised, as indicated by the vertical lines. (C) PKCδ inhibition activates H2AX through JNK. SBcl2 cells were transfected with siRNA targeting JNK1 and JNK2 together (5 nM each) or non-targeting siRNA (10 nM) for 72 hr and subsequently exposed to B106 (0.5 µM) or vehicle (DMSO) for 10 hr. Protein lysates were subjected to immunoblot analysis. Arrows indicate JNK1/2.
Because JNK affects diverse downstream effectors, we next determined whether JNK activation caused by PKCδ inhibition is directly linked to B106-induced H2AX activation. Knockdown of JNK1/2 itself slightly reduced basal phospho-H2AX (pH2AX) expression, indicating that basal phosphorylation of H2AX is regulated by JNK (Lane 2, Figure 5D). B106 exposure robustly induced phosphorylation of H2AX in control siRNA-treated cells (Lane 3, Figure 5D); in comparison, prior downregulation of JNK1/2 protein by siRNA attenuated B106-induced H2AX phosphorylation (Lane 4, Figure 5D). Collectively, these data suggest that PKCδ inhibition directly or indirectly activates MKK4 in cells containing mutated NRAS, which in turn activates JNK1/2 and subsequently H2AX.
H2AX is a critical regulator of caspase-dependent apoptosis induced in response to PKCδ inhibition
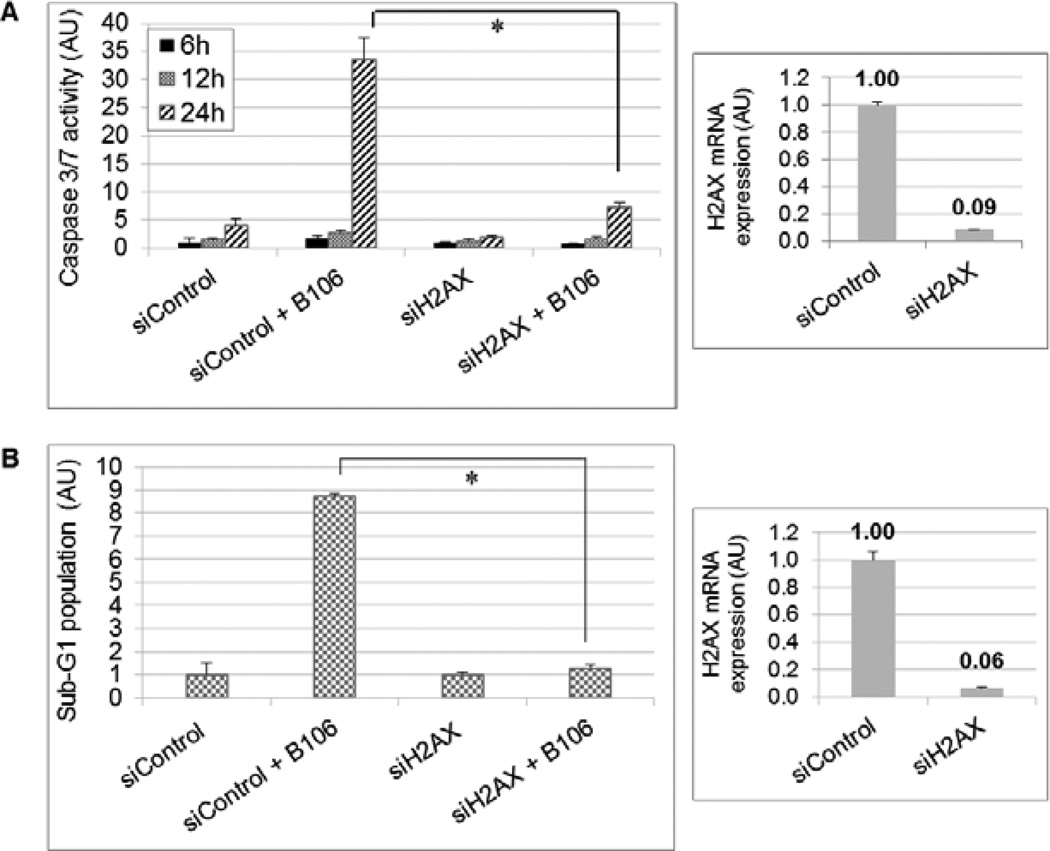
Although phosphorylation of H2AX is best known as a consequence of DNA double-stranded breaks in the DNA-damage response, facilitating repair,28–30 recent studies have demonstrated that phosphorylation of H2AX at Ser 139 resulting from JNK activation actively mediates the induction of apoptosis by inducing DNA fragmentation in UV- or chemotherapy-damaged cells.31–34 Accordingly, the direct involvement of H2AX in apoptotic response to PKCδ inhibition was examined. SBcl2 cells were transfected with siRNA targeting H2AX, or non-targeting siRNA, for 72 hr and then exposed to B106 for 6, 12 or 24 hr, with subsequent assay of caspase 3/7 activation. Downregulation of H2AX prior to B106 treatment greatly decreased the level of caspase 3/7 activation at 24 hr of B106 exposure (Figure 6A).
Figure 6. H2AX is a critical apoptotic regulator in apoptosis induced by PKCδ inhibition.
(A) Activation of caspases 3/7 is mitigated by knockdown of H2AX prior to B106 treatment. SBcl2 cells were transfected with siRNA targeting H2AX or non-targeting siRNA at 5 nM for 72 hours, and subsequently exposed to B106 (0.5 µM) or vehicle for 6, 12 or 24 hr. Caspase 3/7 activity was measured. The average values of triplicates were normalized to those of the vehicle-treated sample at 6 hr between the pairs exposed to the same siRNA. Error bars indicate the standard deviations. P values: * p < 0.005. Downregulation of H2AX at72 hr was confirmed by quantitative PCR.
(B) Induction of DNA fragmentation is mitigated by knockdown of H2AX prior to B106 treatment. SBcl2 cells were transfected with siRNA targeting H2AX, or non-targeting siRNA, at 5 nM for 72 hr, and subsequently exposed to B106 (0.5 µM) or vehicle for 24 hr. The proportion of sub-G1 population was measured by flow cytometry. The average values of duplicates were normalized to those of the vehicle-treated samples between the pairs exposed to the same siRNA. Error bars indicate the standard deviations. P value: * p < 0.0004. Downregulation of H2AX at 96 hr was confirmed by quantitative PCR.
To explore a direct link between H2AX and the execution of apoptosis, PKCδ inhibition-induced DNA fragmentation was examined in the presence or absence of H2AX. SBcl2 cells were transfected with either negative-control siRNA or siRNA targeting H2AX for 72 hr, and then subjected to PKCδ inhibition by exposure to B106 for 24 hr. PKCδ inhibition by B106 treatment increased DNA fragmentation 8.5-fold in the cells transfected with negative control siRNA (Figure 6B). In contrast, PKCδ inhibition by B106 treatment failed to induce DNA fragmentation in the absence of H2AX (Figure 6B), indicating that H2AX is necessary for B106-induced apoptosis (Figure 6B). Collectively, these results suggest that inhibition of PKCδ by B106 treatment triggers caspase-dependent apoptosis through activation of the JNK-H2AX stress-responsive signaling pathway.
BRAF inhibitor-resistant BRAF mutant melanoma lines are susceptible to PKCδ inhibition
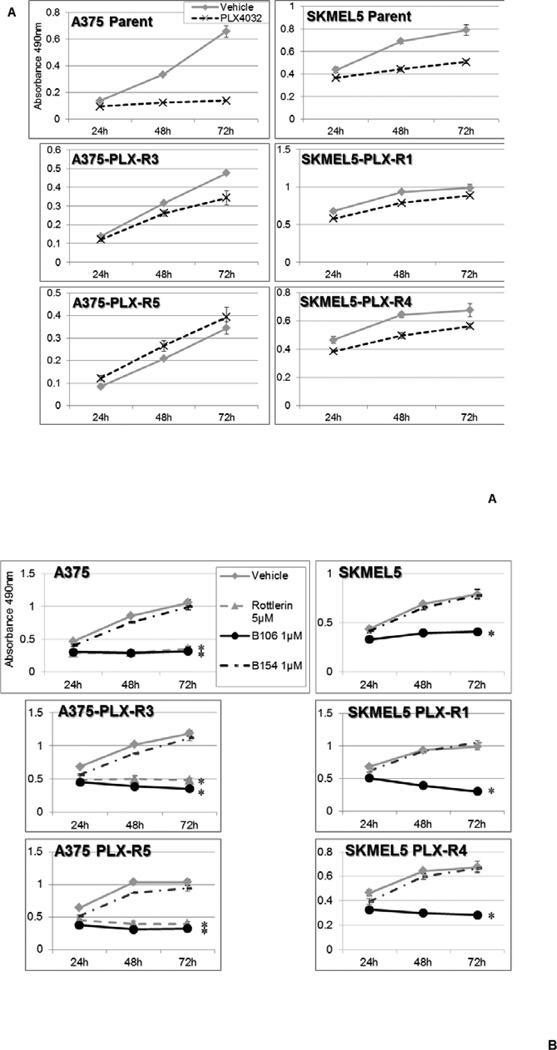
The inevitable development of resistance to the BRAF inhibitor PLX4032 (vemurafenib) in melanomas bearing BRAF mutations remains an ongoing clinical challenge. Several proposed models of PLX4032 resistance involve reactivation of RAS-MEK/ERK mitogenic pathway, induced, for example, by the secondary mutations of NRAS at position 61, or activation of alternative pathways leading to reactivation of ERK signaling, such as IGF1R or AKT.6 Our previous studies have demonstrated the effectiveness of PKCδ inhibitors in the cells with the aberrant CRAF-ERK activation even in the absence of mutations in RAS oncogenes.9–12 We therefore investigated whether PKCδ inhibition could be similarly effective in those BRAF-mutant melanoma cells that have become refractory to a BRAF inhibitor (PLX4032). We generated BRAF-V600E mutant melanoma cell sub-lines resistant to PLX4032 by continuously exposing A375 and SKMEL5 cells to PLX4032, with gradually increasing concentrations of the drug over weeks. Resistance to PLX4032 was verified by comparing their sensitivity to the drug with that of their parental cells (Figure 7A). PLX-R derivative lines from both A375 and SKMEL5 grew in the presence of concentrations of PLX4032 which were cytotoxic to the parental cells. Sequencing revealed that these resistant cell lines retained wild-type NRAS alleles at position 61. The resistant cell sublines derived from both the A375 or SKMEL5 parent lines acquired distinct aberrant alterations in RAS pathway signaling that may be responsible for their resistance (increased activation of ERK1,2 in the resistant A375 lines, and increased CRAF in the resistant SKMEL5 lines). All of these PLX4032-resistant lines were susceptible to cytotoxicity induced by PKCδ inhibitors at concentrations comparable to the NRAS-mutant melanoma lines (Figure 7B). The parental cell lines A375 and SKEML5 (both BRAF-V600E mutant) were also susceptible to PKCδ inhibition (Figure 7B); this finding is consistent with our previous report that cells with aberrant activation/mutation of RAF signaling, and consequent activation of this RAS effector pathway (even in the presence of normal RAS alleles) require PKCδ activity for survival.9–12
Figure 7. PKCδ inhibitors suppress growth of PLX4032-resistant BRAF mutant melanoma cells.
(A) Establishment of PLX4032-resistant cell sub-lines. To establish PLX4032 resistant cell lines, two individual melanoma cell lines with BRAF mutations, A375 and SKMEL5, were continuously exposed to increasing concentrations of PLX4032 up to 10 µM (A375) and 2 µM (SKMEL5). To confirm resistance to PLX4032, the viability of PLX4032-resistant cells and their parental cells was measured by MTS assay during treatment with PLX4032 at 1 µM.
(B) PKCδ inhibitors suppress survival of PLX4032-resistant cells. Two PLX4032-resistant cell sub-lines derived from A375 (Left) and SKMEL5 (Right) cells were exposed to rottlerin (5 µM) or B106 (1 µM) for 24, 48 or 72 hr and MTS assays were performed at each time point. DMSO and B154 (1 µM) served as a vehicle control and a negative compound control, respectively. Each point represents the average of triplicates and error bars indicate the standard deviations. P values (*) were calculated between DMSO (vehicle control) and rottlerin 5 µM, or DMSO and B106, 1 µM in each cell line at 72 hr (p < 0.0002).
PKCδ as a therapeutic target in melanomas with NRAS mutations or BRAF inhibitor resistance
Somatic point mutations of RAS genes at codons 12, 13, and 61 are the most common dominant oncogenic lesions in human cancer,2, 3 making aberrant RAS signaling an important therapeutic target. Inhibition of PKCδ preferentially inhibits the growth of cancer cell lines with genomic mutations in KRAS or HRAS genes, or oncogenic activation of KRAS proteins.9–12, 35, 36 While initially characterized as a specific synthetic lethal interaction between PKCδ and RAS, further work disclosed that aberrant activation of certain RAS effector pathways, PI3K-AKT and CRAF-MEK, would also confer sensitivity to PKCδ inhibition.9–12 Importantly, PKCδ was demonstrated to be non-essential for the survival and proliferation of normal cells and animals,21 suggesting that a therapeutic approach targeting PKCδ would likely spare normal cells, but inhibit the proliferation of tumor cells whose survival depends on PKCδ activity. This report underlines the potential of PKCδ-targeted therapy as a cancer-specific therapy targeting melanoma with NRAS mutations. Cell proliferation and clonogenic assays demonstrated that inhibition of PKCδ suppressed cell growth in multiple melanoma cell lines with NRAS mutations, as well as in PLX4032-resistant cell lines. The cell lines with NRAS mutation that were used in this study had different amino acid substitutions of NRAS codon 61, suggesting the effect of PKCδ inhibitors does not depend on a specific NRAS mutation for their activity. Similarly, PKCδ inhibition was effective in the PLX4032-resistant cell lines tested herein, regardless of the differences in their apparent resistance mechanisms, further supporting the potential of this approach. Constitutive MEK/ERK signaling appears to mediate the majority of acquired resistance to BRAF inhibitors,6 and we have previously reported that aberrant activation of the MEK/ERK arm of the RAS signaling pathway is sufficient to render cells susceptible to PKCδ inhibition, even in the absence of activating mutations of RAS alleles.9–11 Furthermore, we have recently demonstrated that cancer “stem-like” cells (CSCs) derived from a variety of human tumors, including melanomas, are susceptible to PKCδ inhibition.12
The novel PKCδ inhibitor B106, which showed 1000-fold selectivity against PKCδ over PKCα in preliminary in vitro kinase assays, was active at nanomolar concentrations, ten times lower than for rottlerin. These results in cell culture systems suggest the potential of the newest PKCδ inhibitors as targeted agents, although the in vivo efficacy of B106 is yet to be determined. The hydrophobicity of B106 molecule and its rapid metabolism, requiring continuous infusion to generate a pharmacodynamic signal, makes it unsuitable for testing in tumor xenograft models.
Induction of apoptosis is one of the most desirable mechanisms for cytotoxic therapeutic action. The stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), a downstream targets of PKCδ, is activated in response to cellular stresses, including genotoxic stresses.37 Many chemotherapeutic agents employ the JNK pathway for their cytotoxic activity.38, 39 This study demonstrates that PKCδ inhibition activates the JNK pathway through MMK4 to mediate caspase-dependent apoptosis. Consistent with our findings, a recent report demonstrated that knockdown of PKCδ induced apoptosis with elevated phosphorylation of JNK in NIH-3T3 cells stably transfected with HRAS.35 Among the known downstream effectors of JNK, a series of recent reports proposed an active role for phospho-H2AX in apoptosis.31–34 PKCδ inhibition evoked phosphorylation of H2AX subsequent to JNK activation, positioning H2AX phosphorylation downstream of JNK after PKCδ inhibition. Collectively, these results demonstrate the importance of H2AX as an active apoptotic mediator, providing functional evidence showing it to be a necessary component of apoptosis initiated by PKCδ inhibition.
The concept of targeting cancer therapeutics towards specific mutations or aberrations in tumor cells which are not found in normal tissues has the potential advantages of high selectivity for the tumor and correspondingly low secondary toxicities. We have previously demonstrated that knockdown of PKCδ, or its inhibition by previous generations of small molecules, was not toxic to non-transformed primary murine and human cell lines, primary human endothelial cells, or to tumor lines without aberrant activation of the RAS signaling pathway, at concentrations which are profoundly cytotoxic to melanoma lines bearing NRAS mutations (0.5–2.5 µM).. Herein we show that human primary melanocytes are not affected by B106.9–11 In addition, continuous local infusion of B106 at 5 µM concentrations is not cytotoxic to dermal and subdermal tissues in mice. Derivatives of the 3rd generation PKCδ inhibitor B106 are being generated, using structure function analysis of the 36 compounds in that cohort and medicinal chemistry to enhance drug-like properties, to facilitate future in vivo studies. Collectively, our studies suggest that PKCδ suppression may offer a promising tumor-specific option for a subpopulation of melanomas for which we have currently a limited number of effective therapeutics.
Supplementary Material
Acknowledgements
We thank R. Spanjaard and A. Singh (Boston University School of Medicine, Boston, MA) for generously providing cell lines. This study was supported by the Melanoma Research Alliance, National Cancer Institute grants CA112102, CA164245, and CA141908, Department of Defense grants PC100093 and CA110396, a research award from the Scleroderma Foundation, the Raymond and Beverly Sackler Fund for the Arts and Sciences, and the Karin Grunebaum Cancer Research Foundation (DVF); and the Colorado State University Cancer Super Cluster and Phoenicia Biosciences, Inc. (RMW).
Footnotes
Potential Conflicts of Interest:
DVF and RMW have applied for a patent covering some of the structures disclosed in this manuscript. The other authors declare no other conflicts of interest.
Supporting Information Available:
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Inamdar GS, Madhunapantula SV, Robertson GP. Targeting the MAPK pathway in melanoma: why some approaches succeed and other fail. Biochem. Pharmacol. 2010;80:624–637. doi: 10.1016/j.bcp.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takashima A, Faller DV. Targeting the RAS oncogene. Expert. Opin. Ther. Targets. 2013 doi: 10.1517/14728222.2013.764990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, Kim KB, Weber JS, Hersey P, Long GV, Lawrence D, Ott PA, Amaravadi RK, Lewis KD, Puzanov I, Lo RS, Koehler A, Kockx M, Spleiss O, Schell-Steven A, Gilbert HN, Cockey L, Bollag G, Lee RJ, Joe AK, Sosman JA, Ribas A. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin. Oncol. 2013;31:1767–1774. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 7.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 8.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 9.Xia S, Forman LW, Faller DV. Protein Kinase C{delta} is required for survival of cells expressing activated p21RAS. J Biol. Chem. 2007;282:13199–13210. doi: 10.1074/jbc.M610225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S, Chen Z, Forman LW, Faller DV. PKCdelta survival signaling in cells containing an activated p21Ras protein requires PDK1. Cell Signal. 2009;21:502–508. doi: 10.1016/j.cellsig.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Forman LW, Miller KA, English B, Takashima A, Bohacek RA, Williams RM, Faller DV. The proliferation and survival of human neuroendocrine tumors is dependent upon protein kinase C-delta. Endocr. Relat Cancer. 2011;18:759–771. doi: 10.1530/ERC-10-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Forman LW, Williams RM, Faller DV. Protein kinase C delta inactivation inhibits the proliferation and survival of cancer stem cells in culture and in vivo. BMC. Cancer. 2014 doi: 10.1186/1471-2407-14-90. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu T, Chen L, Du W, Tsuji T, Chen C. Synthetic Lethality Induced by Loss of PKC delta and Mutated Ras. Genes Cancer. 2010;1:142–151. doi: 10.1177/1947601909360989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Symonds JM, Ohm AM, Carter CJ, Heasley LE, Boyle TA, Franklin WA, Reyland ME. Protein kinase C delta is a downstream effector of oncogenic K-ras in lung tumors. Cancer Res. 2011;71:2087–2097. doi: 10.1158/0008-5472.CAN-10-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basu A, Pal D. Two faces of protein kinase Cdelta: the contrasting roles of PKCdelta in cell survival and cell death. ScientificWorldJournal. 2010;10:2272-84:2272–2284. doi: 10.1100/tsw.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonne GK, Masoumi KC, Lennartsson J, Larsson C. Protein kinase Cdelta supports survival of MDA-MB-231 breast cancer cells by suppressing the ERK1/2 pathway. J. Biol. Chem. 2009;284:33456–33465. doi: 10.1074/jbc.M109.036186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Wang X, Zhou Y, Evers BM. PKCdelta-mediated regulation of FLIP expression in human colon cancer cells. Int. J. Cancer. 2006;118:326–334. doi: 10.1002/ijc.21373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudot AD, Jeandel PY, Mouska X, Maurer U, Tartare-Deckert S, Raynaud SD, Cassuto JP, Ticchioni M, Deckert M. The tyrosine kinase Syk regulates the survival of chronic lymphocytic leukemia B cells through PKCdelta and proteasome-dependent regulation of Mcl-1 expression. Oncogene. 2009;28:3261–3273. doi: 10.1038/onc.2009.179. [DOI] [PubMed] [Google Scholar]
- 19.Clark AS, West KA, Blumberg PM, Dennis PA. Altered protein kinase C (PKC) isoforms in non-small cell lung cancer cells: PKCdelta promotes cellular survival and chemotherapeutic resistance. Cancer Res. 2003;63:780–786. [PubMed] [Google Scholar]
- 20.Mauro LV, Grossoni VC, Urtreger AJ, Yang C, Colombo LL, Morandi A, Pallotta MG, Kazanietz MG, Bal de Kier Joffe ED, Puricelli LL. PKC Delta (PKCdelta) promotes tumoral progression of human ductal pancreatic cancer. Pancreas. 2010;39:e31–e41. doi: 10.1097/MPA.0b013e3181bce796. [DOI] [PubMed] [Google Scholar]
- 21.Leitges M, Mayr M, Braun U, Mayr U, Li C, Pfister G, Ghaffari-Tabrizi N, Baier G, Hu Y, Xu Q. Exacerbated vein graft arteriosclerosis in protein kinase Cdelta-null mice. J. Clin. Invest. 2001;108:1505–1512. doi: 10.1172/JCI12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soloff RS, Katayama C, Lin MY, Feramisco JR, Hedrick SM. Targeted deletion of protein kinase C lambda reveals a distribution of functions between the two atypical protein kinase C isoforms. J Immunol. 2004;173:3250–3260. doi: 10.4049/jimmunol.173.5.3250. [DOI] [PubMed] [Google Scholar]
- 23.Mochly-Rosen D, Das K, Grimes KV. Protein kinase C-an elusive therapeutic target? Nat. Rev. Drug Discov. 2012;11:937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou WH, Choi DS, Zhang H, Mu D, McMahon T, Kharazia VN, Lowell CA, Ferriero DM, Messing RO. Neutrophil protein kinase Cdelta as a mediator of stroke-reperfusion injury. J Clin. Invest. 2004;114:49–56. doi: 10.1172/JCI21655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 26.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinasedead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, Bode AM, Dong Z. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584:3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol. Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee B, Kessinger C, Kobayashi J, Chen BP, Chen DJ, Chatterjee A, Burma S. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair (Amst) 2006;5:575–590. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan FM, Shao Y, Mayberry MM, Aplin AE. Hyperactivation of MEK-ERK1/2 signaling and resistance to apoptosis induced by the oncogenic B-RAF inhibitor, PLX4720, in mutant N-RAS melanoma cells. Oncogene. 2011;30:366–371. doi: 10.1038/onc.2010.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jane EP, Pollack IF. Enzastaurin induces H2AX phosphorylation to regulate apoptosis via MAPK signalling in malignant glioma cells. Eur. J Cancer. 2010;46:412–419. doi: 10.1016/j.ejca.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Tseng M, Perdreau SA, Rossi F, Antonescu C, Besmer P, Fletcher JA, Duensing S, Duensing A. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67:2685–2692. doi: 10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]
- 34.Wen W, Zhu F, Zhang J, Keum YS, Zykova T, Yao K, Peng C, Zheng D, Cho YY, Ma WY, Bode AM, Dong Z. MST1 promotes apoptosis through phosphorylation of histone H2AX. J Biol. Chem. 2010;285:39108–39116. doi: 10.1074/jbc.M110.151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu T, Chen L, Du W, Tsuji T, Chen C. Synthetic Lethality Induced by Loss of PKC delta and Mutated Ras. Genes Cancer. 2010;1:142–151. doi: 10.1177/1947601909360989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Symonds JM, Ohm AM, Carter CJ, Heasley LE, Boyle TA, Franklin WA, Reyland ME. Protein kinase C delta is a downstream effector of oncogenic K-ras in lung tumors. Cancer Res. 2011;71:2087–2097. doi: 10.1158/0008-5472.CAN-10-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim. Biophys. Acta. 2007;1773:1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee LF, Li G, Templeton DJ, Ting JP. Paclitaxel (Taxol)-induced gene expression and cell death are both mediated by the activation of c-Jun NH2-terminal kinase (JNK/SAPK) J Biol. Chem. 1998;273:28253–28260. doi: 10.1074/jbc.273.43.28253. [DOI] [PubMed] [Google Scholar]
- 39.Koyama T, Mikami T, Koyama T, Imakiire A, Yamamoto K, Toyota H, Mizuguchi J. Apoptosis induced by chemotherapeutic agents involves c-Jun N-terminal kinase activation in sarcoma cell lines. J Orthop. Res. 2006;24:1153–1162. doi: 10.1002/jor.20176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.