Abstract
Purpose.
To investigate the role of DJ-1 in Nrf2-regulated antioxidant defense in corneal endothelial cells (CECs) at baseline and in response to ultraviolet A (UV-A)–induced oxidative stress.
Methods.
DJ-1–deficient CECs were obtained by transfection of an immortalized normal human corneal endothelial cell line (HCECi) with DJ-1 small interfering RNA (siRNA) or by isolation of CECs from ex vivo corneas of DJ-1 knockout mice. Levels of reactive oxygen species (ROS), protein carbonyls, Nrf2 subcellular localization, Nrf2 target genes, and protein interaction between Keap1/Nrf2 and Cul3/Nrf2 were compared between normal and DJ-1–deficient CECs. Oxidative stress was induced by irradiating HCECi cells with UV-A, and cell death and levels of activated caspase3 and phospho-p53 were determined.
Results.
DJ-1 siRNA-treated cells exhibited increased levels of ROS production and protein carbonyls as well as a 2.2-fold decrease in nuclear Nrf2 protein when compared to controls. DJ-1 downregulation led to attenuated gene expression of Nrf2 and its target genes HO-1 and NQO1. Similar levels of Nrf2 inhibitor, Keap1, and Cul3/Nrf2 and Keap1/Nrf2 were observed in DJ-1 siRNA-treated cells as compared to controls. Ultraviolet A irradiation resulted in a 3.0-fold increase in cell death and elevated levels of activated caspase3 and phospho-p53 in DJ-1 siRNA-treated cells compared to controls.
Conclusions.
Downregulation of DJ-1 impairs nuclear translocation of Nrf2, causing decreased antioxidant gene expression and increased oxidative damage. The decline in DJ-1 levels leads to heightened CEC susceptibility to UV-A light by activating p53-dependent apoptosis. Targeting the DJ-1–Nrf2 axis may provide a potential therapeutic approach for enhancing antioxidant defense in corneal endothelial disorders.
Keywords: corneal endothelial cells, DJ-1, Nrf2, oxidative stress, ultraviolet A light
Downregulation of DJ-1 impairs nuclear translocation of Nrf2 causing decreased antioxidant gene expression and increased oxidative damage. Decline in DJ-1 levels leads to heightened corneal endothelial cell susceptibility to UV-A light by activating p53-dependent apoptosis.
Introduction
Corneal endothelium is a monolayer of cells situated on the inner surface of the cornea. The primary function of the corneal endothelium is to maintain corneal transparency and normal visual acuity by controlling leakage of solutes and nutrients from the aqueous humor into corneal layers while water is being pumped from the stroma into the aqueous humor. Human corneal endothelial cells (CECs) have minimal proliferative and repair capacity in vivo, and, therefore, cell loss due to aging and diseases is permanent. Fuchs' endothelial corneal dystrophy (FECD) is the leading cause of endogenous corneal endothelial degeneration characterized by abnormal corneal endothelial morphology, decreased cell density, and abnormal extracellular matrix formation.1
Corneal endothelial cells have the second highest aerobic metabolic rate in the eye—second only to retinal photoreceptors.2 This feature, combined with their lifelong exposure to direct sunlight, make CECs especially prone to oxidative stress. There is growing evidence that oxidative stress plays a critical role in the CEC apoptosis and corneal endothelial degeneration seen in FECD pathogenesis.3–8 In FECD, CECs exhibit a decreased expression of many antioxidants, including glutathione S-transferase, aldehyde dehydrogenase 3A1, peroxiredoxins (Prdx), superoxide dismutase 2, and thioredoxin reductase 1.3,6,9,10 Downregulation of antioxidants in FECD leads to increased production of reactive oxygen species (ROS) and subsequent activation of cell apoptosis.4 Proximal promoter sequence analysis of downregulated antioxidant enzymes in FECD6 reveals a consensus sequence, antioxidant responsive element (ARE), which is the main regulatory site of antioxidant expression.
One of the key transcription factors that binds ARE is nuclear factor erythroid 2–related factor2 (Nrf2).11 Nrf2 is a basic leucine zipper transcription factor that is ubiquitously expressed in a wide range of tissues and cell types. Studies using Nrf2 knockout (KO) mice have demonstrated that Nrf2 and its target genes represent an important antioxidant defense system against a range of chemical and pathologic toxicity.12,13 Different mechanisms of Nrf2 activity and regulation have been proposed at multiple levels, including transcription, translation, and posttranslational modification.14–17 It is commonly accepted that under normal physiological conditions, Nrf2 localizes in the cytoplasm, and its activity is sequestered by Kelch-like ECH-associated protein (Keap1) through the Cullin3-based E3 ligase complex (Keap1/Cul3). This process mediates ubiquitination and degradation, thus keeping Nrf2 at a low production level in the cytoplasm. Conversely, under oxidative stress conditions, ROS and electrophiles bind to sulfhydryl residues of Keap1, leading to the suppression of Nrf2 degradation. Subsequently, released cytoplasmic Nrf2 translocates into the nucleus, where it binds to ARE-bearing sequence and promotes the transcription of antioxidant enzymes, such as heme oxygenase-1 (HO-1), glutamate cysteine ligase, peroxiredoxins, NAD(P)H: quinone oxidoreductase 1 (NQO1), and glutathione S-transferase.
DJ-1, encoded by the PARK7 gene, is a member of the DJ-1/PfpI protein superfamily, with a molecular weight of ∼20 kDa, and is ubiquitously expressed in most cell types and tissues. It has been intensively studied because of its connection to parkinsonism18,19 and cancer.20,21 DJ-1 is a multifunctional protein. Under oxidative stress, it plays critical antioxidant defense roles by several molecular processes. In addition to directly regulating some antioxidant gene expression,22 DJ-1 functions as an atypical ROS scavenger peroxiredoxin-like peroxidase through the oxidization of conserved cysteine residue (Cys106),23 and a defender against UV- and oxidative stress–induced cell death by suppressing proapoptotic factors.24 It is proposed that DJ-1 acts as a stabilizer of Nrf2 by preventing the Keap1/Nrf2 association14,21 or Nrf2 nuclear export.25 However, these observations are challenged by a report that activation of the Nrf2–ARE pathway is independent of DJ-1.26
Ultraviolet A (UV-A, 320–400 nm) has a longer wavelength than ultraviolet B (UV-B, 280–320 nm) and ultraviolet C (UV-C, 100–280 nm). In a natural environment, UV-A constitutes more than 95% of the solar UV radiation that reaches the surface of the earth.27,28 Ultraviolet A wavelengths are not well absorbed by native DNA and instead damage the cells indirectly by producing ROS. Ultraviolet A–induced ROS initiates cell apoptosis, which is a complex event involving different pathways.29–31 Apoptosis caused by UV-A–induced oxidative stress has been shown to result from activation of the tumor suppressor gene p53.30,32 DJ-1 is a potent inhibitor of the oxidative stress-induced, cell death signaling pathway. Under UV-B and UV-C stress, DJ-1 knockdown is determined to be highly susceptible to the UV-induced activation of the apoptotic signaling cascade and cell death.24,33 However, the role of DJ-1 in response to UV-A–induced oxidative stress remains unclear.
Previously, we have found that FECD CECs exhibit decreased levels of Nrf2 protein and HO-1 mRNA as well as increased levels of oxidative DNA marker 8-OHdG, as compared to normal CECs, indicating that Nrf2 deficiency may contribute to oxidant–antioxidant imbalance, causing oxidative DNA damage and apoptosis over the course of FECD development. Interestingly, when accompanied by Nrf2 deficiency, DJ-1 decreases at both mRNA and protein levels in FECD CECs.5
In this study, we aimed to determine the role of DJ-1 in Nrf2-regulated antioxidant defense in CECs as it relates to the pathogenesis of FECD. We analyzed the effect of DJ-1 downregulation on oxidative damage, Nrf2 nuclear translocation, and its target antioxidant gene expression. To mimic the oxidative stress received from daily natural sunlight, we used UV-A irradiation to physically challenge CECs, followed by comparison of the cellular responses between DJ-1 downregulated cells and DJ-1 wild-type (WT) cells. This study presented evidence that reduction in DJ-1 impairs nuclear translocation of Nrf2, causing decreased antioxidant gene expression and heightened cell susceptibility to UV-A–induced oxidative stress. These findings provide not only the role of DJ-1 in Nrf2-regulated antioxidant defense in corneal endothelial cells, but also an oxidative stress model for studying the development of FECD.
Materials and Methods
Human Corneal Endothelial Cell Culture
Normal human corneal endothelial cell line (HCECi), immortalized by infection with an amphotropic recombinant retrovirus containing human papilloma virus type 16 genes E6 and E7, was a generous gift of May Griffith, PhD (Ottawa Hospital Research Institute, Ottawa, ON, Canada).34 Cells were grown in culture flasks or petri dishes in Chen's medium35 at 37°C with 5% CO2.
Small Interfering RNAs Knockdown DJ-1 in HCECi Cells
HCECi cells were seeded in 6-well plates or 35-mm petri dishes, with a density of 2 to 3 × 105 cells per well, in Chen's medium without antibiotics. The next day, the cells were transfected with 25 nM or 50 nM DJ-1 siRNA (Santa Cruz Biotechnology, Santa Cruz, CA, USA) in Opti-MEM reduced serum medium for 6 hours, using transfection reagent Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) followed by replacement of the medium with Chen's medium. Twenty-five nanomolar or 50 nM control siRNA (Santa Cruz Biotechnology) and non-siRNA treatments were used as controls. At 3 days post transfection, cells were imaged by a phase-contrast microscope (Eclipse TS100; Nikon, Tokyo, Japan) and then harvested for further experiments. Cell viability was measured by Trypan blue staining (Invitrogen) using an automated cell counter (Countess; Invitrogen). DJ-1 knockdown efficiency was examined by real-time reverse transcription–polymerase chain reaction (RT-PCR) probing with DJ-1 TaqMan primer (Applied Biosystems, Foster City, CA, USA) and by Western blot analysis with an anti–DJ-1 antibody (1:1000; Novus Biologicals, Littleton, CO, USA).
Detection of ROS and Protein Carbonyl
Three days after siRNA transfection, as described above, cells were washed with warm Hank's balanced salt solution (HBSS; Invitrogen) and then loaded with 25 μM 5-(and -6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA; Invitrogen) for 30 minutes, at 37°C. Cells were harvested by a scraper, and this was followed by washing with warm HBSS by centrifugation at 500g for 5 minutes. Cells were then resuspended in 100 μL warm HBSS and transferred to a flat-bottomed black 96-well plate. The percentage of carboxyl-H2DCF–stained cells was analyzed by measuring their fluorescence (485/520 nm), using a fluorescence microplate reader (BioTek-Synergy 2; BioTek Instruments, Inc., Winooski, VT, USA). Relative fluorescence units were normalized to the cell number. Protein carbonyl content was assessed by using an OxiSelect Protein Carbonyl ELISA kit (Cell Biolabs, Inc., San Diego, CA, USA), according to the manufacturer's guidelines. Briefly, samples and BSA standards (10 μg/mL) were loaded into wells of a 96-well protein-binding plate and left to adsorb overnight, at 4°C, followed by washing with PBS and then by adding dinitrophenylhydrazine (DNPH) working solution. Wells were incubated with anti-DNP antibody for 1 hour after blocking. Wells were next exposed for 1 hour to a horseradish peroxidase (HRP)–conjugated secondary antibody and then developed with substrate solution. Absorbance at 450 nm was read by using an absorbance microplate reader (SpectraMax 34; Molecular Devices, Silicon Valley, CA, USA). The average absorbance was calculated from sample triplicates.
Subcellular Fractionation
HCECi cells were harvested at 72 hours post siRNA transfection. Cytosolic and nuclear extracts were sequentially isolated by using a nuclear/cytosol fractionation kit (BioVision, Milpitas, CA, USA), according to manufacturer's instructions.
Ultraviolet A Irradiation
Cells were irradiated by fluorescent UV-A broadband lamps (320–340 nm, PUVA 180; Herbert Waldman, Werk Fur Lichtechnik Schwenningen, Germany) with the fluence of 10 J/cm2. Briefly, at 48 to 72 hours post DJ-1 siRNA transfection, cells were washed and kept in warm HBSS under UV-A light without a petri dish lid, in a biosafety laminar hood. Ultraviolet A–irradiated cells were incubated in OPTI-MEM base medium at 37°C with 5% CO2 for 24 hours or collected immediately. Controls were taken by keeping the cells in the laminar flow hood under identical conditions with the only exception being lack of UV-A exposure.
Cell Viability Assay
Cytotoxicity was measured by evaluating lactate dehydrogenase release according to manufacturer's instructions (CytoTox 96 Non-radioactive Cytotoxicity Assay; Promega, Madison, WI, USA). In brief, 3 days post DJ-1 siRNA transfection, 50 μL cell culture medium was transferred to a 96-well plate after centrifugation, and 50 μL substrate was added to each well and mixed. The mixture was incubated for 30 minutes at room temperature, in the dark, followed by addition of 50 μL stop solution to each well. The plate was read at 490 nm by using an absorbance microplate reader (SpectraMax 34).
Apoptosis Assay
Activated caspase3 was detected by Western blot analysis with anti–cleaved caspase3 (Asp175) (5A1) Rabbit mAb (Cell Signaling Technology, Danvers, MA, USA) and by the EnzChek Caspase3 Assay Kit No. 2 (Molecular Probes; Invitrogen), following the manufacturer's instructions. Briefly, after treatment, cells were washed in cold PBS and lysed. Fluorescence products of the substrate Z-DEVD-rhodamine 110 generated by activated caspase3 in the cell extracts were measured by using a fluorescence microplate reader (BioTek-Synergy 2) with excitation/emission at 496/520 nm.
Immunoprecipitation
Three days after siRNA transfection, the cell culture medium was removed and cells were rinsed with ice-cold PBS. Whole cell lysates were harvested by scraping the petri dish after addition of 100 μL 1× RIPA cell lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/mL leupeptin; Cell Signaling Technology) supplemented with a protease and phosphatase inhibitor cocktail (Thermo Scientific, Pittsburgh, PA, USA) immediately before use. Scraped cells from five petri dishes were pooled, sonicated on ice for 5 seconds for three times (Sonifier 250; Branson, Danbury, CT, USA) and centrifuged at 14,000g at 4°C for 10 minutes. Supernatants were collected for Pierce Bicinchoninic Acid (BCA) protein assay (Thermo Scientific). The Nrf2 was immunoprecipitated by using rabbit polyclonal anti-Nrf2 antibody (H300) (sc13032; Santa Cruz Biotechnology). Five hundred micrograms of protein from the cell lysates was incubated with 2.5 μg rabbit polyclonal anti-Nrf2 by gentle rocking overnight at 4°C to pull down Nrf2. Protein A magnetic beads (Cell Signaling Technology) were prewashed by adding 40 μL of the bead slurry to a clean tube containing 500 μL 1× cell lysis buffer followed by vortexing and placing the tube in a magnetic separation rack for 20 seconds. Once the solution was clear, the supernatant was carefully removed and the immunocomplex solution was transferred to the prewashed magnetic bead pellets, followed by incubation for 30 minutes with gentle rocking at room temperature. The magnetic bead pellets were washed 3 times with 500 μL cell lysis buffer by placing the tubes in the magnetic separation rack for 20 seconds and carefully removing the supernatant. The bead pellets were suspended with 30 μL 4× Laemmli sample buffer containing 1% β-mercaptoethanol and vortexed. The samples were heated at 95°C for 5 minutes and centrifuged at 14,000g for 1 minute. The supernatants were collected for Western blot analysis with rabbit polyclonal anti-Nrf2 (H300) (1:100, sc13032; Santa Cruz Biotechnology), goat polyclonal anti-Cul3 (1:100, sc-8556; Santa Cruz Biotechnology), and mouse monoclonal anti-Keap1 (1:400, MAB3024; R&D Systems, Minneapolis, MN, USA) for detecting Nrf2, Cul3, and Keap1, respectively. To test the immunoprecipitation specificity of rabbit polycolonal anti-Nrf2 antibody, 2.5 μg normal rabbit IgG (sc 3888; Santa Cruz Biotechnology) used as IgG control was added to 500 μg non–siRNA-treated cell lysates followed by immunoprecipitation–Western blot (IP-WB) as described above. Ten micrograms of whole cell lysates was used for examining the input for DJ-1, Nrf2, Cul3, and Keap1 by Western blot analysis as described later.
Experimental Animals
DJ-1 KO mice with C57BL/6 background and C57BL/6 WT mice (The Jackson Laboratory, Bar Harbor, ME, USA) were used for this study. All animal experiments were approved by the Institutional Animal Care and Use Committee and adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Real-Time RT-PCR
Total RNA was extracted from CECs by using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Total RNA from dissected mouse corneal endothelium with Descemet membrane was extracted by using RNeasy Micro Kit (Qiagen). Complementary DNA was prepared by reverse transcription with a commercially available kit (Promega) in a MyCycler Thermal cycler (Bio-Rad, Hercules, CA, USA), according to manufacturer's protocol. TaqMan primers and probes for DJ-1, Nrf2, Keap1, HO-1, NQO1, and Prdx2 genes, as well as for the endogenous control β2-microglobulin, were obtained from Applied Biosystems. Real-time RT-PCR reactions were set up with Probe Fast master mix (Kapa Biosystems, Woburn, MA, USA) and performed in Mastercycler Realplex2 (Eppendorf, Hamburg, Germany). Each gene was detected in duplicate and the assay repeated three times. For data analysis, the comparative threshold cycle method was performed by using non-siRNA treatment as the calibrator.
Western Blot Analysis
Whole cell extracts from HCECi cells or mouse CECs were lysed with 1× RIPA buffer (Cell Signaling) supplemented with protease and phosphatase inhibitor cocktail (Pierce; Thermo Scientific), followed by brief sonication (Sonifier 250). Protein concentrations of nuclear and cytosolic extracts from HCECi cells were determined by Pierce BCA protein assay (Thermo Scientific). Proteins were separated in 10% or 12% Bis-Tris NuPAGE gels (Invitrogen) and electroblotted to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% dry nonfat milk in PBS for 1 hour and then incubated overnight at 4°C with the following primary antibodies: goat polyclonal anti–DJ-1 (1:1000; Novus Biologicals); rabbit polyclonal anti-Nrf2 for IP-WB (1:100; Santa Cruz Biotechnology); mouse monoclonal anti-Nrf2 (1:400; R&D Systems); mouse monoclonal anti-Keap1 (1:400; R&D Systems); mouse monoclonal anti–phospho-p53 (Ser15) (1:1000; Cell Signaling Technology); rabbit monoclonal anti–cleaved caspase3 (1:1500; Cell Signaling Technology); rabbit polyclonal anti–glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH, 1:4000; Santa Cruz Biotechnology); and mouse anti–β-actin (1:4000; Sigma-Aldrich Corp., St. Louis, MO, USA). Blots were washed with TTBS (50 nM Tris, pH 7.5, 0.9% NaCl2, and 0.1% Tween-20) and exposed for 1 hour to HRP-conjugated secondary antibody: donkey anti-goat IgG 1:2500 for DJ-1 blots; goat anti-mouse IgG 1:1000 for Nrf2 (R&D Systems) and Keap1 blots; anti-rabbit IgG 1:1000 for Nrf2 blots (Santa Cruz Biotechnology), 1:2000 for phospho-p53 (Ser 15) and GAPDH blots, and 1:4000 for cleaved caspase3 blots. After washing in TTBS, antibody binding was detected with a chemiluminescent substrate (Thermo Scientific). Densitometry was analyzed with ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA), and protein content was normalized relative to GAPDH or β-actin. Experiments were repeated a minimum of three times. Results were averaged, and standard errors of the mean were calculated.
Statistical Analysis
Statistical analysis was carried out by using a two-tailed t-test, and P < 0.05 was considered to be statistically different. Data were expressed as the mean ± SEM.
Results
Knockdown of DJ-1 in HCECi Cells
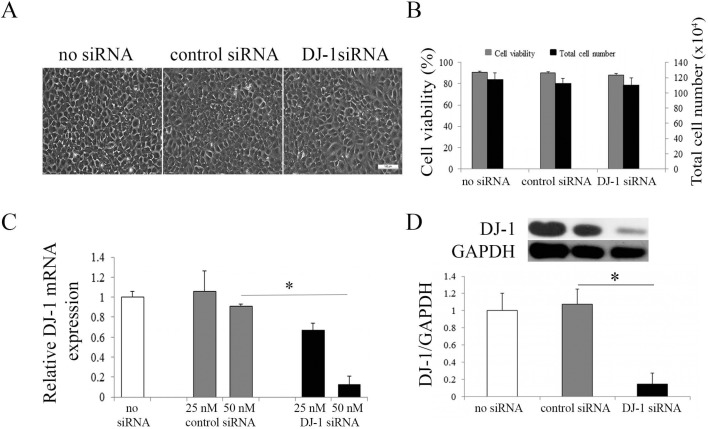
Our previous study has shown significantly decreased mRNA and protein levels of DJ-1 in FECD CECs as compared to normal CECs.5 To further investigate the role of DJ-1 in HCECi cells, we knocked down DJ-1 mRNA and protein levels by siRNA with a lipofectamine transfection method. HCECi cells were treated with DJ-1 siRNA, control siRNA, whereas non–siRNA-treated cells were used as controls. After 72 hours of transfection, siRNA did not cause cellular toxicity, as observed by normal endothelial cell morphology (Fig. 1A). No difference was detected in cell viability and total cell number among DJ-1 siRNA-treated cells and controls (Fig. 1B). Real-time RT-PCR revealed that transfection with 25 nM DJ-1 siRNA resulted in approximately 35% knockdown efficiency, while 50 nM DJ-1 siRNA resulted in approximately 87% knockdown efficiency, at mRNA levels (Fig. 1C). In accordance with mRNA levels, a significant reduction (85%) in DJ-1 protein level was observed in 50 nM DJ-1 siRNA-treated cells as compared to controls (P < 0.05, Fig. 1D).
Figure 1.
Knockdown of DJ-1 in HCECi cells. (A) Representative phase-contrast microcopy images of HCECi transfected with DJ-1 siRNA. Three days post DJ-1 transfection, HCECi cells showed the normal endothelial cell morphology seen in non–siRNA- and control siRNA-treated cells. Scale bar: 100 μm. (B) Trypan blue assay showed similar cell survival and similar total cell numbers in DJ-1 siRNA and control treatments. (C) Real-time RT-PCR analysis of DJ-1 mRNA levels in non–siRNA-treated, control siRNA-treated, and DJ-1 siRNA-treated HCECi cells with 25 nM or 50 nM siRNA transfection. Results were expressed as fold-changes and normalized to β2-microglobulin. Data showed DJ-1 mRNA levels were significantly decreased in 50 nM DJ-1 siRNA-treated HCECi cells as compared to controls. (D) Representative Western blot (upper panel) of DJ-1 protein levels in non–siRNA-treated, control siRNA-treated, and DJ-1 siRNA-treated HCECi cells with 50 nM siRNA transfection. Averaged densitometric analysis (lower panel) showed that DJ-1 protein levels were significantly reduced in DJ-1 siRNA-treated HCECi cells as compared to controls. GAPDH was used for normalization of protein loading. *P < 0.05.
Downregulation of DJ-1 Increases ROS Production and Protein Oxidative Modification
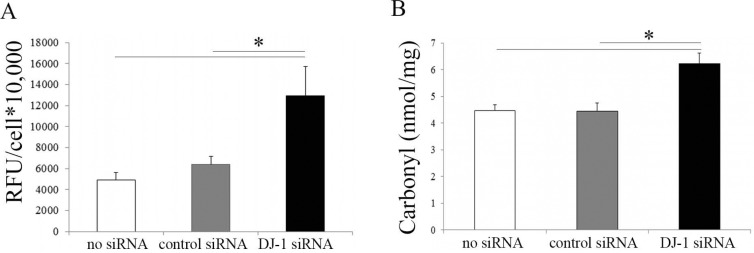
DJ-1 plays a protective role against ROS-induced apoptosis.36,37 In previous studies, we have reported that FECD corneal endothelium displays increased levels of oxidative DNA damage as compared to normal tissue samples.4,6 To test the hypothesis that DJ-1 downregulation leads to increased oxidative damage in HCECi cells, in the current study, we investigated ROS production and protein carbonyl levels in DJ-1 knockdown HCECi cells. The DCFDA intracellular ROS assay showed that DJ-1 siRNA-treated HCECi cells produced a significantly higher level of ROS than did control siRNA-treated and non–siRNA-treated cells (P < 0.05, Fig. 2A). Protein carbonyl is an irreversible oxidation product that has been widely used as a biomarker of oxidative damage to proteins.38 The ELISA results showed that carbonyl levels increased by 1.4-fold in DJ-1 siRNA-treated HCECi cells as compared to controls (P < 0.05, Fig. 2B), indicating that there was an increase in oxidation in DJ-1 downregulated HCECi cells.
Figure 2.
DJ-1 downregulation increases ROS production and oxidative damage. (A) ROS production of non–siRNA-treated, control siRNA-treated, and DJ-1 siRNA-treated HCECi cells. The DCFDA assay showed that ROS levels increased in DJ-1 siRNA-treated cells as compared to controls. (B) ELISA analysis of protein carbonyl content of DJ-1 siRNA and control treatments. Results showed increased protein carbonyls in DJ-1 siRNA-treated HCECi cells as compared to controls, indicating elevated oxidative damage in DJ-1 downregulated HCECi cells. *P < 0.05.
Downregulation of DJ-1 Causes Decreased Nuclear Translocation of Nrf2 and Nrf2-Regulated Antioxidant Gene Expression
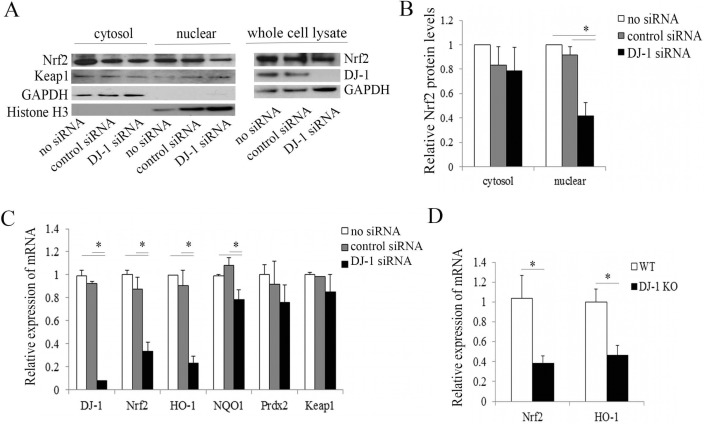
Nuclear translocation is essential for Nrf2 and subsequent antioxidant regulation. In some cell types, DJ-1 has been reported as a stabilizer of Nrf2, enabling Nrf2 to translocate from cytoplasm to the nucleus and upregulate the transcription of antioxidant and oxidative stress response genes.14 Previously, we have reported the decline in DJ-1 and decreased Nrf2 nuclear translocation in FECD.5 To investigate the effect of DJ-1 downregulation on Nrf2 translocation, we analyzed the levels of Nrf2 in the cytoplasm and nucleus in DJ-1 siRNA-treated HCECi cells by Western blot. Despite similar levels of cytoplasmic Nrf2, nuclear Nrf2 protein levels decreased by 2.2-fold in DJ-1 siRNA-treated HCECi cells as compared to non-siRNA and control siRNA groups (P < 0.05, Figs. 3A, 3B). To further examine the Nrf2–ARE pathways in response to impaired Nrf2 nuclear translocation caused by DJ-1 disruption, we quantified the mRNA expression of Nrf2 and its target genes including HO-1, NQO1, Prdx2, and Keap1, through real-time RT-PCR in DJ-1 siRNA-treated HCECi cells. The results showed that downregulation of DJ-1 led to a 2.6-fold decrease in Nrf2, with a 3.9-fold and a 1.4-fold decrease in Nrf2 target genes HO-1 and NQO1 mRNA, respectively (P < 0.05, Fig. 3B). Similar mRNA levels of Keap1 were observed in both DJ-1 siRNA-treated cells and controls (Fig. 3C). To corroborate in vitro observations in HCECi cells, we evaluated Nrf2 and its target gene expression in the corneal endothelium obtained from DJ-1 KO mice. Real-time RT-PCR analysis of mouse CECs obtained from DJ-1 KO mice revealed significant decreases in mRNA levels of Nrf2 and HO-1 (P < 0.05, Fig. 3D) as compared to WT. Interestingly, gene expression levels of Prdx2 did not show significant change among DJ-1 siRNA treatments and controls in normal human CECs (Fig. 3C).
Figure 3.
DJ-1 downregulation decreases Nrf2 nuclear translocation and its target gene expression. Nrf2 subcellular localization. (A) Representative Western blot of Nrf2 protein levels in the cytosolic and nuclear fractions of non–siRNA-treated, control siRNA-treated, and DJ-1 siRNA-treated HCECi cells. (B) Averaged densitometric analysis of Nrf2 protein levels in cytosolic and nuclear fractions from three independent experiments showed that nuclear Nrf2 protein levels decreased in DJ-1 siRNA-treated cells as compared to controls. GAPDH and histone H3 were used for normalization of cytosolic and nuclear protein loading, respectively. The mRNA levels of Nrf2 and its target genes were analyzed by real-time RT-PCR in HCECi cells (C) and in mouse CECs (D). Results were expressed as fold-changes and normalized to β2-microglobulin. *P < 0.05.
Downregulation of DJ-1 Does Not Change Protein Association Between Nrf2 and Either Cul3 or Keap1
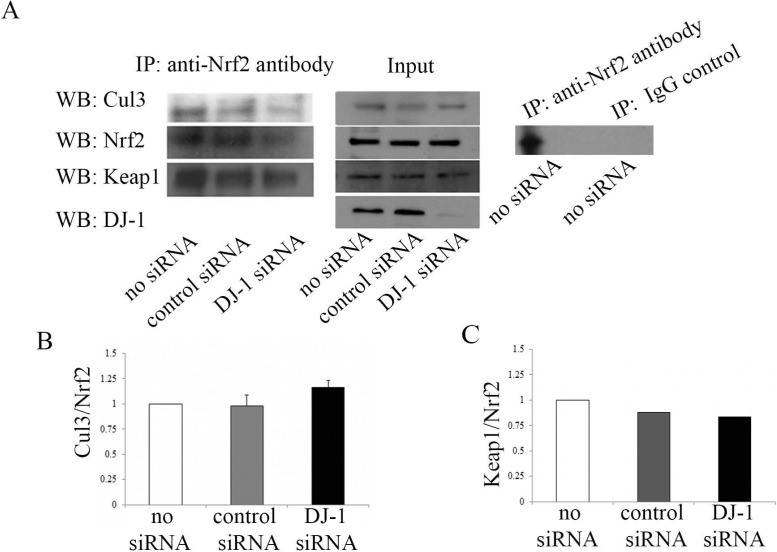
Nrf2 reportedly is retained in cytoplasm by Keap1 through active recruitment of Cul3 and subsequent proteosomal degradation. To understand the underlying mechanism for the decrease in Nrf2 and its target gene expression in DJ-1 downregulated HCECi cells, and the potential link to the Keap1/Cul3 pathway, Nrf2 protein was immunoprecipitated with anti-Nrf2 antibody from DJ-1 siRNA-treated cell lysates, followed by immunoblotting with anti-Cul3 antibody and anti-Keap1 antibody. The IP-WB analysis demonstrated that Nrf2 is associated with Cul3 and Keap1 (Fig. 4A, left panel; see full blot for each IP-WB in Supplementary Fig. S1A). Similar levels of Cul3/Nrf2 were observed in DJ-1 siRNA-treated, control siRNA-treated, and non–siRNA-treated cells (Figs. 4A, 4B). Likewise, similar levels of Keap1/Nrf2 were observed for both DJ-1 siRNA-treated and control cells (Figs. 4A, 4C). Western blot analysis of input did not detect a difference in Nrf2, Cul3, and Keap1 in DJ-1 siRNA-treated, control siRNA-treated, and non–siRNA-treated cells before IP (Fig. 4A, middle panel). In addition, IP-WB analysis of antibody immunoprecipitation specificity showed that the Nrf2 band (∼65 kDa) and IgG heavy chain band (∼50 kDa) were detected in anti-Nrf2 antibody pull-down, while only IgG heavy chain band (∼50 kDa) was detected in normal rabbit IgG (IgG control) pull-down (Fig. 4A, right panel; see full blot in Supplementary Fig. S1B, arrow: IgG heavy chain), indicating that rabbit polyclonal anti-Nrf2 antibody immunoprecipitates Nrf2 specifically.
Figure 4.
The effect of DJ-1 downregulation on Nrf2-associated Cul3 and Keap1 levels. Immunoprecipitation–WB analysis of Cul3/Nrf2 and Keap/Nrf2 levels in non–siRNA-treated, control siRNA-treated, and DJ-1 siRNA-treated HCECi cells. (A) Left panel: IP-WB analysis of cell lysates immunoprecipitated with anti-Nrf2 antibody followed by immunoblotting with anti-Nrf2 antibody, anti-Cul3 antibody, and anti-Keap1 antibody. Middle panel: WB analysis of input immunoblotted with anti-Cul3 antibody, anti-Nrf2 antibody, anti-Keap1 antibody, and anti–DJ-1 antibody. Right panel: IP-WB analysis of non–siRNA-treated cells immunoprecipitated with anti-Nrf2 antibody (left lane) or normal rabbit IgG (IgG control, right lane) followed by immunoblotting with anti-Nrf2 antibody. Averaged densitometric analyses showed that DJ-1 decrease does not change Cul3/Nrf2 (B) and Keap1/Nrf2 (C) levels.
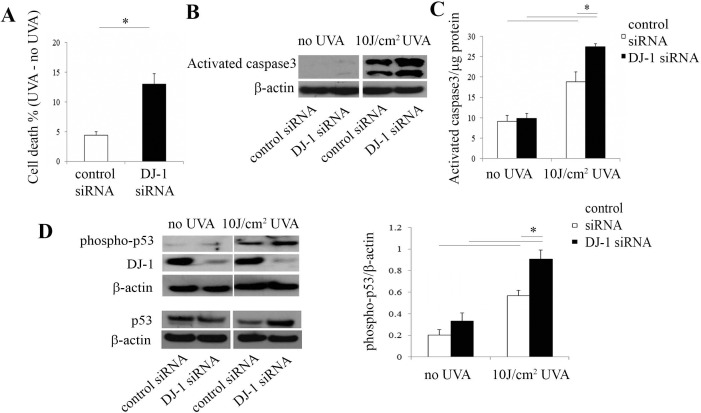
Downregulation of DJ-1 Increases Caspase3 Activation and Phospho-p53 Under UV-A Oxidative Stress
Ultraviolet A light irradiation has been used as an oxidative stressor to induce cell apoptosis39 and Nrf2-mediated antioxidant defense.40 To determine the role of DJ-1 in UV-induced oxidative stress, DJ-1 was downregulated and HCECi cells were irradiated with 10 J/cm2 UV-A light. Cell cytotoxicity assay showed that UV-A irradiation led to a 3.0-fold increase in cell death in DJ-1 siRNA-treated cells as compared to controls (P < 0.05, Fig. 5A). Figure 5B revealed that, after UV-A irradiation, higher levels of activated caspase3 were observed in DJ-1 siRNA-treated cells than in controls (P < 0.05). This finding is consistent with the rhodamine 110 fluorescence–based caspase3 assay (Fig. 5C), which showed that activated caspase3 levels increased by 2.8-fold in DJ-1 siRNA-treated cells and by 2.1-fold in control siRNA-treated cells after UV-A exposure as compared to those with no UV-A–treated cells. Moreover, after UV-A irradiation, the increase was magnified by 1.5-fold in DJ-1 siRNA-treated cells as compared to that in control siRNA-treated cells (P < 0.05, Fig. 5C). These results suggested that downregulation of DJ-1 markedly induced cell apoptosis. We have previously revealed an increase in phospho-p53 levels in Fuchs' dystrophy tissue samples4 and FECDi cells5 as compared to those in normal controls. To further investigate the correlation between phospho-p53 and DJ-1 downregulation, we compared phospho-p53 levels in DJ-1 siRNA-treated and control siRNA-treated cells under UV-A irradiation and no UV-A irradiation. Western blot with anti–phospho-p53 showed that UV-A irradiation caused an approximate 3.0-fold increase in phospho-p53 in DJ-1 siRNA-treated and control siRNA-treated cells as compared to non-UV-A–treated cells. Furthermore, under UV-A irradiation, this effect was augmented with DJ-1 siRNA-treated cells as evidenced by a 1.6-fold increase in phospho-p53 in DJ-1 knockdown cells when compared to controls (P < 0.05, Fig. 5D). These results indicated that DJ-1 downregulation accelerates phospho-53–mediated cell apoptosis.
Figure 5.
Downregulation of DJ-1 increases activated caspase3 and phospho-p53 levels by UV-A irradiation. (A) Cytotoxicity assay by measuring LDH release showed that cell death increased in DJ-1 siRNA-treated HECEi cells as compared to controls. Data were normalized with no UV-A treatment. (B) Representative Western blot of cleaved caspase3 protein levels. (C) Caspase3 activity assay by measuring fluorescence product of Z-DEVD-rhodamine 110. Data were normalized with protein concentrations. (D) Western blot analysis of phospho-p53 and total p53 (left panel), and densitometric analysis (right panel) of phospho-p53 protein levels in control siRNA-treated and DJ-1 siRNA-treated HCECi cells.
Discussion
Previous studies have demonstrated that the decline in Nrf2-regulated antioxidant defense due to diminished Nrf2 nuclear translocation contributes to the oxidant–antioxidant imbalance and resultant cellular and molecular damage seen in FECD.5,6 Along with decreased Nrf2 levels, a significant decrease in DJ-1 is present in both FECD tissue and a FECD cell line at baseline and in response to oxidative stress, as compared to normal corneal endothelium.5 In addition, oxidized and carbonylated forms of DJ-1 have been detected in FECD.5 To determine whether decline in DJ-1 has a role in impaired Nrf2-regulated antioxidant defense, we investigated the effect of DJ-1 downregulation on the oxidant–antioxidant imbalance seen in FECD. DJ-1 has been reported as a stabilizer and positive regulator of Nrf2, aiding Nrf2 in nuclear translocation and antioxidant gene activation.14,21,25 In this study, we reported that DJ-1 knockdown caused increased levels of ROS production and protein carbonyl formation in human CECs, owing to decreased Nrf2 nuclear translocation and its transcriptional target activation. In response to oxidative stress due to UV-A—a physiologic stressor relevant to ocular disorders—the decline in DJ-1 led to activation of p53-dependent apoptosis.
This study detected that, in human CECs, DJ-1 is required for expression of NQO1 and HO-1, the main target genes of Nrf2 transcriptional regulation. In support of these findings, absence of DJ-1 in human lung cells altered Nrf2 target genes including NQO1 expression.14 Similarly, knocking down DJ-1 in human liver cells attenuated NQO1 gene expression.21 These results agreed with the study on mouse primary cortical cultures, which has shown that gene expression levels of NQO1 and HO-1 decreased in DJ-1 KO mice as compared to those in DJ-1 WT mice.26 Interestingly, the Prdx2 gene, although a known Nrf2 target,41 did not result in a significant change in DJ-1 downregulated human CECs. This finding is consistent with GeneChip analysis in lung carcinoma cells, which has not detected that Prdx genes are affected by DJ-1 downregulation.21
In addition to Nrf2 target genes, we observed that Nrf2 mRNA levels were decreased in DJ-1 downregulated human CECs. This finding is supported by reports that DJ-1 deficiency results in a significant reduction of Nrf2 mRNA levels in mouse primary cortical neuronal cultures.26 Contrary to this, one study reports that DJ-1 downregulation in both human and mouse lung cells does not change the Nrf2 mRNA levels,14 while other studies report that knocking down DJ-1 in liver cancer cells does not affect Nrf2 mRNA levels.21 These findings indicate that absence of DJ-1 might affect Nrf2 transcription in a cell-specific manner. The prevailing theory on the mechanism of DJ-1's effect on Nrf2 is that DJ-1enhances Nrf2's protein stability and nuclear localization.14,21 Although Nrf2 has not been shown to be transcriptionally regulated by DJ-1, Nrf2 is known to be its own positive regulator, containing the ARE sequence in its promoter region; thus, decline in Nrf2 protein stability may lead to its own transcriptional downregulation. To investigate the role of DJ-1 in affecting Nrf2 protein levels and stability, we performed subcellular fractionation and detected that, under baseline conditions, there is indeed a decline in Nrf2 nuclear translocation in DJ-1 knockdown CECs. Since nuclear localization of Nrf2 is essential for its transcriptional activation, and we have detected a decline in ARE-dependent gene action with DJ-1 siRNA treatment, we propose that DJ-1 positively regulates Nrf2-mediated antioxidant defense in human CECs by aiding in its nuclear localization. The limitation of our study is that we did not perform ARE activity assays to compare the Nrf2–ARE transcriptional activity between normal and DJ-1 siRNA-treated cells. Since previous reports in lung epithelial cells have not detected any difference in ARE action,14 in DJ-1 knockdown at baseline, further studies are needed to investigate ARE activation in the absence of DJ-1 under oxidative stress, such as UV-A.
To investigate the mechanism of DJ-1 regulation of Nrf2, we analyzed protein levels of Cul3/Nrf2 and Keap1/Nrf2 in DJ-1 siRNA-treated CECs by co-immunoprecipitation. Previous studies have postulated that in the absence of DJ-1, there is an enhanced Keap1/Nrf2 association leading to Cul3-dependent ubiquitination and degradation of Nrf2.14 Our study did not detect a change in the protein interactions between Nrf2 and Cul3 or Keap1 in DJ-1 siRNA-treated cells, suggesting that DJ-1 regulates Nrf2 indirectly through mechanism(s) other than the Keap1–Cul3 ubiquitination–degradation pathway. This finding is consistent with the study showing that DJ-1 does influence the interaction between Nrf2 and Keap1 and does not affect Nrf2 ubiquitination.42 It is possible that DJ-1 regulates Nrf2 by other mechanisms, such as posttranslational modifications of Nrf2 and/or its regulators or interactions with other proteins. For example, CR6-interacting factor 1 (CRIF1) also suppresses Nrf2 protein stability by proteasome-mediated degradation but, unlike Keap1, CRIF1 regulates Nrf2 under both normal and oxidative stress conditions.43
The p53 phosphoprotein acts as a transcriptional factor involving an induction of apoptosis under oxidative stress.44 In this study, we stressed CECs with UV-A, which is a physiological stressor of corneal endothelium in vivo and relevant for study of oxidative stress-induced ocular disorders.45 We detected that UV-A induced phosphorylation of Ser15 on p53 and activated caspase3, as measured by Western blot and fluorescence-based assay designed to detect activated caspase3. Moreover, the UV-A–induced cell death and activation of phospho-p53 and caspase3 were augmented by DJ-1 downregulation. These findings suggest that DJ-1 serves a protective role against UV-A–induced and p53-dependent apoptosis. These results corroborate a previous study in which overexpression of DJ-1 decreases the production of proapoptotic Bcl-2–associated X protein (Bax) and inhibits caspase activation, whereas knockdown of DJ-1 increases Bax levels and accelerates caspase3 activation induced by UV-B exposure.33 In FECD, a decline in DJ-1 correlated with increase in phospho-53, indicating the importance of mechanistic studies on DJ-1–mediated regulation of p53-dependent apoptosis.
The etiology of FECD is a combination of both genetic and environmental factors. Specifically, mutation of transcription factor 4 (TCF4) has shown a strong association with FECD.46–48 Transcription factor 4 regulates a number of cell signaling pathways involved in cell differentiation and survival.49 Knockdown of TCF4 induces apoptosis in neuroblastoma cells.49 Additionally, gene array analysis has shown that TCF4, along with Nrf2, is upregulated during osteoblast differentiation under normal conditions.50 Moreover, under oxidative stress, decreased osteoblastic differentiation is able to be rescued by overexpression of TCF4,51 suggesting the involvement of TCF4 in antioxidant defense. In FECD, expansion of the TGC trinucleotide repeats within the TCF4 gene is found in a very high proportion of patients. These guanine-rich repeats may be particularly susceptible to oxidative damage.48 Given that we have found that the decline in DJ-1 levels in FECD tissue and cell line is accompanied by carbonylation and cysteine oxidation of DJ-1,5 we speculate that, with a defect in TCF4, corneal endothelial cells are especially susceptible to oxidative stress. This leads to the oxidative modification and degradation of DJ-1, which in turn causes the decrease in Nrf2-mediated antioxidant defense and results in corneal endothelial cell apoptosis.
In summary, we demonstrated that DJ-1 is essential in protecting corneal endothelial cells from ROS-induced macromolecular damage under normal and oxidative stress conditions. DJ-1 positively affects Nrf2 by regulating its nuclear translocation and target gene expression. Downregulation of DJ-1 enhances cellular susceptibility to UV-A–induced activation of p53-mediated cell apoptosis. Based on the positive effect of DJ-1 on Nrf2 seen in vitro, it is likely that deficiency in DJ-1 is an important cause of the decreased Nrf2-mediated antioxidant defense in FECD. Since DJ-1 overexpression enhances nuclear translocation of Nrf2,42 targeting the Nrf2–DJ-1 axis provides a potential therapeutic approach for enhancing Nrf2 levels in oxidative stress-induced corneal endothelium disorders.
Acknowledgments
Supported by National Institutes of Health/National Eye Institute Grant R01 EY20581 and a Research to Prevent Blindness Award (UVJ).
Disclosure: C. Liu, None; Y. Chen, None; I.E. Kochevar, None; U.V. Jurkunas, P
References
- 1. Wilson SE, Bourne WM. Fuchs' dystrophy. Cornea. 1988; 7: 2–18 [PubMed] [Google Scholar]
- 2. Dawson DG, Geroski DH, Edelhauser HF. Corneal endothelium: structure and function in health and disease. In: Brightbill FS, McDonnell PJ, McGhee CNJ, Farjo AA, Serdarevic O, eds Corneal Surgery: Theory, Technique and Tissue. Maryland Heights, MO: Mosby Elsevier; 2009: 57–70 [Google Scholar]
- 3. Buddi R, Lin B, Atilano SR, Zorapapel NC, Kenney MC, Brown DJ. Evidence of oxidative stress in human corneal diseases. J Histochem Cytochem. 2002; 50: 341–351 [DOI] [PubMed] [Google Scholar]
- 4. Azizi B, Ziaei A, Fuchsluger T, Schmedt T, Chen Y, Jurkunas UV. p53-regulated increase in oxidative-stress—induced apoptosis in Fuchs endothelial corneal dystrophy: a native tissue model. Invest Ophthalmol Vis Sci. 2011; 52: 9291–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bitar MS, Liu C, Ziaei A, Chen Y, Schmedt T, Jurkunas UV. Decline in DJ-1 and decreased nuclear translocation of Nrf2 in Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2012; 53: 5806–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jurkunas UV, Bitar MS, Funaki T, Azizi B. Evidence of oxidative stress in the pathogenesis of Fuchs endothelial corneal dystrophy. Am J Pathol. 2010; 177: 2278–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Z, Handa JT, Green WR, Stark WJ, Weinberg RS, Jun AS. Advanced glycation end products and receptors in Fuchs' dystrophy corneas undergoing Descemet's stripping with endothelial keratoplasty. Ophthalmology. 2007; 114: 1453–1460 [DOI] [PubMed] [Google Scholar]
- 8. Ziaei A, Schmedt T, Chen Y, Jurkunas UV. Sulforaphane decreases endothelial cell apoptosis in Fuchs endothelial corneal dystrophy: a novel treatment. Invest Ophthalmol Vis Sci. 2013; 54: 6724–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottsch JD, Bowers AL, Margulies EH, et al. Serial analysis of gene expression in the corneal endothelium of Fuchs' dystrophy. Invest Ophthalmol Vis Sci. 2003; 44: 594–599 [DOI] [PubMed] [Google Scholar]
- 10. Jurkunas UV, Rawe I, Bitar MS, et al. Decreased expression of peroxiredoxins in Fuchs' endothelial dystrophy. Invest Ophthalmol Vis Sci. 2008; 49: 2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994; 91: 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Z, Wei Y, Gong J, et al. NRF2 plays a protective role in diabetic retinopathy in mice. Diabetologia. 2014; 57: 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osburn WO, Wakabayashi N, Misra V, et al. Nrf2 regulates an adaptive response protecting against oxidative damage following diquat-mediated formation of superoxide anion. Arch Biochem Biophys. 2006; 454: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malhotra D, Thimmulappa R, Navas-Acien A, et al. Decline in NRF2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, DJ-1. Am J Respir Crit Care Med. 2008; 178: 592–604 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Zhang J, Dinh TN, Kappeler K, Tsaprailis G, Chen QM. La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol Cell Proteomics. 2012; 11:M111.015032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Apopa PL, He X, Ma Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol. 2008; 22: 63–76 [DOI] [PubMed] [Google Scholar]
- 17. Cheng X, Ku CH, Siow RC. Regulation of the Nrf2 antioxidant pathway by microRNAs: new players in micromanaging redox homeostasis. Free Radic Biol Med. 2013; 64: 4–11 [DOI] [PubMed] [Google Scholar]
- 18. Bonifati V, Rizzu P, Squitieri F, et al. DJ-1 (PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci. 2003; 24: 159–160 [DOI] [PubMed] [Google Scholar]
- 19. Bonifati V, Rizzu P, van Baren MJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003; 299: 256–259 [DOI] [PubMed] [Google Scholar]
- 20. Nagakubo D, Taira T, Kitaura H, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997; 231: 509–513 [DOI] [PubMed] [Google Scholar]
- 21. Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006; 103: 15091–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhong N, Kim CY, Rizzu P, et al. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J Biol Chem. 2006; 281: 20940–20948 [DOI] [PubMed] [Google Scholar]
- 23. Andres-Mateos E, Perier C, Zhang L, et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A. 2007; 104: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hwang S, Song S, Hong YK, et al. Drosophila DJ-1 decreases neural sensitivity to stress by negatively regulating Daxx-like protein through dFOXO. PLoS Genet. 2013; 9: e1003412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng X, Chapple SJ, Patel B, et al. Gestational diabetes mellitus impairs Nrf2-mediated adaptive antioxidant defenses and redox signaling in fetal endothelial cells in utero. Diabetes. 2013; 62: 4088–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gan L, Johnson DA, Johnson JA. Keap1-Nrf2 activation in the presence and absence of DJ-1. Eur J Neurosci. 2010; 31: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pourzand C, Tyrrell RM. Apoptosis, the role of oxidative stress and the example of solar UV radiation. Photochem Photobiol. 1999; 70: 380–390 [PubMed] [Google Scholar]
- 28. de Gruijl FR. Skin cancer and solar UV radiation. Eur J Cancer. 1999; 35: 2003–2009 [DOI] [PubMed] [Google Scholar]
- 29. Vile GF, Tyrrell RM. UVA radiation-induced oxidative damage to lipids and proteins in vitro and in human skin fibroblasts is dependent on iron and singlet oxygen. Free Radic Biol Med. 1995; 18: 721–730 [DOI] [PubMed] [Google Scholar]
- 30. von Thaler AK, Kamenisch Y, Berneburg M. The role of ultraviolet radiation in melanomagenesis. Exp Dermatol. 2010; 19: 81–88 [DOI] [PubMed] [Google Scholar]
- 31. Zhuang S, Kochevar IE. Singlet oxygen-induced activation of Akt/protein kinase B is independent of growth factor receptors. Photochem Photobiol. 2003; 78: 361–371 [DOI] [PubMed] [Google Scholar]
- 32. Kappes UP, Luo D, Potter M, Schulmeister K, Runger TM. Short- and long-wave UV light (UVB and UVA) induce similar mutations in human skin cells. J Invest Dermatol. 2006; 126: 667–675 [DOI] [PubMed] [Google Scholar]
- 33. Fan J, Ren H, Jia N, et al. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J Biol Chem. 2008; 283: 4022–4030 [DOI] [PubMed] [Google Scholar]
- 34. Griffith M, Osborne R, Munger R, et al. Functional human corneal equivalents constructed from cell lines. Science. 1999; 286: 2169–2172 [DOI] [PubMed] [Google Scholar]
- 35. Chen CH, Rama P, Chen AH, et al. Efficacy of media enriched with nonlactate-generating substrate for organ preservation: in vitro and clinical studies using the cornea model. Transplantation. 1999; 67: 800–808 [DOI] [PubMed] [Google Scholar]
- 36. Giaime E, Yamaguchi H, Gautier CA, Kitada T, Shen J. Loss of DJ-1 does not affect mitochondrial respiration but increases ROS production and mitochondrial permeability transition pore opening. PLoS One. 2012; 7: e40501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Billia F, Hauck L, Grothe D, et al. Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc Natl Acad Sci U S A. 2013; 110: 6085–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi J, Sullards MC, Olzmann JA, et al. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem. 2006; 281: 10816–10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valencia A, Kochevar IE. Ultraviolet A induces apoptosis via reactive oxygen species in a model for Smith-Lemli-Opitz syndrome. Free Radic Biol Med. 2006; 40: 641–650 [DOI] [PubMed] [Google Scholar]
- 40. Gao X, Talalay P. Induction of phase 2 genes by sulforaphane protects retinal pigment epithelial cells against photooxidative damage. Proc Natl Acad Sci U S A. 2004; 101: 10446–10451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marinho HS, Real C, Cyrne L, Soares H, Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014; 2: 535–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Im JY, Lee KW, Woo JM, Junn E, Mouradian MM. DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum Mol Genet. 2012; 21: 3013–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kang HJ, Hong YB, Kim HJ, Bae I. CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. J Biol Chem. 2010; 285: 21258–21268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009; 137: 413–431 [DOI] [PubMed] [Google Scholar]
- 45. Mallet JD, Rochette PJ. Wavelength-dependent ultraviolet induction of cyclobutane pyrimidine dimers in the human cornea. Photochem Photobiol Sci. 2013; 12: 1310–1318 [DOI] [PubMed] [Google Scholar]
- 46. Baratz KH, Tosakulwong N, Ryu E, et al. E2-2 protein and Fuchs's corneal dystrophy. N Engl J Med. 2010; 363: 1016–1024 [DOI] [PubMed] [Google Scholar]
- 47. Li YJ, Minear MA, Rimmler J, et al. Replication of TCF4 through association and linkage studies in late-onset Fuchs endothelial corneal dystrophy. PLoS One. 2011; 6: e18044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wieben ED, Aleff RA, Tosakulwong N, et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One. 2012; 7: e49083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Forrest MP, Waite AJ, Martin-Rendon E, Blake DJ. Knockdown of human TCF4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013; 8: e73169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beck GR Jr, Zerler B, Moran E. Gene array analysis of osteoblast differentiation. Cell Growth Differ. 2001; 12: 61–83 [PubMed] [Google Scholar]
- 51. Yang Y, Su Y, Wang D, et al. Tanshinol attenuates the deleterious effects of oxidative stress on osteoblastic differentiation via Wnt/FoxO3a signaling. Oxid Med Cell Longev. 2013; 2013: 351895 [DOI] [PMC free article] [PubMed] [Google Scholar]