Abstract
Purpose.
To determine if specific mitochondrial haplogroups associate with nonproliferative diabetic retinopathy (NPDR) and proliferative diabetic retinopathy (PDR).
Methods.
Deidentified medical records for Caucasian patients with diabetic retinopathy (DR; 153 NPDR and 138 PDR) were obtained from BioVU, Vanderbilt University's electronic, deidentified DNA databank. An independent cohort of Caucasian patients with DR (44 NPDR and 57 PDR) from the Vanderbilt Eye Institute (VEI) was used for validation. We tested for an association between mitochondrial haplogroups and PDR among patients with DR.
Results.
In the BioVU cohort, PDR frequency among Caucasian DR patients differed significantly by mitochondrial haplogroup (P = 0.027). Replication in the VEI cohort confirmed this association (P = 0.0064). In the combined cohort, patients from the common haplogroup H were more likely to have PDR (odds ratio [OR] = 2.0 [95% confidence interval (CI) = 1.3–3.0], P = 0.0012), while patients from haplogroup Uk were less likely to have PDR (OR = 0.5 [95% CI = 0.3–0.8], P = 0.0049). In logistic regression analyses, the addition of diabetes duration, hemoglobin A1c (HgbA1c) levels, and hypertension had no effect on the associations of haplogroups H and Uk with PDR.
Conclusions.
In this study, DR patients from mitochondrial haplogroup H were more likely to have PDR, while DR patients from haplogroup Uk were less likely to have PDR. The association was independent of the major clinical variables affecting PDR. The mitochondrial haplogroups were as strong a risk factor for PDR as were elevated HgbA1c levels.
Keywords: diabetes, diabetic retinopathy, proliferative diabetic retinopathy, genetics, mitochondrial haplogroup, mitochondrial genetics, mitochondrial DNA
Using BioVU, Vanderbilt's deidentified DNA databank, and an independent clinical cohort, the authors demonstrate that among patients with diabetic retinopathy, proliferative retinopathy is positively associated with European mitochondrial haplogroup H and negatively associated with haplogroup Uk.
Introduction
Diabetic retinopathy (DR) is the leading cause of new cases of severe visual impairment in US working-age adults.1 Approximately 14% of US adults have diabetes, and the prevalence is projected to increase to between 21% and 33% by 2050.2 Among US diabetics older than 40 years, approximately one-third have DR, and approximately one-third of these patients have vision-threatening DR.3 Strategies to identify individuals at risk for vision-threatening DR are needed in order to provide early intervention.
Diabetic retinopathy is a disorder of the retinal microvasculature, precipitated by exposure to chronically elevated blood glucose levels.4 Classification of DR is based upon vascular changes detected by fundoscopic examination. Retinal findings associated with nonproliferative diabetic retinopathy (NPDR) include microaneurysms, dot-blot hemorrhages, cotton-wool spots, venous beading, and intraretinal microvascular abnormalities. The more severe proliferative diabetic retinopathy (PDR) is characterized by neovascularization of the retina, optic disc, or iris. These new blood vessels can bleed or contract, leading to vision loss from vitreous hemorrhage or retinal traction, respectively. Diabetic macular edema (DME), which can occur in both NPDR and PDR, results from increased retinal vascular permeability and is another major cause of vision loss in diabetes. Diabetic retinopathy can also cause vision loss secondary to capillary nonperfusion in the macula or neovascular glaucoma associated with PDR.5
Mitochondrial dysfunction has been implicated in the pathogenesis of diabetes.6 Since mitochondrial DNA (mtDNA) is inherited maternally, the common mtDNA variants occur in patterns that define haplogroups generally specific to continental populations (i.e., separate sets of European, Asian, and African haplogroups). Subtle variations in mitochondrial function may be accentuated under stress conditions7 and may be the cause for the association of mitochondrial haplogroups with a wide range of disease phenotypes, including diabetes8,9 and DR.10,11 We studied a group of patients with DR from BioVU, Vanderbilt University's deidentified DNA databank, in order to discern whether common mitochondrial haplogroups are associated with severity of DR. We then validated the association in an independent cohort of patients recruited from the Vanderbilt Eye Institute (VEI).
Materials and Methods
Ethics Statement
The clinical case-control study was approved by the Vanderbilt University Human Research Protection Program. Research adhered to the tenets of the Declaration of Helsinki and was conducted in accordance with Health Insurance Portability and Accountability Act regulations. For the VEI cohort, written informed consent was obtained from all participants before study enrollment. BioVU uses an “opt-out” consent model, based on an opinion from the federal Office of Human Research Protection that discarded blood samples can be used for biomedical research without prospective consenting of each individual if the clinical data are deidentified. All BioVU projects are reviewed by the Vanderbilt University Human Research Protection Program and are classified as nonhuman subjects' research before study initiation.
BioVU Patients
BioVU is the Vanderbilt University Medical Center's repository of DNA extracted from discarded blood collected during routine clinical testing and linked to a deidentified copy of the electronic medical record (the Synthetic Derivative, SD).12,13 For deidentification purposes, all dates in each SD record are consistently shifted by a random amount within ±6 months. We identified all Caucasian individuals in the SD with International Statistical Classification of Diseases (ICD)-9 codes for diabetes (250.0, 250.00–250.03) and DR (362.0–362.07) with available genome-wide variant data. To ensure independence from the VEI cohort, the medical record numbers of the VEI cohort (described below) were supplied to the BioVU administrators, and VEI cohort individuals were excluded from the BioVU cohort.
To identify BioVU patients with DR, SD charts were initially screened by using ICD-9 and Current Procedural Terminology (CPT) codes. Synthetic Derivative charts of patients with diabetes mellitus (DM) and DR codes were manually reviewed under the supervision of a fellowship-trained retina specialist (MAB). Diabetes diagnosis was confirmed by presence of a DM ICD-9 code and an internal medicine or endocrinology visit. Diabetic retinopathy diagnosis was confirmed by presence of a DR code and an ophthalmology CPT code. To determine severity of DR (NPDR or PDR), we searched for DR ICD-9 codes occurring on the same day as an ophthalmology CPT code. If NPDR or PDR was coded on the same day as an ophthalmology visit code, the patient's DR severity was determined from that visit. In these cases, ICD-9 code 362.02 was classified as PDR, and ICD-9 codes 362.04, 362.05, and 362.06 (without the presence of 362.02) were classified as NPDR. For ICD-9 code 362.01 (“diabetic retinopathy not otherwise specified”) or for patients without NPDR or PDR codes on the same day as an ophthalmology clinic visit, clinic notes and letters from the SD were reviewed in detail to determine the severity of DR. If we were unable to classify a patient as having NPDR or PDR with these methods, the patient was excluded from the study.
To determine which patients in the BioVU cohort had DME, their SD charts were screened for ICD-9 codes for diabetic macular edema (362.07), cystoid macular edema (362.53), and retinal edema (362.83). The SD charts for all cohort patients with any of these codes were manually reviewed to confirm a diagnosis of DME.
Diabetes type (T1DM or T2DM) was determined by using endocrinology notes when available. If endocrinology notes were not available, diabetes type was determined by reviewing problem lists, reviewing internal medicine notes, or inferring from the patient's medication regimen. Hemoglobin A1c (HgbA1c) for each patient was reported as the median of all values in the SD. Age for each patient was reported as the age at most recent patient encounter or at death. Duration of diabetes was determined by calculating the difference between the age at diagnosis and the age at the most recent patient encounter or at death. A diagnosis of hypertension was determined by the presence of the ICD-9 code 401 for “essential hypertension.” Smoking status (“ever-smoker” or “never-smoker”) was determined by manual chart review. All assessments of the SD were done blinded to the genetic data.
VEI Cohort
Individuals older than 18 years with T1DM or T2DM were recruited from the Retina Division of the VEI. All patients were required to have a diagnosis of DM made by their primary care provider or endocrinologist and to have been prescribed at least one diabetes medication. All patients were diagnosed with DR on the basis of a comprehensive dilated ophthalmologic examination by a fellowship-trained retina specialist, and each patient was classified as having either NPDR or PDR. Nonproliferative diabetic retinopathy was diagnosed from the presence of blot hemorrhages, microaneurysms, cotton-wool spots, or intraretinal microvascular abnormalities, and the absence of signs or history of retinal neovascularization. Proliferative diabetic retinopathy was diagnosed from presence of iris or retinal neovascularization, or evidence of treatment for PDR with laser photocoagulation. Diabetic macular edema was diagnosed from macular edema visible on slit lamp biomicroscopy or history of treatment for DME. Retinopathy status was documented by high-resolution color fundus photography. Medical history was obtained from the electronic medical record, including HgbA1c measurements and hypertension status. Recruitment exclusion criteria for the VEI cohort included the presence of nondiabetic retinopathy (e.g., age-related macular degeneration [AMD], myopic retinopathy, or inherited retinopathy), glaucoma, active uveitis or ocular infection, and ocular surgery within 60 days before enrollment.
At the time of study enrollment, participants in the VEI cohort underwent venipuncture to provide a blood sample and responded to a standardized set of questions regarding disease history and lifetime environmental exposures. From these questions, smoking status was classified as “ever-smoker” or “never-smoker.” Blood was drawn from study participants by using a 21- or 23-gauge butterfly needle. For each participant, approximately 8 mL blood was immediately transferred to a 10 mL K2 EDTA blood collection tube. These samples were delivered to the Vanderbilt Technologies for Advanced Genetics (VANTAGE) Center for DNA isolation and storage.
Genotyping and Mitochondrial Haplogroup Determination
BioVU cohort subjects had been previously genotyped on the Illumina 660W, Illumina 1M, or the Illumina Infinium HumanExome BeadChip (Illumina, San Diego, CA, USA), and the genotyping data had been deposited in BioVU for use in additional research projects such as this one. The Illumina 660W and 1M genotyping chips contain 138 single-nucleotide polymorphisms (SNPs) from the mitochondrial genome, while the Exome chip contains 219 mtDNA SNPs (see the Supplementary Material for the SNP list). The mtDNA SNPs were used to generate a variant list for each BioVU study subject, containing the list of successfully genotyped SNPs for that subject and all variations from the standard mtDNA reference sequence, the rCRS.14 This variant list was used to determine the mitochondrial haplogroup of each BioVU subject, using the HaploGrep software.15,16
For the VEI cohort, a Sequenom pool of 22 mtDNA SNPs was designed to identify the standard European mitochondrial haplogroups.16 These SNPs are listed in the Supplementary Material. DNA samples from the VEI cohort were genotyped by using the Sequenom MassArray Analyzer (Sequenom, San Diego, CA, USA) in the VANTAGE Center. As with the BioVU cohort, a variant list for each subject was then generated and HaploGrep was used to identify the mitochondrial haplogroup of each subject.
Statistics
Tests of variable frequency of PDR or DME with mitochondrial haplogroups were carried out by using χ2 tests on 2 × 3 contingency tables. Odds ratios for an individual haplogroup were calculated from 2 × 2 contingency tables, comparing “haplogroup H” versus “not haplogroup H,” for example. P values in these tests were calculated by χ2 tests with Yates continuity correction, unless otherwise noted in the Results section. Comparisons of continuous variables such as duration of diabetes were carried out with t-tests. Error bars for frequency p were calculated from the sampling error equation 2 × SQRT(p(1 − p)/N), where N is the sample size.
Results
A total of 583 Caucasian individuals from BioVU met the ICD-9 code criteria and had genetic data available. The SDs for these patients were manually reviewed in order to classify each individual as having NPDR or PDR. Approximately 50% of the SD records contained sufficient information to ensure this classification, resulting in 291 patients for analysis (153 NPDR and 138 PDR). Demographic data for the BioVU cohort are presented in Table 1. Sex and smoking status did not differ between NPDR and PDR in this cohort. Frequency of T1DM and median HgbA1c levels were significantly higher in the PDR group. Mean age was significantly lower in the PDR group than in the NPDR group, mainly owing to the greater percentage of T1DM patients in the PDR group. When the BioVU cohort was separated by diabetes type, age was not significantly different between PDR and NPDR in the T1DM patients, but the PDR patients were slightly younger among patients with T2DM. Duration of diabetes was the most significantly different variable between NPDR and PDR patients, with PDR patients having a longer mean duration of diabetes. Demographics for the VEI cohort are shown in Table 2. The VEI cohort had similar demographic data to the BioVU cohort, though the statistics were less significant, likely owing to the smaller sample size.
Table 1.
BioVU Cohort Demographics
|
N |
NPDR |
PDR |
P
Value |
|
| N | 153 | 138 | ||
| Males, % | 291 | 47.1 | 57.2 | 0.10 |
| Ever-smoked, % | 288 | 47.7 | 43.8 | 0.55 |
| T1DM, % | 289 | 17.6 | 42.6 | 4.8E-6 |
| HgbA1c, %* | 273 | 7.4 (6.8–8.2) | 8.1 (7.1–8.9) | 0.0016 |
| Age, y* | 291 | 71 (61–79) | 61 (52–71) | 7E-5 |
| Age T1DM, y* | 85 | 48 (33–59) | 52 (43–58) | 0.55 |
| Age T2DM, y* | 204 | 74 (65–79) | 68 (63–77) | 0.026 |
| Diabetes duration, y* | 232 | 19 (13–26) | 29 (20–39) | 1.7E-7 |
| Diabetes duration T1DM, y* | 75 | 30 (21–34) | 37 (29–43) | 0.094 |
| Diabetes duration T2DM, y* | 157 | 18 (12–24) | 23 (15–30) | 0.0017 |
values calculated by two-tailed Fisher's exact test for dichotomous variables and by two-tailed t-test for continuous variables. IQR, interquartile range.
Median (IQR).
Table 2.
Vanderbilt Eye Institute Cohort Demographics
|
N |
NPDR |
PDR |
P
Value |
|
| N | 44 | 57 | ||
| Males, % | 101 | 59.7 | 70.5 | 0.3 |
| Ever-smoked, % | 101 | 47.7 | 28.1 | 0.06 |
| T1DM, % | 101 | 22.7 | 43.9 | 0.035 |
| HgbA1c, %* | 94 | 8.0 (7.4–9.3) | 8.1 (7.1–8.9) | 0.81 |
| Age, y* | 101 | 62 (46–69) | 52 (45–60) | 0.0092 |
| Age T1DM, y* | 35 | 45 (37–59) | 47 (38–53) | 0.82 |
| Age T2DM, y* | 66 | 63 (57–70) | 57 (50–62) | 0.038 |
| Diabetes duration, y* | 98 | 18 (13–28) | 25 (15–33) | 0.060 |
| Diabetes duration T1DM, y* | 35 | 30 (21–32) | 29 (21–35) | 0.78 |
| Diabetes duration T2DM, y* | 63 | 16 (12–20) | 20 (14–30) | 0.12 |
values calculated by two-tailed Fisher's exact test for dichotomous variables and by two-tailed t-test for continuous variables.
Median (IQR).
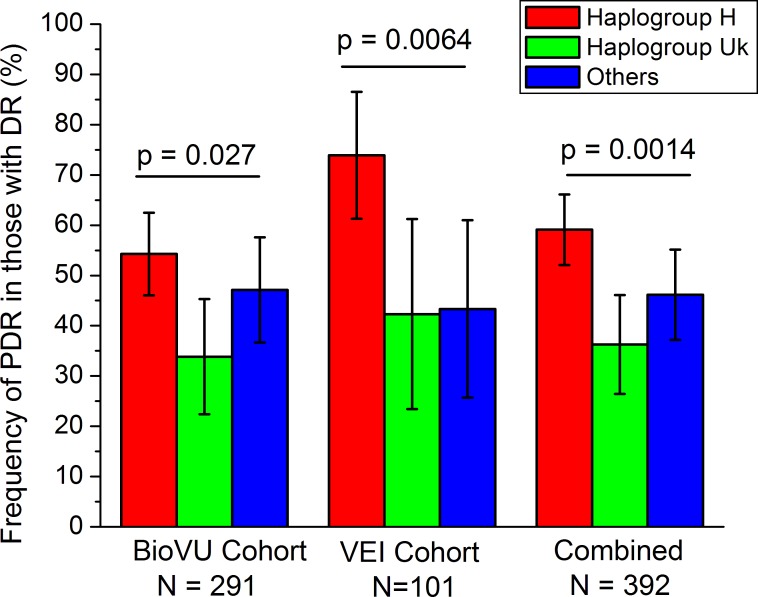
The mitochondrial haplogroup for each subject in the BioVU cohort was determined as described in Materials and Methods. Considering the sample size, analysis of mitochondrial haplogroups was limited to H and Uk (expected to have ∼45% and ∼25% frequency, respectively, in Caucasians). All rarer haplogroups (J, T, I, W, X) were classified as “other.” The proportion of PDR patients among those with DR was determined in these three mitochondrial haplogroups (H, Uk, and “other”; Fig. 1). The proportion of PDR patients was significantly different across these haplogroups (P = 0.027, χ2 test).
Figure 1.
Frequency of PDR within Caucasian patients with DR, split by major European mitochondrial haplogroups. Both the BioVU and the VEI cohorts have statistically significant differences in PDR frequency by mitochondrial haplogroup.
The VEI cohort (Table 2) was used to replicate this association. Mitochondrial haplogroups for the 101 Caucasians in the VEI cohort were determined as described in Materials and Methods. The proportion of PDR patients was significantly different across the tested mitochondrial haplogroups (P = 0.0064, χ2 test) with a similar pattern as that seen in the BioVU cohort (Fig. 1).
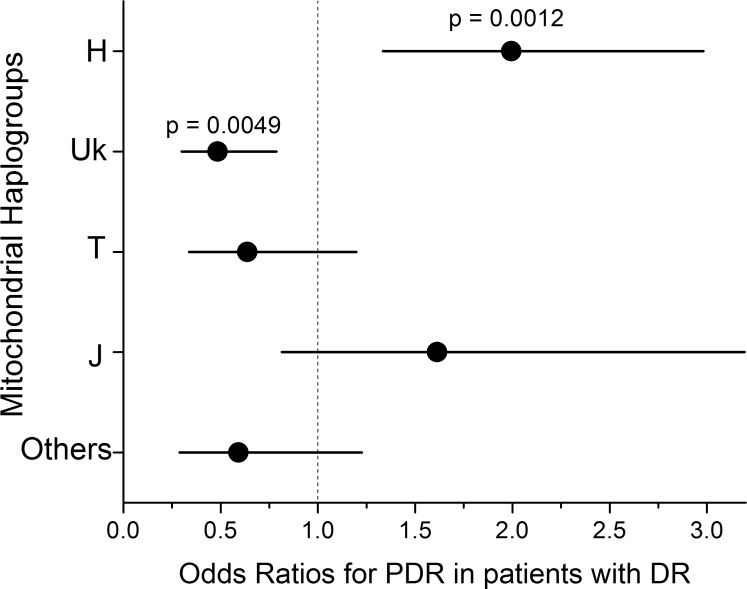
For the combined BioVU and VEI cohorts (n = 392), the association of PDR with European haplogroups was highly significant (P = 0.0014, Fig. 1). In the combined cohort (Fig. 2), patients from haplogroup H were significantly more likely to have PDR (odds ratio [OR] [95% confidence interval (CI)] = 2.0 [1.3–3.0], P = 0.0012 by two-tailed Fisher's exact test), and patients from haplogroup Uk were significantly less likely to have PDR (OR [95% CI] = 0.49 [0.30–0.79], P = 0.0049). The rarer haplogroups T and J (each ∼10% of the population) showed no significant association with PDR.
Figure 2.
Odds ratios and 95% confidence intervals for PDR in patients with DR, measured in the combined BioVU and VEI cohorts. The two most common European mitochondrial haplogroups have significant odds ratios for PDR, but haplogroups T, J, and others (all rarer haplogroups) are not significant.
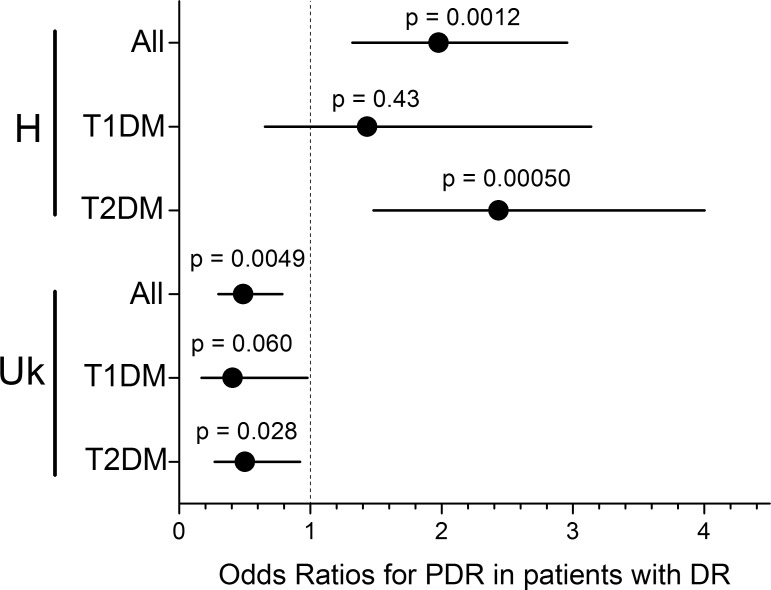
Study subjects were separated by type of diabetes to determine if the PDR associations with mitochondrial haplogroups were present in both T1DM and T2DM. Two subjects were excluded from the BioVU cohort because diabetes type could not be determined from the SD. The H haplogroup association with PDR was highly significant in T2DM subjects (OR [95% CI] = 2.4 [1.5–4.0], P = 0.00050) but was not significant in T1DM subjects (OR [95% CI] = 1.4 [0.7–3.1], P = 0.43) (Fig. 3). The protective Uk association was at a similar strength in both T1DM and T2DM (OR [95% CI] = 0.41 [0.17–0.98] and OR [95% CI] = 0.50 [0.27–0.92], respectively). The Uk association within the T1DM group did not quite meet the 0.05 significance threshold (P = 0.060 by two-tailed Fisher's exact test), while the larger T2DM group was significant (P = 0.028). Since the odds ratio for the Uk association is farther from unity in the T1DM group compared to the full group or the T2DM group, the lack of significance in the T1DM group is likely due to a smaller sample size.
Figure 3.
Odds ratios for PDR in DR patients broken down by diabetes type. For mitochondrial haplogroup H, the susceptibility to PDR is strongest and only significant in the T2DM patients, not the T1DM patients. Both the odds ratio and the P value improve in the T2DM subgroup compared to the combined group, indicating that this association is likely driven by the T2DM patients. In contrast, the protective effect in the Uk haplogroup is similar in both T1DM and T2DM patients, indicating that the same protective effect is occurring in both patient groups.
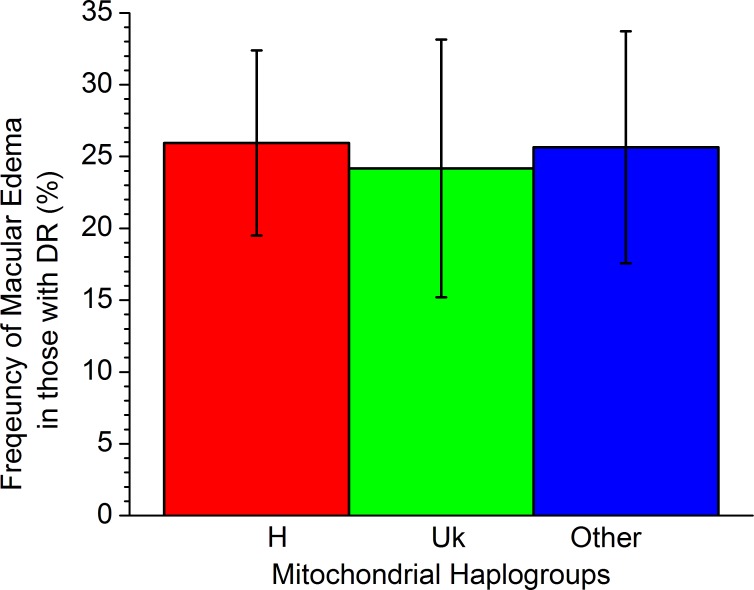
Since DME is an additional form of severe DR that can result in significant vision loss, we examined both the BioVU and VEI cohorts for an association of the mitochondrial haplogroups with DME. The percentage of DR patients with DME (Fig. 4) was almost identical among haplogroups H, Uk, and “other” (P = 0.95), indicating that there is no association of these mitochondrial haplogroups with DME.
Figure 4.
Frequency of macular edema separated by major European mitochondrial haplogroups in the combined cohort. No significant difference by haplogroup occurs for macular edema (P = 0.95).
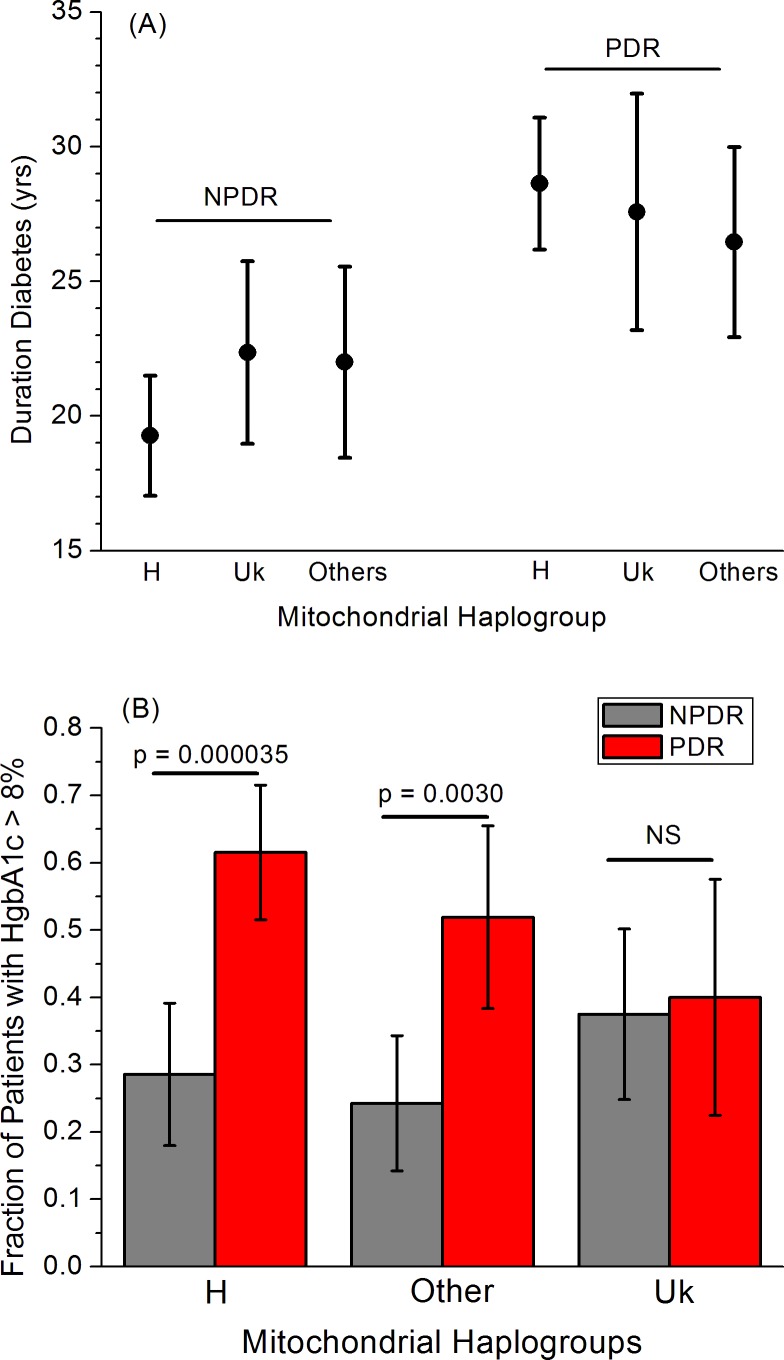
The major clinical variables that may influence progression from NPDR to PDR are duration of diabetes and HgbA1c level (as a measure of diabetes control).1 We assessed these values from both the BioVU and VEI cohorts (see Materials and Methods) and compared them among the H, Uk, and “other” haplogroups for both NPDR and PDR patients (Fig. 5). As expected,17,18 duration of diabetes showed a clear difference between the NPDR and PDR patients, with PDR patients having a longer mean diabetes duration. There was no significant difference among the different mitochondrial haplogroups in duration of diabetes for either NPDR or PDR patients (Fig. 5A). Complications of diabetes, including retinopathy, increased for patients with HgbA1c > 8%.19 The proportion of patients with HgbA1c > 8% was strongly and significantly higher in PDR than NPDR patients for haplogroup H (29% ± 10% in NPDR versus 62% ± 7% in PDR, P = 0.000035) and the “other” group (24% ± 10% in NPDR versus 52% ± 14% in PDR, P = 0.0030), but in haplogroup Uk, this proportion was essentially identical (38% ± 13% in NPDR versus 40% ± 18% in PDR, P = 0.90) (Fig. 5B).
Figure 5.
Major clinical variables for PDR separated by mitochondrial haplogroups. (A) Duration of diabetes had no significant variation by haplogroup for NPDR patients (P = 0.10 for H versus not-H, P = 0.33 for Uk versus not-Uk) or for PDR patients (P = 0.34 for H versus not-H, P = 0.90 for Uk versus not-Uk). (B) The proportion of patients with HgbA1c levels > 8% was significantly elevated in PDR compared to NPDR patients in both haplogroup H and the other minor haplogroups, but was nearly identical in the Uk haplogroup (P = 0.5).
To determine whether the mitochondrial haplogroup associations with PDR were independent of the major clinical variables, we carried out multivariate logistic regressions on the combined cohort, including mitochondrial haplogroup (either H or Uk), HgbA1c > 8% (yes/no), and duration of diabetes as variables. Hypertension was also included in the regression since it has been shown in some studies to affect DR severity.20 For haplogroup H (Table 3), the association with PDR in the multivariate regression (OR = 2.1 [95% CI = 1.3–3.4]) was nearly identical to that in the univariate association test (OR = 2.0 [95% CI = 1.3–3.0], Fig. 2). For haplogroup Uk (Table 4), the protective association with PDR strengthened slightly in the multivariate regression (OR = 0.41 [95% CI = 0.23–0.73]) compared to the univariate test (OR = 0.49 [95% CI = 0.30–0.79]). In both cases, the mitochondrial haplogroup association with PDR was shown to be independent of diabetes duration, HbgA1c level, and hypertension. The most significant covariate for PDR in this study was diabetes duration (Tables 3, 4). The mitochondrial haplogroup associations with PDR were of similar effect size and significance as the HgbA1c level.
Table 3.
Haplogroup H Multivariate Logistic Regression Analysis
|
Risk Factors |
Odds Ratio |
95% CI |
P
Value |
| Haplogroup H, yes/no | 2.1 | 1.3–3.4 | 0.0027 |
| Diabetes duration, y | 1.054 | 1.032–1.078 | 2.5e-06 |
| Median HgbA1c > 8%, yes/no | 2.0 | 1.2–3.3 | 0.0046 |
| Hypertension, yes/no | 0.8 | 0.4–1.8 | 0.67 |
Table 4.
Haplogroup Uk Multivariate Logistic Regression Analysis
|
Risk Factors |
Odds Ratio |
95% CI |
P
Value |
| Haplogroup Uk, yes/no | 0.41 | 0.23–0.73 | 0.0027 |
| Diabetes duration, y | 1.055 | 1.033–1.080 | 2.8e-06 |
| Median HgbA1c > 8%, yes/no | 2.1 | 1.3–3.3 | 0.0033 |
| Hypertension, yes/no | 0.9 | 0.4–1.8 | 0.70 |
Discussion
In this study we demonstrated that common European mitochondrial haplogroups are associated with DR severity. We found that, among patients who have DR, PDR is positively associated with the presence of haplogroup H and negatively associated with the presence of haplogroup Uk. These associations of PDR with mitochondrial haplogroups were initially found in the BioVU cohort and subsequently replicated in the independent VEI clinical cohort. This replication supports the validity of this association.
There are patients with very longstanding diabetes who either never develop DR or whose DR progresses very slowly, regardless of glycemic control.21 This suggests that genetic or environmental factors could also influence DR development and severity. Variants in several genes, including aldolase reductase (ALR2),22,23 erythropoietin (EPO),24,25 and vascular endothelial growth factor (VEGF),26,27 have been demonstrated in some studies to be associated with the presence or progression of DR, but genetic risk factors for DR have not yet been clearly determined.28 In particular, information regarding genetic associations from studies comparing PDR to NPDR is limited.
Mitochondrial DNA variations have been linked with ophthalmologic diseases such as glaucoma29 and AMD,30,31 but the mechanisms of these associations are not well understood. Mitochondria are responsible for the production of most of a cell's energy through oxidative phosphorylation, producing reactive oxygen species (ROS) as a byproduct. Hyperglycemia can increase oxidative stress by generating ROS directly or disrupting cellular redox balance via increased polyol pathway influx, formation of advanced glycation end products, activation of protein kinase C, and disturbance of the hexosamine biosynthesis pathway.32 Additionally, diabetes has been shown to activate matrix metalloproteinase-9, which is thought to directly damage mitochondria and contribute to angiogenesis.33,34 The ability of mitochondria to handle this increased oxidative stress may be critical to the rate of DR progression. Multiple studies have demonstrated variability in bioenergetics among the different mitochondrial haplogroups,35–39 and this may be a key mechanism by which particular haplogroups are more susceptible to advanced forms of DR.
Poor glycemic control is a critical factor influencing progression to the sight-threatening forms of DR, and PDR is typically seen in patient with higher HgbA1c levels. The lack of the expected difference in high HgbA1c levels between PDR and NPDR in haplogroup Uk patients may indicate that the Uk patients are somehow protected against the harmful effects of poor diabetes control. The absence of any association of the mitochondrial haplogroups with DME (Fig. 4) suggests that the effect of the haplogroups is more likely to be influencing DR via angiogenesis than through changes in vascular permeability.
Increasing duration of diabetes and poor glycemic control are major risk factors for development and progression of DR.17,18 Both of these factors were significant covariates for PDR in this study as well. Elevated blood pressure20 may also contribute to PDR, although the evidence is somewhat mixed.40 We did not find hypertension to be a significant covariate for PDR in this study. The associations of haplogroups H and Uk with PDR were just as strong (in terms of odds ratio and P value) as was elevated HgbA1c level, indicating that the mitochondrial haplogroup may be a clinically relevant variable in PDR in Caucasians.
Separating patients by diabetes type (Fig. 3) showed that the susceptibility of the H haplogroup patients to PDR was even stronger in patients with T2DM (OR = 2.4 [95% CI = 1.5–4.0]), but that haplogroup H had a decreased odds ratio and was not significantly associated with PDR in the T1DM patients. The protective effect against PDR in the Uk haplogroup was consistent across both T1DM and T2DM patients (OR = 0.41 [95% CI = 0.17–0.98], P = 0.06 for T1DM and OR = 0.50 [95% CI = 0.27–0.92], P = 0.028 for T2DM). Therefore, the susceptibility of patients from haplogroup H to PDR may act through a mechanism specific to T2DM, while the protective effect of haplogroup Uk may be through a mechanism that is common to both T1DM and T2DM.
The strengths of this study include the use of two independent cohorts obtained in different manners to test and validate the association between mitochondrial haplogroups and DR severity. We demonstrated the feasibility of using BioVU, a university-wide deidentified database, to identify patients with well-classified DR, with the caveat that manual review of the SD charts provided the most precise possible phenotyping. The VEI cohort was consented and ascertained in the Retina Division clinic with full access to fundus photography and the medical record. This model allows the formation of distinct discovery and validation cohorts from a single institution in a timely manner.
The study was limited by the constraints inherent in any deidentified database search. The ICD-9 and CPT codes are used for billing purposes and are often not sufficient to fully characterize clinical phenotypes. Clinic notes and other pertinent information are not available for all patients. The common presence of vague DR codes (e.g., 362.01, diabetic retinopathy not otherwise specified) in the SD did not allow us to fully characterize diabetes phenotypes in all patients meeting initial screening criteria. Sample size constraints led us to group the less common mitochondrial haplogroups into a single “other” group for the initial analysis, and a larger cohort will be necessary to allow detailed investigation of these haplogroups as well.
In this study, we demonstrated a significant association between European mitochondrial haplogroups and sight-threatening PDR. The effect of this association was similar to that of HgbA1c level in this cohort, suggesting that mitochondrial haplogroup analysis deserves further evaluation as a potential biomarker for DR severity. Increased understanding of the biologic basis for these haplogroup associations may identify new targets for intervention during the treatment of DR.
Acknowledgments
Supported by National Eye Institute Grant EY22618 (MAB), the International Retinal Research Foundation (MAB), Core Grant P30 EY08126 to Vanderbilt University, Clinical & Translational Science Awards (CTSA) Award No. UL1TR000445 from the National Center for Advancing Translational Sciences, and an unrestricted departmental grant to Vanderbilt University from Research to Prevent Blindness. Genome-wide genotyping was funded by National Institutes of Health Grants RC2GM092618 from National Institute of General Medical Sciences (Office of the Director) and U01HG004603 from National Human Genome Research Institute & National Institute of General Medical Sciences. The authors alone are responsible for the content and writing of the paper.
Disclosure: C.B. Estopinal, None; I.M. Chocron, None; M.B. Parks, None; E.A. Wade, None; R.M. Roberson, None; L.G. Burgess, None; M.A. Brantley Jr, None; D.C. Samuels, None
References
- 1. Fong DS, Aiello L, Gardner TW, et al. Retinopathy in diabetes. Diabetes Care. 2004; 27 (suppl 1): S84–S87 [DOI] [PubMed] [Google Scholar]
- 2. Boyle JP, Thompson TJ, Gregg EW, Barker LE, Williamson DF. Projection of the year 2050 burden of diabetes in the US adult population: dynamic modeling of incidence, mortality, and prediabetes prevalence. Popul Health Metr. 2010; 8: 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kempen JH, O'Colmain BJ, Leske MC, et al. The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol. 2004; 122: 552–563 [DOI] [PubMed] [Google Scholar]
- 4. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010; 376: 124–136 [DOI] [PubMed] [Google Scholar]
- 5. Antonetti DA, Klein R, Gardner TW. Diabetic retinopathy. N Engl J Med. 2012; 366: 1227–1239 [DOI] [PubMed] [Google Scholar]
- 6. Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005; 307: 384–387 [DOI] [PubMed] [Google Scholar]
- 7. D'Aquila P, Rose G, Panno ML, Passarino G, Bellizzi D. SIRT3 gene expression: a link between inherited mitochondrial DNA variants and oxidative stress. Gene. 2012; 497: 323–329 [DOI] [PubMed] [Google Scholar]
- 8. Feder J, Ovadia O, Blech I, et al. Parental diabetes status reveals association of mitochondrial DNA haplogroup J1 with type 2 diabetes. BMC Med Genet. 2009; 10: 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liou C-W, Chen J-B, Tiao M-M, et al. Mitochondrial DNA coding and control region variants as genetic risk factors for type 2 diabetes. Diabetes. 2012; 61: 2642–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Achilli A, Olivieri A, Pala M, et al. Mitochondrial DNA backgrounds might modulate diabetes complications rather than T2DM as a whole. PLoS One. 2011; 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kofler B, Mueller EE, Eder W, et al. Mitochondrial DNA haplogroup T is associated with coronary artery disease and diabetic retinopathy: a case control study. BMC Med Genet. 2009; 10: 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010; 3: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008; 84: 362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999; 23: 147 [DOI] [PubMed] [Google Scholar]
- 15. Kloss-Brandstatter A, Pacher D, Schonherr S, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011; 32: 25–32 [DOI] [PubMed] [Google Scholar]
- 16. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009; 30: E386–E394 [DOI] [PubMed] [Google Scholar]
- 17. Klein R, Klein BE, Moss SE, Davis MD, DeMets DL. The Wisconsin epidemiologic study of diabetic retinopathy, III: prevalence and risk of diabetic retinopathy when age at diagnosis is 30 or more years. Arch Ophthalmol. 1984; 102: 527–532 [DOI] [PubMed] [Google Scholar]
- 18. Klein R, Klein BE, Moss SE, Davis MD, DeMets DL. The Wisconsin epidemiologic study of diabetic retinopathy, II: prevalence and risk of diabetic retinopathy when age at diagnosis is less than 30 years. Arch Ophthalmol. 1984; 102: 520–526 [DOI] [PubMed] [Google Scholar]
- 19. Shamoon H, Duffy H, Fleischer N, et al. The effect of intensive treatement of diabetes on the development and progression of long-term complications in insulin-dependent diabetes-mellitus. N Engl J Med. 1993; 329: 977–986 [DOI] [PubMed] [Google Scholar]
- 20. Matthews DR, Stratton IM, Aldington SJ, Holman RR, Kohner EM. Risks of progression of retinopathy and vision loss related to tight blood pressure control in type 2 diabetes mellitus: UKPDS 69. Arch Ophthalmol. 2004; 122: 1631–1640 [DOI] [PubMed] [Google Scholar]
- 21. Keenan HA, Costacou T, Sun JK, et al. Clinical factors associated with resistance to microvascular complications in diabetic patients of extreme disease duration: the 50-year medalist study. Diabetes Care. 2007; 30: 1995–1997 [DOI] [PubMed] [Google Scholar]
- 22. dos Santos KG, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I. The −106CC genotype of the aldose reductase gene is associated with an increased risk of proliferative diabetic retinopathy in Caucasian-Brazilians with type 2 diabetes. Mol Genet Metab. 2006; 88: 280–284 [DOI] [PubMed] [Google Scholar]
- 23. Petrovic MG, Peterlin B, Hawlina M, Petrovic D. Aldose reductase (AC)n gene polymorphism and susceptibility to diabetic retinopathy in Type 2 diabetes in Caucasians. J Diabetes Complications. 2005; 19: 70–73 [DOI] [PubMed] [Google Scholar]
- 24. Abhary S, Burdon KP, Casson RJ, Goggin M, Petrovsky NP, Craig JE. Association between erythropoietin gene polymorphisms and diabetic retinopathy. Arch Ophthalmol. 2010; 128: 102–106 [DOI] [PubMed] [Google Scholar]
- 25. Tong Z, Yang Z, Patel S, et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A. 2008; 105: 6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abhary S, Burdon KP, Gupta A, et al. Common sequence variation in the VEGFA gene predicts risk of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2009; 50: 5552–5558 [DOI] [PubMed] [Google Scholar]
- 27. Churchill AJ, Carter JG, Ramsden C, et al. VEGF polymorphisms are associated with severity of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2008; 49: 3611–3616 [DOI] [PubMed] [Google Scholar]
- 28. Kuo JZ, Wong TY, Rotter JI. Challenges in elucidating the genetics of diabetic retinopathy. JAMA Ophthalmol. 2014; 132: 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abu-Amero KK, Gonzalez AM, Osman EA, Larruga JM, Cabrera VM, Al-Obeidan SA. Mitochondrial DNA lineages of African origin confer susceptibility to primary open-angle glaucoma in Saudi patients. Mol Vis. 2011; 17: 1468–1472 [PMC free article] [PubMed] [Google Scholar]
- 30. Kenney MC, Hertzog D, Chak G, et al. Mitochondrial DNA haplogroups confer differences in risk for age-related macular degeneration: a case control study. BMC Med Genet. 2013; 14: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. SanGiovanni JP, Arking DE, Iyengar SK, et al. Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS One. 2009; 4 (5): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005; 54: 1615–1625 [DOI] [PubMed] [Google Scholar]
- 33. Kowluru RA, Zhong Q, Santos JM. Matrix metalloproteinases in diabetic retinopathy: potential role of MMP-9. Expert Opin Investig Drugs. 2012; 21: 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santos JM, Tewari S, Lin JY, Kowluru RA. Interrelationship between activation of matrix metalloproteinases and mitochondrial dysfunction in the development of diabetic retinopathy. Biochem Biophys Res Commun. 2013; 438: 760–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006; 38: 1261–1268 [DOI] [PubMed] [Google Scholar]
- 36. Mueller EE, Brunner SM, Mayr JA, Stanger O, Sperl W, Kofler B. Functional differences between mitochondrial haplogroup T and haplogroup H in HEK293 cybrid cells. PLoS One. 2012; 7: e52367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kenney MC, Chwa M, Atilano SR, et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim Biophys Acta. 2014; 1842: 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kenney MC, Chwa M, Atilano SR, et al. Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PLoS One. 2013; 8: e54339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kenney MC, Chwa M, Atilano SR, et al. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet. 2014; 23: 3537–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chew EY, Ambrosius WT, Davis MD, et al. Effects of medical therapies on retinopathy progression in type 2 diabetes. N Engl J Med. 2010; 363: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]