Abstract
We describe a 8,724-nucleotide-long picornavirus genome encoding a single 2,470-aa polyprotein obtained from the feces of a wild mouse. Rosavirus is genetically closest to the double ORF Dicipivirus found in canine feces that is currently the only picornavirus with a second internal ribosome entry site (IRES). Of note, a section of rosavirus’ 5′UTR showed strong sequence and structural conservation with the type II IRES from the Parechovirus and Hungarovirus genera possibly reflecting exchange of genetic modules between genera. Based on genetic distance criteria rosavirus qualifies as prototype of a new genus of the Picornaviridae family.
Keywords: Picornavirus, Feces, Mice, Genus
The study
We previously reported a partial (~3,956 bases) genome of rosavirus (rodent stool associated picornavirus) sequenced together with the genome of another picornavirus named mosavirus (GenBank JF973687) from the stool sample of a single wild canyon mouse (Peromyscus crinitus) collected in California in May 2010 [1]. Using the Illumina MiSeq platform (http://www.illumina.com/systems/miseq.ilmn) and 5′RACE [2] we generated and describe here a near-complete 8,724-nucleotide-long genome of rosavirus A (GenBank JF973686).
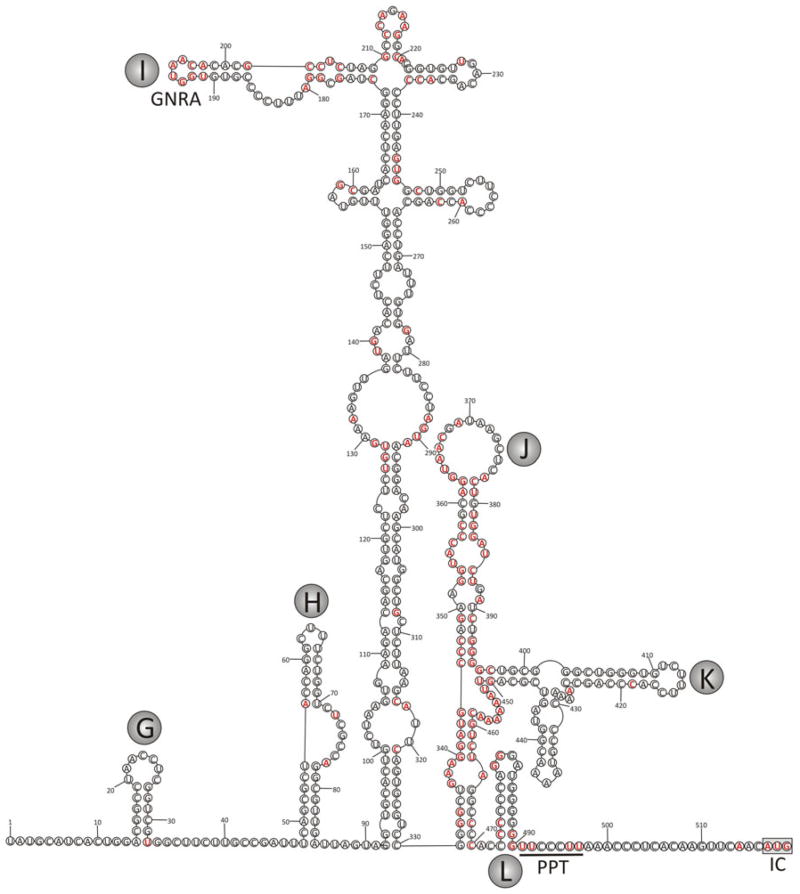
Alignment and RNA structure prediction of the 5′UTR revealed a type II internal ribosomal entry sites (IRES) structure (Fig. 1) similar to that of parechoviruses [3] and hungaroviruses of cattle and sheep [4]. This structural homology and extended regions of substantial nucleotide sequence similarity to picornaviruses in other genera (e.g., 84 and 71 % identity from positions 155–221 and 332–503 between rosavirus and Ljungan virus sequences) reflect a possible example of modular exchange of type II IRESs comparable to that observed for type IV IRES elements in other picornavirus genera [5]. Since the available rosavirus 5′UTR sequence only starts at a position homologous to the G loop of parechovirus [6] it is likely that ~250 bases are missing from the 5′UTR.
Fig. 1.
Proposed RNA secondary structure of the 5′UTR of rosavirus based on minimum free energy folding predictions (MFOLD) and sequence/structural alignment with parechovirus and hungarovirus sequences. Bases conserved with parechoviruses and hungaroviruses are highlighted in red. Stem-loops G-L have been labeled using letters assigned to homologous structures in HPeV [6]. Defined IRES elements GNRA loop, polypyrimidine tract [PPT], and initiation codon [IC] have been labeled
A Kozak sequence, RNNAUGG (AACAUGG), was found at the beginning of the rosavirus ORF [7]. The P1 polypeptide was 869 aa in length sharing the closest aa identity of 35 % with the P1 of canine picodicistrovirus (CPDV) (Table 1) also known as Cadicivirus A, prototype and currently single member of the Dicipivirus genus. CPDV was found in canine feces and contains an unusual 2nd IRES between P1 and P2 [8]. P1 amino acids identities of rosavirus to the next closest picornaviruses were 18 and 14 % to Aichi virus and turdivirus 2 (also known as oscivirus A1), respectively (Table 1). Rosavirus P1 contained the conserved motif GXXXT/S (3GRKDS7) for myristoylation [9]. Similar to CPDV, rosavirus did not have a putative L protein preceding the capsid region [8]. The hypothetical cleavage map of the rosavirus polyprotein was derived from alignments with other picornaviruses and NetPicoRNA prediction [10]. The P1 was hypothetically cleaved at 1A/1B (58D↓S59), 1B/1C (314E↓S315), and 1C/1D (588K↓E589). The 1D protein of rosavirus and CPDV did not have motif [PS]ALXAXETG [8]. The 780-aa P2 polypeptide was hypothetically cleaved at 2A/2B (1102Q↓P1103) and 2B/2C (1312E↓A1313) and shared aa sequence identities of 17, 21, and 18 %—with corresponding polypeptides of CPDV, Aichi virus, and turdivirus 2, respectively. The 2A protein of rosavirus contained an HBox/NC domain [11]. Overall, the rosavirus P3 showed the closest identity (40 %) to CPDV followed by 30 % identity to the P3 of Aichi virus and 31 % to the avian turdivirus 2 (Table 1). Species within a picornavirus genus share >40, >40, and >50 % amino acid identities in P1, P2, and P3 regions, respectively [12], a classification largely supported by discontinuity in pair-wise evolutionary distances distribution among picornaviruses [13, 14]. Since the % aa identities of rosavirus with cogent regions of other picornaviruses are below the within-genus-distance criteria of ICTV, rosavirus is proposed as the prototype genome of the new genus Rosavirus in the Picornaviridae family, pending ICTV approval (http://www.picornaviridae.com/).
Table 1.
Comparison of amino acid sequence identity of rosavirus P1-P3 regions with those of the most closely related picornaviruses
Acknowledgments
We acknowledge NHLBI R01HL105770 and support from BSRI to E.D. We also thank staff at the Division of Communicable Disease Control, California Department of Public Health for sample collection.
Contributor Information
Tung Gia Phan, Blood Systems Research Institute, San Francisco, CA 94118, USA. Department of Laboratory Medicine, University of California at San Francisco, San Francisco, CA 94118, USA.
Nguyen Phung Vo, Blood Systems Research Institute, San Francisco, CA 94118, USA. Pharmacology Department, School of Pharmacy, Ho Chi Minh City University of Medicine and Pharmacy, Ho Chi Minh City, Vietnam.
Peter Simmonds, Roslin Institute, University of Edinburgh, Edinburgh EH25 9RG, UK.
Erik Samayoa, Department of Laboratory Medicine, University of California at San Francisco, San Francisco, CA 94118, USA.
Samia Naccache, Department of Laboratory Medicine, University of California at San Francisco, San Francisco, CA 94118, USA.
Charles Y. Chiu, Department of Laboratory Medicine, University of California at San Francisco, San Francisco, CA 94118, USA
Eric Delwart, Email: delwarte@medicine.ucsf.edu, Blood Systems Research Institute, San Francisco, CA 94118, USA. Department of Laboratory Medicine, University of California at San Francisco, San Francisco, CA 94118, USA.
References
- 1.Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, Zaidi SZ, Delwart E. J Gen Virol. 2009;90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nateri AS, Hughes PJ, Stanway G. J Virol. 2000;74:6269–6277. doi: 10.1128/jvi.74.14.6269-6277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reuter G, Pankovics P, Knowles NJ, Boros A. J Virol. 2012;86:13295–13302. doi: 10.1128/JVI.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simmonds P. Recombination in the evolution of picornaviruses. In: Ehrenfeld E, Domingo E, Roos RP, editors. The Picornaviruses. ASM Press; Washington, DC: 2010. pp. 229–238. [Google Scholar]
- 6.Ghazi F, Hughes PJ, Hyypia T, Stanway G. J Gen Virol. 1998;79(Pt 11):2641–2650. doi: 10.1099/0022-1317-79-11-2641. [DOI] [PubMed] [Google Scholar]
- 7.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo PC, Lau SK, Choi GK, Huang Y, Teng JL, Tsoi HW, Tse H, Yeung ML, Chan KH, Jin DY, Yuen KY. J Virol. 2012;86:2797–2808. doi: 10.1128/JVI.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow M, Newman JF, Filman D, Hogle JM, Rowlands DJ, Brown F. Nature. 1987;327:482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- 10.Blom N, Hansen J, Blaas D, Brunak S. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes PJ, Stanway G. J Gen Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- 12.Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, Zell R. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2012. pp. 855–880. [Google Scholar]
- 13.Lauber C, Gorbalenya AE. J Virol. 2012;86:3905–3915. doi: 10.1128/JVI.07174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauber C, Gorbalenya AE. J Virol. 2012;86:3890–3904. doi: 10.1128/JVI.07173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]