Abstract
RATIONALE
With the increased mass accuracy and resolution in commercialized mass spectrometers, new development on shotgun lipidomics could be expected with increased speed, dynamic range, and coverage over lipid species and classes. However, we found that the major issue by using high mass accuracy/resolution instruments to search lipid species is the partial overlap between the two 13C atom-containing isotopologue of a species M (i.e., M+2 isotopologue) and the ion of a species less a double bond than M (assigned here as L). This partial overlap alone could cause a mass shift of the species L to the lower mass end up to 12 ppm around m/z 750 as well as significant peak broadening.
METHODS
We developed an approach for accurate mass searching by exploring one of the major features of shotgun lipidomics data that lipid species of a class are present in ion clusters where neighboring masses from different species differ by one or a few double bonds. In the approach, a mass-searching window of 18 ppm (from −15 to 3 ppm) was first searched for an entire group of species of a lipid class. Then accurate mass searching of the plus one 13C isotopologue of individual species was used to eliminate the potential false positive.
RESULTS
The approach was extensively validated through comparing with the species determined by the multi-dimensional MS-based shotgun lipidomics platform. The newly developed strategy of accurate mass search enables identifying the overlapped L species and acquiring the corresponding peak intensities.
CONCLUSIONS
We believe that this novel approach could substantially broaden the applications of high mass accurate/resolution mass spectrometry for shotgun lipidomics.
Keywords: accurate mass searching, 13C isotopologue, double bond overlapping effect, shotgun lipidomics, ultra high mass resolution mass spectrometry
Lipidomics, as a newly established research field, is rapidly spreading.[1–7] There exist two approaches for lipidomics analysis, i.e., liquid chromatography-mass spectrometry (LC-MS) based methods and direct infusion based platforms.[3, 8] The latter now is generally referred to as shotgun lipidomics in the field.[3, 9] There exists a dogma in the field that if mass accuracy and resolution of a mass spectrometer is high enough, individual molecular species of a lipid class can be accurately searched through comparison with a theoretical lipid database and its peak intensity or peak area can be extracted from the mass spectral datasets acquired with such an instrument. Such a database can be readily constructed based on the elementary composition as described previously.[10] Therefore, quantitative analysis of individual lipid species can be achieved, at least partially, through accurate mass searching from the mass spectra acquired with those instruments, although ultimate identification of the identities of individual species only can be achieved by tandem MS analysis.
To such reasoning, there are a few questions to be answered. For example, what is the minimal mass resolution of an instrument required for such a purpose and if currently commercially-available instruments did not meet the requirement, whether could we develop an approach to achieve such a goal with a relatively lower mass resolution instrument? OrbiTrap mass analyzers with a mass resolution of over 100,000 at m/z 400 and sub-ppm mass accuracy could offer robust practical solutions for lipidomic analysis in a top-down lipidomic strategy, the aforementioned questions have not been addressed.[11–16] In the reported studies, the researchers used a mass window of ± 3 ppm to search mass spectral datasets acquired with an OrbiTrap mass spectrometer possessing a mass accuracy of 3 ppm.[11]
Herein, we presented a study in which we investigated the requirement of mass resolution for the aforementioned purpose. We found that at this moment, the mass resolution of the majority of the commercially available mass spectrometers is not high enough to totally resolve the overlaps present in the analysis of lipids by shotgun lipidomics. We discovered that the major issue is the partial overlap between the two 13C atom-containing isotopologue of a species M (i.e., M+2 isotopologue) and the ion of a species less a double bond than M (assigned here as L) (referred to as the double bond overlapping effect hereafter). This partial overlap leads to at least two problems including peak broadening and peak shift. The former results in inaccurate quantitative analysis if the peak intensity is used to compare to that of an internal standard for quantification. The latter makes an accurate mass search difficult. It should be pointed out that this issue was not problematic in the case of unit mass resolution mass spectrometry where these ions are completely overlapped each other and the intensity of the overlapped peak is additive. Therefore, the intensity of each individual ion can be extracted after de-isotoping.[17–19]
We simulated this overlap along with the existence of other isotopic atoms including 2H (D), 18O, 15N, 34S, and 37Cl under a variety of experimental conditions and different mass resolution of instruments. We recognized that this partial overlap alone could cause a mass shift of the species L to the lower mass end up to 12 ppm (at m/z 750 where the majority of lipid ions are detected) depending on the molar ratio of the species M and L and the mass resolution of the instrument used. However, if a mass-searching window of −15 to +3 ppm (a 12 ppm mass shift plus ± 3 ppm mass accuracy) was used for accurate mass searching, a substantial number of false positive and non-specific hits could be included. Therefore, such direct mass searching of a high accurate mass spectral dataset for shotgun lipidomics analysis definitely leads to a complicated outcome. This overlapping effect was discussed previously.[11, 13] However, since the double bond overlapping effect mainly affects low abundant species and relatively high resolution MS (the full width at half maximum (FWHM) 100,000 at m/z 400) was used in those reports, the question what is the minimal mass resolution of an instrument required for direct accurate mass searching was not answered.
It should be emphasized that LC-MS offers tremendous advantages to resolve isobaric/isomeric lipid species.[20–22] However, comparing to direct infusion of organic extracts of biological samples (generally referred as to shotgun lipidomics), comprehensive analysis of lipids in a large scale by LC-MS is always time-consuming, prohibiting the high throughput analysis of a cellular lipidome. Moreover, while separation of isobaric/isomeric lipid species is achievable by employing chromatographic technique, resolving all of molecular species of a class is not straightforward.
Based on these findings in the spectra after direct infusion, we developed an approach for accurate mass searching by exploring one of the major features of lipidomics data that molecular species of a lipid class are present in ion clusters where neighboring masses from different species differ by one or a few double bonds. In the approach, we first opened the mass-searching window to 18 ppm (from −15 to +3 ppm) to search an entire group of species of a lipid class; these species contain an identical number of carbon atoms. Then accurate mass searching of the plus one 13C isotopologue was used to eliminate the potential false positive since the overlaps of the plus one 13C isotopologue with other 13C isotopologues are minimal and these overlaps if present are very different from the overlaps between M+2 13C isotopologues and L ions. The intensity of the plus one 13C isotopologue could also be used to calculate the intensity of L ions based on their natural abundance for the purpose of quantification. This new mass searching approach was validated through comparing with the species determined by multi-dimensional MS-based shotgun lipidomics (MDMS-SL),[3, 10] which is a well-developed technology for global lipid identification and quantification directly from lipid extracts of biological samples. We believe that this novel approach could lead to achieving our goal that high throughput analysis of individual lipid species could be rapidly conducted through accurate mass searching followed by targeted tandem MS analysis by high mass accurate/resolution MS without extremely high resolution requirement.
EXPERIMENTAL
Materials
All phospholipid species used either as internal standards or for mixing experiments were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). All the solvents were obtained from Burdick and Jackson (Honeywell International Inc., Muskegon, MI). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Preparation of Phosphatidylcholine Mixtures and Lipid Extracts from Biological Samples
The stock solutions of di14:1, 16:0–18:1, and 16:0–18:0 phosphatidylcholine (PC) species were separately prepared in CHCl3/MeOH (1:1, v/v). The concentration of each stock solution was quantified individually by comparison with the internal standard di14:1 PC using electrospray ionization (ESI) MS (ESI-MS). The mixtures of the PC species were prepared from individual PC species stock solutions. The serial ratios in the mixtures were obtained by fixing the concentration of one PC species while varying the other(s). For our convenience to display the overlapped ion peaks and process the mass shifts, these ratios were given as those between 16:0–18:0 PC and M+2 13C isotopologues of 16:0–18:1 PC.
C57BL/6J wild type male mice (3 months of age) were originally purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in a full barrier facility with a 12-h light/dark cycle and maintained on standard chow (Diet 5053; Purina Inc., St. Louis, MO). All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Studies Committee at the Sanford-Burnham Medical Research Center at Lake Nona, Orlando, FL. Mice were sacrificed by asphyxiation with CO2 and followed by cervical dislocation. The mouse tissues were excised quickly and perfused with ice-cold PBS to remove blood, blotted with Kimwipes (Kimberly-Clark, Roswell, GA) to remove excess buffer, and then immediately freeze-clamped at the temperature of liquid N2. All of tissue samples were stored at −80 °C until lipid extraction and analysis. Lipid extracts of mouse tissue samples were prepared as described previously.[23] The lipid extracts were finally flushed with nitrogen, capped, and stored at −20 °C for ESI-MS analyses.
MS analysis of PC mixtures and lipid extracts of biological samples
High mass accuracy/resolution MS analyses of lipids were performed by using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an automated nanospray apparatus (Advion Bioscience, Ithaca, NY). The NanoMate apparatus was controlled by Chipsoft 8.3.1 software. An ionization voltage of 1.2 kV and a gas pressure of 2.0 psi were employed for the MS analyses. All mass spectra were recorded under Xcalibur 2.2.48 software with an AGC value of 1 × 106 with an injection time up to 120 ms in the full MS mode of an average of 10 s, in which the mass resolution of the analyzer was set at 280,000 (this resolution was kindly offered by Thermo Fisher Scientific as a prototype instrument control software upgrade) or 140,000 (m/Δm, FWHM at m/z 200), corresponding to the resolution of 150K or 75K, respectively, at around m/z 750, where the majority of lipid species are detected, as indicated in this paper.
MDMS-SL was performed by using a TSQ VANTAGE mass spectrometer (Thermo Fisher Scientific, San Jose, CA) as previously described [10, 24]. Individual PC mixtures or lipid extracts of biological samples were diluted to approximately 50 pmol/µL in CHCl3/MeOH/isopropanol (1:2:4, v/v/v) with a 4% LiOH/MeOH solution (200 time dilution from the saturated solution) or specific prior to infusion with the NanoMate device (same as above mentioned).
Simulation of ion overlaps under different mass resolutions
We assume that all of the mass spectrometric peaks follow the Gaussian function:
| (1) |
where e is the Euler’s number (e ≈ 2.71828…); a is the full height of the peak; b is the x-position of the center of the peak (herein the m/z value of the peak); and c controls the width of the peak.
The parameter c relates to FWHM of the peak according to
| (2) |
The resolution, R, of MS is defined as
| (3) |
Therefore, through the combination of equations (1) to (3), the relation between the peak Gaussian shape and the resolution R is
| (4) |
The Gaussian function of each possibly overlapped peak from double bond, 13C, D, 18O, 15N, and so on, was calculated individually. Depending on their natural abundance and the concentrations of the species, which determine the a value for each isotope, those individual Gaussian functions from the components of the overlapped peak at the same x value were added up together to calculate the simulated overlapped MS peaks.
Method and procedures of accurate mass search
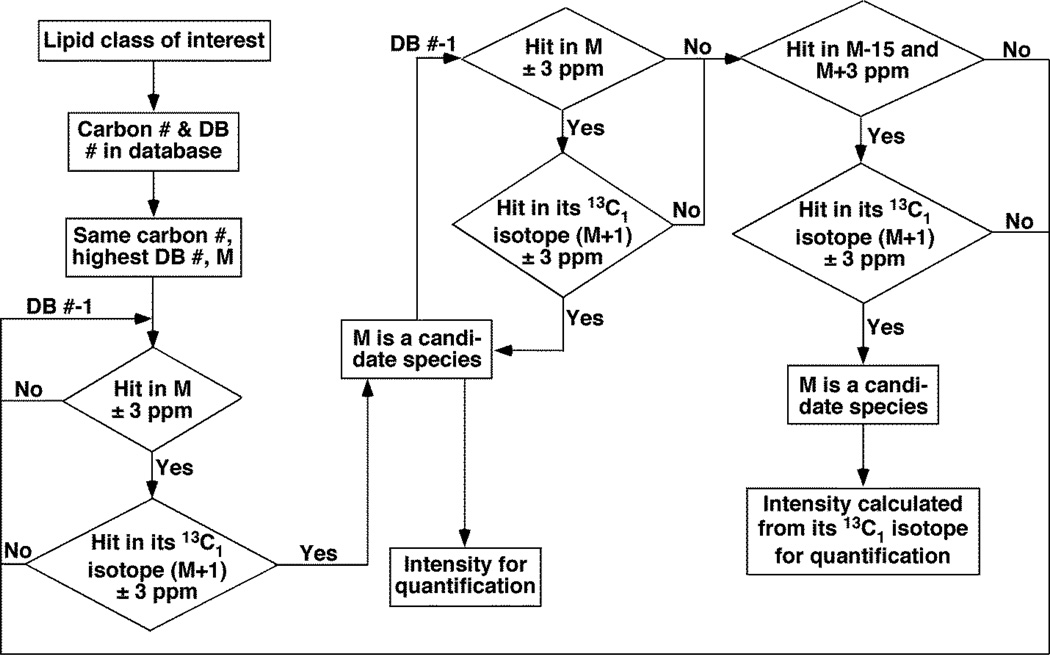
First, a database of interested lipid classes was built up. The database includes the accurate ion masses of the entire possibly known natural existing lipid species, which were grouped according to their numbers of carbon atoms and double bonds in the acyl chains. As shown in Figure 1, the search of the interested lipid class started from the species with the lowest possible carbon number and the highest possible double bond number with a mass window of ± 3 ppm of the theoretical mass of the interesting species. If a hit was found, the presence of its M+1 isotopologue (with one more 13C isotope) was also checked within the mass searching window of ± 3 ppm. Depending on the existence of its isotopologue peak and the intensity ratio between M and M+1 peaks, this species M was validated or discarded. If the species was not present, then the above search was repeated for the next species with the same number of carbon and one less double bond until one species was found. The intensity of this species could be used for quantification by comparing to the internal standard. Next, the same searching process was repeated for the next species with one less double bond until a species was missing. Then, the mass searching window was opened up for this same species from ± 3 ppm to between −15 and +3 ppm. If the species L existed in this expanded window, its L+1 isotopologue (with one more 13C) should be present in its theoretical mass range of ± 3 ppm. If this isotopologue did not exist, this potential candidate species was discarded. If this species was validated with the presence of its L+1 isotopologue, the intensity of the L+1 peak was used to calculate the real intensity of L ions for its quantification. Then, the next species with one less double bond was searched by using the same procedure until the number of the double bond was zero. Similarly, the next group of lipid species with an additional carbon atom and the highest possible double bond number was searched by using the same strategy.
Figure 1.
A flow chart shows a novel accurate mass searching approach for identification of candidate lipid species in high mass accuracy/resolution shotgun lipidomics.
RESULTS AND DISCUSSION
Recognition of the mass shift resulted from the double bond overlapping effect analyzed by high mass accuracy/resolution shotgun lipidomics
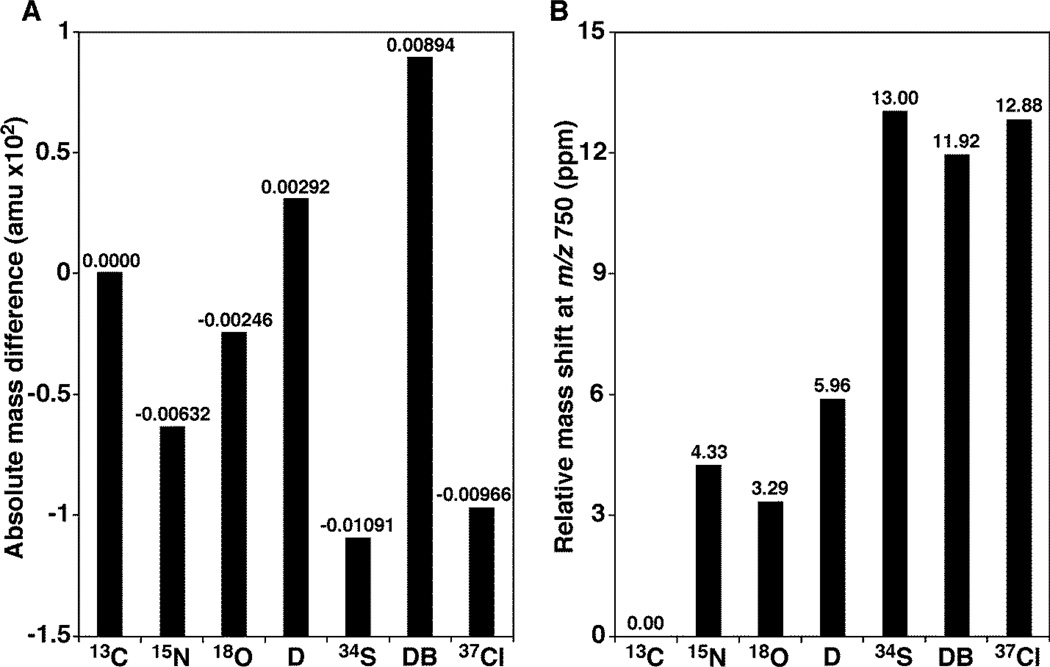
When we calculated the mass defects of natural elements relative to 13C atom (Figure 2), we recognized the partial overlap of the two 13C atom-containing isotopologue (i.e., M+2 isotopologue) of a species M with the ion of a species L less a double bond than M (i.e., the double bond overlapping effect). Although isotopologues resulting from chlorine-37 and sulfur-34 could also overlap with the M+2 13C isotopologue, chlorine (which can be easily probed due to the natural abundance of 37Cl at 31.9784%) and sulfur-containing lipids are less common than C, H, O, N, and P-containing lipids, particularly in the mammalian system.[25] Therefore, the double bond overlapping effect would be the major and most dominant effect contributing to the mass shift and complicating the isotopic patterns of lipid species in high mass resolution shotgun lipidomics. The figure indicated that, if the mass level of species L (one double bond less) is much less than that of species M+2 isotopologue, the differential mass shifts of the species L could be as high as 12 ppm at m/z 750 where the majority of lipid ions are detected nearby by ESI-MS. More complication of this overlapping effect was its dependency on the mass resolution of the employed instrument and the molar ratio of the overlapped ions (see below).
Figure 2.
The double bond overlapping effects complicate the isotopic patterns of lipid species analyzed by high mass accuracy/resolution shotgun lipidomics. Panel A showed the relative mass defects of different elements to one or two 13C mass. D represents deuterium relative to one 13C atom, as well as 15N. Meanwhile, 18O, 34S, and 37Cl are relative to two 13C atoms. DB represents the mass defects of one less double bond (i.e., addition of two protons) relative to two 13C atoms. Panel B indicated the differential mass shifts at ppm of isotopologues containing these isotopes relative to 13C atom(s) at m/z 750 where the majority of lipid ions are detected nearby by ESI-MS.
Mass shift resulted from the double bond overlapping effect depends on the mass resolution of an instrument and the molar ratio of the overlapped ions
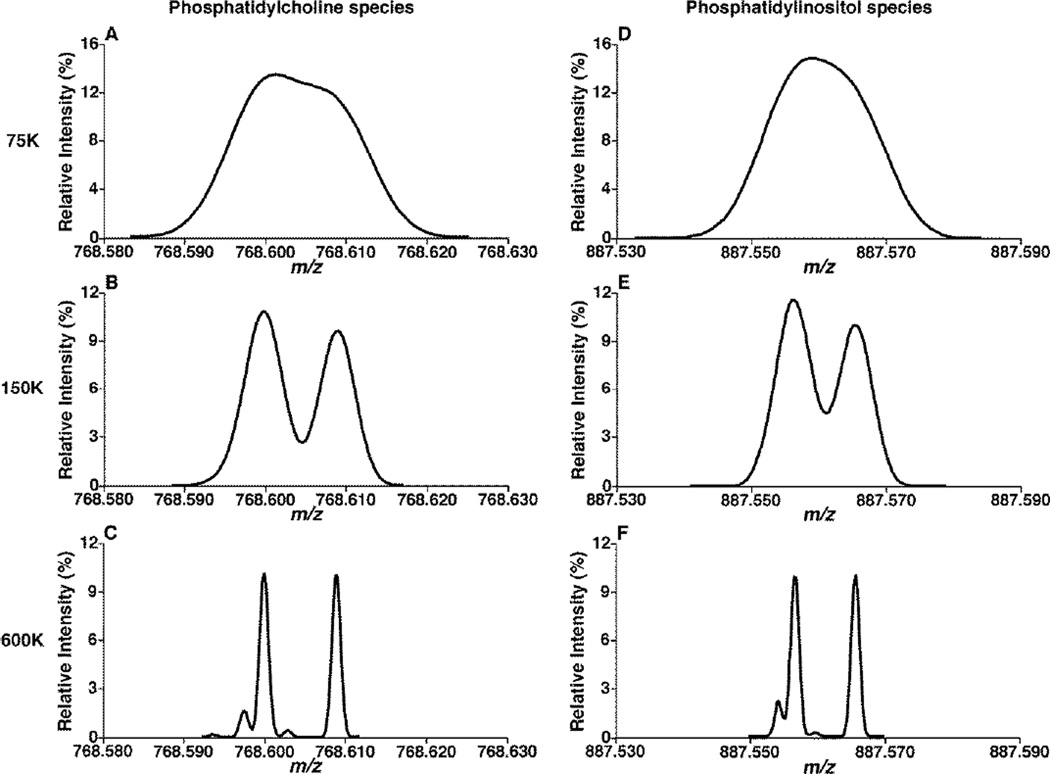
To demonstrate the double bond overlapping effect-induced mass shift, we simulated the expanded mass spectra displaying the ion peaks of the M+2 13C isotopologue and the ion of the species L containing one double bond less than M at an equal intensity ratio for the species of PC and phosphatidylinositol (PI) classes (Figure 3). We found the resolution of these ion peaks depended on the mass resolution of the instrument. Specifically, the two ions essentially collapsed into a single, broad peak at an instrumental mass resolution of 75K (which corresponds to the mass resolution of the Q-Exactive mass spectrometer setting at 140K at this mass region) or less (Figure 3A and D). Although these ions were partially resolved at the mass resolution of 150K (Figure 3B and E), the m/z values of the ion peaks of the M+2 13C isotopologue and the ion of the species L were not affected as much as at the mass resolution of 75K by the totally overlapping. All of the isotopologues ion peaks coming from 13C, D, 14N, etc. could only be totally resolved with an instrumental mass resolution of 600K at this mass range, which is unachievable at the current time (Figure 3C and F).
Figure 3.
Simulation of the mass spectra displaying the M+2 13C isotopologue and the ion (L) which contains one double bond less than the species M with different instrumental mass resolution. An equal intensity ratio of the M+2 isotopologue and the L ion from the classes of both phosphatidylcholine (Panels A, B and C) and phosphatidylinositol species (Panels D, E and F) was used. The mass spectra of these ions with instrumental mass resolution of 75K (Panels A and D), 150K (Panels B and E), and 600K (Panels C and F) were simulated as described in the subsection of “Materials and Methods”.
Obviously, due to this overlap, the apex of this collapsed peak was shifted to the middle of these ions if the instrumental mass resolution is at 75K or less. The location of the apices of the collapsed peaks could shift from the ion of L when the species M is absent to the place of 11.92 ppm downside relative to the ion of L when the species L is absent. This location of the peak apex entirely depends on the intensity (i.e., molar) ratio of the species L and the M+2 isotopologue, as shown by the simulation of the mass spectra with different molar ratio of M+2 vs. L (Figure S1).
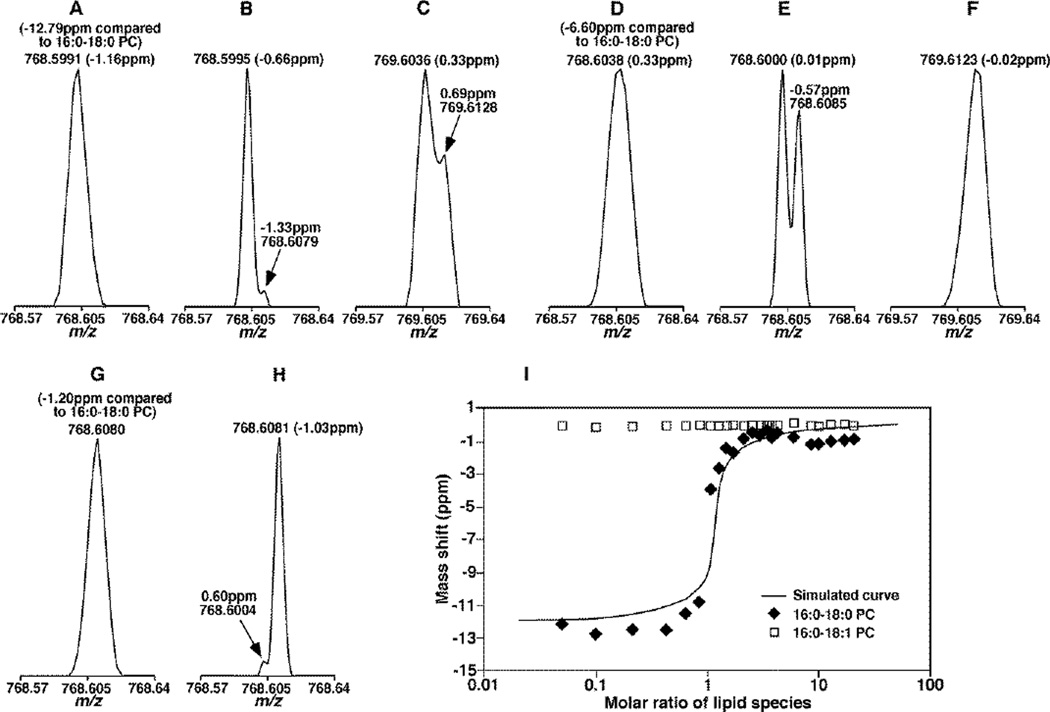
To demonstrate the dependence of the overlapping ion peak on the molar ratio of the L and M+2 ions, we prepared a series of mixtures containing three standard PC species of which the difference between the fatty acyl chains of two species was one double bond (i.e., 16:0–18:1 PC (M) and 16:0–18:0 PC (L)) and the other PC species (di14:1 PC) was used to lock the mass. We determined the mass shift of the apex of the overlapping peak of M+2 13C isotopologue and L ion as varied with the molar ratios of M+2 and L ions by MS with the mass resolution of 75K (Figure 4, panels A, C, D, F and G) and 150K (Figure 4, panels B, E and H). With the instrument resolution at 75K, when the amount of M+2 13C isotopologue (13C2-16:0–18:1 PC) is higher than L ion (16:0–18:0 PC), the collapsed peak shifted to the m/z of 13C2-16:0–18:1 PC and produced −12.79 ppm mass shift comparing to 16:0–18:0 PC peak (Figure 4A). When the intensity ratio of the two ions was close to 1, the collapsed peak was located at the middle of their m/z values with a mass shift of −6.60 ppm comparing to 16:0–18:0 PC peak (Figure 4D). When the amount of 16:0–18:0 PC is higher than 13C2-16:0–18:1 PC, the m/z position of 16:0–18:0 PC was not significantly affected with a mass shift of −1.20 ppm, a little higher than the instrumental specification, 1 ppm (Figure 4G). At the instrument resolution at 150K, the two ion peaks of 13C2-16:0–18:1 and 16:0–18:0 PC could be partially separated under different molar ratios with mass shifts confined in the range of ± 3 ppm (Figure 4B, E and H). At this resolution, the double bond overlapping effects for most of lipids were basically resolved, and direct mass searching with the theoretical m/z could be used. Essentially identical to the simulation, the mass shifts of the overlapping ion peaks with the molar ratio from 1:20 to 20:1 (16:0–18:0 PC vs.13C2-16:0–18:1) at the mass resolution of 75K depended on the molar ratios of the mixtures under experimental conditions (Figure 4, panel I).
Figure 4.
Determination of the mass shift with standard phosphatidylcholine (PC) species resulted from the double bond overlapping effect in a molar ratio dependent manner. Mixtures of standard 16:0–18:0 and 16:0–18:1 PC species were prepared and analyzed by using a Q-Exactive mass spectrometer at the resolution of 75K (panel A, C, D, F, G and I) and 150K (panel B, E and H) as described in the subsection of “Materials and Methods”. Mass spectra of the mixtures were comprised of 10:1 (panel A to C), 1:1 (panel D to F), and 1:10 (panel G and H) ratios of the M+2 13C isotopologue of 16:0–18:1 PC vs. 16:0–18:0 PC ions. For comparison, the corresponding peaks of M+3 13C isotopologue of 16:0–18:1 PC vs. L+1 13C isotopologue of 16:0–18:0 PC were also shown in Panels C and F as indicated. These spectra demonstrated both the overlapping peak shape and the mass shift due to the overlapping of M+2 and L, M+3 and L+1 species. Panel I showed the determined locations of ions corresponding to 16:0–18:1 (open square) and 16:0–18:0 (solid square) PC species relative to their theoretical values of 766.593257 and 768.608907 Da, respectively, and the simulated mass shift of 16:0–18:0 PC relative to 768.608907 Da. Lock mass of 680.483706 Da (i.e., di14:1 PC species) was used during mass spectral acquisition. The variation of the mass shift corresponding to 16:0–18:0 PC was observed as changed in the total concentration of the PC mixtures.
We also found out that the peak resolution is dependent on the total concentration of the lipids species (Figure 2S). The lower total concentration shows better resolution between overlapped peaks. Therefore, analytes with relative low concentration are always preferred in order to show better resolution and avoid lipid species aggregation as well.
As demonstrated above, when the amounts of M+2 13C isotopologue were higher than or close to those of L ion, the mass shifts of L peak could be much bigger than the manufacture’s specification for non-interfered signals, varying from 3 ppm (the mass shift we have seen without overlapping effects with a mass lock) to as high as −15 ppm (3 ppm instrument shift plus 12 ppm (the maximal mass shift due to the double bond overlapping)) at the mass resolution of 75K. However, under the same condition, it was found that the peaks with one more 13C isotopologue, (i.e. M+3 13C isotopologue and L+1 13C isotopologue) showed much less overlapping effects and better resolution due to the substantial reduction of the M+3 13C isotopologue peak intensity and the lower concentration of both of the isotoplogues species (Figure 4C and F). The mass shift of 13C-16:0–18:0 PC was 0.69 ppm when the concentration of 16:0–18:1 PC was 100 times higher than 16:0–18:0 PC, and the isotope peak of 13C-16:0–18:0 PC was dominant and showed a mass shift of −0.02 ppm when the concentration of 16:0–18:1 PC was 10 times higher than 16:0–18:0 PC. Therefore, the peaks corresponding to the L+1 13C isotopologue could be used to assist the accurate mass search at the mass resolution of 75K, and its intensity could assist to derive the intensity of L ions for quantification after identification.
Direct accurate mass searching by using instrumental mass accuracy leads to the substantial miss of species
The mass shift due to the double bond overlapping effect alone apparently also complicates the direct accurate mass searching for identification of lipid species, thereby leading to a significant false negative result. For example, the Q-Exactive mass spectrometer possesses the maximal mass shift up to ± 3 ppm according to our experience that most peaks shift in this range, which is higher than the manufacturer’s specification. However, when accurate mass searching with a mass window of ± 3 ppm was conducted (Table S1A), the hit species included those which were abundant or not overlapped with other interference peaks or M+2 13C isotopologues of the identical class after direct infusion. The searched species also included the species which were not listed in the identification by MDMS-SL (Table 1), which could be the interference peaks from other classes of compounds. The missing species searched by the ± 3-ppm mass window were mainly caused by the double bond overlapping effects, which led to the loss of the species with less double bonds and same carbon numbers (marked with # in Table 1).
Table 1.
Comparison of PC species identified by accurate mass searching using the developed strategy vs. determined by MDMS-SL in a mouse myocardium sample. Species marked with # showed the false negative hits with a searching window of ± 3 ppm, which were missing in Table S1A. “D”, “P” and “A” denote diacyl, plasmenyl, and plasmanyl PC species, respectively.
| Detected by accurate mass search | Detected by MDMS-SL | |||
|---|---|---|---|---|
| PC species | Theoretic m/z | Determined m/z | Intensity | Content (nmol/mg protein) |
| D30:0 | 712.54631 | 712.54557 | 12201.7 | |
| A32:0 | 726.59834 | 726.59877 | 4838.1 | |
| D32:2 | 736.54631 | 736.54650 | 61338.9 | 0.03 |
| D32:1 | 738.56196 | 738.56272 | 75574.5 | 0.20 |
| D32:0 | 740.57761 | 740.57833 | 404423 | 1.29 |
| P34:2 | 748.58269 | 748.58298 | 17969.7 | |
| P34:1 | 750.59834 | 750.59933 | 24749.1 | 0.06 |
| P34:0 | 752.61399 | 752.61480 | 13768.9 | 0.07 |
| D34:4 | 760.54631 | 760.54635 | 147996.7 | 0.87 |
| D34:3 | 762.56196 | 762.56321 | 90457.1 | 0.17 |
| D34:2 | 764.57761 | 764.57838 | 1564914.1 | 4.38 |
| D34:1 | 766.59326 | 766.59452 | 1186745.4 | 3.83 |
| #D34:0 | 768.60891 | 768.60013 | 124198.7 | 0.10 |
| P36:4 | 772.58269 | 772.58302 | 62224.2 | 0.28 |
| P36:3 | 774.59834 | 774.59878 | 17975.4 | 0.07 |
| P36:2 | 776.61399 | 776.61465 | 1237 | 0.01 |
| D36:6 | 784.54631 | 784.54639 | 18062.3 | |
| D36:4 | 788.57761 | 788.57827 | 1693068 | 4.13 |
| D36:3 | 790.59326 | 790.59494 | 676811.9 | 2.19 |
| D36:2 | 792.60891 | 792.61014 | 692685.2 | 2.08 |
| D36:1 | 794.62456 | 794.62623 | 202559 | 0.93 |
| P38:5 | 798.59834 | 798.59931 | 164545.4 | 0.10 |
| P38:4 | 800.61399 | 800.61519 | 42164.2 | 0.01 |
| P38:2 | 804.64529 | 804.64738 | 3758 | |
| P38:0 | 808.67659 | 808.67001 | 25.4 | |
| D38:7 | 810.56196 | 810.56313 | 54220.2 | 0.16 |
| D38:6 | 812.57761 | 812.57849 | 3496274 | 10.63 |
| D38:5 | 814.59326 | 814.59435 | 848389 | 2.46 |
| D38:4 | 816.60891 | 816.60993 | 1224517.4 | 3.46 |
| #D38;3 | 818.62456 | 818.61533 | 203659.4 | 0.75 |
| D38:2 | 820.64021 | 820.64089 | 20896.4 | 0.13 |
| P40:7 | 822.59834 | 822.60008 | 33477.2 | 0.13 |
| P40:6 | 824.61399 | 824.61535 | 36966.5 | 0.15 |
| P40:5 | 826.62964 | 826.62963 | 7231.2 | 0.11 |
| P40:4 | 828.64529 | 828.64033 | 93 | |
| D40:8 | 836.57761 | 836.57849 | 689543.3 | 2.21 |
| D40:7 | 838.59326 | 838.59503 | 389864.3 | 1.23 |
| D40:6 | 840.60891 | 840.60984 | 2019087 | 6.42 |
| D40:5 | 842.62456 | 842.62321 | 336970.3 | 1.09 |
| #D40:4 | 844.64021 | 844.63793 | 76823.4 | 0.11 |
| P42:7 | 850.62964 | 850.63145 | 141.4 | |
| D42:6 | 868.64021 | 868.64153 | 26286.1 | |
| D44:12 | 884.57761 | 884.57938 | 1708.5 | |
| D44:4 | 900.70281 | 900.70030 | 111.8 | |
In order to avoid the missing species caused by the double bond overlapping effects, a mass-searching window from −15 (−3 plus −12 ppm due to double bond overlapping effects) to +3 ppm of each theoretical mass should be used to search all the species. Unfortunately, accurate mass searching with such a wide mass window could result in a significant number of false positive hits (Table S1B). Much more peaks than that identified by MDMS-SL techniques were hit by this wide mass window, and those hits include most of interference peaks and some very low abundant PC species due to much higher sensitivity of Q-Exactive than TSQ mass spectrometer.
A novel accurate mass searching approach for identification of candidate species of a lipid class in high accurate mass shotgun lipidomics
To avoid the aforementioned issues (i.e., both false negative and positive hits), we developed a novel mass searching approach for high accurate mass shotgun lipidomics to identify the candidate species of a lipid class of interest.
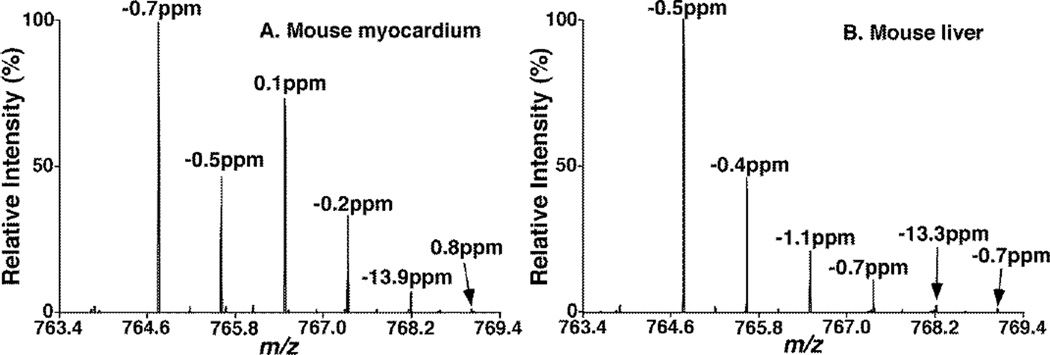
During the accurate mass search at the mass resolution of 75K, we found that the effects of double bond overlapping were much less for the peaks with 13C3 isotopologue (i.e. M+3) and the 13C isotopologue ions with one double bond less (i.e. L+1) in biological samples (Figure 5). For both the mouse myocardium and liver samples, 34:2 PC and 34:1 PC species had abundant amounts and their peaks showed mass shift in the range of ± 3 ppm. However, the intensity of the 34:0 PC peak was weak, and the double bond overlapping effect led to a mass shift of −13.9 ppm in the mouse myocardium (Figure 5A) and −13.3 ppm in the mouse liver sample (Figure 5B). These shifts were in the range of −15 ppm to 3 ppm, consistent with what we expected. On the other hand, their isotopologue peaks of 13C-34:0 PC in these mouse tissues showed little shifts at 0.8 and −0.7 ppm, respectively. Therefore, the plus one isotopologue peak of the species with one less double bond (i.e., L+1) could be used to assist the accurate mass search when this species has low mass level and is affected by the double bond overlapping at the mass resolution of 75K.
Figure 5.
Mass shift of individual peak of 34:2, 34:1 and 34:0 PC species in biological samples. Lipid extracts of mouse myocardium (A) and mouse liver (B) were prepared as described in the subsection of “Materials and Methods” and analyzed by using a Q-Exactive mass spectrometer with a mass resolution of 75K. All the corresponding ion peaks including their 13C isotopologues were indicated.
In the method, we searched the candidate species as a group in which all the species contained an identical number of carbon atoms, but with various numbers of double bonds. We first searched a species M of the group containing a possibly highest number of double bond in the group with a mass searching window ± 3 ppm of the theoretical mass of the interesting species. If a hit was found, we then checked the presence of its M+1 isotopologue also with the mass searching window of ± 3 ppm. If this species was validated with the presence of its M+1 isotopologue and the correct intensity ratio between the two isotope peaks according to the natural abundance within a ± 10% variation range, the intensity of this species M peak could be used for quantification. If its M+1 isotopologue did not exist or the variation of the intensity ratio is out of the range of ± 10%, this species was discarded. Then the search was repeated for the next species with one less double bond until one species was validated by the above criteria. Then, the same search was continued for species with one less double bond until the next species was missing in the mass window of ± 3 ppm. However, if the species L existed in the window between −15 and +3 ppm and the presence of its L+1 isotopologue (with one more 13C) was also validated in the mass range of ± 3 ppm with its intensity equal to or lower than the calculated value from the intensity of the searched L species with a 10% variation, this potential candidate species L was kept, and the intensity of its L+1 isotopologue was used to calculate the intensity of L species for quantification by comparing to the internal standard. If there were multiple candidates for L in the mass range between −15 and +3 ppm (Table S1B), the peak with the correct isotope intensity ratio to the searched L+1 within a ± 10% variation was recorded as the species L. The same searching procedure was repeated for the species with one less double bond until the acyl chains are saturated for this group. It was followed by the same search strategy on the group with more carbon numbers starting with the highest possible double bond until saturated acyl chains. We found that the majority of the false positive ions were eliminated after searching the presence of the M+1 isotopologues while the false negative results were avoided after considering the double bond overlapping effect.
We performed analysis of numerous lipid extracts from biological samples by using this approach and compared the obtained results to those identified by MDMS-SL (e.g., Table 1). We found that all the species identified by MDMS-SL were present in the dataset obtained from the accurate mass searching, indicating the elimination of false negative results. We also found that there existed many extra species in the low abundance in comparison to those identified by MDMS-SL (Table 1). This difference was likely resulted from the better sensitivity and dynamic range possessed by the high mass accuracy/resolution mass spectrometer (Q-Exactive) than that possessed by the nominal mass resolution instrument (TSQ), but these searched species still need to be confirmed by tandem MS spectra.
Finally, we could extend four points from the current study. First, MS/MS could be used to distinguish the double-bond overlapped ion peaks, and based on the acyl chain fragment ions, the overlapped peak could be M+2, L or both ions. However, since the two 13C isotopes in M+2 could be at the head group or the acyl chains, the intensities of acyl chain fragment peaks could not be used for quantification. Second, for identifying all of the searched species, MS/MS analyses still need to be performed in addition to the survey scan. All ion fragmentation (AIF) scans for a few mass ranges could be used to identify the searched species under the help of this novel approach to pick up the characteristic fragment ions for identification of class specified head groups and acyl chains. The composition of individual isomers or the acyl chain compositions/distributions could be calculated by their intensity ratios determined from the MS/MS analysis. Third, it would be more accurate to extract peak areas than to directly measure the peak heights due to the presence of peak broadening resulted from the double bond overlapping effect in high mass accurate shotgun lipidomics. Fourth, although accurate quantification is beyond the scope of this study and needs vigorous determination of linear dynamic range, repeatability, limit of quantification, etc., the dataset of the extracted peak areas has indeed set up a foundation for quantification of those candidate species determined through high accurate mass searching, particularly in combination with the deconvolution of 13C isotopologue patterns which were already conducted in our high accurate mass searching approach.
CONCLUSIONS
The double bond overlapping effect dominates the full scan mass spectra in high mass accurate shotgun lipidomics. We provided herein the evidence from both simulation and experimental data that (1) direct accurate mass searching of high resolution mass spectra could result in a number of false negative hits when a mass window of ± 3 ppm (in our experiences, most of peaks shift in this range with a mass lock) was used for searching and (2) substantial numbers of false positive species would be included if the mass searching window was opened to −15 to 3 ppm which included the mass shift due to the double bond overlapping. This complication has never been resolved[11, 13] and this report is the first to address this issue. At around m/z 750 range where the majority of lipid ions are detected by ESI-MS, with a mass resolution of 75K, although the double bond overlapping effects totally collapse the peaks of the M+2 isotopologue species and the L species (less a double bond than M) together and shift the peak of L species as high as −15 ppm, the new developed strategy of accurate mass search could still identify the collapsed L species peaks and acquire the corresponding peak intensities, which opens the door for lipidomics research by utilizing high mass accuracy/resolution mass spectra with relative low resolution requirements. Another point worth mentioning is that the actual resolution of the Q-Exactive is mass dependent. Lipids with lower m/z, such as nonesterified fatty acids and lysophospholipids, show better resolution at the same settings in Q-Exactive and could fit in this strategy as well or only need lower instrument resolution. However, lipids with higher m/z or doubly charges, such as cardiolipin and phosphatidylinositol phosphates, could require instruments with higher mass resolution to achieve this mass searching strategy. We also extended to point out that the double bond overlapping effect could also complicate the high mass accurate MS-based LC-MS efforts of lipidomics/metabolomics due to the difficult resolution of species differing only by one or two double bonds without extra efforts.
Supplementary Material
Acknowledgement
The authors give the special thanks to Drs. David Peake and Markus Kellmann for facilitating the collaboration between the Sanford-Burnham and Thermo Fisher Scientific, and Dr. Jens Grote for upgrading the prototype instrument control software. This work was partly supported by National Institute of General Medical Sciences Grant R01 GM105724 and intramural institutional research funds.
REFERENCES
- 1.Shevchenko A, Simons K. Lipidomics: Coming to Grips with Lipid Diversity. Nat. Rev. Mol. Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 2.Gross RW, Han X. Lipidomics at the Interface of Structure and Function in Systems Biology. Chem. Biol. 2011;18:284–291. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X, Yang K, Gross RW. Multi-Dimensional Mass Spectrometry-Based Shotgun Lipidomics and Novel Strategies for Lipidomic Analyses. Mass Spectrom. Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanksby SJ, Mitchell TW. Advances in Mass Spectrometry for Lipidomics. Annu. Rev. Anal. Chem. 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 5.Han X. Lipidomics: Developments and Applications. J. Chromatogr. B. 2009;877:2663. doi: 10.1016/j.jchromb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis EA. Lipidomics Joins the Omics Evolution. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2089–2090. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X. Neurolipidomics: Challenges and Developments. Front. Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie WW, Han X. Lipid Analysis: Isolation, Separation, Identification and Lipidomic Analysis. Bridgwater, England: The Oily Press; 2010. [Google Scholar]
- 9.Han X, Gross RW. Shotgun Lipidomics: Multi-Dimensional Mass Spectrometric Analysis of Cellular Lipidomes. Expert Rev. Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 10.Yang K, Cheng H, Gross RW, Han X. Automated Lipid Identification and Quantification by Multi-Dimensional Mass Spectrometry-Based Shotgun Lipidomics. Anal. Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwudke D, Hannich JT, Surendranath V, Grimard V, Moehring T, Burton L, Kurzchalia T, Shevchenko A. Top-Down Lipidomic Screens by Multivariate Analysis of High-Resolution Survey Mass Spectra. Anal. Chem. 2007;79:4083–4093. doi: 10.1021/ac062455y. [DOI] [PubMed] [Google Scholar]
- 12.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A. A Novel Informatics Concept for High-Throughput Shotgun Lipidomics Based on the Molecular Fragmentation Query Language. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwudke D, Schuhmann K, Herzog R, Bornstein SR, Shevchenko A. Shotgun Lipidomics on High Resolution Mass Spectrometers. Cold Spring Harb. Perspect. Biol. 2011;3:a004614. doi: 10.1101/cshperspect.a004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuhmann K, Almeida R, Baumert M, Herzog R, Bornstein SR, Shevchenko A. Shotgun Lipidomics on a Ltq Orbitrap Mass Spectrometer by Successive Switching between Acquisition Polarity Modes. J. Mass Spectrom. 2012;47:96–104. doi: 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho M, Sampaio JL, Palm W, Brankatschk M, Eaton S, Shevchenko A. Effects of Diet and Development on the Drosophila Lipidome. Mol. Syst. Biol. 2012;8:600. doi: 10.1038/msb.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papan C, Penkov S, Herzog R, Thiele C, Kurzchalia T, Shevchenko A. Systematic Screening for Novel Lipids by Shotgun Lipidomics. Anal. Chem. 2014;86:2703–2710. doi: 10.1021/ac404083u. [DOI] [PubMed] [Google Scholar]
- 17.Han X, Gross RW. Quantitative Analysis and Molecular Species Fingerprinting of Triacylglyceride Molecular Species Directly from Lipid Extracts of Biological Samples by Electrospray Ionization Tandem Mass Spectrometry. Anal. Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 18.Eibl G, Bernardo K, Koal T, Ramsay SL, Weinberger KM, Graber A. Isotope Correction of Mass Spectrometry Profiles. Rapid Commun. Mass Spectrom. 2008;22:2248–2252. doi: 10.1002/rcm.3591. [DOI] [PubMed] [Google Scholar]
- 19.Liebisch G, Lieser B, Rathenberg J, Drobnik W, Schmitz G. High-Throughput Quantification of Phosphatidylcholine and Sphingomyelin by Electrospray Ionization Tandem Mass Spectrometry Coupled with Isotope Correction Algorithm. Biochim. Biophys. Acta. 2004;1686:108–117. doi: 10.1016/j.bbalip.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Hermansson M, Uphoff A, Kakela R, Somerharju P. Automated Quantitative Analysis of Complex Lipidomes by Liquid Chromatography/Mass Spectrometry. Anal. Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- 21.Ogiso H, Suzuki T, Taguchi R. Development of a Reverse-Phase Liquid Chromatography Electrospray Ionization Mass Spectrometry Method for Lipidomics, Improving Detection of Phosphatidic Acid and Phosphatidylserine. Anal. Biochem. 2008;375:124–131. doi: 10.1016/j.ab.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 22.Retra K, Bleijerveld OB, van Gestel RA, Tielens AG, van Hellemond JJ, Brouwers JF. A Simple and Universal Method for the Separation and Identification of Phospholipid Molecular Species. Rapid Commun. Mass Spectrom. 2008;22:1853–1862. doi: 10.1002/rcm.3562. [DOI] [PubMed] [Google Scholar]
- 23.Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Alterations in Myocardial Cardiolipin Content and Composition Occur at the Very Earliest Stages of Diabetes: A Shotgun Lipidomics Study. Biochemistry. 2007;46:6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han X, Yang K, Gross RW. Microfluidics-Based Electrospray Ionization Enhances Intrasource Separation of Lipid Classes and Extends Identification of Individual Molecular Species through Multi-Dimensional Mass Spectrometry: Development of an Automated High Throughput Platform for Shotgun Lipidomics. Rapid Commun. Mass Spectrom. 2008;22:2115–2124. doi: 10.1002/rcm.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins and Membranes. Amsterdam: Elsevier Science B.V.; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.