Abstract
Background
We conducted a meta-analysis to assess the association between polymorphisms of GSTM1 null genotype and coronary artery disease (CAD) risk.
Material/Methods
Published literature from PubMed, EMBASE, and China National Knowledge Infrastructure (CNKI) were retrieved before March 2014. All studies reporting adjusted odds ratios (ORs) and 95% confidence intervals (CIs) of CAD risk were included.
Results
A total of 13 case-control studies, including 5453 cases and 5068 controls, were collected. There was a significant association between GSTM1 null genotype and CAD risk (adjusted OR=1.26; 95% CI, 1.11–1.43; I2=3%). When stratified by ethnicity, a significantly elevated risk was observed in whites. In the subgroup analysis according to disease type, a significantly increased myocardial infarction (MI) risk was observed. Subgroup analysis of smoking status showed an increased CAD risk in smokers.
Conclusions
Our results indicate that GSTM1 null genotype is associated with an increased CAD risk.
MeSH Keywords: Coronary Artery Disease; Glutathione Transferase; Meta-Analysis as Topic; Polymorphism, Genetic
Background
Coronary artery disease (CAD) is the leading health problem worldwide and is the leading cause of mortality in the United States. The role of DNA oxidative stress in the pathogenesis of atherosclerosis and its association with increased production of reactive oxygen species has been well established [1]. The mutagenic activities of cigarette smoke chemicals can cause DNA adducts in target tissues and oxidative modification and progression of atherosclerotic lesions. The glutathione S-transferases (GSTs) are a gene superfamily of phase II metabolic enzymes that detoxify free radicals, particularly in tobacco smoke [2]. GSTM1 has been mapped to the GST mu gene cluster on chromosome 1p13.3. One variant in GSTM1 have been identified – a deletion. This inactive form of GSTM1 (null genotype) causes lower detoxification, which may be a risk factor for CAD. The relationship between GSTM1 null genotype and risk of CAD has been studied for more than 10 years. Several studies found GSTM1 null genotype to be a risk factor in CAD, but other studies showed no association between this polymorphism and risk of CAD. These studies reached inconsistent conclusions [3–15], probably due to the relatively small sample sizes. Since individual studies are usually underpowered in detecting the effect of low-penetrance genes, in this study we conducted a meta-analysis to investigate the association between GSTM1 null genotype and the risk for CAD.
Material and Methods
Publication search
We conducted a literature search before February 2014 in PubMed, EMBASE, and Chinese National Knowledge Infrastructure (CNKI) databases without restrictions. Combination of the following terms were applied: ‘coronary heart disease’ OR ‘coronary artery disease’ OR ‘myocardial infarction’ OR ‘acute coronary syndrome’ OR ‘ischemic heart disease’ OR ‘cardiovascular disease’ OR ‘major adverse cardiac event’ OR ‘CHD’ OR ‘CAD’ OR ‘MI’ OR ‘ACS’ OR ‘IHD’ OR ‘MACE’; ‘Glutathione S-transferases’ OR ‘GSTM1; ‘polymorphism’ OR ‘variant’ OR ‘genetic’ OR ‘mutation’. We also conducted a manual search to find other articles based on references identified in the individual articles.
Inclusion criteria and data extraction
We included articles if they met all the following criteria: (1) evaluation of GSTM1 polymorphism and CAD risk, (2) using a case-control design, and (3) adjusted odds ratios (ORs) with 95% confidence intervals (CIs) were reported.
Data were extracted by 2 authors independently. In case of conflicting evaluations, an agreement was reached following a discussion; if agreement could not be reached, another author was consulted to resolve the debate. The following information was extracted from each study: first author, year of publication, ethnicity, age, sex, disease type, sample size, smoking status, covariates, adjusted Ors, and the corresponding 95% CIs of CAD risk.
Statistical analysis
For the GSTM1 gene, we estimated the risk of the null genotype on CAD compared with the non-null genotypes in the recessive model (null verses heterozygous + wild type). The strength of the association between the GSTM1 gene and CAD risk was measured by ORs with 95% CIs. The ORs with corresponding 95% CIs from individual studies were pooled using random-effects models. Heterogeneity between the studies was quantified using the Cochran Q test in combination with the I2 statistic, which represents the percentage of variability across studies that is attributable to heterogeneity rather than to chance. Heterogeneity among studies was considered significant when P was less than 0.1 for the Q-test or when the I2 value was greater than 50%. Subgroup analyses were stratified by ethnicity, disease type, smoking status, and covariates of adjustment. Cumulative meta-analysis was performed. Sensitivity analysis was further performed by excluding single studies sequentially to assess the impact of the individual study on the pooled estimate. Funnel plots and Egger’s regression test were undertaken to assess the potential publication bias [16]. Data analysis was performed using STATA 12 (StataCorp LP, College Station, Texas, USA).
Results
Study characteristics
We ultimately identified a total of 13 articles reporting the relationship between GSTM1 null genotype and CAD risk [3–15]. A total of 5453 cases and 5068 controls were included in this meta-analysis. Table 1 summarized the main characteristics of those included studies. There were 8 case-control studies from white populations and 5 case-control studies from Asian populations.
Table 1.
Characteristics of the case-control studies included in this meta-analysis.
| First author | Year | Race | Age | Sex | Type | Smoking | Case | Control | Covariate |
|---|---|---|---|---|---|---|---|---|---|
| Li | 2000 | White | 54 | Mixed | CAD | Mixed* | 400 | 890 | Age, sex, LDL, HDL, hypertension and diabetes |
| Masetti | 2003 | White | 61 | Mixed | CAD | Mixed* | 308 | 122 | Sex, dyslipidemia, hypertension, diabetes, and family history |
| Girisha | 2004 | Asian | 47 | Mixed | CAD | Mixed* | 59 | 132 | Age, sex, smoke, alcohol, diet, cholesterol, triglyceride, HDL, LDL, VLDL, apoprotein B |
| Tamer | 2004 | White | 51 | Mixed | CAD | Mixed* | 148 | 247 | Smoke |
| Cornelis | 2007 | White | 58 | Mixed | MI | Mixed | 2042 | 2042 | Smoke, waist-to-hip ratio, income, physical activity, history of diabetes and hypertension, intake of alcohol, and energy-adjusted saturated fat and folate |
| Manfredi | 2007 | White | 56 | Mixed | CAD | Smoker | 165 | 53 | Age, sex, dyslipidemia, hypertension, diabetes, and family history |
| Kim | 2008 | Asian | 60 | Mixed | CAD | Mixed* | 582 | 110 | Age, sex, hypertension, diabetes, body mass index, and lipid profile |
| Wang | 2008 | Asian | 62 | Mixed | CAD | Mixed* | 277 | 277 | Age, sex, smoke, dyslipidemia, hypertension, diabetes, body mass index |
| Kariž | 2012 | White | 62 | Mixed | MI | Mixed | 206 | 257 | Age, sex, smoke, BMI, duration of diabetes and lipid parameters |
| Lakshmi | 2012 | Asian | 68 | Mixed | CAD | Mixed | 352 | 282 | Age, sex, diabetes |
| Taspinar | 2012 | White | 62 | Mixed | CAD | Mixed* | 132 | 151 | Age, sex, smoke, diabetes and family history |
| Cora | 2013 | Caucasian | 62 | Mixed | MI | Mixed* | 324 | 296 | Age, sex, smoke, diabetes, hypertension, family history and lipid profile |
| Yeh | 2013 | Asian | 64 | Mixed | CAD | Mixed | 458 | 209 | Age, sex, smoke, alcohol use, diabetes mellitus, levels of serum total cholesterol and high-density lipoprotein cholesterol |
Information of smoking status can be extracted.
CAD – coronary artery disease; MI – myocardial infarction; LDL – low-density lipoprotein; HDL – high-density lipoprotein; VLDL – very low-density lipoprotein; BMI – body mass index.
Quantitative data synthesis
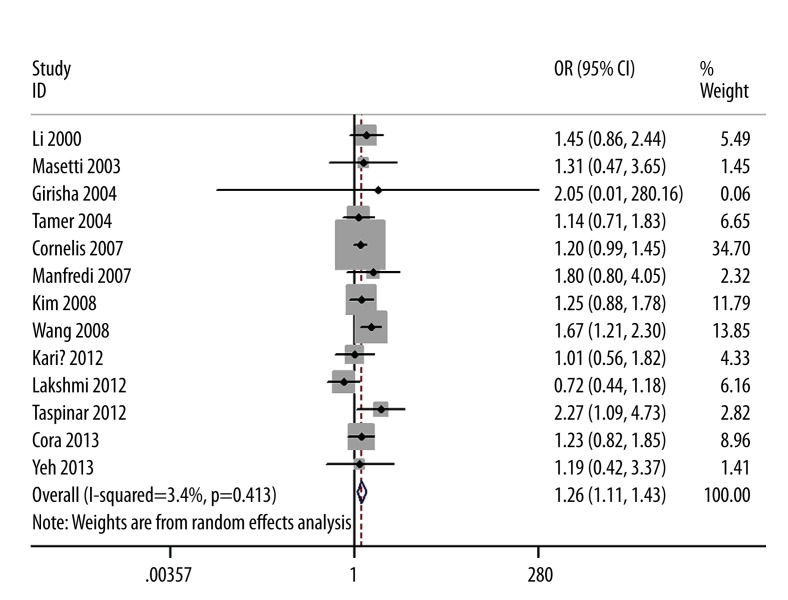
The evaluations of the association between GSTM1 polymorphism and CAD risk are summarized in Table 2. The null genotype of GSTM1 was associated with a significantly increased risk of CAD when compared with present genotype (adjusted OR=1.26; 95% CI 1.11–1.43; I2=3%; Figure 1). When stratified by ethnicity, a significantly elevated risk was observed in whites (OR=1.22; 95% CI 1.06–1.41; I2=0%) but not in Asians (OR=1.20; 95% CI 0.85–1.71; I2=50%). In the subgroup analysis according to disease type, a significantly increased myocardial infarction (MI) risk was observed (OR=1.19; 95% CI 1.01–1.40; I2=0%). Subgroup analysis of smoking status showed that increased risks were found in smokers (OR=1.97; 95% CI 1.59–2.44; I2=4%) but not in non-smokers (OR=1.20; 95%CI, 0.96–1.51; I2=0%). When we limited the meta-analysis to studies that controlled for confounders such as age, sex, smoking, diabetes, hypertension, family history, and dyslipidemia, a significant association between GSTM1 null genotype and CAD risk remained.
Table 2.
Results of meta-analysis.
| Test of association | Heterogeneity | |||
|---|---|---|---|---|
| OR (95% CI) | P value | P value | I2 (%) | |
| Overall | 1.26 (1.11–1.43) | <0.01 | 0.41 | 3.0 |
| Asian | 1.20 (0.85–1.71) | 0.30 | 0.09 | 50.0 |
| Caucasian | 1.22 (1.06–1.41) | <0.01 | 0.97 | 0.0 |
| MI | 1.19 (1.01–1.40) | 0.04 | 0.85 | 0.0 |
| Smoker | 1.97 (1.59–2.44) | <0.01 | 0.40 | 4.0 |
| Non-smoker | 1.20 (0.96–1.51) | 0.11 | 0.63 | 0.0 |
| Adjusted for | ||||
| Age and sex | 1.31 (1.07–1.59) | <0.01 | 0.23 | 23.0 |
| Smoke | 1.29 (1.12–1.48) | <0.01 | 0.50 | 0.0 |
| Diabetes and hypertension | 1.31 (1.15–1.50) | <0.01 | 0.69 | 0.0 |
| Family history | 1.46 (1.07–1.99) | 0.02 | 0.50 | 0.0 |
| Dyslipidemia | 1.25 (1.03–1.51) | 0.03 | 0.32 | 13.0 |
MI – myocardial infarction.
Figure 1.
Forest plot for the association between CAD risk and GSTM1 null genotype.
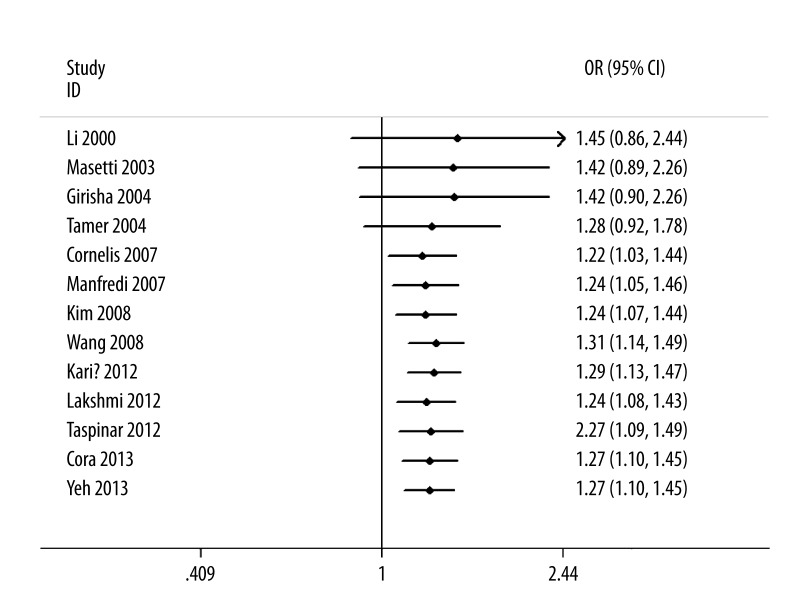
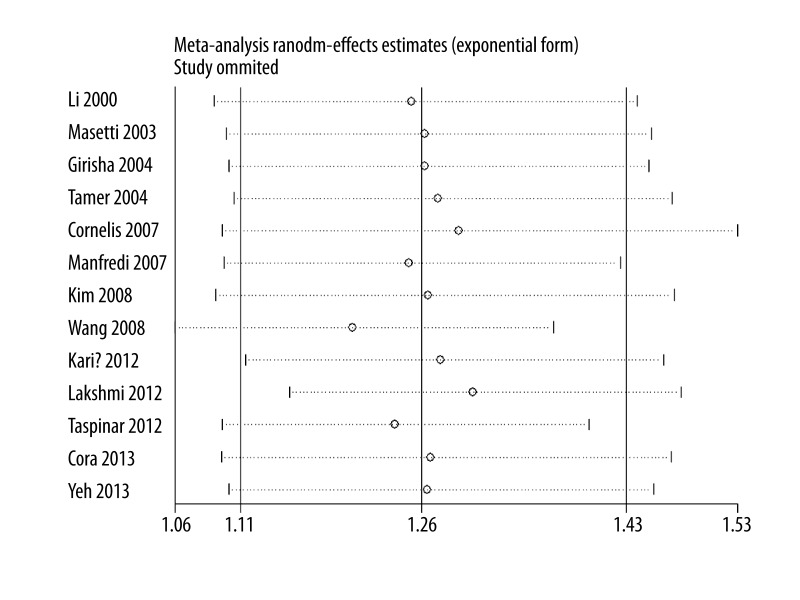
As shown in Figure 2, significant associations were evident with each addition of more data over time. The results showed that the pooled ORs tended to be stable. A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs, and the corresponding pooled ORs were not materially altered (Figure 3).
Figure 2.
Cumulative meta-analysis for the association between CAD risk and GSTM1 null genotype.
Figure 3.
Sensitivity analysis for the association between CAD risk and GSTM1 null genotype.
Funnel plot and Egger’s test were performed to assess the publication bias of the literature. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (data not shown). Egger’s test did not find evidence of publication bias (P=0.68).
Discussion
The present meta-analysis, including 5453 cases and 5068 controls from 13 case-control studies, explored the associations of GSTM1 null genotype with CAD risk. We demonstrated that this polymorphism is significantly associated with an increased CAD risk. Subgroup analyses stratified by ethnicity showed whites, but not Asians, with GSTM1 null genotype had increased CAD risk. It was possible that different lifestyles, diets, and environments may account for this discrepancy. In the MI subgroup, we found that this polymorphism was associated with MI risk. Cigarette smoking is a pro-inflammatory stimulus and an important risk factor for CAD. Some studies have explored the interaction between GSTM1 genotype and smoking habits. Our results showed a significant association among smokers but not in non-smokers, suggesting that even the same variant in the same gene may have a different effect on the pathogenesis and occurrence of CAD in different individuals.
Previous studies have shown that individuals with GSTM1 null genotype have a decreased capacity to detoxify certain carcinogens. Thus, impaired GSTM1 function may lead to serious DNA damage. Toxic molecules produce DNA adducts that contribute to the development of atherosclerosis. Evidence indicates that the interaction of DNA adducts with DNA may trigger pathogenic pathways in the cell. Significant correlation was found between DNA adduct levels, which are accepted as a biomarker of exposure to environmental carcinogens, and atherogenic risk factors. Higher DNA adduct levels were detected in individuals with severe CAD [17,18] and in atherosclerotic plaques [19]. Therefore, it is biologically plausible that the GSTM1 null genotype may increase risk of CAD.
Our study has some strengths. First, it was the first meta-analysis to report the adjusted ORs between GSTM1 null genotype and CAD risk. Second, the methodological issues for meta-analysis (e.g., subgroup analysis, cumulative meta-analysis, and sensitivity analysis) were well investigated. Third, when we limited the meta-analysis to studies that controlled for age and sex, smoking, diabetes and hypertension, family history, dyslipidemia, the significant positive association was only marginally altered. Finally, we did find significant heterogeneity and publication bias in this meta-analysis.
Some limitations in this meta-analysis should be addressed. First, the number of studies included in our meta-analysis remained small. Thus, publication bias may exist, although the funnel plots and Egger’s linear regression tests indicated no remarkable publication bias. Second, lack of the original data of the eligible studies limited the evaluation of the effects of the gene-environment interactions in CAD development. Third, no prospective studies have addressed this association between GSTM1 null genotype and CAD risk, and all included studies followed a retrospective case-control design. Thus, owing to the limitations of case-control design, we cannot exclude the possibility of undetected bias.
Conclusions
This meta-analysis supports an association between GSTM1 null genotype and risk of CAD. Prospective studies are suggested to further ascertain the relationship between GSTM1 null genotype and genetic predisposition to CAD.
Footnotes
Source of support: Self financing
Declaration of interest statement
The authors declare that they have no competing interests.
References
- 1.Harrison D, Griendling KK, Landmesser U, et al. Role of oxidative stress in atherosclerosis. Am J Cardiol. 2003;91:7–11. doi: 10.1016/s0002-9149(02)03144-2. [DOI] [PubMed] [Google Scholar]
- 2.Jana S, Mandlekar S. Role of phase II drug metabolizing enzymes in cancer chemoprevention. Curr Drug Metab. 2009;10:595–616. doi: 10.2174/138920009789375379. [DOI] [PubMed] [Google Scholar]
- 3.Li R, Boerwinkle E, Olshan AF, et al. Glutathione S-transferase genotype as a susceptibility factor in smoking-related coronary heart disease. Atherosclerosis. 2000;149:451–62. doi: 10.1016/s0021-9150(99)00483-9. [DOI] [PubMed] [Google Scholar]
- 4.Masetti S, Botto N, Manfredi S, et al. Interactive effect of the glutathione S-transferase genes and cigarette smoking on occurrence and severity of coronary artery risk. J Mol Med (Berl) 2003;81:488–94. doi: 10.1007/s00109-003-0448-5. [DOI] [PubMed] [Google Scholar]
- 5.Girisha KM, Gilmour A, Mastana S, et al. T1 and M1 polymorphism in glutathione S-transferase gene and coronary artery disease in North Indian population. Indian J Med Sci. 2004;58:520–26. [PubMed] [Google Scholar]
- 6.Tamer L, Ercan B, Camsari A, et al. Glutathione S-transferase gene polymorphism as a susceptibility factor in smoking-related coronary artery disease. Basic Res Cardiol. 2004;99:223–29. doi: 10.1007/s00395-004-0465-8. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis MC, El-Sohemy A, Campos H. GSTT1 genotype modifies the association between cruciferous vegetable intake and the risk of myocardial infarction. Am J Clin Nutr. 2007;86:752–58. doi: 10.1093/ajcn/86.3.752. [DOI] [PubMed] [Google Scholar]
- 8.Manfredi S, Federici C, Picano E, et al. GSTM1, GSTT1 and CYP1A1 detoxification gene polymorphisms and susceptibility to smoking-related coronary artery disease: a case-only study. Mutat Res. 2007;621:106–12. doi: 10.1016/j.mrfmmm.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Kim SJ, Kim MG, Kim KS, et al. Impact of Glutathione S-Transferase M1 and T1 Gene Polymorphisms on the Smoking-Related Coronary Artery Disease. J Korean Med Sci. 2008;23:365–72. doi: 10.3346/jkms.2008.23.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LS, Tang JJ, Tang NP, et al. Association of GSTM1 and GSTT1 gene polymorphisms with coronary artery disease in relation to tobacco smoking. Clin Chem Lab Med. 2008;46:1720–25. doi: 10.1515/CCLM.2008.353. [DOI] [PubMed] [Google Scholar]
- 11.Kariž S, Nikolajević Starčević J, Petrovič D. Association of manganese superoxide dismutase and glutathione S-transferases genotypes with myocardial infarction in patients with type 2 diabetes mellitus. Diabetes Res Clin Pract. 2012;98:144–50. doi: 10.1016/j.diabres.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Lakshmi SV, Naushad SM, Saumya K, et al. Role of CYP1A1 haplotypes in modulating susceptibility to coronary artery disease. Indian J Biochem Biophys. 2012;49:349–55. [PubMed] [Google Scholar]
- 13.Taspinar M, Aydos S, Sakiragaoglu O, et al. Impact of genetic variations of the CYP1A1, GSTT1, and GSTM1 genes on the risk of coronary artery disease. DNA Cell Biol. 2012;31:211–18. doi: 10.1089/dna.2011.1252. [DOI] [PubMed] [Google Scholar]
- 14.Cora T, Tokac M, Acar H, et al. Glutathione S-transferase M1 and T1 genotypes and myocardial infarction. Mol Biol Rep. 2013;40:3263–67. doi: 10.1007/s11033-012-2401-6. [DOI] [PubMed] [Google Scholar]
- 15.Yeh HL, Kuo LT, Sung FC, et al. GSTM1, GSTT1, GSTP1, and GSTA1 genetic variants are not associated with coronary artery disease in Taiwan. Gene. 2013;523:64–69. doi: 10.1016/j.gene.2013.02.052. [DOI] [PubMed] [Google Scholar]
- 16.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:6296–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Flora S, Izzotti A, Walsh D, et al. Molecular epidemiology of atherosclerosis. FASEB J. 1997;11:1021–103. [PubMed] [Google Scholar]
- 18.Van Schooten FJ, Hirvonen A, Maas LM, et al. Putative susceptibility markers of coronary artery disease: association between VDR genotype, smoking and aromatic DNA adduct levels in human right atrial tissue. FASEB J. 1998;12:1409–17. doi: 10.1096/fasebj.12.13.1409. [DOI] [PubMed] [Google Scholar]
- 19.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–9. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]