Abstract
Separase, an enzyme that cleaves the chromosomal cohesin during mitosis, is overexpressed in a wide range of human epithelial cancers of breast, bone and prostate [Meyer et al, 2009, Clin Cancer Res 15(8): 2703-2710] [1]. Overexpression of Separase in animal models results in aneuploidy and tumorigenesis. We have examined the expression and localization of Separase protein in adult and pediatric glioblastoma and normal brain specimens. Immunofluorescence microscopy and Western blot analysis showed significant overexpression of Separase in all adult and a subset of pediatric glioblastoma cells. Tumor status and patient survival strongly correlate with the mislocalization of Separase into the nucleus throughout all stages of the cell cycle. Unlike exclusively nuclear localization in mitotic control cells, glioblastoma samples have a significantly higher number of resting (interphase) cells with strong nuclear Separase staining. Additionally, patient survival analysis demonstrated a strong correlation between overexpression of Separase protein in adult glioblastoma and a high incidence of relapse and reduced overall survival. These results further strengthen our hypothesis that Separase is an oncogene whose overexpression induces tumorigenesis, and indicate that Separase overexpression and aberrant nuclear localization are common in many tumor types and may predict outcome in some human malignancies.
Keywords: Separase, Espl1, GBM
Introduction
Cohesin, an evolutionarily conserved protein complex consisting of four proteins, Smc1, Smc3, Rad21 and Stag1 or Stag2 (SA1/SA2), plays a crucial role in chromosomal segregation. The importance of chromosomal cohesion and separation in tumorigenesis has become increasingly evident ([2-5]. Mutations of cohesin subunits have been reported in various human tumors. For example, STAG2 is mutated in 20% of glioblastoma cases [3], and multiple components of the cohesin complex, including STAG2, RAD21, SMC1 and SMC3, in different myeloid neoplasms [6]. The cohesin complex holds two newly replicated sister chromatids together from S-phase until metaphase producing the accurate segregation of sister chromatids into two daughter cells. At the beginning of anaphase, Separase, an endopeptidase, is activated and cleaves the cohesin subunit Rad21 (also known as Scc1 or Mcd1 in budding yeast), which then releases sister chromatids with consequent chromosome disjunction. Because Separase activity is critical to the smooth progression to cytokinesis, it is very tightly regulated via several mechanisms to ensure timely cleavage of cohesin Rad21 during metaphase to anaphase transition [7-9]. Separase, normally a cytoplasmic protein, is segregated from the nucleus until mitosis. Following the breakdown of the nuclear envelop, at the metaphase to anaphase transition, Separase enzyme is activated after its inhibitory chaperone Securin is degraded by anaphase promoting complex/cyclosome (APC/c)-mediated ubiquitination and proteasome targeted degradation [10-14]. Activated Separase then resolves the cohesed chromatids by cleaving the Rad21 subunit of the cohesin ring. Via an additional mechanism, Separase activity is inhibited by Cdk1/cyclin B-mediated phosphorylation of Separase at S1126 and Thr1346 residues [15-17]. Despite these innate control mechanisms, overexpression and constitutive nuclear localization of Separase has been reported in many human tumors, including breast, prostate and osteosarcoma [1]. Overexpression of Separase in animal models has been shown to cause chromosomal missegregation and aneuploidy [1, 18] resulting in tumorigenesis [19] . However, the detailed landscape of Separase overexpression in other human tumors and the mechanisms of Separase overexpression-driven tumorigenesis have not been fully investigated.
Here we report that Separase overexpression and mislocalization to non-mitotic nuclei is a marker for more aggressive and high-grade adult and pediatric glioblastomas and can be considered as a potential predictor of progression-free survival and relapse in these patients. This study underscores the importance of further investigation into targeting Separase for the early detection and treatment of Separase-overexpressing tumors including glioblastoma.
Materials and Methods
Tissue arrays
Arrays for adult glioblastoma and normal brain specimens were obtained from the MD Anderson Cancer Center tissue repository using an IRB approved protocol. Scoring was done by taking pictures on a Nikon eclipse E800 microscope of three randomly selected areas from any given tissue, which were analyzed by two different investigators, the second being uninformed of the origin of the sample and the first scoring result. In each field, at least 100 cells were counted and evaluated for the magnitude of Separase expression determining the propensity (on a scale of 0-5) and the intensity (on a scale of 0-3) scores for a total of 300 cells per tissue spot as previously described [20]. Briefly, the level of expression for Separase was quantified using the Allred score, [20] used widely for setting the staining threshold and quantification of immunohistochemical staining of cancer specimens. After reviewing the entire immunostained slide, a proportion score (PS) is assigned representing the estimated proportion of positive staining target cells (0 = none; 1 > 1/100th; 2 = 1/100th to 1/10th; 3 = 1/10th to 1/3rd; 4 = 1/3rd to 2/3rd; 5 > 2/3rd). Then an intensity score (IS) is assigned representing the estimated average intensity of positive target cells (0 = none; 1 = weak; 2 = intermediate; 3 = strong). The PS and IS are then added to obtain a total score (TS; i.e., the Allred Score), ranging from 0 to 8. This system is fast, easy to learn, and highly reproducible.
Tumor specimen
A set of anonymized human glioblastoma tissues (both adult and pediatric) with matched normal brain tissues were obtained from the tissue repository of the Methodist Hospital and Texas Children's Cancer and Hematology Center using appropriate IRB approved protocols. All tumors were harvested from the patient by a neuropathologist and quality-controlled to ensure the nature and the neoplastic cell composition and histology in each specimen. Each tumor specimen contained at least 85% tumor cells. All tissues in this study were obtained after institutional review board-approved informed consents.
Immunofluorescence microscopy
Human tissue arrays and paraffin-embedded samples were baked for 2 h at 60°C and deparaffinized in xylene for 2 × 10 min, respectively. Following stepwise rehydration in 100%, 95%, 80%, 75%, and 30% ethanol for 10 min each and 2 × 5 min in deionized water, antigen retrieval was done in a pressure cooker at 121°C for 20 min in buffer (14.5 mL of 0.1 mol/L citric acid monohydrate + 61.5 mL of 0.1 mol/L sodium citrate in 750 mL double-distilled H2O at pH 6.0). Nonspecific binding was blocked with 10% normal goat serum in phosphate buffered saline (PBS) at room temperature for 3 h before incubation with primary anti-Separase antibody (Abnova, Taiwan, Cat# ESPL1-6H6) and normal mouse IgG as control antibody overnight at 4°C in 10% normal goat serum/PBS in a humidified chamber. After rinsing with PBS on a shaker for 4 × 15 min, incubation with secondary antibody (goat anti-mouse Alexa Fluor 488 or 594) was done in 10% normal goat serum/PBS in a humidified chamber at room temperature for 1.5 h. To detect the proliferation status of the cells in the tissue array, a set of slides were counterstained with Ki-67 antibody (Vector Labs) followed by a secondary goat anti-rabbit Alexa Fluor antibody (Molecular Probes) for detection. After PBS rinse on a shaker 3 × 10 min, slides were mounted using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) and kept at -20°C if not immediately imaged on a Nikon eclipse E800 microscope as previously described [1]. In brief, the images were processed using background subtraction to remove shading due to non-uniform illumination and inhomogeneous staining effects and using color compensation to minimize the effects of spectral bleed-through among the three-color channels (red, green, and blue)
Statistical analysis
Immunofluorescence data for the normal brain and glioblastoma samples were analyzed for the presence of Separase and Ki-67. For each sample, the average expression levels and localization in the 100 cells evaluated in each of the three randomly selected microscopy spots were calculated. The magnitude of expression was evaluated using either PS and IS or using the combined TS. Due to the high degree of correlation between PS and IS in the observed data, combining the two into TS did not have a significant effect on the inferences drawn from the analysis. Due to the binary nature of the scoring, a Fisher's exact test was used to analyze statistical significance of the distribution of Separase expression. A log-rank (Mantel-Cox) test was used to calculate significance in overall and progression-free survival in patients corresponding to their Separase levels.
Results
Immunofluorescence analysis of tumor tissue for Separase expression and localization
Paraffin-embedded tissue sections of human adult glioblastoma and normal brain sections were used to assay levels of Separase expression and its cellular localization (Fig. 1). All sections were pathologically confirmed to be glioblastoma with varying features of the disease including high levels of tumor hemorrhage and microvascular proliferation (Fig.1, A, B), and tumor necrosis (Fig. 1, C) compared to normal brain (Fig. 1, D). Using Separase antibody, we performed immunofluorescence microscopy on individual patient tumor tissue sections as well as tumor tissue microarrays with appropriate normal brain tissue controls (Fig. 2). The slides were also counterstained with Ki-67 antibody to distinguish Separase staining in non-proliferating cells versus proliferating cells. Separase expression was scored according to the Allred scoring system with a PS ranging from 0 to 5 and an IS from 0 to 3 [20]. Adding the PS and IS yielded a TS used for statistical analysis of the data. A 2×2 contingency result was used to run the Fisher's exact test to estimate the statistical significance of the distribution of Separase expression. The p-value estimated was 1.546E-18 implying extreme significance of Separase expression in tumor versus normal brain tissue (Supplemental Table 1).
Figure 1. H&E staining of glioblastoma sections from human patients.
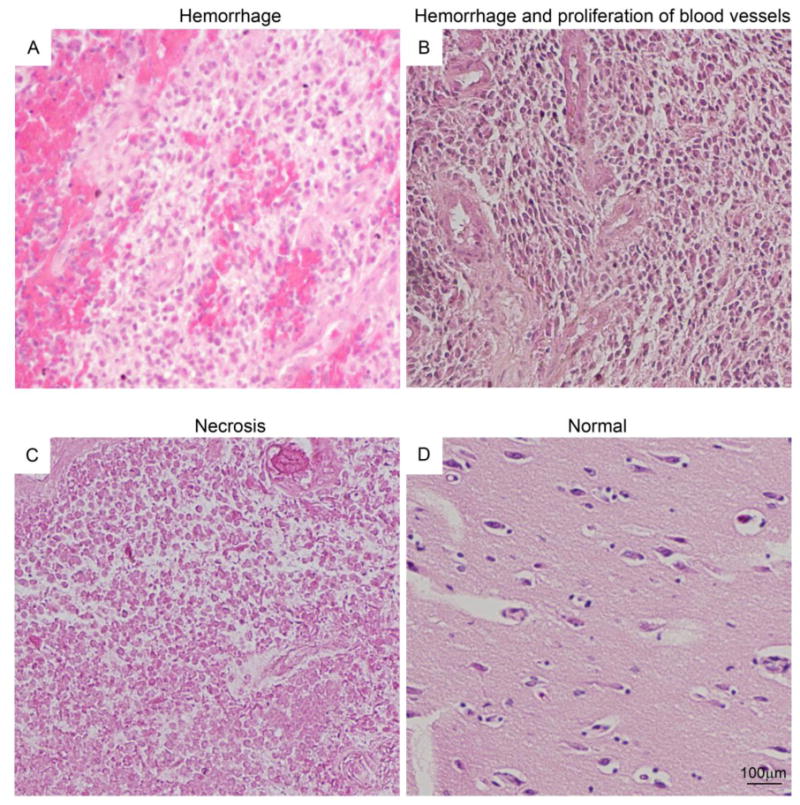
Glioblastoma samples show large areas of hemorrhage (A), proliferating blood vessels (B) and necrotic regions (C) compared to a normal brain (D).
Figure2. Immunofluorescence staining shows overexpression of Separase in glioblastoma tissue sections.
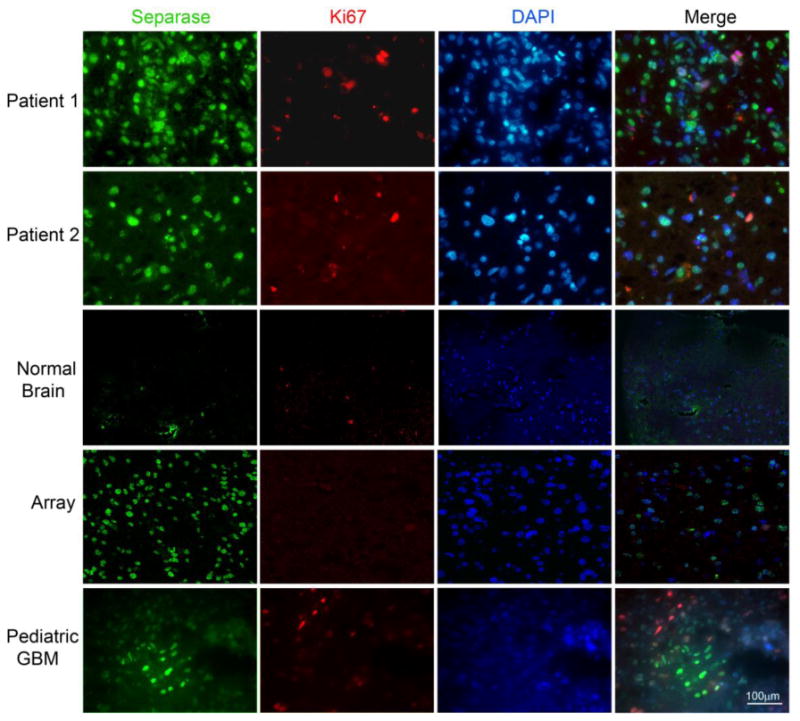
Separase staining (green) showed a higher percentage of positive cells, not all of which were proliferative (shown by Ki-67 staining in red). Normal brain sections stained for the same markers showed only background level of staining. Pediatric glioblastoma shows clusters of positive Separase expression in non-proliferating cells (bottom panel).
Immunofluorescence data for the glioblastoma and normal brain arrays were analyzed for Separase and Ki-67 and their mutual predictability pattern, if any. The results of Separase and Ki-67 expression were coded into a contingency table (Supplemental Table 2). To estimate the extent of association between them, the marginal distributions were calculated; the phi coefficient was estimated at 0.254, implying that the correlation between the two markers in this data set was low, but statistically significant (p-value = 0.024). Binary logistic regression run on these data did not imply any statistical significance (G-test = 0.072, p-value = 0.788). Repeating the same tests on the smaller data set of individual patient samples with survival data, (n=10, Supplemental Table 3), yielded a phi coefficient of 0.654, indicating a higher correlation between Separase and Ki-67 in this set of samples. Binary logistic regression analysis of these data was found to be significant (G-test = 5.487, p-value = 0.019). Ki-67 was not detected in samples with normal Separase expression. When Separase was overexpressed, Ki-67 was detected in 70.5% of adult glioblastoma specimens and in 71.4% of samples with survival data. While our smaller dataset is not sufficiently powered to make conclusions about mutual predictability between Separase and Ki-67, the contingency tables summarizing the relationship between these genes follow the same distribution. To demonstrate this, we carried out a numerical simulation, selecting 10,000 samples of size 10 from the larger sample of 38 glioblastoma tumors. Our 10-sample dataset included 5 samples with overexpressed Separase and where Ki-67 was detected by staining, while randomly chosen same-size datasets from our larger 38-sample dataset had an average of 5.1 samples that were both positive for Separase overexpression and Ki-67, with a standard deviation of 1.1. The 0.13 standard-deviation distance between results derived from our 10-sample dataset and expectation resulting from the analysis of our 38-sample dataset suggests that the distributions are identical with respect to mutual predictability between Separase and Ki-67.
To evaluate Separase misexpression, we scored Separase expression stratified by localization to cytoplasm, nucleus, or both compartments. Separase was primarily localized to the nuclei in glioblastoma cells with cytoplasmic expression observed in only a small fraction of the samples (Fig. 2, Patient 1,2). In normal brain tissue no Separase expression was observed. Expression of Separase protein was also tested in a small set of three pediatric glioblastomas obtained as xenografts. Unlike adult glioblastoma, pediatric glioblastoma had small clusters of cells with nuclear Separase expression (Fig. 2, bottom panel).
Western Blot Analysis
To examine levels of Separase expression in glioblastoma tissue, Western blotting was performed on protein extracted from tumors. For comparison, normal brain white and grey matter was used. Our results showed that Separase was significantly overexpressed (up to thirty-fold increase) in several of the glioblastoma patient samples compared to normal-like white matter (NLWM) from brain used as a control (Fig. 3, A-D). Using both Western blot and immunofluorescence within this small set of samples, Separase expression correlated with progression-free survival; higher Separase levels correlated with a shorter total survival time (p=0.0003 log-rank Mantel-Cox test) and a shorter progression-free survival (Fig. 3, B, p=0.0003 log-rank Mantel-Cox test).
Figure 3. Western Blot shows overexpression of Separase protein in primary adult glioblastoma tissues (n=23) (A-D) that correlates with decreased overall survival (E) and decreased time for progression-free survival (F) of patients.
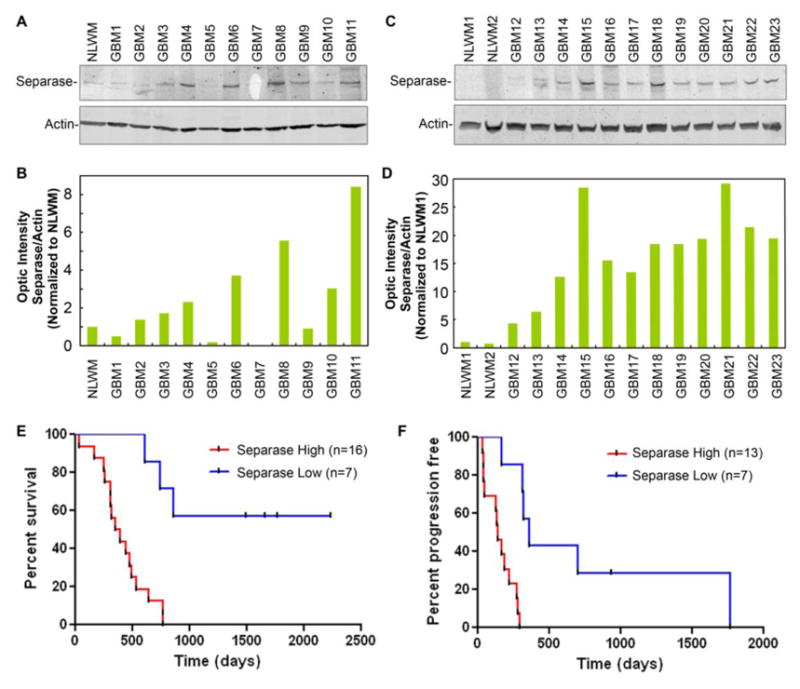
(p= 0.0003 for overall survival and p= 0.0003 for progression-free survival). Quantification of Western blotting data shows a 2-30 fold increase in Separase levels in glioblastoma specimens (B, D).
Primary Cell Analysis
To investigate the significance of Separase localization in glioblastoma cells further, primary cells obtained from the tumors used in the studies described above in Figure 3 (n=7 patients and two established glioblastoma cell lines, Daoy and U87) were briefly cultured (2-3 days at passage 0 or 1). Cells were then fixed and stained for Separase, Phospho-Histone H3 (PH3), a commonly used marker for mitotic cells, GFAP (glial fibrillary acidic protein) and Nestin (two markers for neuroprogenitor cells) and VEGF (vascular endothelial growth factor). Separase was frequently found to be localized in non-PH3 positive (non-mitotic) nuclei of primary cells in these tumors and also co-localized in the nucleus as well as cytoplasm with GFAP, a marker for glioblastomas with a precursor (stem cell-like) signature as well as with Nestin (a marker for a progenitor population of glial cells) (Fig. 4). Finally, overexpression of VEGF and intracellular VEGF-positive connections were frequently observed in Separase overexpressing glioblastoma primary cells compared to established glioblastoma cell lines such as like U87 (Fig. 4).
Figure 4. Immunofluorescence images show overexpression of Separase protein in primary glioblastoma cells.
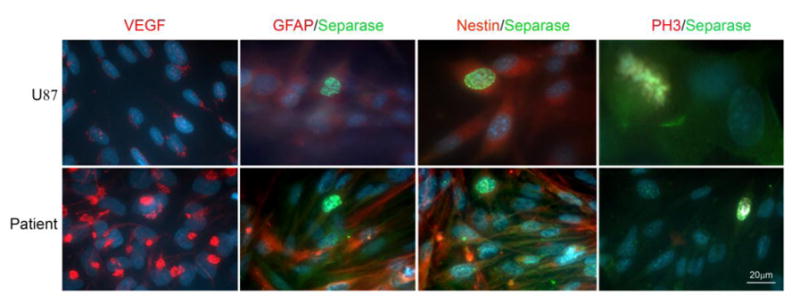
Separase is expressed in nuclei and cytoplasm of cells that are also positive for GFAP and Nestin. Separase expression is detected in nuclei of non-mitotic (PH3 negative) primary glioblastoma cells. VEGF expression is also abundant in these glioblastoma cells.
Analysis of the Cancer Genome Atlas (TCGA) glioblastoma data [21] indicates that ESPL1 transcripts, that encode Separase protein, are overexpressed across all molecular categories of adult glioblastoma (Fig. 5, B). However, ESPL1 expression is significantly higher when IDH1 is mutated and not significantly differentially expressed (p<0.05) (Fig. 5, C). There is also a significant correlation between ESPL1 transcript levels and overall patient survival, as shown by Kaplan-Meier survival curves that show that probability of reduced survival in REMBRANDT [22] patients with up-regulated ESPL1 mRNA expression is highly significant (p=3.81923E-5) (Fig. 5,A).
Figure5.
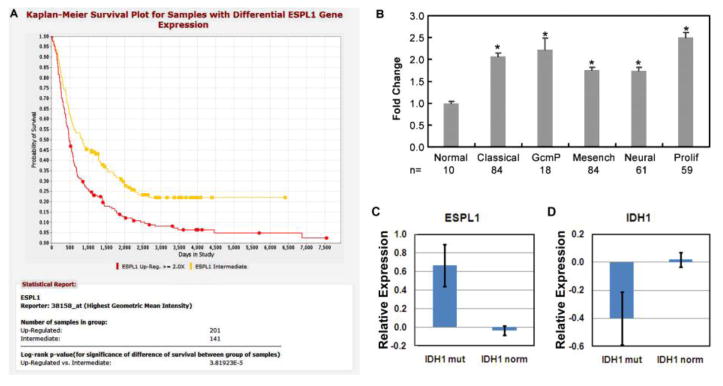
Kaplan-Meier survival plot analysis of human patients overexpressing the ESPL1 gene encoding Separase protein shows a significantly reduced survival corresponding to Separase expression (A, NCI Rembrandt data set). ESPL1 is also found to be significantly overexpressed in several subsets of glioblastoma (B, TCGA data set), including classical, proneural G-CIMP (GcmP), Mesenchymal (Mesench), Neural and Proliferative (Prolif) compared to normal brain. ESPL1overexpression correlates with increased IDH1 mutation status in GBMs (p<0.05) while IDH1 is not significantly differentially expressed in any GBM subtype (mRNA levels of mutated and normal IDH1 remain the same).(C, TCGA and REMBRANDT data set).
Discussion
The role of Separase in human brain tumors has not been investigated. Here we have examined the expression levels and cellular localization of Separase in a set of 23 individual adult and three pediatric glioblastoma patients as well as in a tissue microarray consisting of 38 individual adult glioblastoma and 38 normal brain sections. We found that Separase protein is overexpressed and constitutively mislocalized in ∼70% of the glioblastoma specimens tested. Additionally the level of Separase protein expression as well as its nuclear localization in non-mitotic cells correlated with patient outcomes. This result is consistent with our finding in tumor transcriptomes of gliomas (Fig. 5), breast, prostate and bone cancers [1] of strong positive correlation between Separase mRNA expression and tumor grade and a strong negative correlation with disease-free and overall survival.
Separase overexpressing glioblastoma primary cell lines showed a higher expression of VEGF, suggesting an increased potential for invasiveness. Separase expression was found in adult glioblastoma cells both in the cytoplasm and nucleus and was frequently co-localized with the expression of GFAP and Nestin, two markers for neural progenitor cells [23, 24] .
Similar to our previous study [1], addressed the question of whether stronger Separase staining in tumor tissue correlates solely with increased proliferation status of the tumor, we counterstained glioblastoma tissue array samples and cultured cells with the proliferation marker Ki-67 and Separase. Most tumor tissues showed that high Separase expression is constitutively nuclear regardless of the proliferative status of the cells. Additional studies using a stable Separase overexpressing HeLa clone and a Tet-inducible diploid, non-tumorigenic mouse mammary epithelial cell line, FSK3, indicated that Separase nuclear localization correlates with its overexpression irrespective of proliferative status [1, 18], suggesting that overexpression of Separase may contribute to its aberrant localization to nucleus.
The mechanistic significance of nuclear Separase localization is unclear, but there are several possible explanations. First, it is possible that the normal mechanism of active nuclear exclusion of Separase may be overwhelmed by Separase overexpression. Second, export of Separase from the nucleus of proliferating tumor cells may be inefficient owing to Separase overexpression. Third, a high Separase expression level and its nuclear localization may poise cells for division. Finally, it is known that cohesin is recruited to damaged sites along chromosomes during repair, and it is removed by Separase following DNA repair [25, 26]. Hence, nuclear retention of Separase in proliferating tumor cells could result in premature removal of cohesin, a process normally occurring only after the repair process is complete. Premature cohesion removal would enhance mutation defects in the tumor DNA-damage response.
Although Separase enzyme activity is tightly regulated during cell-cycle progression, its transcriptional expression remains constant. How overexpression and nuclear localization are connected or contribute to tumor formation/progression is not fully understood. In mouse mammary epithelial cells, transcriptional regulation of Separase expression is regulated by estrogen and progesterone, and Separase expression is further facilitated by loss of p53 [27]. The observation that Separase overexpression and loss or mutations of p53 are strongly correlated in breast cancers might not be coincidental. These findings strengthen the hypothesis that misregulation of sister chromatid cohesion and segregation and resultant aneuploidy could be a strong driving force for tumorigenesis and/or tumor progression. Incidentally, glioblastoma is a highly aneuploid cancer with frequent mutations in p53 [28]. A role of Separase in this disease has not been previously investigated and future studies present the possibility of using this protein as a diagnostic marker for the disease.
To correlate Separase expression to IDH1 mutations in GBMs, we have analyzed the status of IDH1 (mutant vs. WT) in relation to ESPL1 expression from the TCGA and REMBRANDT datasets. We found that ESPL1 gene expression is significantly increased in IDH1 mutated GBMs (p= <0.05). As IDH1 is also mutated in the G-CIMP (glioma CpG island methylator phenotype) subtypes of human GBMs, this may also imply that ESPL1 expression is significantly increased in G-CIMP GBMs. These correlations further strengthen the case that Separase is an important predictor and marker for adult GBM outcome.
The average age of the adult glioblastoma patients in this study was 57 years and 6 years for the pediatric patients. The finding that Separase was overexpressed in both adult and pediatric glioblastoma, makes this a potential diagnostic marker for both populations, even though previous reports have confirmed that the pathways in pediatric glioblastomas are significantly different from adult glioblastomas. Further studies with expanded populations are needed to confirm these initial findings. Also determining the effect of pharmacologic suppression or direct targeting of overexpressed Separase in these tumors could add relevant information regarding the contribution of Separase overexpression to glioblastoma growth and/or progression. In summary, the present study investigated the profile of Separase in glioma patients. Our analysis revealed that Separase is overexpressed as well as mislocalized in glioblastomas compared to its nearly insignificant levels in normal brain. This overexpression of Separase is strongly associated with patients' prognosis in glioma and can be developed in the future as a molecular target for GBM diagnosis and treatment. These results also provide important information that can be used to examine underlying mechanisms and potential therapeutic strategies for glioma patients.
Supplementary Material
Acknowledgments
This study was supported by grants awarded to D. Pati from the National Cancer Institute (1RO1 CA109478 and 1RO1 CA109330).
Footnotes
Conflict of interest: Malini Mukherjee, Tiara Byrd, Vita S. Brawley, Kevin Bielamowicz, Xiao-Nan Li, Fatima Merchant, Saurabh Maitra, Pavel Sumazin, Greg Fuller, Yvonne Kew, David Sun, Suzanne Z. Powell, Nabil M. Ahmed, Nenggang Zhang, and Debananda Pati declare that they have no conflict of interest.
References
- 1.Meyer R, Fofanov V, Panigrahi A, Merchant F, Zhang N, Pati D. Overexpression and mislocalization of the chromosomal segregation protein separase in multiple human cancers. Clin Cancer Res. 2009;15:2703–2710. doi: 10.1158/1078-0432.CCR-08-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Tomaszewski JM, McKay MJ. Can corruption of chromosome cohesion create a conduit to cancer? Nat Rev Cancer. 2011;11:199–210. doi: 10.1038/nrc3018. [DOI] [PubMed] [Google Scholar]
- 3.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagemann C, Weigelin B, Schommer S, Schulze M, Al-Jomah N, Anacker J, Gerngras S, Kuhnel S, Kessler AF, Polat B, et al. The cohesin-interacting protein, precocious dissociation of sisters 5A/sister chromatid cohesion protein 112, is up-regulated in human astrocytic tumors. Int J Mol Med. 2011;27:39–51. doi: 10.3892/ijmm.2010.551. 10.3892/ijmm.2010.551. [DOI] [PubMed] [Google Scholar]
- 5.Pati D. Oncogenic activity of separase. Cell Cycle. 2008;7:3481–3482. doi: 10.4161/cc.7.22.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 7.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 8.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 9.Haering CH, Nasmyth K. Building and breaking bridges between sister chromatids. Bioessays. 2003;25:1178–1191. doi: 10.1002/bies.10361. [DOI] [PubMed] [Google Scholar]
- 10.Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 11.Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 13.Hornig NC, Knowles PP, McDonald NQ, Uhlmann F. The dual mechanism of separase regulation by securin. Curr Biol. 2002;12:973–982. doi: 10.1016/s0960-9822(02)00847-3. [DOI] [PubMed] [Google Scholar]
- 14.Waizenegger I, Gimenez-Abian JF, Wernic D, Peters JM. Regulation of human separase by securin binding and autocleavage. Curr Biol. 2002;12:1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- 15.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Andreu-Vieyra CV, York JP, Hatcher R, Lu T, Matzuk MM, Zhang P. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells. PLoS Biol. 2008;6:e15. doi: 10.1371/journal.pbio.0060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorr IH, Boos D, Stemmann O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol Cell. 2005;19:135–141. doi: 10.1016/j.molcel.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, Zhao YJ, Li XN, Cai WW, El-Naggar AK, et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci U S A. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee M, Ge G, Zhang N, Edwards DG, Sumazin P, Sharan SK, Rao PH, Medina D, Pati D. MMTV-Espl1 transgenic mice develop aneuploid, estrogen receptor alpha (ERalpha)-positive mammary adenocarcinomas. Oncogene. 2013 doi: 10.1038/onc.2013.493. 10.1038/onc.2013.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, Gage FH. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morshead CM, Garcia AD, Sofroniew MV, van Der Kooy D. The ablation of glial fibrillary acidic protein-positive cells from the adult central nervous system results in the loss of forebrain neural stem cells but not retinal stem cells. Eur J Neurosci. 2003;18:76–84. doi: 10.1046/j.1460-9568.2003.02727.x. [DOI] [PubMed] [Google Scholar]
- 25.Nagao K, Adachi Y, Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430:1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 26.McAleenan A, Clemente-Blanco A, Cordon-Preciado V, Sen N, Esteras M, Jarmuz A, Aragon L. Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature. 2013;493:250–254. doi: 10.1038/nature11630. 10.1038/nature11630. [DOI] [PubMed] [Google Scholar]
- 27.Pati D, Haddad BR, Haegele A, Thompson H, Kittrell FS, Shepard A, Montagna C, Zhang N, Ge G, Otta SK, et al. Hormone-induced chromosomal instability in p53-null mammary epithelium. Cancer Res. 2004;64:5608–5616. doi: 10.1158/0008-5472.CAN-03-0629. [DOI] [PubMed] [Google Scholar]
- 28.Milinkovic V, Bankovic J, Rakic M, Milosevic N, Stankovic T, Jokovic M, Milosevic Z, Skender-Gazibara M, Podolski-Renic A, Pesic M, et al. Genomic instability and p53 alterations in patients with malignant glioma. Exp Mol Pathol. 2012;93:200–206. doi: 10.1016/j.yexmp.2012.05.010. 10.1016/j.yexmp.2012.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


