Abstract
Deciphering the signaling pathways that govern stimulation of naïve CD4+ T helper cells by antigen-presenting cells via formation of the immunological synapse is key to a fundamental understanding of the progression of successful adaptive immune response. The study of T cell – APC interactions in vitro is challenging, however, due to the difficulty of tracking individual, nonadherent cell pairs over time. Studying single cell dynamics over time reveals rare, but critical, signaling events that might be averaged out in bulk experiments, but these less common events are undoubtedly important for an integrated understanding of a cellular response to its microenvironment. We describe a novel application of microfluidic technology that overcomes many limitations of conventional cell culture and enables the study of hundreds of passively sequestered hematopoietic cells for extended periods of time. This microfluidic cell trap device consists of 440 18 μm×18 μm×10 μm PDMS, bucket-like structures opposing the direction of flow which serve as corrals for cells as they pass through the cell trap region. Cell viability analysis revealed that more than 70% of naïve CD4+ T cells (TN), held in place using only hydrodynamic forces, subsequently remain viable for 24 hours. Cytosolic calcium transients were successfully induced in TN cells following introduction of chemical, antibody, or cellular forms of stimulation. Statistical analysis of TN cells from a single stimulation experiment reveals the power of this platform to distinguish different calcium response patterns, an ability that might be utilized to characterize T cell signaling states in a given population. Finally, we investigate in real-time contact and non-contact-based interactions between primary T cells and dendritic cells, two main participants in the formation of the immunological synapse. Utilizing the microfluidic traps in a daisy-chain configuration allowed us to observe calcium transients in TN cells exposed only to media conditioned by secretions of lipopolysaccharide-matured dendritic cells, an event which is easily missed in conventional cell culture where large media-to-cell ratios dilute cellular products. Further investigation into this intercellular signaling event indicated that LPS-matured dendritic cells, in the absence of antigenic stimulation, secrete chemical signals that induce calcium transients in TN cells. While the stimulating factor(s) produced by the mature dendritic cells remains to be identified, this report illustrates the utility of these microfluidic cell traps for analyzing arrays of individual suspension cells over time and probing both contact-based and inter-cellular signaling events between one or more cell populations.
Introduction
T cells play a central role in cellular mediated adaptive immune responses because they are highly involved in recognizing and responding to foreign or dangerous antigens. Complete activation of naïve T cells (TN), resulting in clonal expansion, differentiation, and development of effector functions, relies upon contact-based (i.e., receptor/ligand) interactions with antigen-presenting cells (APCs) such as dendritic cells (DCs) in conjunction with signals from secreted molecules. Proper functional differentiation and migration of effector T cells to sites of inflammation, in response to antigen stimulation, rely on a complex array of chemokine, cytokine, and other chemical signaling agents.1 These chemical signals are both autocrine and paracrine in nature, and often are intended to act locally. For example, ligation of the T-cell receptor by an antigen-primed APC results in the secretion of IL-2, also known as T cell growth factor, which then acts in an autocrine fashion as part of a positive feedback loop by binding the IL-2 receptor (IL-2R) on the surface of the T cell from which it was produced. This enhances T cell activation and promotes clonal expansion. Dendritic cells are known to produce cytokines such as IL-12, which influence the differentiation of the T cells they activate. While these and other signaling pathways that govern the adaptive immune response are among the most intensely studied, the intricacies of many signaling mechanisms remain unclear, perhaps in part due to the lack of suitable analytical tools available which can effectively study the complex interactions between non-adherent, hematopoietic cells in a physiologically relevant manner.
Conventional cell culture methodologies involve analyzing bulk responses from millions of T cells which may average out rare signaling events important in the progression of a successful immune response.2 However, monitoring individual hematopoietic cells is not a trivial venture, largely because they are non-adherent. Methods such as antibody coatings or planar lipid bilayers incorporating T cell receptor ligands,3;4 which initiate intracellular signaling pathways by restricting non-adherent T cells to a known physical location, have been employed with limited success. However, these techniques are generally challenging to set up and do not faithfully capture the earliest signaling interactions between TN and DCs during the formation of the immune synapse. Furthermore, the large ratio of media volume to cell volume in ordinary cell culture leads to the dilution of cellular secretions and increases distance between cells, thereby diluting the autocrine and paracrine signals that are so important to the immune response.
The interrogation of T cells, either from a patient or from culture, is commonly done through fluorescence-activated cell sorting (FACS), a powerful technology that is capable of yielding single cell data and is currently the gold standard for single cell signaling analysis in immunological research. But FACS cannot provide any information on the temporal cellular signaling dynamics in a single cell.5 There is a need for a new type of experimental methodology that enables the analysis of signaling dynamics in individual T cells over time. We believe microfluidic technology serves this purpose.
The use of microfluidic devices for cellular applications has increased over the past ten years due to a promise of lower costs for production and reagents, tighter control of experimental parameters, faster reaction times, and reduced diffusion times compared to traditional cell culture techniques.2;6;7 The latter feature entertains the possibility of interrogating cellular activities that occur over time scales too fast to observe with conventional techniques.8 Flexible and relatively easy fabrication enables incorporation of complicated microfluidic designs, including valves, pumps, and switches which enhance the researcher’s ability to manipulate the cellular microenvironment.9–11 For this study, the most important aspect of microfluidics is the ability to study cells at densities that are much closer to those encountered in tissue and lymph nodes than is the case for conventional suspension cell culture. Furthermore, culturing cells in a constant flow-through environment is arguably more physiological than maintaining cells in a relatively large, stagnant pool of media, as is done in conventional cell culture.
The ability to maintain mammalian cells for extended periods of time within microfluidic devices has been demonstrated in adherent cell lines.12–14 Cells readily adhere to a glass or plastic surface (sometimes functionalized to promote adhesion) and can be studied individually or en masse. However, methods for adhering non-adherent cells to a surface (antibodies or adhesion proteins) will induce undesired intracellular signaling pathways that alter the state of the cell.
One solution to this problem is to use suction to hold a cell in a “trap,” which has been demonstrated to be very effective with cardiac myocytes.15–17 However, T cells, one of the main focuses of this research, are extremely small compared to cardiac myocytes, with diameters of TN cells ranging between 5–7 μm. The width of the suction channels would ideally be much smaller than the diameter of the cell, but this aspect ratio is nearing the resolution limit for common microfabrication facilities. Furthermore, T cells are highly deformable and sensitive to pressure. Suction easily lyses these cells, and those that remain intact may be subject to apoptosis (data not shown). Thus, for analysis of TN cells, a passive trapping process would be most suitable.
A truly passive trapping mechanism was developed by Wheeler et al.18 A unique device design incorporating on-chip valves allowed high precision control of fluid flow such that individual T cells could be directed into a bucket-shaped chamber. Once trapped, the intricate hydrodynamic controls allowed the cell to be directly addressable by dyes, stimulants, or lysing agents. However, the design was arranged such that only a single cell was trapped and interrogated during an experiment, which is not conducive to collecting data from a large population of T cells. In order to capture the heterogeneity in T cell responses, it is desirable to have many (hundreds to thousands) T cells that are individually trapped and fluidically addressable within a single microfluidic platform.
Here we report the development and validation of a novel microfluidic platform, the multitrap nanophysiometer, which addresses these needs. In contrast to microphysiometers, which constrain hundreds of thousands of cells to volumes of several microliters for physiological measurements,19–24 nanophysiometers constrain small numbers of cells in nanoliter volumes.25;26 We demonstrate the ability not only to utilize this platform to passively capture primary human CD4+ TN cells using only hydrodynamic forces, but also to sustain the cells within the microfluidic device for more than 24 hours, a significant accomplishment when dealing with non-transformed, non-proliferating cells that have short half-lives of three to five days in conventional suspension cell culture conditions. Despite these difficulties, primary cells were chosen because their activities would more closely model in vivo responses compared to immortalized T cell lines.
In addition, we show that in this microfluidic environment we are able to stimulate calcium signaling in the primary TN cells using chemical, antibody, and cellular methods. Data collection and analysis can be completely automated to sort the resulting calcium traces. The multitrap nanophysiometer enables the analysis and classification of individual cells based on their dynamic, phenotypic response to stimuli and requires the use of only a tiny sample volume, such as might be obtained by a finger stick blood sample.27 Thus, this technology could also be considered complementary to flow cytometry, which requires larger numbers of cells for analysis.
A schematic of the microfluidic multitrap nanophysiometer is shown in Figure 1. This platform is a self-contained unit fabricated of optically transparent, bio-inert PDMS bonded to glass and designed to maintain non-adherent cells in a microfluidic environment. This particular trap design was based conceptually on a platform used by Prokop et al.,14 but modified to accommodate long-term maintenance of small, non-adherent T cells. Cells are introduced into one of the inlet ports using computer-controlled microsyringe pumps.27 As cells flow through the trap region, they are passively trapped in one of 440 18 μm × 18 μm × 10 μm PDMS, bucket-like structures, each with a volume of ~ 3 pL. For the device shown, the overall dimensions of the trap region are 2 mm × 2 mm. It is important to note that the height of the device is 10 μm throughout to prevent stacking of cells in the visual field. The volume of the region containing the traps is ~20 nL, hence the use of the term nanophysiometer. Fluid flow rate is optimized such that cells remain in traps and receive necessary nutrients while shear stress and waste accumulation are minimized. This provides a method of quickly sequestering cells into individual compartments without using any type of adhesive agents, such as the normally used cell surface antibodies, poly-l-lysine, or fibronectin, which can themselves induce unwanted intracellular signaling cascades. Furthermore, only relatively small quantities of cells (aliquots as small as 10,000 cells), media (~100s of microliters for long-term experiments), and other reagents are required to obtain data on hundreds of single cells in parallel, resulting in decreased labor and material costs. The small size of the device enables imaging of the entire cell trap region during automated, high-magnification microscopy.
Figure 1.
Details of the multitrap nanophysiometer and experimental setup. (A) Schematic illustrating the standard experimental setup for the cell trap device. (B) Photograph of the device after the PDMS structure is bonded to a glass coverslip. (C) A schematic overview of the device. (D) Close-up schematic of the cell trap region showing the design and arrangement of individual cell traps. (E) Time lapse imaging of a single representative cell trap within the microfluidic device, demonstrating that traps are easily loaded with cells within minutes of introduction.
Materials & Methods
Experimental Setup
All experiments were performed using an Axiovert 200 microscope (Carl Zeiss), which was equipped to facilitate long-term maintenance of cells during experiments, as shown in Figure 1D. A plexiglass incubator enclosed the microscope such that the temperature was consistently maintained at 37°C by circulating warm air generated by an Air-Therm heater (WPI, Sarasota, FL). The multitrap nanophysiometer chip was enclosed within a 60 mm Petri dish (Fisher Scientific, Hampton, NH) with holes machined in the bottom to allow microscope viewing, as well as slots in the top for polyetheretherketone (PEEK) tubing. In addition, a port was drilled in the side of the dish to allow a tube connected to a gas supply to deliver a constant flow of 5% CO2 and 95% air to the Petri dish.
To minimize evaporation, 3 to 4 layers of 55 mm filter paper (Whatman, Florham Park, NJ) saturated with distilled H20 surrounded the device inside the Petri dish. The motorized microscope stage, image acquisition, focus, and filter settings were controlled using Metamorph™ software (Molecular Devices, Downington, PA). Syringe pump flow rates were also under computer control, utilizing software developed by VIIBRE researchers.28 Images were obtained with the electrically cooled CoolSNAP HQ CCD camera (Photometrics, Tucson, AZ).
Cell Culture
Primary human CD4+ T cells and dendritic cells (derived from human monocytes) were obtained following isolation and purification from human blood donors.29 Primary CD4+ T cells were thawed from liquid nitrogen aliquots (5×105 cells/vial) 24 hours prior to use and cultured using sterile technique in RPMI 1640 cell culture media (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (ATCC, Manassas, VA) and 10 μg/ml Ciprofloxacin (Mediatech, Herndon, VA). Dendritic cells were derived by incubating primary human CD14+ monocytes with 20 ng/ml IL-4 and 100 ng/ml GM-CSF for 4–6 days.29;30 Mature dendritic cells (mDC) were generated following 24-hour incubation with 10 ng/ml lipopolysaccharide (LPS).29;30 All cells were incubated in a sterile environment maintained at 37°C, complete with 5% CO2 and 95% humidity. DCs were normally maintained at a concentration of 5×105 cells per milliliter of media, but for one experiment this was increased to 5×106 cell per milliliter of media in an attempt to obtain supernatants containing a higher concentration of DC secretions.
Microfluidic Cell Loading
Cell loading followed methods developed by our lab and described in greater detail elsewhere.31 Cells were centrifuged to a pellet at 5000 rpm for 1 minute in a 1.5 ml Eppendorf tube (Fisher Scientific, Hampton, NH) which was then mounted on a glass slide with cellophane tape for microscope viewing. Once the pellet was located using the microscope, the opposite end of 50 μm I.D. PEEK tubing (Upchurch, Oak Harbor, WA), connected to a 50 μl gas-tight syringe (Hamilton, Reno, NV), was mounted on a computer-controlled syringe pump (PicoPumps, Harvard Apparatus, Holliston, MA) and guided to the center of the cell pellet. Cells were then aspirated directly into the PEEK tubing using the syringe pump set to withdraw fluid at a flow-rate of 0.5 μl/min. The tubing was then inserted into a loading channel of a device pre-cleared with sterile media. Once connected to the device, the pump controlling the cell channel was set to a forward flow rate of 100 nl/min for passive loading of cells into the traps as they traverse the device. Figure 1E shows a series of images obtained of a single cell trap during the cell loading process. In this trap, six cells were captured within one minute of cell introduction. Unless otherwise noted, the media used to perfuse the device and the cells contained within consisted of RPMI 1640 supplemented with 10% FBS and 10 μg/ml ciprofloxacin.
Cell Viability Assay
Approximately 5×104 primary human CD4+ T cells were loaded with a cytosolic calcium indicator dye by incubating the cells in a solution of five μM rhod-2 AM (Invitrogen, Carlsbad, CA) for 30 minutes at 37°C. Cells were then loaded into the microfluidic device as described previously. The perfusing media was supplemented with CaCl2 to 1.8 mM (Sigma, St. Louis, MO) to mimic normal in vivo calcium levels.32 A second syringe was prepared containing normal cell culture media supplemented with 1 nM Yopro-1 viability indicator (Invitrogen, Carlsbad, CA). Using Metamorph, the microscope was programmed to obtain DIC (bright field), FITC (Yopro-1), and TRITC (rhod-2) images for five fields of view spanning the cell trap region every 30 minutes for 24 hours. Pump control software developed by VIIBRE researchers was configured to switch from the perfusing media to media containing Yopro-1 every two hours for five minutes. Unless otherwise noted, the flow rate was maintained at 100 nl/min.
Cytosolic Calcium Assay
Release of intracellular calcium stores, an early signaling event in T-cell activation, was induced chemically, with antibody-coated beads, or by co-incubation with antigen-presenting cells. In all cases, naïve primary CD4+ T cells were first loaded with rhod-2 calcium indicator by incubation in a 5 μM solution for 30 minutes at 37°C. Cells were then loaded into a microfluidic device (pre-cleared with media to remove air bubbles) and perfused with normal culture media supplemented with CaCl2 to 1.8 mM. Using Metamorph, the microsope was programmed to obtain baseline bright field (DIC or phase-contrast) and TRITC (rhod-2 fluorescence) images for four to five fields of view (FOV) spanning the entire cell trap region of the microfluidic device every 30 seconds.
Chemical Stimulation
A solution consisting of plain culture media supplemented with 1.8 mM CaCl2, 20 ng/ml IL-2, and 8 μM ionomycin, a known calcium ionophore,33 was introduced into an adjacent entrance port. Calcium release from the T-cell endoplasmic reticulum, followed by calcium flux across the cell membrane, was stimulated chemically upon switching perfusate flow completely to the media containing ionomycin.
Bead Stimulation
Protein A coated microspheres (Bangs Laboratories, Fishers, IN) were conjugated to anti-CD3 and anti-CD28 antibodies (R & D Systems, Minneapolis, MN). Antibodyconjugated beads were loaded in the same manner as cells (aspiration from a centrifuged pellet) and introduced into the device directly following cell trapping. Image acquisition was begun upon bead introduction and continued for the duration of the experiments as described above.
Dendritic Cell Stimulation
Lipopolysaccharide (LPS), a protein found in the membrane of gram-negative bacteria, was used to promote maturation of primary human dendritic cells (DCs) by culturing these cells in 100 ng/ml LPS for 24 hours. Prior to loading, the DCs were incubated with 5 μg/ml staphylococcal enterotoxin B (SEB), a superantigen that binds to the outer portion of the major histocompatibility complex (MHC), which can then trigger the T-cell receptor (TCR ) upon contact with T cells, for thirty minutes at 37°C. DCs were then rinsed with fresh media to remove all unbound SEB. The resuspended cells were then centrifuged to a pellet for cell loading as described above. TN were trapped first, and then DCs were introduced to the devices. Image acquisition began upon introduction of DCs.
Daisy-chain Configuration
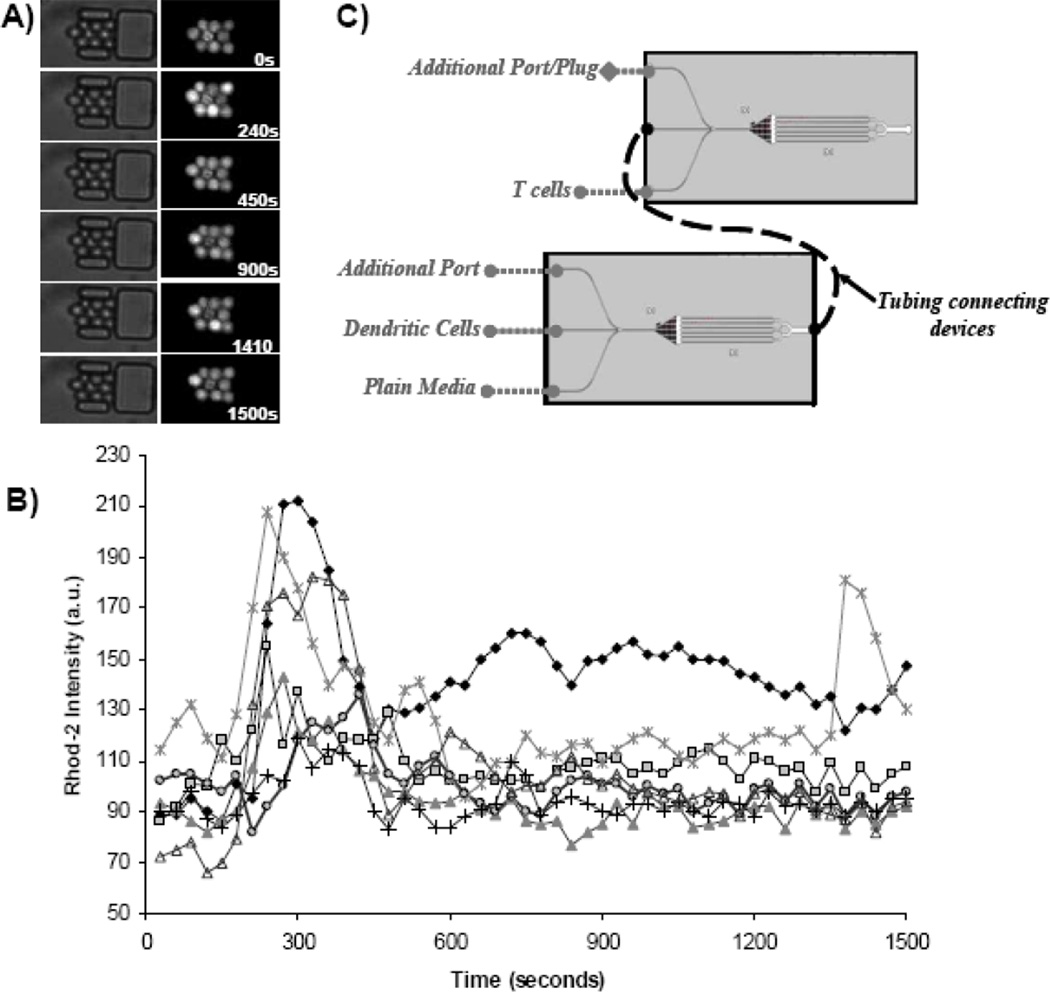
Two separate devices were prepared as described above, except for the replacement of the Petri dish surrounding the device with a chambered coverglass (Nunc, Fisher Scientifc, Hampton, NH). Both devices were secured to a custom-machined stage insert capable of accommodating several devices. Filter paper saturated with water and CO2 were added as before. One device was loaded with naïve CD4+ T cells only. The other device was loaded with the TN stimulus of interest (e.g., mDC). Background data were collected every 30 seconds on both devices running on completely separate pump systems. To connect the two devices, the tubing connecting the syringe pump delivering plain media to the TN cells-only device was cut and the end attached to that device was inserted into the exit port of the stimulus device, such that the TN in the downstream device were exposed to the effluent of the cells in the upstream stimulus device. Figure 5 illustrates the daisy-chain setup for studying intercellular signaling. Image acquisition was restarted immediately following reconnection and continued for at least one hour.
Figure 5.
Observation of intercellular signaling between mature DCs and naïve CD4+ T cells (TN). Using the device configurations illustrated in (C) we were able to observe non-contact-based stimulation of T cells by mature, SEB-pulsed DCs. TN are trapped in microfluidic cell traps connected downstream of a nanophysiometer containing mature DCs. Bright field images of T cells beginning upon daisy-chain connection (zero seconds) are shown in the left of (A) with the corresponding fluorescence (rhod-2 AM ester) images shown on the right. The calcium transients of the corresponding cells are shown in (C) where each individual graph represents a single cell. Cytosolic calcium transients are clearly seen in TN within minutes of daisy-chain connection.
Image Storage and Analysis
Due to the large quantity of information that can be extracted from a single experiment when monitoring hundreds of cells with a variety of fluorescent markers over an extended period of time, it was both desirable and necessary to develop an automated analysis program. Metamorph was used to collect images and store them in a stacked format to the hard drive. Analysis of image data was done primarily using the ImageJ software package (XXX CITE SOURCE), with the Wright Cell Imaging Facility bundled plugins and Excel™ (Microsoft), along with custom-written macros. First, phase contrast and fluorescence images obtained at a single time point were aligned and re-indexed using a modified routine written by Philippe Thévenaz.34 This algorithm calculates the X and Y translation of each phase contrast (bright field) image relative to a reference image of the same series and saves the data to a file. The resulting translations were applied to the phase contrast and fluorescent images together to ensure that proper alignment between each image at a given time point was maintained. The particle analyzer routines embedded with the WCIF ImageJ bundle were used to find the positions of cells in the resulting images, and the mean gray values at these positions were recorded to file.
Excel macros were used to segment the data files into trap regions based on the XY positions recorded by the ImageJ particle analyzer. A typical field of view contains up to 85 traps that contain one to four cells which are to be tracked during the analysis. Segmentation into trap regions is easily accomplished since all the microfluidic traps are spaced very precisely. Segmentation of individual trap zones into individual cells is more complex. Briefly, the first slice which occurred that contained the maximum number (Nmax) of putative cells for the time series was selected as a cardinal slice. The positions of each of the cells were stored in a 2×Nmax array. Every time point was then sequentially compared to the cardinal time point, and the XY positions of the cardinal time point were updated and replaced by the XY positions of each time point weighted by observation. In this way, if a cell is repeatedly found near one particular position (even if it is not found in the cardinal slice), it is weighted heavily as a probable cell position in the final analysis. Similarly, if a spurious cell or debris is located at a single time point it is weighted less heavily and is less likely to appear in the final analysis. Fluorescence intensities of cells found using this algorithm were automatically stored in an Excel spreadsheet for further analysis.
Cluster Analysis of Calcium Data
The extracted tracings were automatically classified or clustered into groups by their responses to a given stimulus. Statistical cluster analysis, or unsupervised learning, is a set of statistical methods used to identify meaningful or informative subgroups within a dataset. For this study, we used hierarchical clustering to identify scales of similarity between individual T cells and groups of T cells. A distance metric must be specified to measure the pair-wise similarity between T cells. Each cell's time series is treated as a vector in a multidimensional space, and for this study we used the Pearson correlation metric, which is well suited for measuring differences in shape between two curves.35;36
The clustering algorithm works by creating a pair-wise distance matrix between all cells and a cluster is created by merging the pair of cells with the smallest separating distance. A new distance matrix then is created with the unclustered cells and the merged cells, and the process is repeated. The location of the merged cluster can be defined in different ways, including single linkage, complete linkage, and average linkage (the average of the cell vectors). We use complete linkage, though all methods give similar clusters.36
FACS simulation
To emphasize the differences between single cell studies in FACS scanners and our parallel measurements in the multitrap nanophysiometer, we used the calcium transients from data collected on ionomycin-stimulated T cells, using the microfluidic platform to simulate FACS data that might have been collected. The microfluidic data were collected from 447 cells once every 30 seconds. For the first of the 57 frames in the image sequence, we randomized the order of the 447 observed calcium levels (measured as rhod-2 fluorescence) for that frame to create the first 447 sequential time points in our simulated FACS signal. The second image frame, with a new randomization, produced the next 447 FACS time points. This was repeated for each of the 57 frames and resulted in a random sampling of calcium fluorescence levels at a rate of approximately 15 samples per second for 1700 seconds.
Results & Discussion
Cell Environment and Viability
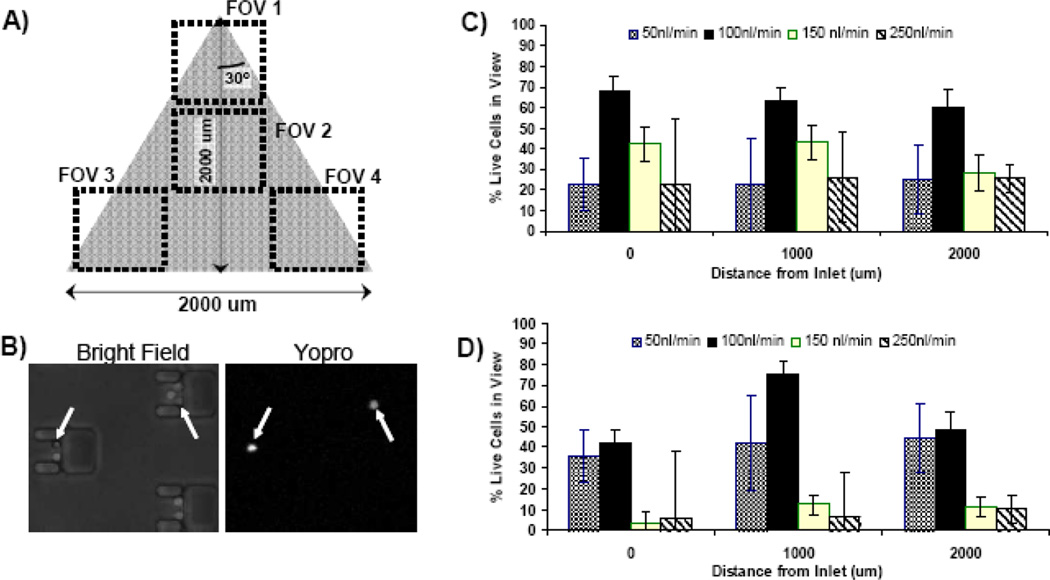
In order to reliably utilize our microfluidic cell traps for cellular assays it is important to first validate that T cells are stably maintained within the microfluidic device over time. Basic environmental conditions established in conventional cell culture settings, such as infusion of CO2, humidity control, and regulation of temperature to 37°C, were all addressed by various components of our experimental setup (See Figure 1). In vivo, T cells naturally migrate in the constant-flow environment of blood and lymph, but the flow rate to which they are exposed in vitro can have a profound impact on viability. Perfusion rates too low can lead to accumulation of waste and, due to the small volume of the device (~20 μl), depletion of nutrients. Higher flow rates will, of course, result in high shear stress, which might induce apoptosis or even cause cell lysis. Thus, while investigating long-term viability within our multitrap nanophysiometer, we analyzed a range of flow rates to determine the optimal conditions for maximal T-cell survivability.
Two widely used markers of T cell activation are the secretion of IL2 and expression of IL-2Rα (CD25), which require translocation of various transcription factors to the nucleus, followed by transcription, translation, and expression of the proteins.14;37 Peak expression of CD25 can occur as late as 24 hours following stimulation, and thus our goal was to maintain primary T cells for at least this long within the microfluidic cell traps. Cell viability was assessed every two hours by transient (5 min) exposure of the cells to Yopro-1, a dye that fluorescently labels dead cells. In addition, we included a cellular acitivity measurement at the conclusion of the 24-hour experiment by perfusing the device with media containing ionomycin and monitoring calcium flux via rhod-2 fluorescence. This activity measurement allows us to gauge how well the live cells (unstained by the Yopro-1 viability dye) retain normal T cell functionality following this long-term incubation.
Results from the viability experiments are shown in Figure 2C. A range of flow rates was evaluated, and it was determined that 100 nl/min is optimal for promoting long-term cell viability in this device with an average of 68% of cells alive after 24 hours. This flow rate was also best in terms of cellular activity measurements, with up 75% of live cells exhibiting a measurable calcium response upon exposure to ionomycin after 24 hours in the middle region of the device (Figure 2D). While these viability data may seem low compared to cell lines in culture, it is important to remember that primary TN cells naturally rely heavily upon external signaling factors to avoid apoptosis. Although the in vivo half life of naïve CD4+ T cells can be years, typical in vitro lifetimes are on the order of 3–5 days, which can only be prolonged to weeks in the presence of survival factors such as cytokines IL-7.38;39 We did not utilize IL-7 in our experiments. Thus, the survival rates within the microfluidic device are only slightly lower than normal cell culture without survival factors. We continue to investigate ways to optimize our experimental conditions, including optimizing nutrient composition of the media and oxygen levels,40 in order to enhance cell survival, which will be of great importance for future long-term studies. However, for the proof-of-principle experiments discussed here, the cells were subject to relatively short durations (approximately 6 hours) within the microfluidic devices, which were extremely well tolerated, with greater than 90% of the cells remaining viable.
Figure 2.
Cell viability analysis within the multitrap nanophysiometer. (A) Schematic illustrating the four fields of view that were imaged every 20 minutes. (B) An example of images obtained during the viability experiments. Cells exhibiting fluorescence due to Yopro-1 staining are dead. (C) Viability as measured by percentage of cells alive after 24 hours with respect to both flow rate and position within the device. (D) Cellular activity measured as percentage of cells that respond to ionomycin exposure with a measurable cytosolic calcium transient following 24 hours within the device. Results are shown with respect to both flow rate and position within the device.
T Cell Stimulation
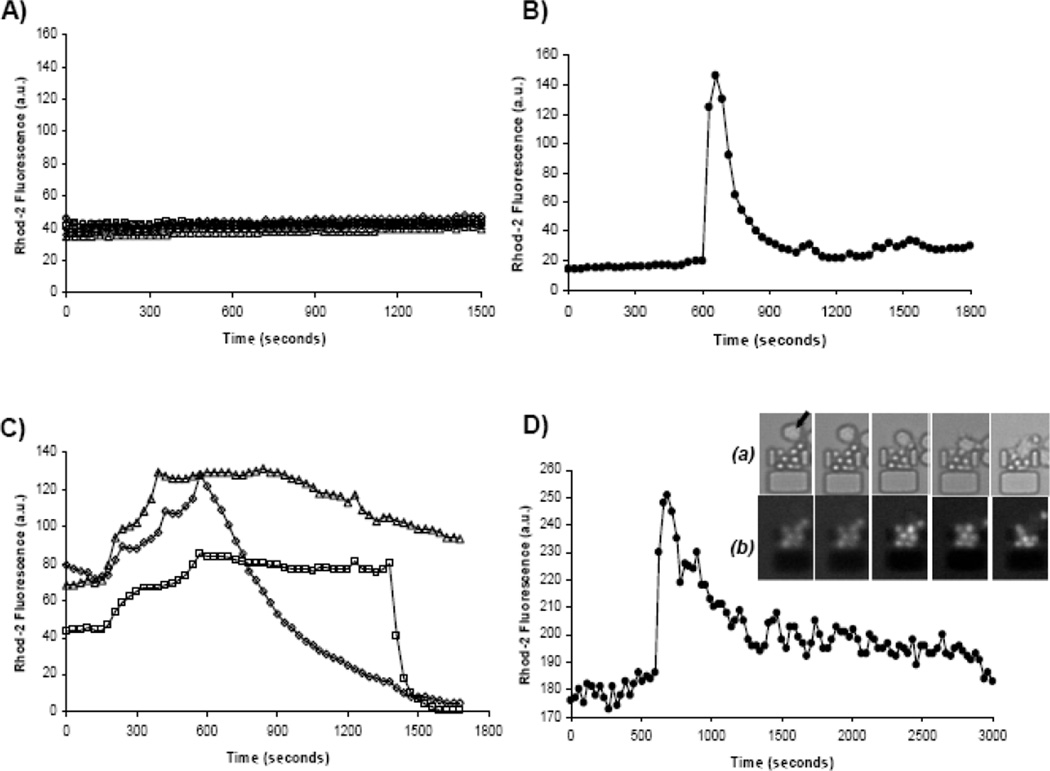
Next, we wished to demonstrate the ability to activate the primary T cells within the device, using an increase in cytosolic calcium levels as an early indicator of stimulation. We employed three well-established methods of activation: chemical stimulation with ionomycin, antibody stimulation with anti-CD3- and anti-CD28-coated microspheres, and cellular stimulation using SEB-pulsed, LPS-matured, primary human dendritic cells. Quantitative results of these experiments are shown in Figure 3. In each case we were successful in activating the primary CD4+ T cells as illustrated by the cytosolic calcium transients observed using rhod-2 AM ester calcium indicator.
Figure 3.
Examples of cytosolic calcium transients, as indicated via rhod-2 calcium dye, in single primary T cells exposed to (A) plain media, (B) antibody-coated microspheres, (C) ionomycin, and (D) mature, stimulated dendritic cells. Each trace represents a single cell. Three traces are shown in (C) to illustrate the three typical calcium transient patterns observed when stimulating T cells with ionomycin. Inset relative to graph D shows example of (a) naïve T cells and mature dendritic cells (indicated in first frame by arrow) interacting within a single trap. The corresponding fluorescence images are shown in (b) as the cellular interactions stimulate calcium fluxes in TN cells (pre-loaded with rhod-2 calcium indicator).
The results of the control experiment shown in Figure 3A clearly show that rhod-2 loaded cells did not exhibit any cytosolic calcium increases when exposed to plain media. Introduction of antibody-coated beads (Figure 3B), ionomycin (Figure 3C), and mDCs (Figure 3D) all resulted in strong calcium responses. The cytosolic calcium transients induced by antibody-coated beads and DCs are also consistent with traces found in literature.41;42
Real-time Cell-Cell Interactions
Another key strength of this platform is the ability to generate and study cell-cell interactions in real time. Figure 3D and the corresponding inset clearly show pulsing of calcium transients within the TN cells as they interacted with DCs within the device. Observation of the TC/DC interactions revealed a marked similarity to that reported in studies employing intravital microscopy of murine lymph node in that DCs exhibited multiple extensive protrusion and often would migrate (in many cases opposing flow) towards and “sample” multiple T cells over the course of an experiment.43;44 Cellular interactions, particularly those of TN and DCs, are central to the progression of immune response but are not easily reproduced in vitro. The large media-to-cell-volume ratios in conventional cell culture make studying cellular interactions such as the formation of immunological synapse difficult because of the time required for diffusion of cells through the media. It is practically impossible to capture the earliest signaling events initiated by cell-cell contact without restricting cell motion via antibodies or other adhesion proteins, which as discussed previously can trigger intracellular signaling events. Through sequential loading of two or more cell types, the microfluidic cell traps easily capture multiple types of cells passively, quickly, and much more predictably than current methods. Because the entire trap region is continuously surveyed by the computer-controlled microscope, the interactions are recorded from the beginning. The capacity to monitor hundreds of cell couplings and manipulate their environment with relative ease has great potential to contribute to advancing immunological studies.
Stochastic Analysis of Cell Signaling Dynamics for Individual Cells Versus Cell Ensembles
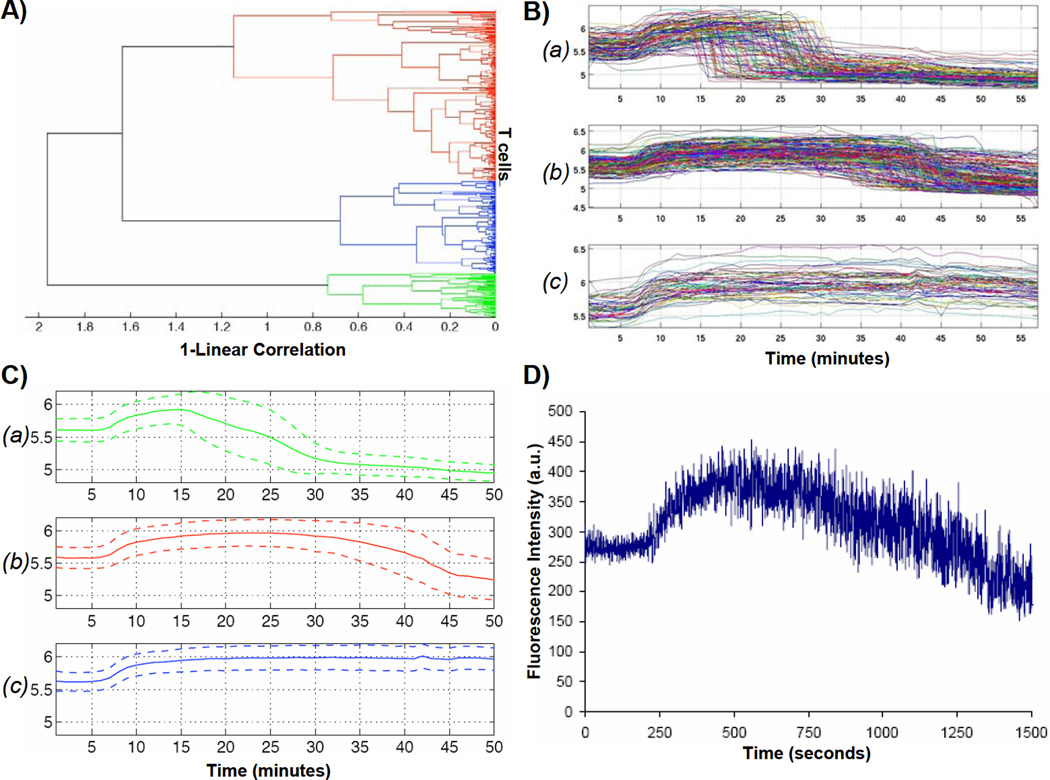
Ionomycin stimulation of T cells resulted in calcium transients whose shape and duration differed from more physiological forms of stimulation. This illustrates a key strength of our microfluidic approach in that we were able to observe the individual cell responses to ionomycin from hundreds of individual cells and clearly identify three distinct populations of responses in a single experiment (as shown in representative curves in Figure 3C). Automated ImageJ and Excel routines were used to analyze fluorescence images of T cells challenged with ionomycin. The analysis found 809 raw fluorescence traces which included 69 duplicates, 195 truncated curves with insufficient data, and 88 flat-lines or unresponsive traces found by calculation of the data trend. Out of this data, 457 unique traces (e.g., calcium transients from 457 individual T cells) were found and subsequently analyzed using automated clustering analysis, the results of which are illustrated in Figure 4. A dendrogram plot (Figure 4A) illustrates the distinct clusters of T cells comprising the range of calcium responses to ionomycin as determined by the clustering algorithm. At the far right, each horizontal dendrogram line begins with an individual T cell, and clusters are constructed by assembling the most similar pairs of cells or resultant clusters of cells. Each vertical line in the dendrogram connects a pair of T cells or cluster of T cells. The location of the vertical line projected onto the horizontal axis quantifies the dissimilarity between the pair. Dissimilarity is calculated by 1-P, where P is the Pearson correlation between calcium time-series profiles of the corresponding T cells. Vertical dendrogram lines further to the left connect T cells or clusters of T cells that are more dissimilar. By inspection of the dendrogram in Figure 4A one can observe that there are three major subgroups of cellular calcium responses to ionomycin. In Figure 4B, we created cluster profiles by plotting the 457 individual T cell time-series profiles according to their group assignment.
Figure 4.
Clustering analysis results of calcium response from ionomycin-stimulated T cells within the nanophysiometer. (A) Dendrogram representing the distance in N-dimensional space between green, red, and blue type curves, where N is the number of time points. (B) Profiles of the green, red, and blue subgroups a, b, and c, respectively, that clearly illustrate three distinct response profiles to ionomycin stimulation that would not have been apparent using FACS analysis. (C) Average responses of the profiles shown in B. Dotted lines on top and bottom indicate maximum deviation from the mean. (D) Simulated FACS data computed from the calcium data set in (B), demonstrating that FACS analysis will conceal the three distinct response patterns illustrated in (C).
The distinguishing feature of these subgroups is the fall-time, or time it takes the intensity to return to baseline following the initial intensity rise following exposure to ionomycin. Cluster A is composed of T cells exhibiting a marked increase in fluorescence upon ionomycin exposure followed by a sharp drop in fluorescence that falls to below initial baseline fluorescence levels, often within a single data point (30 seconds). Cells within cluster B respond to ionomycin with a similar increase in fluorescence intensity, but both the time spent at peak fluorescence intensity and the time required to return to baseline intensity levels are extended. Finally, cells in cluster C exhibit an increase in fluorescence intensity but remain at the peak intensity for the duration of the experiment. We theorize that the cells in cluster A are undergoing non-apoptotic cell death because of the dramatic and often complete loss of fluorescence signal. This is perhaps due to the initial state of the cell, environmental stress, ionomycin exposure, or a combination thereof. In vivo, there is a complex array of biological variables that can influence the ability of a given T cell to respond to stimulation. Factors such as, but not limited to, receptor configuration, duration of interaction, cytokine/chemokine exposure, configuration of the microniche within an individual trap, and timing all contribute to the signaling state of the cell and, thus, determine the outcome of T cell activation and differentiation.44–46;46–51 We believe this heterogeneity of response to ionomycin identified after a single experiment both highlights an important biological modifier, which is essential to fully understanding and fine-tuning adaptive immune responses, and demonstrates the power of microfluidic technologies to resolve single cell events. This type of temporal information on single cells and small cell clusters, which is required to probe the relationship between cellular environment and stimulation, is very difficult to obtain using conventional cell culture techniques or FACS analysis.
To emphasize this point, Figure 4D contains a prediction of the data had these cells been analyzed using FACS. The same data set used in Figure 4A and B was used to simulate FACS data that might have been collected with these cells undergoing the same stimulation. Beneath the noise of cell variation, we see a single monotonic increase and decrease in calcium. Not surprisingly, the heterogeneity of the underlying populations is completely masked. While our 447 cells sampled every 30 seconds correspond to a very slow FACS rate of 15 cells per second, the image in Figure 4D shows that we did not observe any evidence of the three distinct cell populations, independent of our cell sorting rate, since a faster FACS with more samples would simply produce a finer scale replica of Figure 4D. Cell-to-cell variations, even in relatively homogenous subsets such as these naïve CD4+ T cells, can have a large impact upon their response to a given stimuli. The contrast between Figures 4A–C and Figure 4D illustrates additional information available from time-series measurements on a modest number of individual cells, as compared to single-pass FACS measurements on much larger cell populations. Hence, the multitrap nanophysiometer is an extremely useful companion to traditional FACS analysis for the study of the temporal activation dynamics of T cells.
Intercellular Signaling
As we have mentioned, a variety of chemical signaling factors can modify the state of T cell activation. Even products secreted by the T cells acting in both an autocrine and paracrine manner are critical to this activation process.52 In the majority of experiments, the precise composition of biological modifiers the T cells are exposed to during activation is not known, despite careful formulation of cell culture media and supplements. It is in general very difficult to determine what cells are producing in response to various stimuli and how these are affecting adjacent cells. This problem is also present in the microfluidic cell traps we present here. However, while in traditional cell culture methods large media volumes rapidly dilute cellular signaling agents, thus rendering it very difficult to monitor individual suspension cells over time, our multitrap nanophysiometer allows us to control this dilution with ease, providing us with an important, readily controlled parameter. By configuring the content of the media supplying the device, we can begin to probe the cause and effect of various chemical signaling agents upon T cell signaling. The greatly reduced media-to-cell-volume ratio creates a more physiologically relevant model, as neither stimuli nor cellular products are diluted as significantly as in conventional suspension cell culture.
Our first evidence of the value of the reduced media-to-cell-volume ratio in studying autocrine and paracrine signaling was the observation of calcium transients in TN cells that were downstream of DC-TN cell pairs, but not in direct contact with a DC. In the following set of experiments, we took advantage of the capabilities of our microfluidic devices to investigate further intercellular signaling events between two cell populations in real time. Utilizing a daisy-chain configuration of multiple microfluidic cell trap devices, we were able to effectively monitor the effects of DC secretions upon naïve T cells.
We used two identical microfluidic devices to trap and study two distinct cell populations. After collecting baseline data, the two devices were connected as shown in Figure 5C, such that the exit port of the first device was connected to an inlet port of the second device. In this arrangement the cells in the second device are exposed to all the signaling factors produced and secreted by the cells in the upstream device. The earliest experiments involved the first device containing mDCs and TN cells trapped together while the second (downstream) device contained only TN cells. Calcium transients in the TN cells were observed in both devices despite the absence of DC contact with the TN cells in the second device. Figure 5 illustrates the non-contact-dependent calcium transients observed in the TN cells trapped in the second device, which occurred within minutes of connecting the daisy-chain.
Once we verified that there was a stimulatory factor transmitted in a non-contact-dependent manner, we sought to better characterize its source. This involved evaluating both populations of cells in the upstream device, as well as chemicals used in the experiment, such as SEB and LPS, that were being transferred to the downstream device in the media. A comparison of control data with experimental data is shown in Figure 6. Graphs A – C clearly demonstrate that calcium transients are not induced from plain media, SEB, or LPS. Figure 6D illustrates calcium transients in the downstream device with mDCs pre-pulsed with SEB in the upstream device. The stimulation of calcium release in the downstream cells revealed that the DCs were in fact the source stimulating the T cells.
Figure 6.
Monitoring cytosolic calcium levels in naïve T cells exposed to (A) plain media (RPMI 1640+ 10% FBS+10 mg/ml Cipro+20 ng/ml IL2), (B) plain media supplemented with 5 mg/ml LPS, (C) plain media supplemented with 5 nM SEB, and (D) daisy-chained to a device upstream which contained mature DCs only. Each line represents a single cell analyzed over the entirety of the experiment. T cells were incubated with 5 mM rhod-2 AM-ester prior to the experiments. No calcium activity is observed in response to plain media or LPS. Some calcium activity is observed with the addition of SEB, as expected, but the relative intensity of this response is quite small compared to that shown in graph D. Within seconds to minutes, T cells respond with distinct cytosolic calcium transient to the effluent from mature DCs. It is clear that the DCs are secreting a signaling factor that stimulates this calcium release in T cells. Inset (E) shows the average of the fluorescence signals plotted in graph D, with the SEM indicated by the upper and lower lines.
Table 1 summarizes the experiments conducted to further characterize the source of the stimulatory factor. We evaluated the stimulation ability of mDCs pulsed with SEB and paired with TN cells, mDCs pulsed with SEB, mDCs only (no SEB), naïve DCs, SEB alone, LPS alone, and finally, the supernatant taken from mDCs (no SEB) incubated in a cell culture dish for 24 hours at a concentration of 5×105 and 5×106 DCs per milliliter of media (LPS concentration held constant). The analysis revealed that mDCs secrete the stimulating factor and that neither pulsing with SEB nor incubation with TN cells was necessary for production. In addition, the fact that the supernatant from mDCs cultured at 5×105 cells/ml was unable to elicit a calcium response suggests that the stimulatory factor not secreted in abundance. This hypothesis was supported by the fact that concentrating DCs tenfold (essentially reducing the media volume to cell ratio) resulted in supernatant that induced transient calcium flux in TN cells.
Table 1.
Summary of results from intercellular signaling study. The left column describes the configuration of cells within the first device. The right column indicates whether or not a calcium signal was detected in the naïve CD+ T cells housed in the downstream daisy-chained device. These results indicate that maturation of DCs by LPS alone was sufficient to induce DCs to produce products that stimulate calcium fluxes in naïve T cells, but only at low media-to-cell-volume ratios.
| Device 1: TN cell stimulus | Device 2: Ca2+ Signal in TN Cells? |
|---|---|
| LPS-matured DCs + SEB + TN cells | Yes |
| LPS-matured DCs + SEB | Yes |
| LPS-matured DCs | Yes |
| Immature DCs | No |
| T cells only | No |
| SEB only | No |
| LPS only | No |
| Supernate from LPS-Matured (24 hours) DCs in culture (5×105 cells/mL) | No |
| Supernate from LPS-Matured (24 hours) DCs in culture (5×106 cells/mL) | Yes |
After we determined that factors secreted by mDCs stimulated calcium transients in the TN cells, we proceeded to compare this to contact-based stimulation and determine whether or not exposure to DC effluent has any effect upon subsequent DC contact. To examine this question, mDCs were daisy-chained upstream of TN cells as in previous experiments. After allowing the TN in the second device to be exposed to the effluent from the first device containing DC, the daisy-chain was disconnected and a bolus of SEB-pulsed mDCs were injected into the second device. We then compared the calcium response of cells exposed to the secreted factor followed by direct DC contact to those stimulated by DC contact alone. The results are shown in Figure 7. We found that cells initially exposed to DC products were subsequently able to be stimulated by DC contact, although the average calcium flux was lower compared to DC contact alone. This may indicate that exposure to DC products somehow diminishes the signaling strength of subsequent cellular contact. It is also possible that the two stimulatory events could accumulate in the overall scheme of activation.
Figure 7.
Comparison of calcium flux magnitudes in naïve CD4+ TN exposed to DC contact only, products of mature DCs (using daisy-chain configuration), or DC contact following one-hour exposure to products produced by mature DCs. Error bars indicate +/− one standard deviation.
These results raise interesting questions into the nature of these early, pre-synaptic communications between DCs and TN. One could clearly envision DC products having chemotactic properties, as mDCs ready for antigen presentation would like to traffic and/or stimulate TN trafficking to lymph nodes. Increased T cell motility and sampling of nearby mDCs would also be desirable in the early stages of immune response. The concentration of DC products is relatively small, suggesting that it is meant to act locally to support synapse formation in vivo to propagate the signals. Another possibility is that mDCs continually produce small amounts of signaling products which accumulate over time. It is also conceivable that these initial signals “prime” neighboring T cells to initiate cell-to-cell contact to begin “sniffing” signals from the DCs to form antigen-specific immunological synapses.
Taken together these results illustrate the ability of our multitrap nanophysiometers to maintain concentrations of signaling molecules at physiologically relevant levels. Essentially, the daisy-chaining of microfluidic cell traps provides a method to generate conditioned media, but the undiluted effects are observable in real time and without the interference of feeder cell or intermediate filtering stages. This can be of great utility to detect and monitor locally produced and acting agents that are known to be critical for initiating, fine-tuning, and propagating biological processes, such as those that occur in immune responses and stem cell differentiation. This setup can also easily be expanded to contain any number of microfluidic devices in series, the inputs and outputs of which are easily configurable to suit the complexity of the experiment.
Conclusion
In this report we have demonstrated a novel microfluidic platform for studying both cellcell interactions and intercellular signaling events of non-adherent cells. The novelty of our multitrap nanophysiometers lies in the ability to passively trap and maintain hundreds of nonadherent cells in parallel, easily and quickly dictate the physical location of cells, and obtain temporal data on any given cells. The devices operate with a low media-to-cell-volume environment such that cellular secretions are not diluted and their effects may be monitored in real time. Due to its small dimensions, the device requires very few cells and reagents to obtain large amounts of data. We successfully demonstrated the ability to passively trap non-adherent, primary CD4+ T cells from samples with less than fifty thousand cells while sustaining their viability for more than 24 hours. Furthermore, we successfully induced cytosolic calcium transients in trapped TN cells using chemical, antibody, and cellular forms of activation. Utilizing a daisy-chain configuration of multiple microfluidic devices, we were able to probe intercellular communications between DCs and TN cells in real time with minimal dilution. These experiments show that DCs matured with LPS, but in the absence of antigen, release low concentrations of a chemical signal that stimulates cytosolic calcium release in TN cells. Future experiments will continue to investigate both intercellular and intracellular signaling events surrounding the immune synapse. Taken together, these results establish the multitrap nanophysiometer as a tool well suited for the study of T cell signaling in that it enables the collection of important information on individual cell dynamics which current, widely utilized technologies are unable to provide. We hope that the demonstration of the capabilities of this microfluidic platform will inspire more interest in the use of this type of technology in biological research.
Acknowledgments
The authors wish to thank Derya Unutmaz and Michelle Tseng for providing the primary cells as well as their invaluable assistance and advice on cell culture. In addition, we would like to thank Walter Georgescu for the use of the computer guided pump controller program he developed. We are indebted to E. Duco Jansen for the use of cell culture facilities, Allison Price for her generous help with this manuscript, and Don Berry, Phil Samson, and Dmitry Markov for assistance with illustrations. This research was supported in part by the Vanderbilt Institute for Integrative Biosystems Research and Education (VIIBRE), Air Force Office of Sponsored Research (AFOSR) grants FA95500410364 and FA95500510349, National Institutes of Health grant U01 A1061223, the Systems Biology and Bioengineering Undergraduate Research Experience (SyBBURE), and the Simons Center for Systems Biology at the Institute for Advanced Study, which provided JPW with an environment that enabled completion of the manuscript.
References
- 1.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Annual Review of Immunology. 2000;18:767. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Wikswo JP, Prokop A, Baudenbacher F, Cliffel D, Csukas B, Velkovsky M. IEE.Proc.Nanobiotechnol. 2006;153:81. doi: 10.1049/ip-nbt:20050045. [DOI] [PubMed] [Google Scholar]
- 3.Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD. Proc.Natl.Acad.Sci.U.S.A. 2007;104:20296. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310:1191. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 5.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Science. 2005;308:523. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell P. Nat Biotechnol. 2001;19:717. doi: 10.1038/90754. [DOI] [PubMed] [Google Scholar]
- 7.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 8.Voldman J. Curr.Opin.Biotechnol. 2006;17:532. doi: 10.1016/j.copbio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 10.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annu.Rev.Biomed.Eng. 2001;3:335. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 11.Whitesides GM. Nature. 2006;442:368. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 12.Di CD, Wu LY, Lee LP. Lab Chip. 2006;6:1445. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 13.Kim L, Toh YC, Voldman J, Yu H. Lab Chip. 2007;7:681. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 14.Prokop A, Prokop Z, Schaffer D, Kozlov E, Wikswo J, Cliffel D, Baudenbacher F. Biomed.Microdevices. 2004;6:325. doi: 10.1023/B:BMMD.0000048564.37800.d6. [DOI] [PubMed] [Google Scholar]
- 15.Klauke N, Smith G, Cooper JM. Lab Chip. 2007;7:731. doi: 10.1039/b706175g. [DOI] [PubMed] [Google Scholar]
- 16.Klauke N, Smith GL, Cooper JM. Anal.Chem. 2007;79:1205. doi: 10.1021/ac061547k. [DOI] [PubMed] [Google Scholar]
- 17.Werdich AA, Lima EA, Ivanov B, Ges I, Anderson ME, Wikswo JP, Baudenbacher FJ. Lab Chip. 2004;4:357. doi: 10.1039/b315648f. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, Liao YH, Farrell K, Manger ID, Daridon A. Anal.Chem. 2003;75:3581. doi: 10.1021/ac0340758. [DOI] [PubMed] [Google Scholar]
- 19.Parce JW, Owicki JC, Kercso KM, Sigal GB, Wada HG, Muir VC, Bousse LJ, Ross KL, Sikic BI, McConnell HM. Science. 1989;246:243. doi: 10.1126/science.2799384. [DOI] [PubMed] [Google Scholar]
- 20.Beeson C, Rabinowitz J, Tate K, Gutgemann I, Chien YH, Jones PP, Davis MM, McConnell HM. J.Exp.Med. 1996;184:777. doi: 10.1084/jem.184.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arimilli S, Deshpande SV, Nag B. J.Immunol.Methods. 1998;212:49. doi: 10.1016/s0022-1759(98)00010-6. [DOI] [PubMed] [Google Scholar]
- 22.Arimilli S, Howard MC, Nacy CC, Deshpande SV. J.Cell.Biochem. 2000;77:409. [PubMed] [Google Scholar]
- 23.Eklund SE, Kozlov E, Taylor DE, Baudenbacher F, Cliffel DE. ch. Chapter 16. In: Rosenthal S, Wright D, editors. NanoBiotechnology Protocols. Vol. 303. Totawa, NJ: Humana Press; 2005. pp. 209–223. [Google Scholar]
- 24.Eklund SE, Snider RM, Wikswo J, Baudenbacher F, Prokop A, Cliffel DE. J.Electroanal.Chem. 2006;587:333. [Google Scholar]
- 25.Werdich A, Lima EA, Ivanov B, Ges I, Wikswo JP, Baudenbacher FJ. Lab Chip. 2004;4:357. doi: 10.1039/b315648f. [DOI] [PubMed] [Google Scholar]
- 26.Ges IA, Ivanov BL, Werdich AA, Baudenbacher FJ. Biosens.Bioelectron. 2007;22:1303. doi: 10.1016/j.bios.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Seale KT, Faley S, Chamberlain J, Wikswo JP. Lab Chip. 2008 , Submitted. [Google Scholar]
- 28.Georgescu W, Jourquin J, Estrada L, Anderson ARA, Quaranta V, Wikswo JP. Lab Chip. 2008;8:238. doi: 10.1039/b716203k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oswald-Richter K, Torres VJ, Sundrud MS, VanCompernolle SE, Cover TL, Unutmaz D. J.Virol. 2006;80:11767. doi: 10.1128/JVI.00213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eger KA, Sundrud MS, Motsinger AA, Tseng M, Kaer LV, Unutmaz D. PLoS.ONE. 2006;1:e50. doi: 10.1371/journal.pone.0000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seale KT, Faley SL, Wikswo JP. In Preparation. 2007 [Google Scholar]
- 32.Navarrete C, Sancho R, Caballero FJ, Pollastro F, Fiebich BL, Sterner O, Appendino G, Munoz E. J Pharmacol.Exp.Ther. 2006;319:422. doi: 10.1124/jpet.106.108209. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Hermann TE. J Biol Chem. 1978;253:5892. [PubMed] [Google Scholar]
- 34.Thevenaz P, Unser M. Microsc.Res.Tech. 2007;70:135. doi: 10.1002/jemt.20393. [DOI] [PubMed] [Google Scholar]
- 35.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc.Natl.Acad.Sci.U.S.A. 1998;95:14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKinney BA, Reif DM, Ritchie MD, Moore JH. Appl.Bioinformatics. 2006;5:77. doi: 10.2165/00822942-200605020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnement MR, Faley SL, Wikswo JP, Rosenthal SJ. IEEE Trans.Nanobioscience. 2006;5:268. doi: 10.1109/tnb.2006.886573. [DOI] [PubMed] [Google Scholar]
- 38.Rathmell JC, Farkash EA, Gao W, Thompson CB. J.Immunol. 2001;167:6869. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 39.Vivien L, Benoist C, Mathis D. Int.Immunol. 2001;13:763. doi: 10.1093/intimm/13.6.763. [DOI] [PubMed] [Google Scholar]
- 40.Sahaf B, Atkuri K, Heydari K, Malipatlolla M, Rappaport J, Regulier E, Herzenberg LA, Herzenberg LA. Proc.Natl.Acad.Sci.U.S.A. 2008;105:5111. doi: 10.1073/pnas.0712363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donnadieu E, Bismuth G, Trautmann A. J.Biol.Chem. 1992;267:25864. [PubMed] [Google Scholar]
- 42.Donnadieu E, Cefai D, Tan YP, Paresys G, Bismuth G, Trautmann A. J Immunol. 1992;148:2643. [PubMed] [Google Scholar]
- 43.Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, Nussenzweig MC. Nat Immunol. 2004;5:1243. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 44.Miller MJ, Safrina O, Parker I, Cahalan MD. J Exp.Med. 2004;200:847. doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viola A, Lanzavecchia A. Science. 1996;273:104. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 46.Yang and Parkhouse. Immunology. 1998;93:26. doi: 10.1046/j.1365-2567.1998.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Curtsinger JM, Schmidt CS, Mondino A, Lins DC, Kedl RM, Jenkins MK, Mescher MF. J Immunol. 1999;162:3256. [PubMed] [Google Scholar]
- 48.Friedman RS, Jacobelli J, Krummel MF. Nat Immunol. 2006;7:1101. doi: 10.1038/ni1384. [DOI] [PubMed] [Google Scholar]
- 49.Kupfer A, Kupfer H. Semin Immunol. 2003;15:295. doi: 10.1016/j.smim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Bode U, Wonigeit K, Pabst R, Westermann J. Eur.J.Immunol. 1997;27:2087. doi: 10.1002/eji.1830270837. [DOI] [PubMed] [Google Scholar]
- 51.Pajusto M, Ihalainen N, Pelkonen J, Tarkkanen J, Mattila PS. Eur.J.Immunol. 2004;34:2771. doi: 10.1002/eji.200324761. [DOI] [PubMed] [Google Scholar]
- 52.Smith KA. Science. 1988;240:1169. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]