Abstract
Two syndromic cognitive impairment disorders have very similar craniofacial dysmorphisms. One is caused by mutations of SATB2, a transcription regulator, and the other by heterozygous mutations leading to premature stop codons in UPF3B, encoding a member of the nonsense-mediated mRNA decay complex. Here we demonstrate that the products of these two causative genes function in the same pathway. We show that the SATB2 nonsense mutation in our patient leads to a truncated protein that localizes to the nucleus, forms a dimer with wild-type SATB2 and interferes with its normal activity. This suggests that the SATB2 nonsense mutation has a dominant negative effect. The patient’s leukocytes had significantly decreased UPF3B mRNA compared to controls. This effect was replicated both in vitro, where siRNA knockdown of SATB2 in HEK293 cells resulted in decreased UPF3B expression, and in vivo, where embryonic tissue of Satb2 knock-out mice showed significantly decreased Upf3b expression. Furthermore, chromatin immunoprecipitation demonstrates that SATB2 binds to the UPF3B promoter, and a luciferase reporter assay confirmed that SATB2 expression significantly activates gene transcription using the UPF3B promoter. These findings indicate that SATB2 acts as an activator UPF3B expression through binding to its promoter. This study emphasizes the value of recognizing disorders with similar clinical phenotypes to explore underlying mechanisms of genetic interaction.
Introduction
Cognitive impairment occurs in approximately 2–3% of the population (Ropers and Hamel 2005). Whether syndromic or non-syndromic, it can be caused by genetic and/or environmental factors. There are at least 1,870 phenotypic entries with cognitive impairment as a feature in the OMIM database (searched May 2012). We identified an individual with profound mental delay, a jovial personality and craniofacial dysmorphism including a long thin face, facial asymmetry, cleft palate, anterior open bite and a pointed chin (MIM 119540); this person had a nonsense mutation in the Special AT-rich Sequence Binding 2 (SATB2) gene (Leoyklang et al. 2007). Subsequently, other individuals with syndromic cognitive impairment showing similar craniofacial features, including a long thin face, facial asymmetry, high-arched palate and a prominent chin (MIM 300676), were reported to have mutations in the UPF3 regulator of nonsense transcripts homolog B (UPF3B) gene (Tarpey et al. 2007).
SATB2 (MIM 608148) resides on chromosome 2q32–q33, contains 11 exons, and spans 191-kb. It encodes a 733-amino acid protein containing a Pfam-B_10016 or SATB domain required for dimerization (residues 57–231), two CUT domains with a DNA-binding motif (residues 352–437 and 482–560), a domain for DNA binding (residues 614–677) and a nuclear localization signal (NLS; residues 613–616) (FitzPatrick et al. 2003) (Fig. 1a). SATB2 specifically binds to genomic nuclear matrix attachment regions (MAR) and participates in transcription regulation and chromatin remodeling (Apostolova et al. 2010; Chung et al. 2010; Savarese et al. 2009). Satb2 knock-out mice have craniofacial abnormalities and defects in osteoblast differentiation and function (Dobreva et al. 2006). Human SATB2 defects cause cognitive deficits, craniofacial dysmorphism, behavioral changes and osteoporosis (Leoyklang et al. 2007; Tegay et al. 2009; Urquhart et al. 2009; Rosenfeld et al. 2009). Our patient harbored a heterozygous nonsense c.715C>T (p.R239X) mutation in exon 6 of SATB2. Previous expression studies showed that the mutant SATB2 mRNA was present (Leoyklang et al. 2007) and was expected to produce a truncated protein that retained the SATB2 dimerization domain, but lost the DNA binding motifs; this could exert a dominant negative effect.
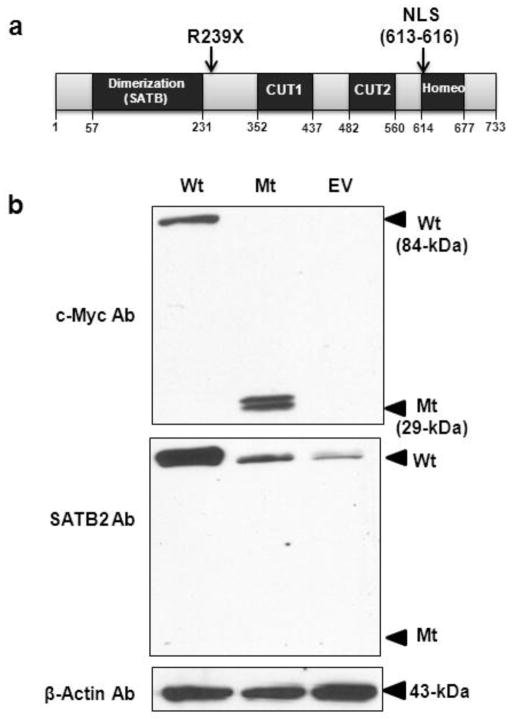
Fig. 1. Protein domains of SATB2 and expression of the wild-type and truncated SATB2 proteins.
(a) SATB2 protein outline. SATB2 contains 733 amino acids, including an N-terminal dimerization domain (residues 57–231), two CUT domains with DNA-binding motifs (residues 352–437 and 482–560), a DNA-binding domain (residues 614–677), and a nuclear localization signal (NLS; residues 613–616). The position of our patient’s nonsense mutation, p.R239X, is indicated.
(b) Immunoblot analysis. HEK293 cells transfected with the wild-type SATB2/myc-His plasmid (Wt; 84 kDa), the truncated SATB2/myc-His plasmid (Mt; 29 kDa), or with the empty pcDNA 3.1/myc-His plasmid (EV). Upper panel: Probing with anti c-Myc antibodies (Ab) showed SATB2 bands at the expected molecular weights. Middle panel; Reprobing the membrane with antibodies to the C-terminus of SATB2 showed abundant expression of SATB2 in the Wt SATB2 transfected cells, and less SATB2 expression in the Mt SATB2 and EV transfected cells. Lower panel: Reprobing the same membrane with anti β-actin antibodies demonstrated equal protein loading in all lanes.
UPF3B (MIM 300298) resides on chromosome Xq25-q26, contains 11 exons and spans 18.9 kb (Serin et al. 2001). It encodes a protein containing a region necessary for interaction with UPF2 (residues 30 – 255) (Serin et al. 2001), a region for binding to UPF2 (residues 52 – 57) (Kadlec et al. 2004), an EJC-binding domain (residues 418–432) (Buchwald et al. 2010), and a region for interaction with RBM8A (residues 430 – 447) (Gehring et al. 2003). The UPF3B protein is a member of the nonsense mediated mRNA decay (NMD) complex that rids eukaryotic cells of aberrant mRNAs containing premature termination codons. These are discriminated from true termination codons by downstream cis elements, such as exon-exon junctions (Lykke-Andersen et al. 2000). No Upf3B mutant mice have been reported. Human UPF3B defects cause cognitive deficits and craniofacial dysmorphisms (Tarpey et al. 2007). Since the two syndromic cognitive impairment entities had similar craniofacial dysmorphisms, we explored whether the two underlying genes could function in the same signaling pathway.
We noticed that the severe clinical features of our SATB2 mutated patient significantly overlapped with those of patients carrying UPF3B mutations. Our patient is so far the only reported person with a nonsense mutation in SATB2; other SATB2 deficient patients had either small interstitial deletions leading to haploinsufficiency (Rosenfeld et al. 2009) or a missense mutation with unknown pathomechanism (Rauch et al. 2012). Another group of patients had chromosomal aberrations involving 2q32-q33 which may also affect other genes (Urquhart et al. 2009; Rosenfeld et al. 2009; Tegay et al. 2009; FitzPatrick et al. 2003). We examined the molecular mechanism, i.e., haploinsufficiency versus a dominant negative effect, leading to the phenotypic abnormalities of our patient. And we subsequently explored potential genetic interactions between SATB2 and UPF3B.
Methods
Plasmid constructs
Wild-type and truncated (expressing amino acid 1-238, mimicking our patient’s p.R239X mutation) SATB2 cDNA were amplified from the IRAMp95E224Q clone (SATB2 cDNA inserted in pCR-Blunt II-TOPO vector; RZPD, Berlin, Germany) using primers shown in Supplementary Table 1. The PCR products were subcloned into pGEM-T Easy (Promega, Madison, WI), and transferred into the EcoRI-SfuI sites of pcDNA™3.1/myc-His version C (Invitrogen, Carlsbad, CA), resulting in a myc-His tagged C-terminus of the SATB2 translated proteins.
Cell culture, transfections and subcellular fractionations
Human Embryonic Kidney 293 (HEK293) cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with penicillin-streptomycin-glutamine (PSG) and 10% fetal bovine serum (Invitrogen). 2 × 106 HEK293 cells were transfected with 2 μg of wild-type (Wt) or truncated (Mt) SATB2/myc-His plasmids or the empty vector (pcDNA™3.1/myc-His) by electroporation, using an Amaxa Nucleoporator, Q001 program, nucleofector reagent kit V, according to the manufacturer’s instructions (Lonza group Ltd., Basel, Switzerland). After transfections (24 hr), cells were harvested and separated into one portion for whole cell extracts and one portion for nuclear-cytoplasmic extractions using a NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific, Waltham, MA). All extracts were immunoblotted as described below.
Immunoblotting
SATB2-transfected cells were harvested and lysed in 250 μl lysis buffer (50 mM Tris, 300 mM NaCl, 0.5% Triton, and 5 mM EDTA) in the presence of protease inhibitors (Complete Mini; Roche Applied Science, Mannheim, Germany). Total protein concentrations were measured by the DC Protein assay (Bio-Rad Laboratories, Hercules, CA). Total protein extracts (25 μg) were mixed with 2x loading dye, Laemmli Sample buffer (Bio-Rad Laboratories), boiled at 95° C for 5 min, and loaded onto 4–12% Tris-Glycine gels (Invitrogen), followed by electroblotting onto nitrocellulose membranes (Invitrogen). The membranes were probed with one of the following primary mouse monoclonal antibodies: anti-c-Myc (sc-40, Santa Cruz Biotechnology, Santa Cruz, CA), anti-SATB2 raised against a recombinant protein corresponding to the C-terminal region of human SATB2 (Sc-81376, Santa Cruz Biotechnology), anti β-actin (Sigma Aldrich, St. Louis, MO), or anti-α-tubulin (CP06, Calbiochem EMD Millipore, Billerica, MA). Detection was performed with either horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibodies for Enhanced Chemiluminescence (ECL) detection (Amersham Biosciences, Piscataway, NJ) (Fig. 1) or with anti-mouse secondary antibodies were IRDye800CW-conjugated for Li-Cor Odyssey Infrared detection (Li-Cor Biosciences, Lincoln, NE) (Figs. 2 and 3). Before reprobing with other primary antibodies, membranes were stripped with Restore™ Western Blot Stripping Buffer (Thermo Fisher Scientific Inc. Rockford, IL).
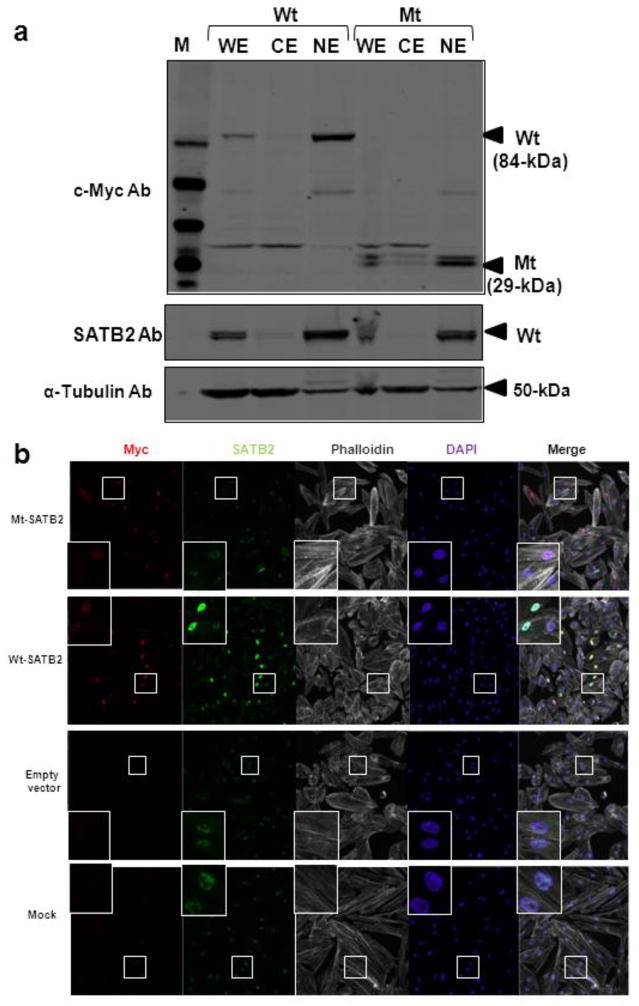
Fig. 2. Nuclear localization of the wild-type and truncated SATB2.
HEK293 cells were transfected with the Wt (lanes 2–4) or Mt (lanes 5–7) SATB2/myc-His plasmids.
(a) Immunoblotting of subcellular fractionated cell extracts with anti c-Myc antibodies (Ab) showed that both the Wt and Mt SATB2 proteins were present in the whole cell extracts (WE) and nuclear extracts (NE) and absent from cytoplasmic extracts (CE), upper panel. The membrane was reprobed with antibodies to the C-terminus of SATB2, middle panel and α-tubulin as loading control, lower panel.
(b) Immunofluorescence of normal human fibroblasts transfected with Wt or Mt SATB2/myc-His plasmids. Cells were stained with rabbit anti c-Myc (red) or mouse anti-SATB2 (green) antibodies, the phalloidin (staining F-actin) to outline cell boundaries (white), and the nuclear dye DAPI (blue). Merged images showed nuclear localization of the Wt and Mt proteins. Note that mock transfected cells, empty vector transfected cells and cells transfected with Mt SATB2/myc-His showed endogenous SATB2 upon SATB2 antibody staining.
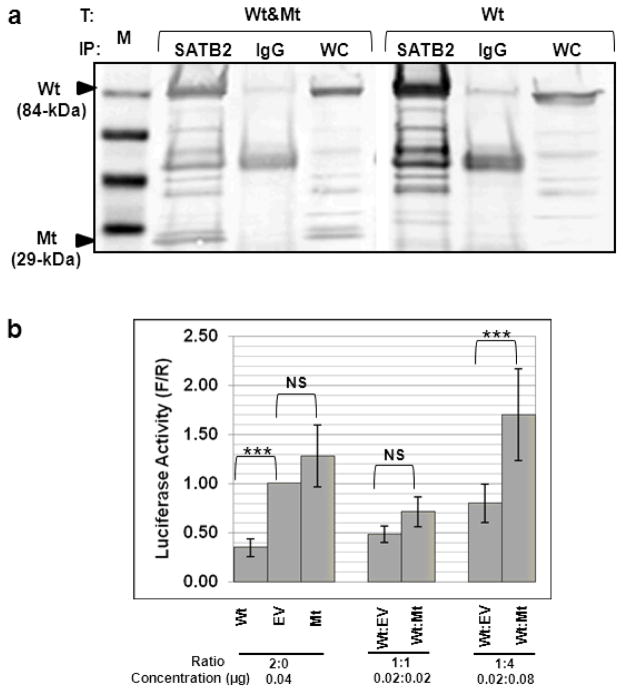
Fig. 3. Interaction between the wild-type and truncated SATB2.
(a) HEK293 cells were co-transfected (T) with a combination of the Wt and Mt SATB2/myc-His plasmids (Wt&Mt), or with only the Wt SATB2/myc-His plasmid (Wt). Cell extracts were immunoprecipitated (IP) with anti-SATB2 or mouse IgG (negative control) antibodies. All IP-fractions and whole cell extracts (WC) were immunoblotted with c-Myc antibodies. The Mt protein was detected in cells transfected with both Wt&Mt SATB2 plasmids (WC fraction) and precipitated with SATB2 antibodies (SATB2 fraction), indicating interaction of the Wt SATB2 with the Mt SATB2. Cells transfected with the Wt plasmid only did not show the 29 kDa protein band, as expected. M = SeeBlue® Plus2 Pre-Stained Standard (Invitrogen).
(b) Firefly (F) luciferase activities (from the MAR SATB2 reporter plasmid) shown as fold of Renilla (R) luciferase activity (from normalization plasmid) (F/R, Y-axis) in cells co-transfected with different ratios of the Wt or Mt SATB2/myc-His plasmid, or the empty pcDNA3.1/myc-His vector (EV). Transfection of the Wt SATB2 repressed MAR-luciferase activity (likely due to repression by SATB2 dimers) compared to EV or Mt transfections. Co-transfections of the Wt with either EV or Mt SATB2 decreased repression (likely due to decreased or aberrant SATB2 dimer formation). The activity of each combination of plasmids was assayed in three independent experiments and the means and standard errors were calculated (***, ANOVA, P< 0.001).
Immunofluorescence
Normal human fibroblasts were cultured in DMEM supplemented with PSG and 10% fetal bovine serum (Invitrogen). 4×105 cells were transfected with 2 μg of the wild-type or truncated SATB2/myc-His plasmids by electroporation, using an Amaxa Nucleoporator, U023 program, nucleofector reagent kit NHDF, according to the manufacturer’s instructions (Lonza group Ltd., Basel, Switzerland). The transfected cells were transferred into Lab-Tek®II Chamber Slides, 4 wells, 4.0 cm2 growth area per well (Nunc, Thermoscientific Roskilde, Denmark). The cells were fixed 24 hr post-transfection with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in phosphate buffered saline (PBS)/2 mg/ml bovine serum albumin (BSA)/1 mM NaN3, followed by blocking with 0.05% Tween-20 in PBS/2 mg/ml BSA/1 mM NaN3. Cells were stained with a combination of rabbit anti-Myc (Cell Signaling Technology, Inc, Danvers, MA) and mouse anti-SATB2 (Santa Cruz Biotechnology) antibodies followed by secondary anti-rabbit Alexa Fluor 555 (Invitrogen) and secondary anti-mouse Alexa Fluor 488 (Invitrogen). Cells were co-stained with Phalloidin (Invitrogen) that stained cytosolic F-actin and the nuclear dye DAPI (Vector Laboratories, Burlingame, CA). Cells were imaged with a Zeiss510META confocal laser-scanning Microscopy (Carl Zeiss, Jena, Germany).
Co-immunoprecipitation (co-IP)
HEK293 cells were transfected with either a combination of Wt and Mt SATB2/myc-His plasmids, or only the Wt plasmid. Cells were harvested 48 hr post-transfection in lysis buffer (150 mM NaCl, 1% Triton-X, 50 mM M Tris-Cl, pH 8.0). Protein extracts were pre-cleared with 1 μg of mouse IgG antibodies (Santa Cruz Biotechnology, Inc.) and 100 μl (1 mg) of Dynabeads M-280 linked to sheep anti-mouse IgG (Invitrogen) in an end-over-end rotating mixer for 1 hr, at 4°C. The pre-cleared proteins were then mixed with 2 μg of SATB2 antibodies or 1 μg mouse IgG antibodies (negative control) and mixed overnight (all mixing occurred in an end-over-end rotating mixer at 4°C). Dynabeads linked to M-280 sheep anti-mouse IgG (Invitrogen) were then added and mixed for 2 hr. After washing the beads three times with cell lysis buffer, immunoprecipitated proteins were eluted from the beads by boiling for 5 min in 2× sodium dodecyl sulfate (SDS) loading buffer and subjected to immunoblotting as described above.
Luciferase reporter assays
COS7 cells were grown in DMEM supplemented with PSG and 10% fetal bovine serum (Invitrogen), and seeded in 12-well plates (Nunc). The cells were transfected 24 hr after seeding (Lipofectamine 2000; Invitrogen) with combinations of the following plasmids. For determining interference between Wt and Mt SATB2 on SATB2 mRNA transcription activity, a firefly luciferase reporter plasmid (0.4 μg) pFos-Luc containing the nuclear matrix attachment region (MAR) recognized by SATB2 (kindly provided by Professor Gergana Dobreva, Max-Planck Institution for Immunobiology, Freiburg, Germany) was co-transfected with either Wt or Mt SATB2/myc-His plasmid or empty pcDNA3.1/myc-His vector (EV). To compensate for endogenous expressed SATB2 in COS7 cells, the cells were also co-transfected with varying ratios of Wt or Mt SATB2/myc-His plasmids: (Wt:Mt ratios of 1:1 and 1:4 (1 portion = 0.02 μg)) or with Wt:EV as a negative control.
For determining transcription factor activity of SATB2 on the UPF3B promoter, a firefly luciferase reporter plasmid was constructed by subcloning the UPF3B promoter from position -1 to -633 (ENSR00000261309) into the KpnI-HindIII sites of pGL4.10 [luc2] (Promega), using primers listed in Supplementary Table 1. This UPF3B promotor-pGL4.10 firefly reporter plasmid (0.4 μg) was then co-transfected with five concentrations of the Wt SATB2/myc-His plasmid (0.004, 0.012, 0.04, 0.12, and 0.4 g) or with the empty pcDNA3.1/myc-His vector (EV) as a control.
All above transfections included 0.04 μg of the pGL4.74 [hRluc/TK] plasmid encoding Renilla luciferase to normalize for transfection efficiency. Both firefly and Renilla luciferase activities were assayed 48 hr post-transfection using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) on a VICTOR3 Multilabel Plate Reader (PerkinElmer, Waltham, MA). All luciferase assays were performed in three independent experiments with each reaction performed in triplicate per experiment. P-values were calculated with the ANOVA test.
SATB2 small interfering RNA (siRNA) transfections
We acquired a SATB2 Trilencer kit, containing 3 SATB2 siRNAs (A, B, C) (SR308179; Origene, Rockville, MD). HEK293 cells were transfected with a combination of 2 specific siRNAs against human SATB2; Set I (siRNA A+B+C) and Set II (siRNA B+C), or a negative control siRNA (Trilencer-27 Universal Scrambled Negative Control siRNA Duplex, SR30004; Origene). For all transfections, the siTran 1.0 system was used according to the manufacturer’s protocols (TT300001; Origene). Transfected cells were cultured in six-well plates. The cells were harvested and analyzed 48 hr post-transfection. For RNA extraction, we used combination of Trizol (Invitrogen) and the RNA-Easy Mini-Kit (QIAGEN) according to the manufacturer’s protocols. RNA was treated with DNA-free DNase (Applied Biosystems, Austin, TX). First-strand cDNA was synthesized with a high-capacity RNA-to-cDNA kit (Applied Biosystems).
Satb2 knock-out mice
Satb2 knockout mice were created as described (Dobreva et al. 2006). Heterozygous knockout mice have no phenotype and homozygous Satb2 mutant mice die shortly after birth (Dobreva et al. 2006). Therefore, embryos from a heterozygous mating were collected at embryonic day (E) 18.5, and kidney tissues (Satb2 and Upf3b genes are endogenously expressed in embryonic kidneys) were isolated and snap frozen. Genotyping of the embryos was performed as described (Dobreva et al. 2006). No wild type embryos were present in this litter, therefore, four heterozygous embryos and one homozygous embryo were used for our gene expression experiment (Fig 4d). RNA was isolated from the embryonic kidney tissues using a combination of Trizol (Invitrogen) and the RNA-Easy Mini-Kit (QIAGEN) according to the manufacturer’s protocols. RNA was treated with DNA-free DNase (Applied Biosystems, Austin, TX). First-strand cDNA was synthesized with a high-capacity RNA-to-cDNA kit (Applied Biosystems).
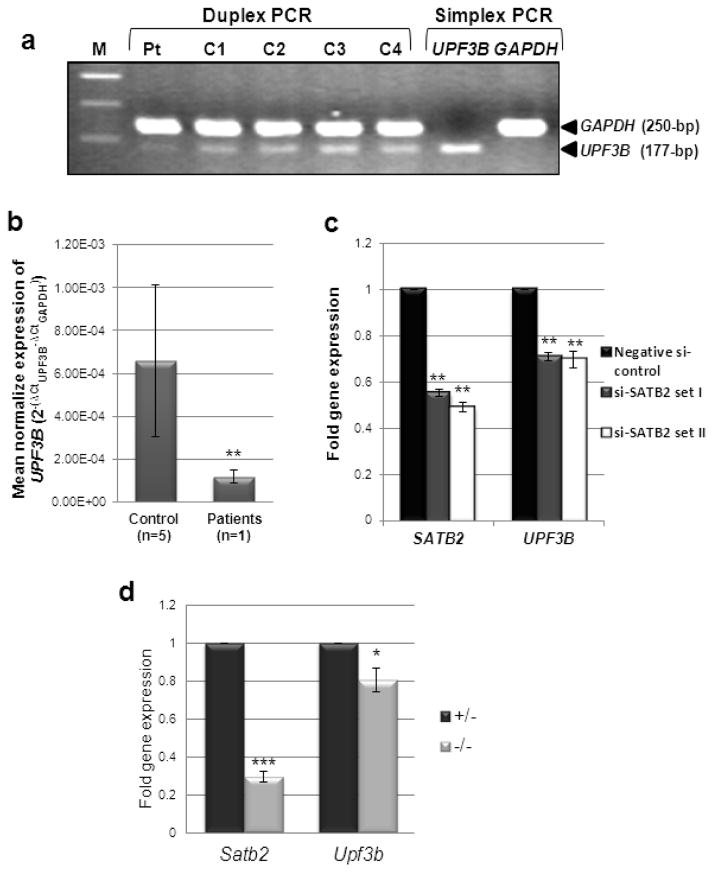
Fig. 4. UPF3B mRNA expression in SATB2-deficient cells.
(a) Duplex RT-PCR of leukocyte cDNA showing lower levels of UPF3B expression in the SATB2 mutated patient (Pt) compared to those in unaffected female (C1 and C4) or male (C2 and C3) controls. GAPDH was served as a normalizing control gene. Simplex PCR of UPF3B and GAPDH in normal cDNA is shown in lanes 7 and 8. Lane 1; 100-bp DNA ladder.
(b) UPF3B mRNA expression by qRT-PCR in leukocyte cDNA of our SATB2 mutated patient is significantly decreased compared to unaffected controls. (**, unpaired t-test, P=0.0008).
(c) HEK293 cells were transfected with two sets (I and II) of siRNAs against SATB2 or a negative siRNA control. SATB2 and UPF3B expression were measured by qRT-PCR and normalized to β-actin expression. Data are represented as means ± standard deviation (Two sample unequal variant t-test, **P< 0.01).
(d) mRNA was extracted from heterozygous and homozygous mutant Satb2 knockout mouse littermate embryonic kidneys (age E18.5). Both Satb2 and Upf3b expression, tested by qRT-PCR, was significantly decreased in mutant (−/−) kidneys compared to that of heterozygous (+/−) littermates. Experiments were performed three times in triplicate. Data are represented as means ± standard deviation. (Two sample unequal variant t-test, *P<0.05, ***P< 0.001).
RNA analysis
Total RNA was isolated from peripheral blood leukocytes of the patient (after obtaining written informed consent) and unaffected controls using a QIAamps RNA blood mini kit (Qiagen, Valencia, CA). mRNA (225 ng) of each sample was subjected to DNase I treatment, followed by reverse transcription into cDNA using ImProm-II TM reverse transcriptase (Promega). cDNAs of our patient (Pt), two male and two female Thai unaffected controls (C1-C4) were subjected to PCR amplification (standard conditions) of UPF3B in simplex or in duplex with GAPDH (normalization control). For primer sequences, see Supplementary Table 2. For quantitative real-time PCR (qRT-PCR) on patient’s cDNA, each reaction contained 100 ng cDNA, 0.2 μM of each primer and UPF3B or GAPDH labeled probes (Supplementary Table 2), and TaqMan® Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA) in a final volume of 20 μl. Reactions were carried out using a StepOnePlus™ V2.1 Real-Time PCR System (Applied Biosystems) for 40 cycles (95°C for 15 s and 60°C for 60 s) following the manufacturer’s recommendations.
For qRT-PCR experiments on the siRNA transfected HEK293 cells, human TaqMan gene expression assays for the SATB2 (Hs00392652), UPF3B (Hs00224875) and ACTB (Hs99999903) genes were used (Applied Biosystems). For qRT-PCR experiments on embryonic mouse kidney mRNA, mouse TaqMan gene expression assays for the Satb2 (Mm00507331), Upf3b (Mm01297389) and B2m (Mm00437762) genes were used (Applied Biosystems). qRT-PCR was performed with100 ng cDNA on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The cycling conditions were as above. Data were analyzed with the comparative CT method (ΔΔCT), which measures relative gene expression (Livak and Schmittgen 2001). All experiments were performed three times in triplicate.
Chromatin immunoprecipitation assay (ChIP)
HEK293 cells were co-transfected with the Wt SATB2/myc-His plasmid and the UPF3B-promoter-containing luciferase reporter plasmid (UPF3B promoter-pGL4.10). After 18 hours, cells were harvested and lysed. ChIP assay was performed using the MAGnify™ Chromatin Immunoprecipitation System kit (Invitrogen) according to the manufacturer’s protocols. Protein lysates were immunoprecipitated with a SATB2 antibody (Santa Cruz Biotechnology). A mouse IgG antibody (Santa Cruz Biotechnology) was used as a negative control. Primers specific for the UPF3B promoter were used to amplify DNA fragments (Supplementary Table 1).
Results
Detection Mt SATB2 protein
We previously showed the existence of SATB2 mRNA, despite the presence of a nonsense mutation (c.715C>T; p.R239X), in our patient’s cells (Leoyklang et al. 2007). Here, we studied whether the mutant protein was translated and maintained. We transfected both wild-type (Wt) and truncated (Mt) SATB2/myc-His plasmids into HEK293 cells (a cell type with endogenous expression of both SATB2 and UPF3B), and performed immunoblotting assays. After transfections, both Wt and Mt SATB2 proteins were detected with anti-c-Myc tag antibodies at expected molecular weights of 84 and 29 kDa, respectively (Fig. 1b). SATB2 antibodies against the C-terminus of SATB2 detected abundant Wt SATB2 in transfected cells. Cells transfected with Mt SATB2/myc-His (lacking the C-terminal recognition domain of SATB2 antibodies) or the empty vector (EV) showed a significantly lighter band upon SATB2 antibody staining, representing the endogenous SATB2 in HEK293 cells (Fig. 1b).
Nuclear localization of Mt SATB2 protein
To investigate the localization of Mt SATB2 protein lacking the nuclear localization signal (NLS), subcellular fractionation and immunofluorescence assays were performed in HEK293 cells. Immunoblotting showed that both Wt and Mt SATB2 proteins were present in nuclear extracts (NE) and were not detected in cytosolic fractions (CE) (Fig. 2a). Furthermore, immunofluorescence studies of transfected HEK293 cells also demonstrated localization of both Wt and Mt SATB2 proteins to the nucleus (Fig. 2b).
Interaction of Wt and Mt SATB2
Although Mt SATB2 lacks CUT, NLS and homeodomains, it still contains the dimerization domain (Fig. 1a). Co-IP studies demonstrated that the 29-kDa Mt SATB2 protein was pulled down along with the 84-kDa Wt SATB2 protein using an antibody specific to the SATB2 C-terminus (Fig. 3a). This indicated that Mt SATB2 retained its ability to dimerize and/or interact with Wt SATB2 (Fig. 3a). The Wt SATB2/myc-His construct was used to confirm antibody specificity.
Effect Mt SATB2 on Wt SATB2 mRNA transcription
SATB2 can regulate transcription regulation by binding to genomic nuclear matrix attachment regions (MAR). We determined the effects of Mt and Wt SATB2 proteins on luciferase expression regulated by a MAR sequence (subcloned into the pFos-Luc plasmid). Co-expression of the MAR-pFos-Luc plasmid with the Wt SATB2/myc-His plasmid showed significantly decreased levels of luciferase activity, while co-expression of the MtSATB2/myc-His plasmid exhibited luciferase activity similar to that of the empty vector (EV) (Fig. 3a). These results suggest that the Wt SATB2 protein acts as an inhibitor of MAR-regulated transcriptional activity. With increasing concentrations of the Mt SATB2 protein co-translated with Wt SATB2, the inhibitory effect decreased, with a significant difference between Wt:Mt and Wt:EV at the ratio of 1:4 (Fig. 3b). This suggests that the Mt SATB2 protein has a dominant negative effect on promoter activity of the Wt SATB2 protein, possibly by forming aberrant dimers unable to bind to the MAR sequences.
Decreased UPF3B mRNA expression in the SATB2 mutated patient
To explore whether SATB2 can act as a transcription regulator for UPF3B, we first determined the effect of our patient’s nonsense SATB2 mutation (p.R239X) on UPF3B mRNA expression in his leukocytes. Duplex reverse transcribed (RT) PCR showed decreased UPF3B expression in the patient’s cells compared to that of four unaffected ethnic-matched controls (Fig. 4a). These results were confirmed by qRT-PCR, which revealed significant reduction of UPF3B mRNA levels in the patient’s leukocytes compared to those of five age-, sex-, and ethnicity-matched controls (unpaired t-test, P=0.0008). Experiments were performed twice, in triplicate (Fig. 4b).
SATB2 regulates UPF3B expression
To further explore SATB2 regulation of UPF3B expression, we employed an in vitro (siRNA knockdown of SATB2 in HEK293 cells) and an in vivo (embryonic kidneys of Satb2 knock-out mice) system. Two combinations of SATB2 siRNA were transfected into HEK293 cells. We first confirmed that transfection of SATB2 siRNAs (Set I and Set II) significantly decreased SATB2 expression (Fig. 4c). We then demonstrated that decreased siRNA-induced SATB2 expression in HEK293 cells resulted in significantly decreased UPF3B expression, indicating that UPF3B is likely a downstream target of SATB2.
We next compared embryonic kidney mRNA of heterozygous (+/−) Satb2 knockout mice to that of a homozygous mutant (−/−) littermate (wild type embryos were not available to us). Both Satb2 as well as Upf3b mRNA expression was significantly decreased in mutant embryonic kidney tissue (Fig. 4d). These data confirmed a regulation of Upf3B gene expression by the Satb2 protein in vivo.
SATB2 protein binds and activates the UPF3B promoter
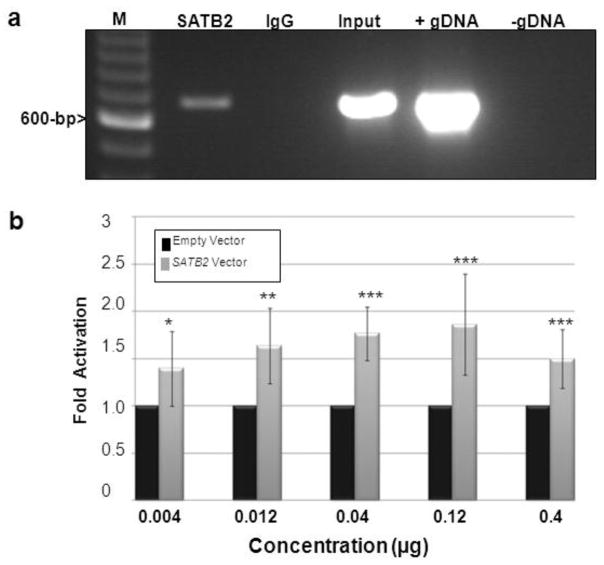
After ChIP with SATB2 antibodies in Wt SATB2/myc-His transfected HEK293 cells, UPF3B promoter DNA sequence was demonstrated to be present in the immunoprecipitates, but was absent from IgG antibody immunoprecipitates (Fig. 5a). These results indicate that SATB2 was able to bind to the UPF3B promoter. We next demonstrated the activating effects of the Wt SATB2 protein on UPF3B promoter activity by luciferase reporter assays. With transfecting increasing concentrations of Wt SATB2/myc-His plasmid at 0.004, 0.012, 0.04, 0.12, and 0.4 g, luciferase activity was augmented by 1.4 (P=0.032), 1.6 (P=0.0022), 1.8 (P=0.00018), 1.9 (P=0.001), and 1.5 (P=0.0013) fold respectively, compared with the empty vector (Fig. 5b), strongly suggesting that Wt SATB2 binds and activates the UPF3B promoter.
Fig. 5. SATB2 protein binds and activates the UPF3B promoter.
(a) Chromatin fragments of Wt SATB2/myc-His transfected HEK293 cells were affinity-purified by either a SATB2 antibody (SATB2, lane 2), or a mouse IgG antibody as a negative control (IgG, lane 3), followed by PCR amplification with primers located in the UPF3B promoter. PCR fragments were run on an agarose gel and visualized with ethidium bromide. Input (lane 4) represents PCR products of bulk chromatin extracts (before immunoprecipitation). Positive (+ control gDNA, lane 5) and negative (− control gDNA, lane 6) controls of PCR preparation were included. M (lane 1):100-bp DNA ladder.
(b) Firefly luciferase activities from the UPF3B promoter reporter plasmid, normalized to Renilla luciferase activity, were measured with co-expression of increasing amounts of the Wt SATB2/myc-His plasmid (gray bars) as indicated. Expression is reported as fold-activation of the SATB2/myc-His transfected cells relative to that of the empty pcDNA3.1/myc-His vector transfected cells (black bars). Experiments were performed three times in triplicate. Data are represented as means ± standard deviation. (ANOVA, *P<0.05, **P< 0.01, ***P< 0.001).
Discussion
SATB2 is a nuclear matrix protein that acts as a molecular node in the transcriptional network that regulates skeletal development (including craniofacial patterning) through regulating transcription factors involved in osteoblast differentiation (Dobreva et al. 2006; Srinivasan et al. 2012). SATB2 has also been shown to be a specific marker of colorectal origin among carcinomas (Magnusson et al. 2011; Eberhard et al. 2012). Here we identify UPF3B, encoding a member of the NMD complex, as a new binding partner of SATB2. This finding associates SATB2 function to the nonsense-mediated mRNA decay pathway.
Based on molecular pathomechanism, SATB2 mutations can be divided into three groups. The first group includes small interstitial deletions spanning only SATB2, leading to haploinsufficiency. Three patients with intellectual disability, microcephaly, growth retardation, thin and sparse hair, feeding difficulties and cleft or high-arched palate had deletions that ranged in size from 173.1 kb to 185.2 kb and spanned a part of SATB2 (Rosenfeld et al. 2009). The second group contains chromosomal aberrations, either large deletions or translocations involving 2q32-q33 which may affect more than one gene. For example, a patient with developmental delay and cleft palate had a 4.5 Mb deletion including the SATB2 gene (Urquhart et al. 2009). Four patients with severely intellectual disability and cleft or high-arched palate shared an 8.1 Mb minimal deleted region of 2q32 (van Buggenhout et al. 2005). Another patient with a diagnosis of Toriello-Carey syndrome had agenesis of the corpus callosum, cleft palate/Robin sequence, cardiac defect, hypotonia, intellectual disability with microcephaly, growth retardation, and facial dysmorphism. He had an apparently balanced de novo translocation between chromosomes 2 and 14 [46,XY,t (2;14)(q33;q22)], with the break on chromosome 2 interrupting the SATB2 gene (Tegay et al. 2009). A female with 46,XX,t(2;10)(q33;q21.2) disrupting SATB2 had developmental delay, cleft palate and behavioral abnormalities (Baptista et al. 2008). The two girls first found to have apparently balanced chromosomal translocations involving chromosome 2q32 disrupting SATB2 also had cleft palate, facial dysmorphism and mild learning disability (Brewer et al. 1999; FitzPatrick et al. 2003). Genes other than SATB2 may contribute to the phenotypes of the patients with chromosomal aberrations. The third group contains patients with intragenic SATB2 (point) mutations. This group includes our patient with the SATB2 p.R239X nonsense mutation (Leoyklang et al. 2007), as well as the recently reported female patient with severe intellectual disability and bifid uvula with SATB2 p.V381G mutation (Rauch et al. 2012). The pathomechanism of these point mutations remains unknown. We investigated whether a dominant negative effect could cause our patient’s phenotypic abnormalities.
We previously showed expression of mutant SATB2 mRNA caused by a nonsense mutation, c.715C>T (p.R239X), in our patient’s leukocytes (Leoyklang et al. 2007). We now demonstrate the presence of the truncated SATB2 protein of 238 residues (29 kDa) (Fig. 1b), which retains its dimerization domain (residues 57–231) but lacks the two functional CUT domains, homeodomain, and NLS domain (residues 613–616) (Fig. 1a). We also showed that the truncated SATB2 was localized to the nucleus, similar to the wild-type SATB2 (Fig. 2). Most proteins destined to the nucleus require a NLS (Schlenstedt 1996). Some proteins lacking the NLS, however, could enter the nucleus by either interacting with another protein containing the NLS (Steidl et al. 2004) or via passive diffusion, if their molecular weights are less than 50 kDa (Macara 2001). Our co-immunoprecipitation assays showed that the truncated SATB2 could bind to the wild-type SATB2 (Fig. 3a), providing an explanation for the presence of the truncated SATB2, lacking the NLS, in the nucleus.
Luciferase assays using a MAR sequence binding domain showed that the truncated SATB2 interfered with the repressive function of the wild-type SATB2 (Fig. 3b). These findings suggested that the truncated SATB2 had a dominant negative effect on the wild-type SATB2, possibly by formation of dimers unable to repress MAR-regulated gene expression, leading to the phenotypic abnormalities in our patient.
Patients with SATB2 mutations including ours had intellectual disability and craniofacial dysmorphisms similar to patients with UPF3B mutations. Patients with UPF3B mutations exhibit attention deficit hyperactivity disorder, childhood onset schizophrenia (Addington et al. 2011), intellectual disability with/without autism (Tarpey et al. 2007), and craniofacial dysmorphisms. Different phenotypes stemming from mutations of the same gene are not uncommon and could be related to different pathomechanisms, residual protein activities, and alleles of modifier genes (Grandis et al. 2008; Lebrun et al. 2011; To-Figueras et al. 2011). Regardless of the underlying mechanisms, some patients with UPF3B mutations had craniofacial dysmorphisms similar to those of patients with SATB2 mutations. It is also known that SATB2 can serve as both an activator and a repressor in different cell types (Dobreva et al. 2003; Ellies and Krumlauf 2006; Dobreva et al. 2006). This led us to investigate whether the products of the two genes functioned in the same pathway.
Hypothesizing that SATB2 is an activator of UPF3B, we first performed expression studies. We found significantly lower UPF3B mRNA levels in leukocytes of our SATB2 patient and in embryonic Satb2 deficient mouse kidney tissue than in unaffected controls (Fig. 4a,b,d). Down-regulation of SATB2 expression by siRNA also led to significantly decreased UPF3B expression (Fig. 4c). Using ChIP, we showed that the SATB2 protein could bind to the UPF3B promoter (Fig. 5a). In addition, the luciferase reporter assays showed that SATB2 could activate transcription of the UPF3B promoter, suggesting transcriptional activation of UPF3B by SATB2 (Fig. 5b). In our patient’s cells, the truncated SATB2 led to decreased UPF3B mRNA, likely causing decreased UPF3B protein. Since UPF3B is a member of the NMD complex, lower UPF3B levels may decrease the ability of cells to remove aberrant mRNAs containing premature termination codons, leaving the truncated SATB2 mRNA in our patient undegraded, creating a vicious cycle potentiating its effects.
There exist other examples of using similar human genetic disease clinical phenotypes to elucidate disease gene functions (van Driel et al. 2006). For example, the distinctive and overlapping features of Noonan syndrome, Costello syndrome and cardiofaciocutaneous (CFC) syndrome (facial appearance, cognitive deficits, heart defects, and musculocutaneous abnormalities) linked these disorders clinically (Aoki et al. 2008). Half of Noonan patients have mutations in the PTPN11 gene, while HRAS germline mutations are present in some patients with Costello syndrome; both genes function in the RAS-RAF-ERK pathway (Aoki et al. 2008). Based upon the observation of the clinical similarities among the three disorders, the KRAS and BRAF genes, encoding proteins functioning in the RAS-RAF-ERK pathway were identified as genes associated with CFC (Niihori et al. 2006). Similarly, the resemblance of two cognitive impairment disorders led us to demonstrate that the SATB2 protein acts upstream of UPF3B to regulate its transcription.
In summary, using a phenotype database to identify a new protein-gene interaction, we investigated two diseases with overlapping clinical features (profound cognitive impairment and palatal and craniofacial abnormalities) to link two different genes (SATB2 and UPF3B). Our evidence has indicated that the SATB2 protein acts as a transcriptional activator of the UPF3B gene.
Supplementary Material
Acknowledgments
We like to thank Drs. Gergana Dobreva and Rudolph Grosschedl (Max Planck Institution for Immunobiology and Epigenetics, Freiburg, Germany) for kindly providing the pfosluc-Wt SATB2 plasmid and Satb2 knockout mice. This study was supported by the Royal Golden Jubilee Ph.D. Program to PL (Grant No PHD/0026/2549); the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA; the Thailand Research Fund; The Integrated Innovation Academic Center, Chulalongkorn University Centenary Academic Development Project (CU56-HR05), and the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (HR1163A).
Abbreviations
- BSA
bovine serum albumin
- ChIP
Chromatin immunoprecipitation
- DMEM
Dulbecco’s modified Eagle medium
- E
embryonic day
- HEK
Human Embryonic Kidney
- hr
hours
- HRP
horseradish peroxidase
- MAR
matrix attachment regions
- min
minutes
- Mt
mutant/truncated
- NLS
nuclear localization signal
- NMD
nonsense mediated mRNA decay
- PBS
phosphate buffered saline
- PSG
penicillin-streptomycin-glutamine
- SATB2
Special AT-rich Sequence Binding 2
- SDS
sodium dodecyl sulfate
- siRNA
small interfering RNA
- UPF3B
UPF3 regulator of nonsense transcripts homolog B
- Wt
wild type
Footnotes
This article contains online supplementary materials, including five Tables.
References
- Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Germain G, Gochman P, Long R, Rapoport JL, Rouleau GA. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16:238–239. doi: 10.1038/mp.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- Apostolova G, Loy B, Dorn R, Dechant G. The sympathetic neurotransmitter switch depends on the nuclear matrix protein Satb2. J Neurosci. 2010;30:16356–16364. doi: 10.1523/JNEUROSCI.3502-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, Maloney V, Thomas NS, Jacobs PA, Crolla JA. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer CM, Leek JP, Green AJ, Holloway S, Bonthron DT, Markham AF, FitzPatrick DR. A locus for isolated cleft palate, located on human chromosome 2q32. Am J Hum Genet. 1999;65:387–396. doi: 10.1086/302498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci U S A. 2010;107:10050–10055. doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Lau J, Cheng LS, Grant RI, Robinson F, Ketela T, Reis PP, Roche O, Kamel-Reid S, Moffat J, Ohh M, Perez-Ordonez B, Kaplan DR, Irwin MS. SATB2 augments DeltaNp63alpha in head and neck squamous cell carcinoma. EMBO Rep. 2010;11:777–783. doi: 10.1038/embor.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17:3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard J, Gaber A, Wangefjord S, et al. A cohort study of the prognostic and treatment predictive value of SATB2 expression in colorectal cancer. Br J Cancer. 2012;106:931–938. doi: 10.1038/bjc.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellies DL, Krumlauf R. Bone formation: The nuclear matrix reloaded. Cell. 2006;125:840–842. doi: 10.1016/j.cell.2006.05.022. [DOI] [PubMed] [Google Scholar]
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, Markham AF, Fantes JA, Bonthron DT. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;1:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Grandis M, Vigo T, Passalacqua M, Jain M, Scazzola S, La Padula V, Brucal M, Benvenuto F, Nobbio L, Cadoni A, Mancardi GL, Kamholz J, Shy ME, Schenone A. Different cellular and molecular mechanisms for early and late-onset myelin protein zero mutations. Hum Mol Genet. 2008;17:1877–1889. doi: 10.1093/hmg/ddn083. [DOI] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- Lebrun AH, Moll-Khosrawi P, Pohl S, Makrypidi G, Storch S, Kilian D, Streichert T, Otto B, Mole SE, Ullrich K, Cotman S, Kohlschutter A, Braulke T, Schulz A. Analysis of Potential Biomarkers and Modifier Genes Affecting the Clinical Course of CLN3 Disease. Mol Med. 2011;17:1253–1261. doi: 10.2119/molmed.2010.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoyklang P, Suphapeetiporn K, Siriwan P, Desudchit T, Chaowanapanja P, Gahl WA, Shotelersuk V. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum Mutat. 2007;28:732–738. doi: 10.1002/humu.20515. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25 (4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson K, de Wit M, Brennan DJ, et al. Satb2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am J Surg Pathol. 2011;35:937–948. doi: 10.1097/PAS.0b013e31821c3dae. [DOI] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380(9854):1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Ballif BC, Lucas A, Spence EJ, Powell C, Aylsworth AS, Torchia BA, Shaffer LG. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS One. 2009;4:e6568. doi: 10.1371/journal.pone.0006568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F, Davila A, Nechanitzky R, De La Rosa-Velazquez I, Pereira CF, Engelke R, Takahashi K, Jenuwein T, Kohwi-Shigematsu T, Fisher AG, Grosschedl R. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 2009;23:2625–2638. doi: 10.1101/gad.1815709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G. Protein import into the nucleus. FEBS Lett. 1996;389:75–79. doi: 10.1016/0014-5793(96)00583-2. [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Leone DP, Bateson RK, Dobreva G, Kohwi Y, Kohwi-Shigematsu T, Grosschedl R, McConnell SK. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci U S A. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl S, Tuncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage AA. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J Mol Biol. 2004;342:515–524. doi: 10.1016/j.jmb.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegay DH, Chan KK, Leung L, Wang C, Burkett S, Stone G, Stanyon R, Toriello HV, Hatchwell E. Toriello-Carey syndrome in a patient with a de novo balanced translocation [46,XY,t(2;14)(q33;q22)] interrupting SATB2. Clin Genet. 2009;75:259–264. doi: 10.1111/j.1399-0004.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- To-Figueras J, Ducamp S, Clayton J, Badenas C, Delaby C, Ged C, Lyoumi S, Gouya L, de Verneuil H, Beaumont C, Ferreira GC, Deybach JC, Herrero C, Puy H. ALAS2 acts as a modifier gene in patients with congenital erythropoietic porphyria. Blood. 2011;118:1443–1451. doi: 10.1182/blood-2011-03-342873. [DOI] [PubMed] [Google Scholar]
- Urquhart J, Black GC, Clayton-Smith J. 4.5 Mb microdeletion in chromosome band 2q33.1 associated with learning disability and cleft palate. Eur J Med Genet. 2009;52:454–457. doi: 10.1016/j.ejmg.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Van Buggenhout G, Van Ravenswaaij-Arts C, Mc Maas N, Thoelen R, Vogels A, Smeets D, Salden I, Matthijs G, Fryns JP, Vermeesch JR. The del(2)(q32.2q33) deletion syndrome defined by clinical and molecular characterization of four patients. Eur J Med Genet. 2005;48:276–289. doi: 10.1016/j.ejmg.2005.05.005. [DOI] [PubMed] [Google Scholar]
- van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA. A text-mining analysis of the human phenome. Eur J Hum Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.