Abstract
Many of the actions of thyroid hormones (THs) occur via TH binding to intracellular receptors. Although it was long thought that THs diffused passively across plasma membranes, it is now recognized that cellular entry is mediated by a variety of membrane transporter proteins. In this study, we identified cDNAs encoding the TH transporters monocarboxylate transferase 8 (mct8) and 10 (mct10) as well as eight distinct organic anion-transporting polypeptide (oatp) proteins from fathead minnow (Pimephales promelas). Analysis of the tissue distribution of transporter mRNAs revealed that mct8 and mct10 transcripts were both abundant in liver, but also present at lower levels in brain, gonad and other tissues. Transcripts encoding oatp1c1 were highly abundant in brain, liver and gonad, and exhibited significant sex differences in the liver and gonad. Treatment of adult male minnows with 3,5,3′-triiodothyronine (T3) or the goitrogen methimazole altered gene transcript abundance for several transporters. Fish given exogenous T3 had reduced mct8 and oapt1c1 mRNA levels in the liver compared to methimazole-treated fish. In the brain, transcripts for mct8, mct10, oatp2b1, and oatp3a1 were each reduced in abundance in fish with elevated T3. As a whole, these results provide evidence that TH status influences the transcriptional dynamics of mct8, mct10 and several Oatp genes including oatp1c1 in teleost fish.
Keywords: monocarboxylate transporter, Mct, gene expression, OATP, plasma membrane, solute carrier organic anion, liver, brain, transport protein, SLC16, Pimephales promelas
1. Introduction
Thyroid hormones regulate a variety of physiological and behavioral functions in teleost fishes including growth and morphological development (Reddy and Lam, 1992; Power et al., 2001; Lema and Nevitt, 2006), reproduction (Jacob et al., 2005; Nelson and Habibi, 2006), and behaviors such as rheotaxis and olfactory imprinting (McCormick et al., 1998; Lema and Nevitt, 2004; Edeline et al., 2005). These diverse functions are regulated in part by variation in the secretion of THs from the thyroid follicles, but also via extrathyroidal conversion of circulating thyroxine (T4) either to the more bioactive 3,5,3′-triiodothyronine (T3) or to less active forms (e.g., rT3, T2) by iodothyroinine deiodinase enzymes in peripheral tissues (Orozco et al., 2012). These peripheral tissues are also the sites where THs induce cellular responses, either by binding to intracellular thyroid hormones receptors (TRs) that function as transcription factors to activate or repress gene transcription, or via interactions with structural membrane proteins (e.g., integrin ανβ3) to have non-genomic cellular actions (Bergh et al., 2005; Yonkers and Ribera, 2009; Cheng et al., 2010).
In mammals, it has recently become clear that the peripheral actions of THs also rely upon the presence of plasma membrane transporters expressed by a target cell or tissue (Bernal, 2005; Visser et al., 2011). Given the lipophilic properties of THs, it was long assumed that THs diffused passively across the lipid bilayer of plasma membranes (e.g., Robbins and Rall, 1960). It is now recognized, however, that transmembrane movement of THs is facilitated by several transporters belonging to the solute carrier (Slc) group of proteins, which includes the monocarboxylate transporter (Mct) and organic anion transporter polypeptide (Oatp) families of plasma membrane transporters (for reviews, see Bernal, 2005; Schweizer et al., 2008; Visser et al., 2011).
Of the Mct family, current evidence suggests that only Mct8 (solute carrier family 16 member 2, or Scl16a2) and Mct10 (solute carrier family 16 member 10, or Scl16a10) appear to function in the transport of THs (Halestrap & Meredith 2004, Visser et al., 2011), while other proteins in the Mct transporter family function in the movement of substrates such as aromatic amino acids, lactate, pyruvate, or ketone bodies across plasma membranes (Halestrap & Price 1999, Halestrap & Meredith 2004). Mct8 proteins from both human and rat show a high specificity for THs and have been documented to transport T4, T3, rT3, and T2 (Friesema et al., 2003; 2006; Visser et al., 2007). A recent study of the transport kinetics of zebrafish (Danio rerio) Mct8 indicated that this transporter likewise functions in TH transport, although the functional specificity of this fish protein may vary from that in mammals (Arjona et al., 2011). While not yet studied in any teleost fish, human Mct10 transports both T3 and T4, shows a greater affinity for T3, and transports aromatic amino acids in addition to THs (Friesema et al., 2008; Visser and Visser, 2013).
The Oatp superfamily of transmembrane proteins mediates sodium-independent cellular transport of a wide variety of amphipathic organic compounds including xenobiotics, hormones, hormonal conjugates, bile salts, and pharmaceutical drugs (e.g., Hagenbuch and Meier, 2003; 2004; Mikkaichi et al., 2004; Hagenbuch and Gui, 2008; Svoboda et al., 2011). While over 160 Oatps have been identified to date from the sequenced genomes of more than 25 species (Hagenbuch and Gui, 2008), many basic questions remain unaddressed about the tissue distribution, expressional regulation and functional differentiation of Oatps, even in humans (Svoboda et al., 2011). It is known, however, that select Oatps function in the transport of hormones or hormone metabolites including T3, T4, and glucuronidated steroid hormones (Pizzagalli et al., 2002; Hagenbuch, 2007; Hagenbuch and Gui, 2008), and recent studies in vertebrates have demonstrated a role for Oatps in hormone-associated contexts ranging from compensating for the endocrine disrupting effects of environmental pollutants (Szabo et al., 2009; Noyes et al., 2013), to changing their expression levels with hormone-dependent cancers (Pressler et al., 2011).
Mammalian Oatp1c1 (also known as Slco1c1 and Oatp14) has a high transport specificity for THs including T3, T4, rT3, and T4 sulfate (Sugiyama et al., 2003; Tohyama et al., 2004; Hagenbuch, 2007; van der Deure et al., 2008; Visser et al., 2011). Other mammalian Oatps including Oatp2b1, Oatp3a1, and Oapt4a1 also transport T4 and, in some cases, T3 or rT3, although often with lower affinity (Fujiwara et al., 2001; Huber et al., 2007; van der Deure et al., 2008; for review, see Visser et al., 2011). It is thus likely that several Oatps contribute to the membrane transport of THs. In teleost fishes, however, almost nothing is known about evolutionary diversity and functional specificity of Oatp transporter proteins. The sole exception to date is one recent study in zebrafish that identified 14 oatp genes belonging to the eight families of Oatp proteins present in tetrapod vertebrates (Popovic et al., 2010), indicating that Oatp transporters can be more diverse in teleost fishes than mammals.
In this study, we identified cDNAs encoding Mct8, Mct10 and eight distinct Oatps from the fathead minnow (Pimephales promelas), a teleost fish commonly employed for studies on the toxic and endocrine disrupting effects of chemicals (Ankley and Villeneuve, 2006). Using real-time quantitative reverse transcription PCR (qRT-PCR) methods, we document the tissue distribution patterns for transcripts encoding these Mct and Oatp transporter proteins and report on relative expressional differences between sexes. We next explored whether circulating thyroid hormone status influences the relative abundance of mct8, mct10, and oatp transcripts in the brain and liver by experimentally modulating circulating TH levels in adult male minnows via oral treatment with exogenous T3 or the goitrogen methimazole. In fish, as in mammals, the liver is the major organ of TH conversion and metabolism and therefore plays a key role in regulating systemic TH levels and effects (e.g., Van der Geyten et al., 1998; Malik and Hodgson, 2002). In the brain and nervous system of vertebrates, THs mediate cellular proliferation and neural differentiation (e.g., Forrest et al., 1996; Lema and Nevitt, 2004; Desouza et al., 2005), and limited evidence from the study of TH transporters in mammalian system indicates that TH status regulates mRNA and protein expression of Oatp1c1 in brain capillary endothelial cells (Sugiyama et al., 2003). We therefore hypothesized that circulating TH status would likewise influence the transcriptional dynamics for oatp1c1, and possibly for mct8 and mct10, in the liver and brain of this teleost fish model.
2. Materials and Methods
2.1. Isolation and sequencing of mct and oatp cDNAs
2.1.1. Total RNA isolation
Liver tissue was dissected from an adult male fathead minnow (Pimephales promelas) purchased from Aquatic BioSystems, Inc. (Fort Collins, CO, USA). The tissue was flash frozen in liquid N2 and stored at −80 °C. Total RNA was extracted using TriReagent (Molecular Research Center, Inc., Cincinnati, OH, USA) with bromochloropropane for phase separation. The resulting RNA was DNase I treated (DNAfree Kit, Ambion) and quantified by spectrophotometery (Nanodrop 2000, ThermoScientific).
First stand cDNA was generated in 20 µL reverse transcription reactions by incubating 5 µg of total RNA template (4.0 µL) with 1.0 µL random primers (random hexadeoxynucleotides; Promega Corp., Madison, WI, USA) at 70 °C for 10 min. Subsequently, the above mixture was combined with 3.0 µL of MgCl2 (25 mM), 1.0 µL deoxynucleotides triphosphates (dNTPs) (100 mM, Promega), 0.25 µL recombinant RNasin® ribonuclease inhibitor (20 U/µL, Promega), 4.0 µL of 5× reaction buffer, 0.5 µL GoScript™ reverse transcriptase, and 5.5 µL of nuclease-free H2O, as per the protocol of the GoScript™ Reverse Transcription System (Promega). RNA was reverse transcribed under a thermal profile of 25 °C for 5 min and 42 °C for 1 h, followed by 70 °C for 15 min to inactivate the reverse transcriptase enzyme.
2.1.2. Isolation and sequencing of partial cDNAs encoding mct8 and mct10
In silico BLAST searches using mammalian mct8 cDNA sequences identified ESTs encoding putative partial mct8 cDNAs from Chinese rare minnow (Gobiocypris rarus; GenBank accession no. EE393294), zebrafish (EH490251), and three-spined stickleback (Gasterosteus aculeatus, DT965355), as well as an unannotated protein sequence from pufferfish (Tetraodon nigroviridis, CAG09736). Nucleotide sequences from the ESTs were aligned, and a nested set of degenerate primers was designed to consensus regions of these aligned sequences. Nucleotide sequences for these degenerate primers were as follows: (forward1) 5'-CTGCTACATGGTGCAACGG-3', (forward2) 5'-ACATGATGCTGGTGAARSACC-3', (reverse3) 5'-AAATAATGMCCCAGGATCACCAGG -3', (reverse2) 5'-GGCTGRAAYGCAAAYGAGGA-3', and (reverse1) 5'-GAGGAGCCGCAGCCRAASA -3' (Integrated DNA Technologies, Inc., Coralville, IA, USA). These degenerate primers were then used in 50 µL PCR reactions containing 25 µL of GoTaq® Polymerase Colorless Master Mix (Promega), 21 µL nuclease-free H2O, 1 µL each of forward and reverse primers (50 µM), and 2 µL of cDNA. PCR products were examined on 1.2% agarose gels with EtBr, and any products of predicted size were cleaned (QIAQuick PCR Kit, Qiagen) and Sanger sequenced (MacrogenUSA, Rockville, MD, USA).
A sequence encoding mct10 from zebrafish (Danio rerio, GenBank accession no. NM_001080028) was located in GenBank (http://www.ncbi.nlm.nih.gov/) and used to BLAST search EST and genomic databases (www.ensemble.org) for additional sequences from teleost fishes. These in silico searches identified an EST encoding a partial cDNAs for mct10 from fathead minnow (DT292022), as well as unannotated ESTs encoding partial mct10 cDNAs from three-spined stickleback (DN712990), Atlantic salmon (Salmo salar, DY715060 and AM402521) and mandarin fish (Siniperca chuatsi, GR474247). An EST was also identified encoding mct10 from Japanese ricefish (medaka, Oryzias latipes, DK098183), which was combined with several genomic sequences for mct10 in this same species obtained from Ensemble. Resulting sequences were aligned using Sequencher v4 software (GeneCodes Corp., Ann Arbor, MI, USA), and gene-specific primers were designed to consensus regions and used to amplify a 615-bp nucleotide partial cDNA of mct10 from fathead minnow. These primers were the following: (forward1) 5'-TCCAGAACGCCTTCGGTATCATGT-3', (forward2) 5'-TTCTTCTGCTCTCCGATCGTCAGT-3', and (reverse) 5'-AAGGCCCATATGCGGTATCCAAGA-3' (Integrated DNA Technologies, Inc.).
2.1.3. Identification of oatp partial cDNA sequences
Deduced protein sequences from previously identified oatp cDNAs from zebrafish (Popovic et al., 2010) were used for in silico BLAST searches to identify EST sequences encoding putative oatp cDNAs from fathead minnow. EST’s identified from these searches were checked against zebrafish sequences to determine open reading frame regions, and then aligned using Sequencher v4 software (GeneCodes Corp.). Partial cDNAs encoding eight Oatps were identified from fathead minnow using this method: oatp1c1 (GenBank accession nos. DT233541 and DT235267), oatp1f1 (DT350917), oatp1f2 (DT350916), oatp2a1 (DT161775), oatp2b1 (DT195991), oatp3a1 (DT287125), oatp4a1 (DT228118, DT173660 and DT118223), and oatp5a1 (DT197132). Nested sets of gene-specific primers were designed for each of these eight oatp cDNAs, and used to amplify and sequence each oatp cDNA to confirm nucleotide composition. Nucleotide sequences for these primers are provided in Table 1 of the Online Supplemental Materials. Thermal cycling conditions for the PCR reactions were as follows: 95 °C for 2 min, 35 cycles of 95 °C for 30 s, 54 °C for 30 s, 72 °C for 1–2 min, and then 72 °C for 2–4 min, followed by a second round of PCR using 95 °C for 2 min, 30 cycles of 95 °C of 30 s, 55 °C for 30 s, and 72 °C for 1–2 min, and then a final extension step of 72 °C for 3–4 min. All PCR products were examined on 1.2% agarose gels, and the resulting PCR products were purified (QIAQuick PCR Kit, Qiagen) and Sanger sequenced (MacrogenUSA).
2.2. Phylogenetic analyses
Deduced amino acid sequences obtained from fathead minnow mct and oatp cDNAs were aligned to all teleost fish sequences of Mct8 and Mct10 proteins or Oatp proteins identifiable on GenBank, as well as select tetrapod sequences. Amino acid sequences for Mct8 and Mct10 were aligned using ClustalX software (Larkin et al., 2007), and an unrooted tree was assembled by the Neighbor-Joining method (Saitou and Nei, 1987) using the p-distance model with pairwise deletion of gaps in MEGA v5 software (Tamura et al., 2011). Confidence values for nodes were determined using bootstrap tests (1000 replicates). For the Oatp phylogeny, a tree was constructed by the maximum likelihood method using the Jones-Taylor-Thornton model (Jones et al., 1992) with all amino acid sites included. Again, 1000 bootstrap replicates were used to determine confidence values for each node.
2.3. Tissue distribution of relative mRNA abundance
Adult fathead minnows (Pimephales promelas) were purchased from Aquatic BioSystems, Inc. (Fort Collins, CO, USA), and maintained in closed system, 208 L tanks under conditions of 24–26 °C, 0.2 ppt salinity, and a 14L:10D photoperiod. Fish were maintained for 30 days prior to commencing any fish sampling. All procedures were approved by the Animal Care and Use Committee of California Polytechnic State University (Protocol no. 1207).
Adult male (n = 3; body mass, 3.96 ± 0.69 g [mean ± SEM]; standard length, 53.87 ± 3.45 mm) and female (n = 3; 1.74 ± 0.24 g; 41.07 ± 2.24 mm) minnows were euthanized in buffered tricaine methansulfonate (MS222), and the brain, gills, gonads, liver, intestine, spleen, kidney, skeletal muscle were dissected. The brain was further subdivided into the forebrain, hypothalamic, optic tectum, cerebellum and hindbrain (medulla oblongata). All tissues were immediately frozen in liquid N2 and stored at −80 °C.
Total RNA was extracted using TriReagent (Molecular Research Center, Inc.), DNase I treated (TURBO DNAfree Kit, Ambion), and quantified using a P300 NanoPhotometer (Implen, Inc., Westlake Village, CA, USA). Total RNA was then reverse transcribed in 30 µL reactions by incubating 15.5 µL of total RNA template (7 µg) with 6.0 µL 5× buffer, 0.25 µL of nuclease-free H2O (Sigma-Aldrich, St. Louis, MO, USA), 4.5 µL MgCl2, 1.5 µL dNTPs, 1.0 µL of random primers, 1.0 µL of GoScript™ reverse transcriptase, and 0.25 µL of RNasin® ribonuclease inhibitor (20 U/µL, Promega). The mixture was incubated under a thermal profile of 25 °C for 5 min followed by 42 °C for 60 min and 70 °C for 15 min (T100 thermal cycler, BioRad).
Primers and Taqman probes for quantitative real-time PCR assays were designed to the cDNAs encoding mct8, mct10, and the eight oatps from fathead minnow. Nucleotide sequences for these primers and probes are provided in Table 1. Quantitative real-time PCRs were conducted in 16 µL reactions, with each reaction containing 4.5 µL nuclease-free water (Sigma), 8.0 µL qPCR Master Mix (iTaq™ Universal SYBR® Green Supermix, BioRad), 0.5 µL of both forward and reverse primer (45 µM), 0.5 µL probe (5 µM), and 2.0 µL of cDNA template. The PCR thermal profile for each reaction was 50 °C for 2 min, 95 °C for 3 min, 45 cycles of 95 °C for 15 s and 60 °C for 1 min, and all assays were run on a 7300 Real Time PCR System (Applied Biosystems, Inc.). For each gene, a standard curve was made from a pool of total RNA from samples representing all treatments and sexes, and each standard was assayed in triplicate. DNA contamination was assessed for each gene by analyzing RNA samples that were not reverse-transcribed, and each qPCR run included two samples without cDNA template to further control for contamination. PCR efficiencies for each transcript assay are provided in Table 1, and correlation coefficients (r2) for all transcripts were always greater than r2 = 0.991. Finally, expression of each gene of interest was expressed as a relative level by dividing the resulting values by the mean values of the forebrain mRNA levels in each gene.
Table 1.
Nucleotide sequences for primer and Taqman probe used in quantitative real-time RT-PCR.
| Transcript | Primer | Nucleotide Sequence (5´ to 3´) |
Amplicon Size (bp) |
% Efficiency (Avg.) |
|---|---|---|---|---|
| mct8 | Forward primer | ACCAGGCGTCTCAGTTTAAAGTG | 72 bp | 97.55% |
| Probe | CATGGGTCGGCGCTCTGGC | |||
| Reverse primer | CCGGCGAGCAGAAGAAGAT | |||
| mct10 | Forward primer | GCTCTCCGATCGTCAGTGTGT | 61 bp | 94.96% |
| Probe | CACGGATCTGCTGGGATGCCG | |||
| Reverse primer | TCCTCCGACAGCCGTGAT | |||
| oatp1c1 | Forward primer | GAGAGACGGTCACCGGAGAA | 67 bp | 93.70% |
| Probe | TCCAAGGCCCTGTTGCTCAAGCC | |||
| Reverse primer | AGGCCACGAGGAACATCTTG | |||
| oatp1f1 | Forward primer | TGACGCCCAGAATACTGTTCAA | 64 bp | 98.17% |
| Probe | AGCTTGTGGCCGGATGCCA | |||
| Reverse primer | CGAGCTTTCCTCTTCCCCTTT | |||
| oatp1f2 | Forward primer | TGCAGAATGTATGACACTGAGACTT | 71 bp | 95.03% |
| Probe | AGGTTCCTGTTTCTGGGACTGG | |||
| Reverse primer | CCTAACACCCGCAGAGAGTTG | |||
| oatp2a1 | Forward primer | TGGCCTTGACAGCCTCTCTT | 65 bp | 93.43% |
| Probe | AGGAACCGTCTCGTCCCTCCAT | |||
| Reverse primer | TCAGCACTGTGTTCCCAACCT | |||
| oatp2b1 | Forward primer | GAAAAATGGTGACGCCAGATG | 65 bp | 96.94% |
| Probe | TTGGAGCTTGGTGGTTGGGA | |||
| Reverse primer | GGAGGGTGGCAGCTATCAAG | |||
| oatp3a1 | Forward primer | CAACCTGGCCCTCATTCTCTTTGT | 168 bp | 98.34% |
| Probe | ATGGCTTTGGGAGCGCTGCTCTCCGCATTA | |||
| Reverse primer | CCCAATTCCTTGCTCCATGTGTCT | |||
| oatp4a1 | Forward primer | TCGCTCATTGCAGGCATCATCTAC | 160 bp | 94.34% |
| Probe | AGCTGTGAGAGCACAGATGTGGGAGGCCAT | |||
| Reverse primer | TGATTGGCAGGTCTCTGTTCTTCC | |||
| oatp5a1 | Forward primer | TTGACCAGAGTGACCCACGATTTG | 169 bp | 92.46% |
| Probe | TGGAGTGGGTTTCTCCTGTGTGCTGTGGCA | |||
| Reverse primer | CTGGACACATCACCTGCACTCTTT | |||
| ef1α | Forward primer | GTTTGAGAAGGAAGCTGCCGAGAT | 100 bp | 98.50% |
| Probe | AGGGCTCCTTCAAATATGCCTGGGTGCT | |||
| Reverse primer | ATCAATGGTGATACCACGCTCAC | |||
| rpl8 | Forward primer | CTGTTGTTGGTGTTGTTGCTGGTG | 95 bp | 94.92% |
| Probe | ACCCATCCTGAAGGCAGGACGTGCATACCA | |||
| Reverse primer | GCAGTTCCTCTTGGCCTTGTACTT | |||
2.4. Effects of thyroid hormone status on mct and oatp mRNA levels
2.4.1. Dietary T3 and methimazole treatments
Exogenous T3 (Sigma-Aldrich) or the goitrogen methimazole (Sigma-Aldrich) were administered orally within a 1:1 ratio mixture of commercial spirulina flake (Aquatic Eco-Systems, Inc., Apopka, FL) and brine shrimp flake (San Francisco Bay Brand, Inc., Newark, CA, USA) feed. Flake food was sprayed with a 20 mL solution containing 19 mL of 95% ethanol and 1 mL of 0.1% NaOH containing either 20 mg T3 (Sigma-Aldrich), 150 mg methimazole (Sigma-Aldrich), or no additional chemical (control).
Adult minnows (n = 16–18 per treatment; body mass: 3.25 ± 0.10 g; standard length: 52.19 ± 0.57 mm; mean ± SEM) were maintained in 113 l tanks under conditions of 25.80 ± 0.01 °C, and 0.2 ppt salinity. Fish were fed 0.05 g flake feed · fish−1 · day−1, over two separate feeding times (11:00–13:00 h and 16:00–19:00 h), for average exposure doses of 20 µg T3 · fish−1 · day−1 and 150 µg methimazole · fish−1 · day−1. After 14 days of treatment, minnows were euthanized (MS222) and body mass (g) and standard length (mm) were recorded. Blood was collected in heparinized capillary tubes from the caudal peduncle, transferred to heparinize microcentrifuge tubes, and centrifuged at 3,000 g for 10 min at 4 °C, and the resulting plasma was stored at −80 °C for subsequent quantification of plasma TH concentrations. The whole brain and liver were dissected, frozen in liquid N2, and then stored at −80 °C.
2.4.2. Quantification of plasma total T4 and T3 concentrations
Plasma from individual male fish of the same treatment tank was pooled (n = 3–6 fish per pool), and circulating total T4 (tT4) and total T3 (tT3) concentrations were quantified by liquid chromatography tandem mass spectrometry (LC/MS/MS) (Wang and Stapleton, 2010) in conjunction with a newly developed extraction method for fish plasma described in detail elsewhere (Noyes, 2013; Noyes et al., submitted). Briefly, THs were extracted by incubating the pooled plasma samples (50 µl) with 50 µl of isotopically labeled hormones 13C12-T4 and 13C6-T3 (10 ng/mL in MeOH; Cambridge Isotope Laboratories, Andover, MA), which were used as internal standards to quantify levels of tT4 and tT3, respectively. Blank controls (deionized water) containing labeled THs alone were extracted alongside plasma samples to correct for trace unlabeled hormone levels (~0.5%) present in the labeled standards. The labeled hormone 13C6-T4 (Isotec, Canton, GA) was added prior to LC/MS/MS analysis to measure recovery of the 13C12-T4 internal standard. The percent recovery of 13C12-T4 was 93.50 ± 6.85 % (mean ± SD). Standard curves for each hormone spanned from 0.05 to 10 ng/mL with r2 values for each curve using a quadratic regression at > 0.99. Method detection limits (MDLs) were defined as three times the standard deviation (mean + 3*SD) of thyroid hormones detected in blanks containing no plasma and were measured at 0.25 ng/mL and 0.16 ng/mL for tT4 and tT3, respectively. The resulting TH values were normalized to the volume of plasma extracted (50 µL). The average intra-assay CV % for this LC/MS/MS method with fathead minnow plasma was calculated to be 4.73% for tT3 and 5.97% for tT4 (Noyes et al., submitted).
2.4.3. Quantitative real-time RT-PCR
Total RNA was extracted using TriReagent (Molecular Research Center, Inc.), DNase I treated (TURBO DNAfree Kit, Ambion), and quantified (P300 NanoPhotometer; Implen, Inc.). Total RNA was then reverse transcribed in 30 µL reactions by incubating 15.5 µL of total RNA template (10 µg/µL) with 6.0 µL 5× buffer, 0.25 µL of nuclease-free H2O (Sigma-Aldrich), 4.5 µL MgCl2, 1.5 µL dNTPs, 1.0 µL of random primers, 1.0 µL of GoScript™ reverse transcriptase, and 0.25 µL of RNasin® ribonuclease inhibitor (20 U/µL, Promega). The mixture was incubated under a thermal profile of 25 °C for 5 min followed by 42 °C for 60 min and 70 °C for 15 min (T100 thermal cycler, BioRad).
Quantitative real-time PCRs were conducted in 16 µL reactions as described above. Nucleotide sequences for primers and probes are shown with average PCR efficiencies for each transcript in Table 1, and r2 for all standard curves were greater than r2 = 0.97. Transcript abundance for ribosomal protein L8 (rpl8) was also quantified for use as the normalizing gene in liver, while elongation factor 1α (ef1α) was used as the normalizing gene in the brain. These transcripts were selected for each tissue because their relative expression levels did not vary across treatment groups (liver rpl8, F2,52 = 0.013, p = 0.987; brain ef1α, F2,47 = 0.898, p = 0.414). For each gene, relative mRNA levels were subsequently calculated using the standard curve and normalized to rpl8 mRNA expression (liver) or ef1α mRNA expression (brain). Finally, expression of each gene of interest was expressed as a relative level by dividing the resulting values by the mean values of the forebrain mRNA levels in each gene.
2.5. Statistical analyses
Tissue distribution data on relative levels of mct and oatp mRNAs was square root transformed to equalize variances. Two-tailed t tests with modified α levels as per the step-down Bonferroni-Holm correction method (Holm, 1979) were then used to examine sex differences in transcript abundance for each transcript within each tissue. This approach of using t tests in combination with step-down modified α levels was chosen to balance our reporting of tissues where clear sex differences in mct or oatp transcript abundance appeared evident in the data with the family-wise type I statistical error rate, given the large number of tissues and transcripts examined.
Plasma TH concentrations in male minnows treated with feeds containing exogenous T3, the goitrogen methimazole, or control vehicle were analyzed using a one-factor ANOVA model followed by Tukey HSD multiple comparisons. Analysis of relative levels of each mct and oatp transcript in the liver and brain was likewise conducted using a one-factor ANOVA model followed by Tukey HSD multiple comparison tests (α = 0.05). For select transcripts, data were square root transformed to homogenize variances prior to running ANOVA models. All data are presented as mean ± SEM.
3. Results
3.1. Sequencing and tissue distribution of mct8 and mct10 cDNAs
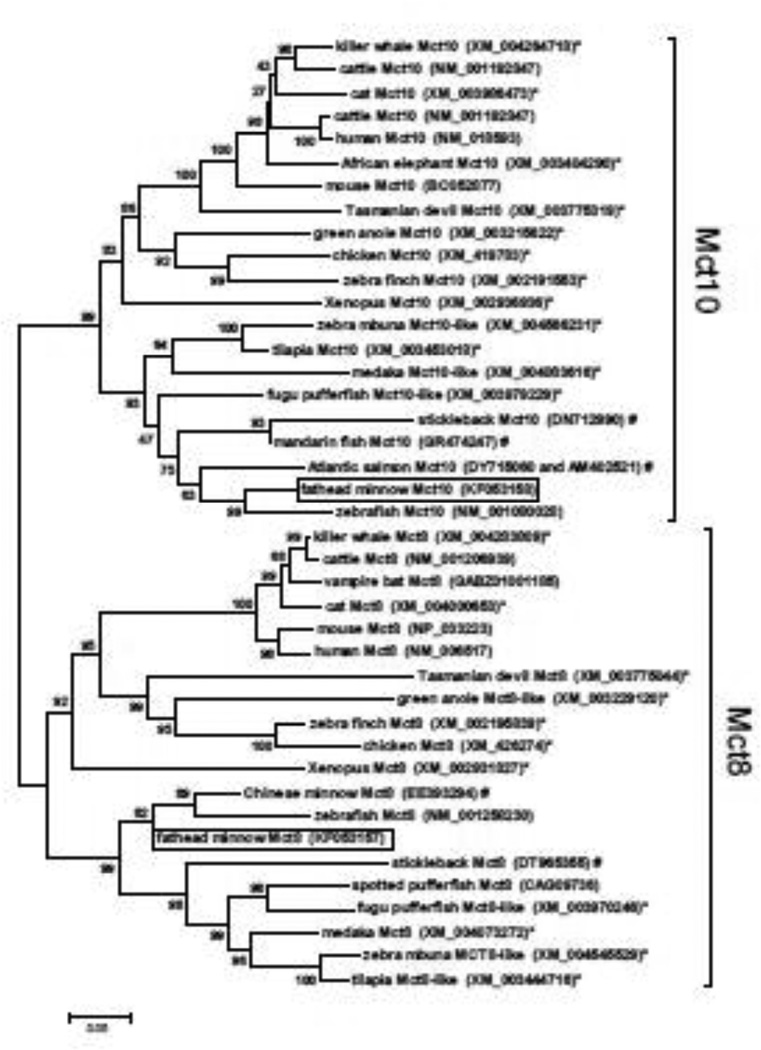
Two partial cDNAs encoding putative mct8 (231 bp nucleotides, GenBank accession no. KF053157) and mct10 (846 bp nucleotides; KF053158) were identified from fathead minnow by RT-PCR using degenerate primers. BLAST analyses (http://blast.ncbi.nlm.nih.gov) confirmed that these two cDNA fragments shared both nucleotide and deduced amino acid sequence identity with mammalian mct8 and mct10 genes, as well as the previously identified mct10 cDNA from zebrafish (NM_001080028). Alignment and phylogenetic analysis of the predicted amino acid sequences of these fathead minnow mct8 and mct10 cDNAs further confirmed their identities as mct8 and mct10 (Fig. 1).
Figure 1.
Phylogenetic tree based on alignment of deduced amino acid sequenced for Mct8 and Mct10 from teleost fishes and other select vertebrates. Tree was assembled using the neighbor-joining method with pairwise deletion, and bootstrap values (1000 replicates) are indicated at each node. Boxes indicate the mct8 and mct10 cDNAs identified from fathead minnow; (*) indicates predicted proteins based on sequenced genomes; (#) indicates partial proteins deduced from unannotated EST sequence. GenBank accession nos. are shown following each taxon designation.
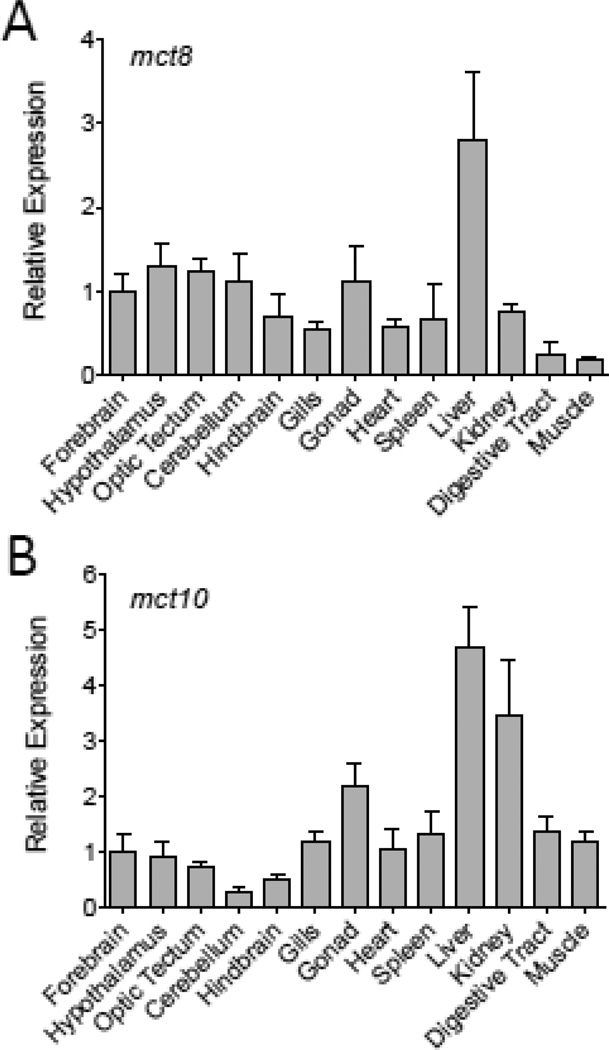
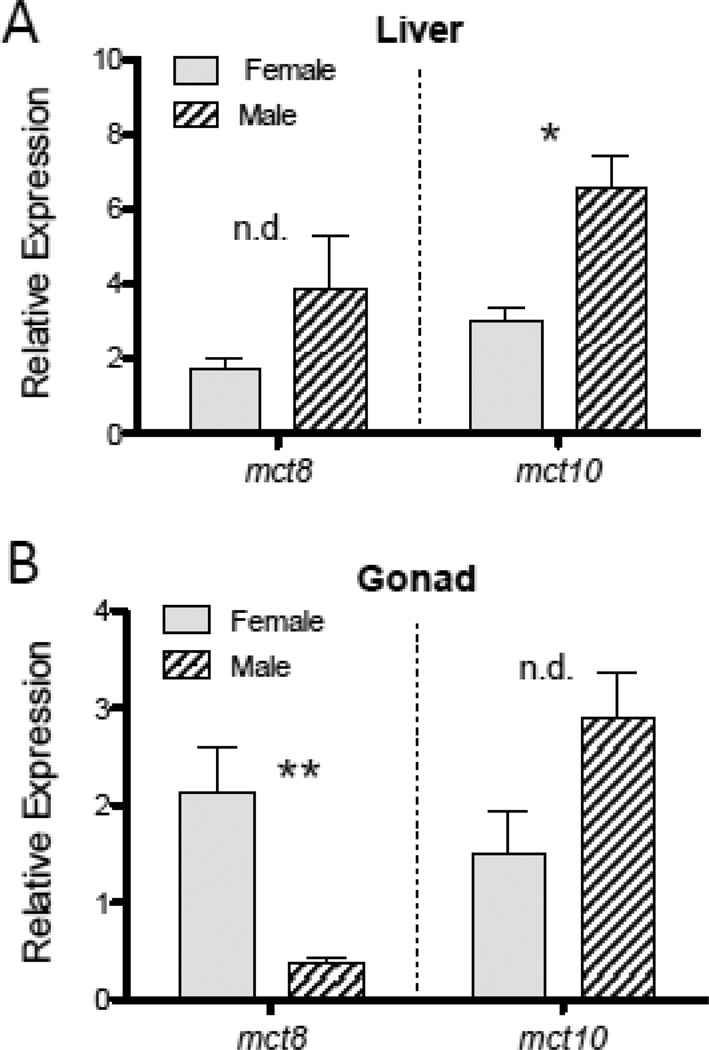
The relative abundance of mct8 and mct10 mRNAs, as indicated by quantitative real-time RT-PCR, was found to vary among tissues (Fig. 2). Both mct8 and mct10 mRNAs were at greatest abundance in the liver, although mct10 transcripts also showed a high abundance in kidney and, to a lesser degree, gonad tissues. The abundance of mct10 transcripts in the liver was observed to be at greater relative levels in males than in females (t = 4.2502, p = 0.0196) (Fig. 3). Sex differences in gene transcript abundance were also observed for mct8 in the gonad, with higher mct8 mRNA levels in ovarian tissues than in testicular tissues (t = −5.1840, p = 0.0226) (Fig. 3).
Figure 2.
Tissue distribution of mct8 (A) and mct10 (B) mRNAs in adult male and female fathead minnows (n = 6). Data values are shown as mean ± SEM. Relative expression values for each transcript are normalized against the mean abundance value of the forebrain tissue.
Figure 3.
Sex differences in the relative abundance of mct8 and mct10 mRNAs observed in the liver (A) and gonad (B) tissues of adult fathead minnows. Data are shown as mean ± SEM, with * indicating p < 0.025, ** indicating p < 0.01, and n.d. indicating ‘no difference’ between sexes. Data values are normalized to the mean abundance value for that same gene transcript in the forebrain. Sample sizes are n = 3 fish per gender.
3.2. Phylogenetic analysis and tissue distribution of oatp mRNAs
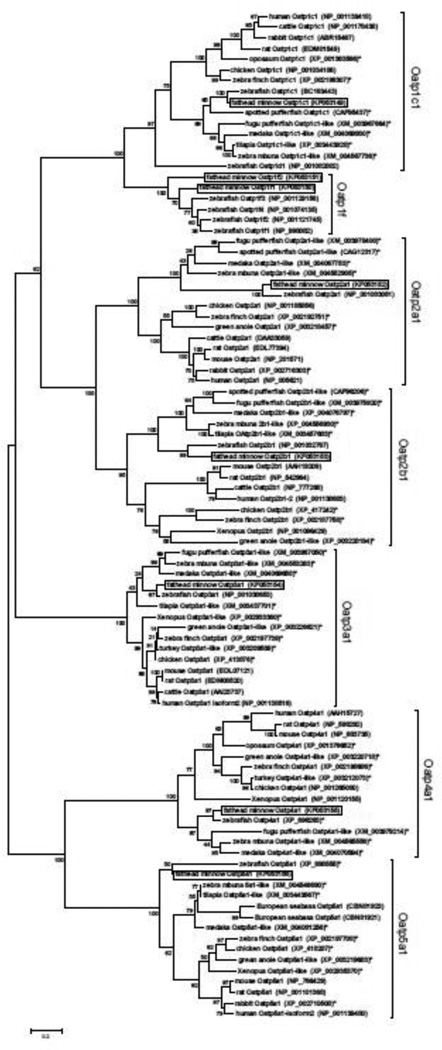
BLAST (http://blast.ncbi.nlm.nih.gov) analyses of available unannotated EST sequences from fathead minnow identified eight putative oatp cDNAs. Gene-specific primers were subsequently designed to each of these cDNAs and used in PCRs to amplify and sequence each cDNA to confirm nucleotide sequence composition. The resulting cDNAs included the following: a partial cDNA of 870 bp nucleotide encoding oatp1c1 (slco1c1; GenBank accession no. FK053149), two partial cDNAs of 589 bp and 311 bp nucleotides encoding putative Oatp1f-type transporters oatp1f1 (slco1f1; FK053150) and oatp1f2 (slco1f2; FK053151), a partial oatp2a1 cDNA of 422 bp nucleotides (slco2a1; FK053152), a 700 bp nucleotide partial cDNA encoding oatp2b1 (slco2b1; FK053153), a partial oatp3a1 cDNA of 464 bp nucleotides (slco3a1; FK053154), a partial cDNA of 1,462 bp nucleotides encoding oatp4a1 (slco4a1; FK053155), and a partial cDNA of 618 bp nucleotides encoding oatp5a1 (slco5a1; FK053156). Phylogenetic analysis of the predicted amino acid sequences of each of these oatp cDNAs from fathead minnow revealed that each cDNA grouped into a separate clade of Oatp genes, with the exception of the oatp1f1 and oatp1f2 cDNAs, which grouped within a single Oatp1f-type clade (Fig. 4). Four distinct full-length Oatp1f-type cDNAs were previously identified from zebrafish, indicating that some Cyprinid fishes may have multiple Oatp1f-type transporters evolved through gene duplication (Popvic et al., 2010).
Figure 4.
Phylogenetic tree from alignment of deduced amino acid sequences for Oatps from fish and select vertebrates. Tree was assembled by maximum likelihood using a Jones-Taylor-Thornton model (Jones et al., 1992) with all sites. Bootstrap values (1000 replicates) are indicated at each node. Boxes denote the eight Oatp cDNAs identified from fathead minnow. Asterisks (*) indicate predicted sequences based on genome projects, and GenBank accession nos. are provided following each taxon name.
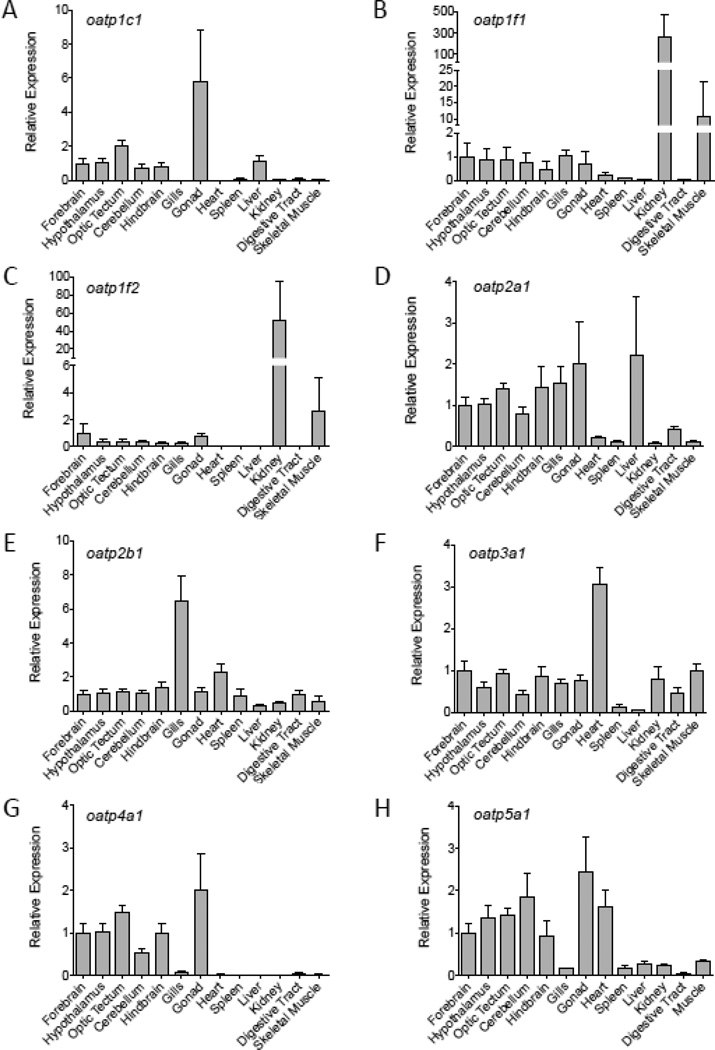
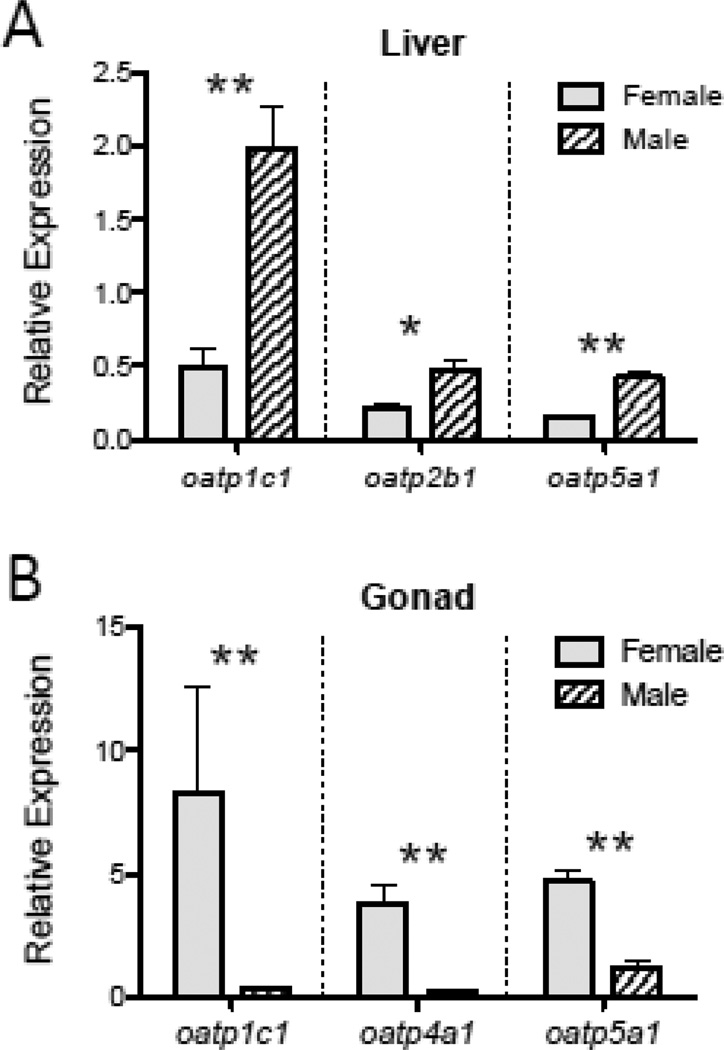
Transcripts encoding oatp anion transporter proteins differed in relative abundance among organs and tissues (Fig. 5). Relative transcript levels of oatp1c1 were greatest in the gonad, liver and brain (Fig. 5A). The high relative abundance of oatp1c1 mRNAs in the gonad appeared to be due to elevated levels of oatp1c1 mRNAs in ovarian tissues, since oatp1c1 mRNAs were on average more than 7 fold greater in the ovary than in the testis (Fig. 6B; t = −12.395, p = 0.0007). A sex difference in oatp1c1 transcript abundance was also observed in the liver, with male minnows exhibiting 4-fold greater oatp1c1 mRNA levels in this tissue compared to females (Fig. 6A; t = 5.331, p = 0.0064).
Figure 5.
Relative oatp mRNA levels among tissues of adult fathead minnows (n = 6). Data values are shown as mean ± SEM. Relative expression values for each transcript are normalized against the mean relative abundance value for that same transcript in forebrain tissue.
Figure 6.
Sex differences in oatp mRNA transcript abundance in the liver (A) and gonadal (B) tissues of adult fish. Data are shown as mean ± SEM, with * indicating p < 0.025, and ** indicating p < 0.01. Data values are normalized to the mean abundance value for that same gene transcript in the forebrain (see Fig. 5). Sample sizes are n = 3 fish per gender.
Relative levels of oatp1f1 and oatp1f2 were observed to be greatest in the kidney (Fig. 5B,C). Transcripts encoding oatp2a1 were most abundant in brain, gill, gonad and liver (Fig. 5D), and while oapt2b1 mRNAs were observed to be at greatest abundance in gill tissue, a significant sex difference in relative levels of oapt2b1 was observed in the liver (Fig. 6A; t = 3.693, p = 0.021). Other Oatp-encoding mRNAs likewise exhibited expressional variation in relative abundance among tissues, with oatp3a1 mRNAs most abundant in cardiac muscle (Fig. 5F), oatp4a1 mRNAs most abundant in brain and gonadal tissues (Fig. 5G), and oatp5a1 transcripts at greatest levels in brain, gonad and heart (Fig. 5H). Transcripts encoding oatp5a1 were observed to be at greater relative levels in the liver of male fish compared to females (Fig. 6A; t = 7.908, p = 0.0015), and both oatp4a1 and oatp5a1 transcripts were at higher abundance in ovarian tissue than in testicular tissue (Fig. 6B; oapt4a1, t = −6.363, p = 0.0231; oatp5a1, t = −7.632, p = 0.0017).
3.3. Thyroid hormone regulation of mct and oatp mRNAs in liver and brain
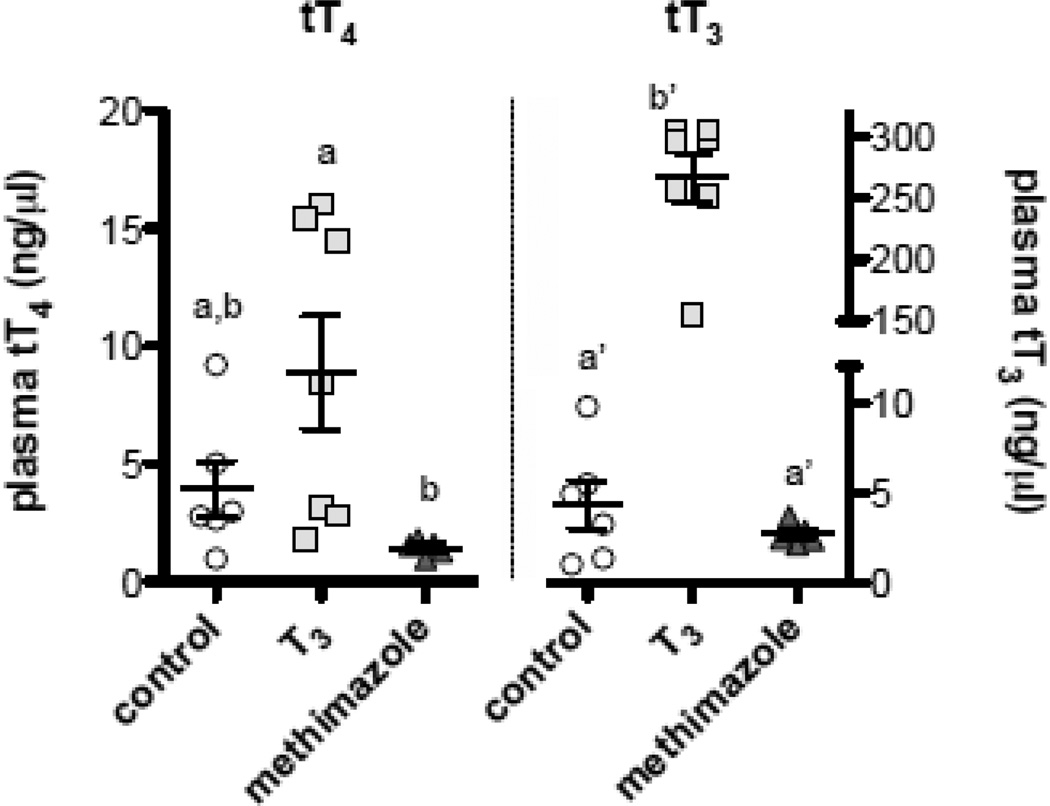
Oral treatment of adult male minnows with either exogenous T3 or the goitrogen methimazole generated variation in plasma concentrations of total T4 (tT4) (F2,15 = 5.255, p = 0.0186), and post hoc pairwise comparisons revealed tT4 to be significantly lower in methimazole-treated fish compared to T3-treated fish (Fig. 7). Plasma concentrations of total T3 (tT3) also differed among treatment groups, and were significantly elevated in males treated orally with exogenous T3 (F2,15 = 291.35, p < 0.0001) (Fig. 7).
Figure 7.
Plasma total T4 (tT4) and total T3 (tT3) concentrations in adult male minnows given dietary treatments of control vehicle, exogenous T3, or the goitrogen methimazole for 14 days. Methimazole depressed tT4, while exogenous T3 significantly elevated plasma tT3 concentrations over control and methimazole-treated fish. Each data point represents the hormone concentration measured from the pooled plasma of n = 3–6 minnows, and bars denote mean ± SEM values for each treatment group. Letters indicate pairwise differences among treatment groups for each hormone (Tukey HSD tests, α = 0.05)
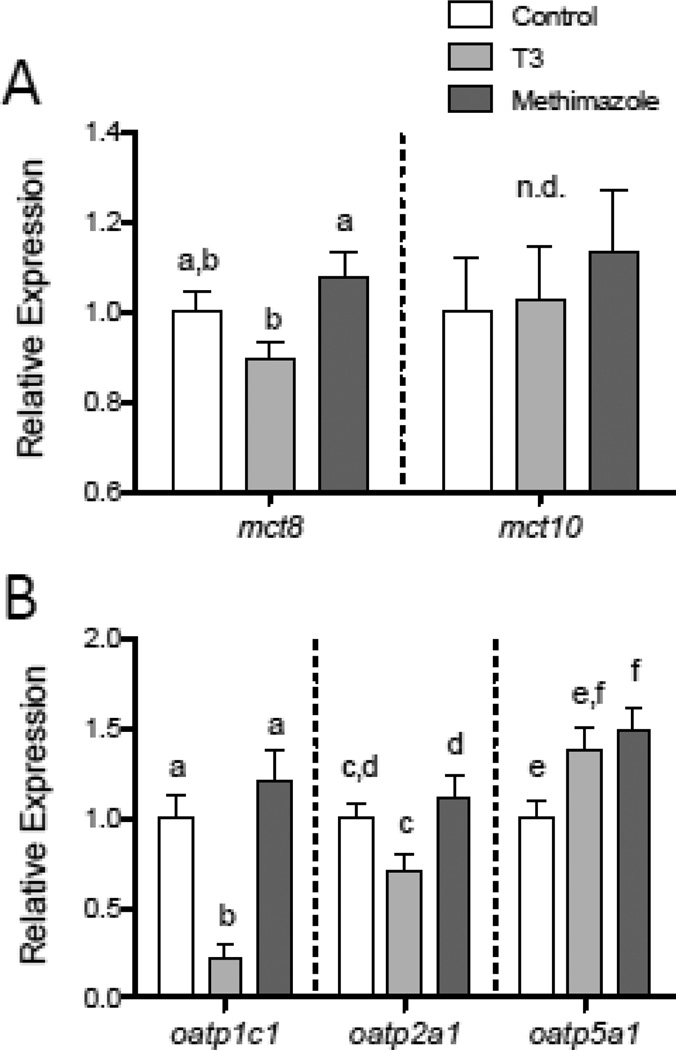
These experimentally-induced changes in circulating THs were found to influence relative mRNA levels for select mct and oatp membrane transporter genes, with the specific effects varying depending on transcript and tissue. In the liver, male fish treated with exogenous T3 had lower relative levels of mct8 transcripts compared to methimazole-treated fish (Fig. 8A; F2,52 = 3.839, p = 0.0279). Experimental modulation of plasma TH levels did not, however, alter liver mct10 mRNA levels (Fig. 8A). In the liver, transcripts encoding oatp1c1 were found to be strongly altered by T3 treatment, with T3-exposed fish showing more than a 60% reduction in oatp1c1 mRNA abundance compared to control or methimazole-treated fish (Fig. 8B; F2,52 = 14.273, p < 0.0001). Transcripts for oatp2a1 were likewise reduced in abundance in the liver tissue of T3-treated males (Fig. 8B; F2,52 = 4.544, p = 0.0152). The relative abundance of oatp5a1 transcripts was also altered by the experimentally-induced change in plasma TH concentrations, although this effect differed qualitatively from that observed for oatp1c1 and oatp2b1 transcripts with methimazole-treated fish exhibiting elevated oatp5a1 mRNA levels compared to control fish (Fig. 8B; F2,52 = 4.549, p = 0.0151).
Figure 8.
Variation in relative levels of mct and oatp transcripts in the liver with experimentally-induced changes in circulating TH concentrations. Transcripts encoding mct8, but not mct10, were reduced in male fathead minnows treated with exogenous T3 for 14 days, compared to fish treated with the goitrogen methimazole. (A). Transcripts encoding oatp1c1 and oatp2a1 were likewise down-regulated in the liver of T3-treated male fish, while oatp5a1 mRNAs became elevated in the liver of both T3- and methimazole-treated (B) in the liver. Letters indicate significant differences between groups for each transcript (Tukey HSD tests, p < 0.05), and n.d. indicates “no difference” among treatment groups. Sample sizes are n = 16–18 fish per treatment.
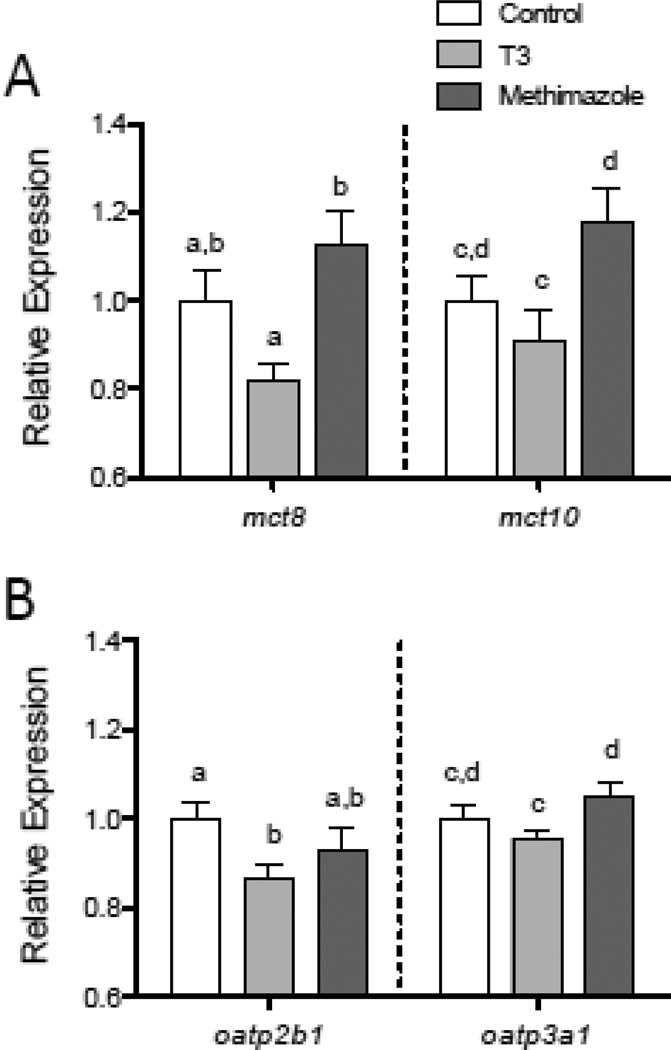
In the brain, both mct8 and mct10 showed changes in relative mRNA abundance in response to experimental alteration of plasma TH concentrations. Minnows treated with T3 had a reduced relative abundance of transcripts encoding mct8 (F2,47 = 6.829, p = 0.0025) and mct10 (F2,47 = 3.819, p = 0.0291) compared to methimazole-treated fish (Fig. 9A). Transcripts encoding oatp2b1 and oatp3a1 also exhibited small (< 10%), but statistically significant reductions in relative abundance in response to T3-treatment (Fig. 9B), with oatp2b1 mRNA levels lower in T3-treated males than control males (F2,47 = 3.346, p = 0.044), and oatp3a1 transcripts reduced in T3-treated fish compared to methimazole-treated fish (F2,47 = 3.504, p = 0.0381).
Figure 9.
Changes in mct and oatp relative mRNA levels in the brain of male fish treated with exogenous T3 or methamizole for 14 days. Transcripts encoding mct8 and mct10 were both found to be lower in the brain of T3-treated fish compared to methimazole-treated fish (A). Transcripts encoding oatp2b1 and oatp3a1 also showed reduced relative mRNA levels in the brain of T3-treated fish (B). Letters indicate significant differences between groups for each transcript (Tukey HSD tests, p < 0.05). Sample sizes are n = 16–18 fish per treatment.
4. Discussion
Only within the last decade or so has it become evident that the movement of THs into target cells occurs not by diffusion, but rather via facilitated transport mediated by plasma membrane transport proteins (Bernal, 2005; Visser et al., 2011). The first TH transport protein was identified in mammals at the molecular level over 15 years ago (Abe et al., 1998), and yet almost nothing is presently known about the diversity, patterns of expression, or regulation of TH membrane transporters in other vertebrates including teleost fishes. In this study, we identified cDNAs encoding mct8, mct10, and eight transporters from the Oatp family of solute carrier proteins in the fathead minnow, and documented the expressional distribution of mRNAs encoding these transporters among organs and tissues. We also began to explore how varying conditions of thyroid hormone status may alter the expression of these transporters by experimentally modulating circulating TH status and then quantifying the effects on relative mRNA levels for these membrane transporters in the liver and brain.
4.1. Distribution of monocarboxylate transporter Mct8 and Mct10 transcripts
Using degenerate primer PCR and in silico searches of available EST libraries from teleost fishes, partial cDNA sequences encoding mct8 and mct10 from the fathead minnow were isolated and sequenced. Phylogenetic analysis of the deduced amino acid sequences from these cDNAs confirmed the identity of these mRNAs and revealed that each respective protein comprised part of clades containing previously identified mct8 and mct10 proteins from mammals, as well as mct8 and mct10 genes recently annotated from the genomes of select fish, birds and reptiles (e.g., zebrafish, Takifugu pufferfish, green anole) and other teleost fish cDNAs newly identified from unannotated EST libraries as part of the current study (Fig. 1). Quantitative real-time RT-PCR (qRT-PCR) analyses of the relative levels for mct8 and mct10 transcripts in fathead minnow tissues revealed that mRNAs for both genes are most abundant in the liver, but are also present at lesser abundances in all organs and tissues examined.
Mammalian Mct8 has likewise been detected in a variety of tissues including brain, liver, kidney, thyroid gland, placenta, and the adrenal glands (Price et al., 1998; Biebermann et al., 2005). Mammalian Mct8 has a high specificity for the bidirectional transport of a variety of THs (T4, T3, rT3 and T2) across cellular membranes (Friesema et al., 2003, 2006), but its role in regulating T3 transport across the blood-brain barrier is perhaps the best evidence that Mct8 plays a key role in target tissue responses to circulating THs (reviewed by Visser et al., 2011). Clinical studies in humans have now linked several mutations in Mct8 to behavioral and cognitive abnormalities including impaired speech development, dystonic movements, low intelligence quotient values, and mental retardation (e.g., Dumitrescu et al., 2004; Schwartz and Stevenson, 2007; Visser et al., 2011; Visser and Visser, 2012). Clinical patients with non-functional Mct8 often also show atypical TH signaling profiles with elevated circulating T3 and reduced total and free T4 and rT3 concentrations suggestive of compensatory responses in either TH secretion or peripheral conversion by iodothyronine deiodinases (Visser et al., 2011). Mammalian Mct10, the most structurally similar Mct protein to Mct8, has also been shown to transport both T3 and T4 (Friesema et al., 2008). Recently, Arjona and coworkers (2011) provided the first characterization of Mct8 from a teleost fish, the zebrafish Danio rerio, and demonstrated that zebrafish Mct8 (zMct8) likewise mediates cellular TH uptake. Similar to our findings here with fathead minnows, mct8 mRNAs are found nearly ubiquitously in adult zebrafish tissues, with greatest expression levels in brain, liver and kidney (Arjona et al., 2011). Transcripts encoding Mct8 also are present in zebrafish during early developmental stages (Arjona et al., 2011), and morpholino-based knockdown and rescue of mct8 in this species provides evidence that zMct8 expression during early life is required for brain and spinal cord development (Vatine et al., 2013). Interestingly, however, the transport specificity of zMct8 appears to vary from mammalian Mct8 in that zMct8 transports T3 with a higher affinity and also shows temperature-dependent variation in TH specificity, transporting only T3 at a temperature of 26 °C, but both T3 and T4 at 37 °C (Arjona et al., 2011). An environmental temperature of 37 °C is much higher than standard laboratory rearing conditions for zebrafish and approaches the maximum recorded temperatures for this species in the wild (Engeszer et al., 2007). Nonetheless, many teleost fishes tolerate fluctuating water temperatures daily or seasonally, and future studies into the temperature dependency of Mct8 function in fish could provide new insights into the dynamics of TH signaling in variable temperature environments.
Our initial comparisons of mct8 and mct10 mRNA levels among male and female minnows also revealed evidence of sex differences in mct transcript abundance in select tissues. Relative transcript abundance for mct10 in the liver was more than 2-fold greater in males than in females, and ovarian mct8 mRNA levels were nearly 2-fold higher than testicular mct8 mRNA levels (Fig. 3). For gonadal tissues, such differences in mct8 mRNA levels are likely associated with differential roles for THs in ovarian and testicular function in fish. THs have been known to influence reproductive function in fish for some time (Cyr and Eales 1996), and there is evidence that thyroid status can affect testicular androgen production and spermatogenesis (Jacob et al., 2005; Swapna et al., 2006). In fathead minnows, ovarian and testicular tissues have previously been shown to vary in the expression of other TH signaling components such as TH receptors α and β. For instance, the relative abundance of TH receptor β transcripts in the testis is associated negatively with the proportion of primary and secondary spermatocytes, but positively with the proportion of spermatozoa (Lema et al., 2009). From this evidence it is clear that patterns of gene expression for proteins involved in TH signaling in the gonad will be highly dependent on variation in cellular composition with oogenesis and spermatogenesis, and additional studies focused on the role of THs in gonadal function in fish are needed. It is also important to note, however, that the small sample sizes of each sex used to assess tissue distributions in the present study points to a need for additional studies to confirm these sex differences in gonadal mct8 mRNA expression. Future studies should use these results as a foundation to confirm and extend the study of sexual variation in Mct expression in teleost fish.
4.2. Diversity and distribution of Oatp transporters
The Oatp family of solute carrier proteins is highly diverse with most proteins in this family transporting a variety of ligands (Hagenbuch and Meier, 2003, 2004; Hagenbuch, 2007; Hagenbuch and Gui, 2008). From the fathead minnow, we identified eight putative oatp cDNAs from seven oatp gene families, including what appears to be two distinct oatp1f cDNAs (Fig. 4). These results provide only the second assessment of Oatp transporter diversity in a teleost fish, as previous work by Popovic and coworkers (2010) identified 14 distinct oatp genes in the zebrafish, including paralogs of oatp3a and oatp5a, and four distinct oatp1f genes. These findings in zebrafish point to the likehood of considerable variation in oatp diversity among teleost fish species, possibly in accordance with taxonomic variation in genome duplication among Actinopterygiian fishes (e.g., Larhammar et al., 2009).
Although only limited comparative data are available at present, the tissue distribution patterns of relative oatp mRNA abundance observed here in adult fathead minnow (Fig. 5) generally corresponds with the pattern observed previously in adult zebrafish (Popovic et al., 2010). Popovic et al. (2010) likewise found oatp2b1 mRNAs to be highly abundant in the gills, and oatp1f transcript most abundant in kidney and at low or undetectable levels in other tissues. Popovic and coworkers (2010) did not, however, target oatp1c1 in the zebrafish, so there is no data available to compare our finding of oatp1c1 mRNAs as abundant in the brain, liver and gonadal tissues of adult fathead minnows. Distinct sex differences were also measured in relative oatp1c1 expression with greater abundance in the liver of males than females (Fig. 6A) as well as elevated levels in the ovary compared to testis (Fig. 6B). Similar patterns of male-female transcript abundance variation were also observed for oatp2b1 and oatp5a1 in the liver and oatp4a1 and oatp5a1 in the gonadal tissues (Fig. 6).
Although beyond the scope of the current study, it is not expected that all Oatps identified here function in the membrane transport of THs in fathead minnows as the functional specificity for substrate transport varies widely among mammalian Oatps (e.g., Hagenbuch and Meier, 2003; Meier-Abt et al., 2006; Hagenbuch, 2007; Svoboda et al., 2011). Rather, several of these Oatps have established alternate functions in mammals; mammalian Oatp2a1, for instance, is a key prostaglandin transporter, and Oatp2b1 is involved in the liver uptake and disposition of several classes of xenobiotics. It is still largely unclear which compounds Oatp5a1 transports, even in mammals (Svoboda et al., 2011). Of the eight oatp cDNAs identified here in the fathead minnow, only oatp1c1, oatp2b1, oatp3a1 and oatp4a1 have been identified as TH membrane transporters in mammalians systems (Mikkaichi et al., 2004; Svoboda et al., 2011; Visser et al., 2011).
Of this group of TH transporting Oatps, mammalian Oatp1c1 has the highest affinity for THs (T4, T3, rT3) (Pizzagalli et al., 2002), although this protein also is involved in the uptake of estrone-3- sulfate and 17β-estradiol-glucuronide (Sugiyama et al., 2003; Hagenbuch, 2007; van der Deure et al., 2008; Visser et al., 2011). In mammalian systems, Oatp1c1 appears to play key roles in TH uptake across the blood-brain barrier as well as by Leydig cells in the testis (Sugiyama et al., 2003; Tohyama et al., 2004; Chu e al., 2008). Our finding of high oatp1c1 mRNA abundance in the brain, gonad and liver may therefore support a similar important of Oatp1c1 in these tissues in fish. In any case, much remains unknown about the function and expressional dynamics of Oatp1c1, even in mammals. For instance, human patients identified with genetic mutations in oatp1c1 continue to have circulating THs in the normal physiological range (van der Deure et al., 2008). And yet, experimentally-induced changes in circulating TH concentrations have been observed to alter Oatp1c1 protein and mRNA levels in endothelial cells in the rat brain (Sugiyama et al., 2003), suggesting that systemic THs may regulate Oatp expression under some circumstances.
4.3. Effects of circulating TH concentrations on mct and oatp mRNA levels
Regulation of TH signaling is known to occur not only via changes in the production and secretion of THs from the thyroid follicles, but also via the up- or down-regulation of iodothyronine deiodinase enzymes or the abundance and type of thyroid hormone receptors in peripheral tissues (Cheng et al, 2010; Orozco et al., 2012). Such peripheral regulation can generate variation among target tissues in sensitivity to circulating hormones (e.g., Lema and Kitano, 2013). Target tissue variation in the type and density of TH membrane transporters may therefore provide an additional mechanism for regulatory control of TH action on peripheral tissues. At present, however, our understanding of TH transporter regulation is limited, and to our knowledge only a few studies have tested whether TH membrane transporters show expressional regulation under controlled conditions where TH titers are modulated experimentally. In one recent study of male New Zealand rabbits manipulated to generate a phenotypic state mimicking illness, ‘ill’ rabbits were found to have elevated mRNA levels of mct8 in liver and mct10 in skeletal muscle; treatment of these rabbits with a combination of exogenous T4 and T3 was observed to down-regulate mct8 gene transcription in liver and mct10 transcription in muscle (Mebis et al., 2009). And, in a separate study of rats, hypothyroidism caused either by thyroidectomy or by treatment with the goitrogen methimazole was found to increase oatp1c1 mRNA levels and protein levels in the endothelial cells lining brain capillaries; exogenous T3-induced hyperthyroidism was observed to have the opposing effect in these same animals (Sugiyama et al., 2003). And, in a recent study of adult fathead minnows similar to our work here, oral dosing of males with either the goitrogen 6-propyl-2-thiouracil (PTU) or the (PBDE) flame retardant decabromodiphenyl ether BDE-209 both resulted in an increase in brain mct8 mRNA levels (Noyes et al., 2013).
In the present study, we tested whether changes in systemic TH status would alter gene transcript abundance for TH transporters in fish tissues by exposing adult male minnows orally for 14 days to either exogenous T3 or methimazole, and then quantifying relative mct and oatp mRNA levels in the liver and whole brain. In the liver, fish with experimentally elevated plasma T3 levels showed a nearly 70% reduction in relative transcript abundance for oatp1c1, and also induced smaller reductions in mct8 and oatp2a1 mRNA levels. In the brain, fish treated with exogenous T3 showed reduced abundances of mct8, mct10 and oatp3a1 compared to methimazole-treated fish. Transcripts encoding oatp2b1 in the brain also varied among treatment groups, with T3-treated fish exhibiting reduced oatp2b1 mRNA levels relative to control fish. Interestingly, these oatp transporter transcripts that we observed responding to T3 or goitrogen treatment correspond with Oatps identified as transporting THs in mammalian systems (e.g., Oatp1c1, Oatp3a1, and Oatp2b1) with the exception of Oatp2a1, which transports prostaglandins in mammals, and Oatp5a1, which has no identified substrates to date (Hagenbuch, 2007; Svoboda et al., 2011).
The direction of abundance changes that we observed in mct or oatp mRNAs generally follows the pattern predicted if these changes were part of a compensatory response, where higher circulating T3 concentrations down-regulate TH transporter expression to reduce TH uptake, and lower circulating THs trigger an up-regulation of transporter gene transcription. With the exception of oatp1c1 down-regulation in the liver of T3-treated fish, however, all other changes in membrane transporter mRNA abundance that we observed varied no more than 15–20% from the levels observed in control or methimazole-treated fish, even under the current experimental scenario where plasma tT3 concentrations exceeded the normal physiological range for this species (e.g., Lema et al., 2008; 2009). This finding may suggest that transcriptional responses of TH transporters to alterations in circulating TH concentrations may be a fairly minor component in the regulation of peripheral tissue sensitivity compared to TH-induced changes in deiodinase or TH receptor expression (Johnson and Lema, 2011). Furthermore, evidence from mutant and gene knockout studies in mammals suggests that changes in TH membrane transporter density itself may trigger compensatory changes in deiodinase enzymes or TH receptor density, as target cells counteract a reduced capacity to uptake THs (e.g., Dumitrescu et al., 2006; Mayerl et al., 2012).
While more studies are needed to understanding the dynamics of TH membrane transporter regulation in modulating tissue sensitivity to THs, our results here indicate that changes in TH transporter gene transcription in response to TH can vary among tissues. Even though our analysis of mct and oatp mRNA tissue distributions indicated that oatp1c1 transcripts are abundant in both brain and liver tissues (Fig. 5A), the strong down-regulation of oatp1c1 mRNAs in the liver – but absence of an oapt1c1 transcriptional response in the brain – of T3-treated minnows provides evidence for differences in TH transport regulation in these tissues. Supporting this finding, we recently observed that oral treatment of adult male fathead minnows with the goitrogen PTU induced a 12-fold increased in oatp1c1 transcript abundance in the liver, but had no effect on oatp1c1 mRNA levels in the brain (Noyes et al., 2013). In this same study, dietary exposure of male minnows to the PBDE flame retardant BDE-209 depressed circulating tT3 and tT4 concentrations and increased liver oatp1c1 mRNA levels nearly 7-fold, but again had no effect on brain oatp1c1 transcript abundance (Noyes et al., 2013). While the functional significance of tissue variation in oatp1c1 transcriptional dynamics remains unclear, the absence of a detectable response of oatp1c1 mRNAs in the brain may indicate a lessened ability for this tissue to modulate TH uptake (e.g., Lema et al., 2008; Noyes et al., 2013). Under normal conditions, the liver is thought to be the primary organ for conversion and metabolism of THs in blood circulation (Van der Geyten et al., 1998; Malik and Hodgson, 2002). Here we found that the effects of plasma TH concentrations on liver oatp1c1 transporter mRNA abundance were particularly large in magnitude, suggesting that regulatory changes in liver Oatp1c1 expression may be a key component in maintaining systemic TH homeostasis in teleost fishes. Even so, large changes in circulating TH concentrations induced either experimentally or via exposure to EDC pollutants could overwhelm the ability of the liver to adjust circulating THs, so that the brain’s ability to regulate TH uptake may be key in determining the impacts of altered TH status on brain development and function (Zoeller et al., 2002; Tan and Zoeller 2007). It is also important to note that the absence of oatp1c1 mRNA changes in the brain of T3-treated fish stands in contrast to the regulatory response of oatp1c1 mRNAs and protein observed in the brain capillary endothelial cells of rats (Sugiyama et al., 2003). This distinction may reflect the differences in TH transport mechanisms across the blood-brain barrier between mammals and fish, and points to the need for future studies to look in more detail at the mechanisms and regulatory dynamics TH membrane transport in teleost fish models.
Supplementary Material
Acknowledgements
The authors thank Rob Brewster and Doug Brewster for methodological assistance via the design and construction of animal facilities for this research. This research was supported by a California State University Program for Research and Education in Biotechnology (CSUPERB) President’s Commission Scholars Award to A.M.M., by an Undergraduate College Based Fee (CBF) research award from California Polytechnic State University to A.M.M., by an U.S. Environmental Protection Agency STAR graduate fellowship (FP-917145010) to P.D.N., by a National Institute of Environmental Health Sciences R01-ES016099 to H.M.S., and by a CSUPERB New Faculty Award to S.C.L. The authors also thank two anonymous referees for comments and suggestions that greatly improved the quality of this manuscript.
Abbreviations
- TH
thyroid hormone
- T3
3,5,3′-triiodothyronine
- T4
thyroxine
- Mct
monocarboxylate transferase
- Oatp
organic anion-transporting polypeptide
- Slc
solute carrier protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Kakyo M, Sakagami H, Tokui T, Nishio T, Tanemoto M, Nomura H, Hebert SC, Matsuno S, Kondo H, Yawo H. Molecular characterization and tissue distribution of a new organic anion transporter subtype (oatp3) that transports thyroid hormones and taurocholate and comparison with oatp2. J. Biol. Chem. 1998;273:22395–22401. doi: 10.1074/jbc.273.35.22395. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Villeneuve DL. The fathead minnow in aquatic toxicology: Past, present and future. Aquat. Toxicol. 2006;78:91–102. doi: 10.1016/j.aquatox.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Arjona FJ, de Vrieze E, Visser TJ, Flik G, Klaren PHM. Identification and functional characterization of zebrafish solute carrier Slc16a2 (Mct8) as a thyroid hormone membrane transporter. Endocrinology. 2011;152:5065–5073. doi: 10.1210/en.2011-1166. [DOI] [PubMed] [Google Scholar]
- Bergh JJ, Lin HY, Lansing L, Mohamed SN, Davis FB, Mousa A, Davis PJ. Integrinανβ3 contains a cell surface receptor site for thyroid hormone that is linked to activation of mitogen-activated protein kinase and induction of angiogenesis. Endocrinology. 2005;146:2864–2871. doi: 10.1210/en.2005-0102. [DOI] [PubMed] [Google Scholar]
- Bernal J. The significance of thyroid hormone transporters in the brain. Endocrinology. 2005;146:1698–1700. doi: 10.1210/en.2005-0134. [DOI] [PubMed] [Google Scholar]
- Biebermann H, Ambrugger P, Tarnow P, von Moers A, Schweizer U, Grueters A. Extended clinical phenotype, endocrine investigations and functional studies of a loss-of-function mutation A150V in the thyroid hormone specific transporter MCT8. Eur. J. Endocrinol. 2005;153:359–366. doi: 10.1530/eje.1.01980. [DOI] [PubMed] [Google Scholar]
- Cheng S-Y, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone action. Endocrine Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Li JY, Boado RJ, Pardridge WM. Blood-brain barrier genomics and cloning of a novel organic anion transporter. J. Cereb. Blood Flow Metab. 2008;28:291–301. doi: 10.1038/sj.jcbfm.9600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DG, Eales JG. Interrelationships between thyroidal and reproductive endocrine systems in fish. Rev. Fish Biol. Fish. 1996;6:165–200. [Google Scholar]
- Desouza LA, Ladiwala U, Daniel SM, Agashe S, Vaidya RA, Vaidya VA. Thyroid hormone regulates hippocampal neurogenesis in the adult rat brain. Mol. Cell. Neurosci. 2005;29:414–426. doi: 10.1016/j.mcn.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao XH, Best TB, Brockmann K, Refetoff S. A novel syndrome combining thyroid and neurological and abnormalities is associated with mutations in a monocarboxylate transporter gene. Am. J. Hum. Genet. 2004;74:168–175. doi: 10.1086/380999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao X-H, Weiss RE, Millen K, Refetoff S. Tissue-specific thyroid hormone deprivation and excess in monocarboxylate transpoerter (Mct) 8-deficient mice. Endocrinology. 2006;147:4036–4043. doi: 10.1210/en.2006-0390. [DOI] [PubMed] [Google Scholar]
- Edeline E, Bardonnet A, Bolliet V, Dufour S, Pierre E. Endocrine control of Anguilla anguilla glass eel dispersal: effect of thyroid hormones on locomotor activity and rheotactic behavior. Horm. Behav. 2005;48:53–63. doi: 10.1016/j.yhbeh.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Engeszer RE, Patterson LB, Rao AA, Parichy DM. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007;4:21–38. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- Forrest D, Erway LC, Ng L, Altschuler R, Curran T. Thyroid hormone receptor β is essential for development of auditory function. Nat Genet. 1996;13:354–357. doi: 10.1038/ng0796-354. [DOI] [PubMed] [Google Scholar]
- Friesema EC, Ganguly S, Abdalla A, Manning Fox JE, Halestrap AP, Visser TJ. Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter. J. Biol. Chem. 2003;278:40128–40135. doi: 10.1074/jbc.M300909200. [DOI] [PubMed] [Google Scholar]
- Friesema EC, Kuiper GG, Jansen J, Visser TJ, Kester MH. Thyroid hormone transport by the human monocarboxylate transporter 8 and its rate-limited role in intracellular metabolism. Mol. Endocrinol. 2006;20:2761–2772. doi: 10.1210/me.2005-0256. [DOI] [PubMed] [Google Scholar]
- Friesema EC, Jansen J, Jachtenberg JW, Visser WE, Kester MH, Visser TJ. Effective cellular uptake and efflux of thyroid hormone by human monocarboxylate transporter 10 (MCT10) Mol. Endocrinol. 2008;22:1357–1369. doi: 10.1210/me.2007-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Adachi H, Nishio T, Unno M, Tokui T, Okabe M, Onogawam T, Suzuki T, Asano N, Tanemoto M, Seki M, Shiiba K, Suzuki M, Kondo Y, Nunoki K, Shimosegawa T, Iinuma K, Ito S, Matsuno S, Abe T. Identification of thyroid hormone transporters in humans: Different molecules are involved in a tissue-specific manner. Endocrinology. 2001;142:2005–2012. doi: 10.1210/endo.142.5.8115. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B. Cellular entry of thyroid hormones by organic anion transporting polypeptides. Best Pract. Res. Clin. Endocrinol. Metabol. 2007;21:209–221. doi: 10.1016/j.beem.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta. 2003;1609:1–18. doi: 10.1016/s0005-2736(02)00633-8. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the OATP/SLC21 family: phylogenetic classification as OATP/SLCO superfamily, new nomenclature and molecular/functional properties. Eur. J. Physiol. 2004;447:653–665. doi: 10.1007/s00424-003-1168-y. [DOI] [PubMed] [Google Scholar]
- Hagenbuch B, Gui C. Xenobiotic transporters of the human organic anion transporting polypeptides (OATP) family. Xenobiotica. 2008;38:778–801. doi: 10.1080/00498250801986951. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, Price NT. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 1999;343:281–299. [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP, Meredith D. The SLC16 gene family – from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Eur. J. Physiol. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- Heuer H, Maier MK, Iden S, Mittag J, Friesema ECH, Visser TJ, Bauer K. The monocarboxylate transporter 8 linked to human psychomotor retardation is highly expressed in thyroid hormone-sensitive neuron populations. Endocrinology. 2005;146:1701–1706. doi: 10.1210/en.2004-1179. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinav. J. Stat. 1979;6:65–70. [Google Scholar]
- Huber RD, Gao B, Sidler Pfändler MA, Zhang-Fu W, Leuthold S, Hagenbuch B, Folkers G, Meier PJ, Stieger B. Characterization of two splice variants of human organic anion transporting polypeptide 3A1 isolated from human brain. Am. J. Physiol. Cell Physiol. 2007;292:C795–C806. doi: 10.1152/ajpcell.00597.2005. [DOI] [PubMed] [Google Scholar]
- Jacob TN, Pandey JP, Raghuveer K, Sreenivasulu G, Gupta AD, Yoshikuni M, Jagota A, Senthilkumaran B. Thyroxine-induced alterations in the testis and seminal vesicles of air-breathing catfish, Clarias gariepinus. Fish Physiol. Biochem. 2005;31:271–274. doi: 10.1007/s10695-006-0035-0. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Lema SC. Tissue-specific thyroid hormone regulation of gene transcripts encoding iodothyronine deiodinases and thyroid hormone receptors in striped parrotfish (Scarus iseri) Gen. Comp. Endocrinol. 2011;172:505–517. doi: 10.1016/j.ygcen.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Larhammar D, Sundstrom G, Dreborg S, Daza DO, Larsson TA. Major genomic events and their consequences for vertebrate evolution and endocrinology. Ann. NY Acad. Sci. 2009;1163:201–208. doi: 10.1111/j.1749-6632.2008.03659.x. [DOI] [PubMed] [Google Scholar]
- Lema SC, Nevitt GA. Evidence that thyroid hormone induces olfactory cellular proliferation in salmon during a sensitive period for imprinting. J. Exp. Biol. 2004;207:3317–3327. doi: 10.1242/jeb.01143. [DOI] [PubMed] [Google Scholar]
- Lema SC, Nevitt GA. Testing an ecophysiological mechanism of morphological plasticity in pupfish and its relevance to conservation efforts for endangered Devils Hole pupfish. J. Exp. Biol. 2006;209:3499–3509. doi: 10.1242/jeb.02417. [DOI] [PubMed] [Google Scholar]
- Lema SC, Dickey JT, Schultz IR, Swanson P. Dietary exposure to 2,2’,4,4’-tetrabromodiphenyl ether (PBDE-47) alters thyroid status and thyroid hormone-regulated gene transcription in the pituitary and brain. Environ. Health Perspect. 2008;116:1694–1699. doi: 10.1289/ehp.11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema SC, Dickey JT, Schultz IR, Swanson P. Thyroid hormone regulation of mRNAs encoding thyrotropin β-subunit, glycoprotein α-subunit, and thyroid hormone receptors α and β in brain, pituitary gland, liver, and gonads of an adult teleost, Pimephales promelas. J. Endocrinol. 2009;202:43–54. doi: 10.1677/JOE-08-0472. [DOI] [PubMed] [Google Scholar]
- Lema SC, Kitano J. Hormones and phenotypic plasticity: implications for the evolution of integrated adaptive phenotypes. Curr. Zool. 2013;59:504–523. [Google Scholar]
- Liu Q, Dou S, Wang G, Li Z, Feng Y. Evolution and functional divergence of monocarboxylate transporter genes in vertebrates. Gene. 2008;423:14–22. doi: 10.1016/j.gene.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Malik R, Hodgson H. The relationship between the thyroid gland and the liver. Quart. J. Med. 2002;95:559–569. doi: 10.1093/qjmed/95.9.559. [DOI] [PubMed] [Google Scholar]
- Mayerl S, Visser TJ, Darras VM, Horn S, Heuer H. Impact of Oatp1c1 deficiency on thyroid hormone metabolism and action in the mouse brain. Endocrinology. 2012;153:1528–1537. doi: 10.1210/en.2011-1633. [DOI] [PubMed] [Google Scholar]
- McCormick SD, Hansen LP, Quinn TP, Saunders RL. Movement, migration, and smolting of Atlantic salmon (Salmo salar) Can. J. Fish. Aquat. Sci. 1998;55:77–92. [Google Scholar]
- Mebis L, Paletta D, Debaveye Y, Ellger B, Langouche L, D’Hoore A, Darras VM, Visser TJ, Van der Berghe G. Expression of thyroid hormone transporters during critical illness. Eur. J. Endocrinol. 2009;161:243–250. doi: 10.1530/EJE-09-0290. [DOI] [PubMed] [Google Scholar]
- Meier-Abt F, Mokrab Y, Mizuguchi K. Organic anion transporting polypeptides of the OATP/SLCO superfamily: identification of new members in nonmammalian species, comparative modeling and a potential transport mode. J. Mem. Biol. 2006;208:213–227. doi: 10.1007/s00232-005-7004-x. [DOI] [PubMed] [Google Scholar]
- Mikkaichi T, Suzuki T, Tanemoto M, Ito S, Abe T. The organic anion transporter (OATP) family. Drug Metab. Pharmacokin. 2004;19:171–179. doi: 10.2133/dmpk.19.171. [DOI] [PubMed] [Google Scholar]
- Morgado I, Campinho MA, Costa R, Jacinto R, Power DM. Disruption of the thyroid system by diethylstilbestrol and ioxynil in the sea bream (Sparus aurata) Aquat. Toxicol. 2009;92:271–280. doi: 10.1016/j.aquatox.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Nelson ER, Habibi HR. Molecular characterization and sex-related seasonal expression of thyroid receptor subtypes in goldfish. Mol. Cell. Endocrinol. 2006;253:83–95. doi: 10.1016/j.mce.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Nelson ER, Habibi HR. Functional characterization and sex-related seasonal expression of thyroid receptor subtypes in goldfish. Mol. Cell. Endocrinol. 2008;253:4702–4709. doi: 10.1016/j.mce.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Noyes PD, Lema SC, Macaulay LJ, Douglas K, Stapleton HM. Low level exposure to the flame retardant BDE-209 reduces thyroid hormone levels and disrupts thyroid signaling in fathead minnows. Environ. Sci. Technol. 2013;47:10012–10021. doi: 10.1021/es402650x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes PD, Lema SC, Roberts SC, Cooper EM, Stapleton HM. Rapid method for the measurement of circulating thyroid hormones in low volumes of teleost fish plasma by LC-ESI/MS/MS. doi: 10.1007/s00216-013-7528-3. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes PD. PhD dissertation. Durham, NC: Duke University; 2013. Polybrominated diphenyl ether (PBDE) flame retardants: accumulation, metabolism, and disrupted thyroid regulation in early and adult life stages of fish. [Google Scholar]
- Orozco A, Valverde-R C, Olvera A, García-G C. Iodothyronine deiodinases: a functional and evolutionary perspective. J. Endocrinol. 2012;215:207–219. doi: 10.1530/JOE-12-0258. [DOI] [PubMed] [Google Scholar]
- Pizzagalli F, Hagenbuch B, Stieger B, Klenk U, Folkers G, Meier PJ. Identification of a novel human organism anion transporting polypeptide as a high affinity thyroxine transporter. Mol. Endocrinol. 2002;16:2283–2296. doi: 10.1210/me.2001-0309. [DOI] [PubMed] [Google Scholar]
- Popvic M, Zaja R, Smital T. Organic anion transporting polypeptides (OATP) in zebrafish (Danio rerio): phylogenetic analysis and tissue distribution. Comp. Biochem. Physiol. A. 2010;155:327–335. doi: 10.1016/j.cbpa.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Power DM, Llewellyn L, Faustino M, Nowell MA, Björnsson BTH, Einarsdottir IE, Canario AVM, Sweeney GE. Thyroid hormones in growth and development of fish. Comp. Biochem. Physiol. C. 2001;130:447–459. doi: 10.1016/s1532-0456(01)00271-x. [DOI] [PubMed] [Google Scholar]
- Pressler H, Sissung TM, Venzon D, et al. Expression of OATP family members in hormone-related cancers: potential markers of progression. PLOS One. 2011;6:e20372. doi: 10.1371/journal.pone.0020372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NT, Jackson VN, Halestrap AP. Cloning and sequencing of four new mammalian monocarboxylate transporter (MCT) homologues confirms the existence of a transporter family with an ancient past. Biochem. J. 1998;329:321–328. doi: 10.1042/bj3290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PK, Lam TJ. Effect of thyroid hormones on morphogenesis and growth of larvae and fry of telescopic-eye black goldfish, Carassius auratus. Aquaculture. 1992;107:383–394. [Google Scholar]
- Robbins J, Rall JE. Proteins associated with the thyroid hormones. Physiol. Revs. 1960;40:415–489. doi: 10.1152/physrev.1960.40.3.415. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schwartz CE, Stevenson RE. The MCT8 thyroid hormone transporter and Allan-Herndon-Dudley syndrome. Best Pract. Res. Clin. Endocrinol. Metab. 2007;21:307–321. doi: 10.1016/j.beem.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer U, Weitzel JM, Schomburg L. Think globally: act locally – new insights into the local regulation of thyroid hormone availability challenge long accepted dogmas. Mol. Cell. Endocrinol. 2008;289:1–9. doi: 10.1016/j.mce.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Sugiyama D, Kusuhara H, Taniguchi H, Ishikawa S, Nozaki Y, Aburatani H, Sugiyamam Y. Functional characterization of rat brain-specific organic anion transporter 15 (Oatp14) at the blood-brain barrier: high affinity transporter for thyroxine. J. Biol. Chem. 2003;278:43489–43495. doi: 10.1074/jbc.M306933200. [DOI] [PubMed] [Google Scholar]
- Svoboda M, Riha J, Wlcek K, Jaeger W, Thalhammer T. Organic anion transporting polypeptides (OATPs): Regulation of expression and function. Curr. Drug Metab. 2011;12:139–153. doi: 10.2174/138920011795016863. [DOI] [PubMed] [Google Scholar]
- Swapna I, Rajasekhar M, Supriya A, Raghuveer K, Sreenivasulu G, Rasheeda MK, Majumdar KC, Kagawa H, Tanaka H, Dutta-Gupa A, Senthikumaran B. Thiourea-induced thyroid hormone depletion impairs testicular recrudescence in the air-breathing catfish, Clarias gariepinus. Comp. Biochem. Physiol. A. 2006;144:1–10. doi: 10.1016/j.cbpa.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Szabo DT, Richardson VM, Ross DG, Diliberto JJ, Kodavanti PR, Birnbaum LS. Effects of perinatal PBDE exposure on hepative phase I, phase II, phase III, and deiodinase 1 gene expression involved in thyroid hormone metabolism in male rat pups. Toxicol. Sci. 2009;107:27–39. doi: 10.1093/toxsci/kfn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SW, Zoeller RT. Integrating basic research on thyroid hormone action into screening and testing programs for thyroid disruptors. Crit. Rev. Toxicol. 2007;37:5–10. doi: 10.1080/10408440601123396. [DOI] [PubMed] [Google Scholar]
- Tohyama K, Kusuhara H, Sugiyama Y. Involvement of multispecific organic anion transporter, Oatp14 (Slc21α14), in the transport of thyroxine across the blood-brain barrier. Endocrinology. 2004;145:4384–4391. doi: 10.1210/en.2004-0058. [DOI] [PubMed] [Google Scholar]
- van der Deure WM, Hansen PS, Peeters RP, Kyvik KO, Friesema EC, Hegedüs L, Visser TJ. Thyroid hormone transport and metabolism by OATP1C1 and consequences of genetic variation. Endocrinology. 2008;149:5307–5314. doi: 10.1210/en.2008-0430. [DOI] [PubMed] [Google Scholar]
- Van der Geyten S, Mol KA, Pluymers W, Kühn ER, Darras VM. Changes in plasma T3 during fasting/refeeding in tilapia (Orechromis niloticus) are mainly regulated through changes in hepatic type II iodothyroinine deiodinase. Fish Physiol. Biochem. 1998;19:135–143. [Google Scholar]
- Vatine GD, Zada D, Lerer-Goldshtein, Tovin A, Malkinson G, Yaniv K, Appelbaum L. Zebrafish as a model for monocarboxyl transporter 8-deficiency. J. Biol. Chem. 2013;288:169–180. doi: 10.1074/jbc.M112.413831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser WE, Visser TJ. Finding the way into the brain without MCT8. J. Clin. Endocrinol. Metabol. 2012;97:4362–4365. doi: 10.1210/jc.2012-3759. [DOI] [PubMed] [Google Scholar]
- Visser WE, Friesema ECH, Jansen J, Visser TJ. Thyroid hormone transport in and out of cells. Trends Endocrinol. Metabol. 2007;19:50–56. doi: 10.1016/j.tem.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Wang DL, Stapleton HM. Analysis of thyroid hormones in serum by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2010;397:1831–1839. doi: 10.1007/s00216-010-3705-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonkers MA, Ribera AB. Molecular components underlying nongenomic thyroid hormone signaling in embryonic zebrafish neurons. Neural Dev. 2009;4:20. doi: 10.1186/1749-8104-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller RT, Dowling ALS, Herzig CTA, Iannacone EA, Gauger KJ, Bansal R. Thyroid hormone, brain development, and the environment. Environ. Health Perspect. 2002;110(suppl. 3):355–361. doi: 10.1289/ehp.02110s3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.