Abstract
Repeated pesticide contaminations of lentic freshwater systems located within agricultural landscapes may affect population evolution in non-target organisms, especially in species with a fully aquatic life cycle and low dispersal ability. The issue of evolutionary impact of pollutants is therefore conceptually important for ecotoxicologists. The impact of historical exposure to pesticides on genetic divergence was investigated in the freshwater gastropod Lymnaea stagnalis, using a set of 14 populations from contrasted environments in terms of pesticide and other anthropogenic pressures. The hypothesis of population adaptive divergence was tested on 11 life-history traits, using Q ST -F ST comparisons. Despite strong neutral differentiation (mean F ST = 0.291), five adult traits or parameters were found to be under divergent selection. Conversely, two early expressed traits showed a pattern consistent with uniform selection or trait canalization, and four adult traits appeared to evolve neutrally. Divergent selection patterns were mostly consistent with a habitat effect, opposing pond to ditch and channel populations. Comparatively, pesticide and other human pressures had little correspondence with evolutionary patterns, despite hatching rate impairment associated with global anthropogenic pressure. Globally, analyses revealed high genetic variation both at neutral markers and fitness-related traits in a species used as model in ecotoxicology, providing empirical support for the need to account for genetic and evolutionary components of population response in ecological risk assessment.
Introduction
Understanding the causes of genetic divergence within species is a central goal in evolutionary biology. Population evolution is also a question of importance in environmental sciences and conservation biology, and, as a consequence, evolutionary principles are increasingly taken into account in the management of biodiversity and ecosystems [1]–[3]. Indeed, evolutionary change can be rapid, especially in the current environmental context. Besides evolutionary textbook cases of insecticide resistance [4], environmental stress may trigger fast evolutionary responses [5], and rapid adaptive responses have been documented in ecological situations involving anthropogenic disturbance [6], [7]. Furthermore, it is noteworthy that the rate of phenotypic change can itself increase in human-disturbed and toxic environments [8], [9]. In spite of this, the potential evolutionary impact of chemicals and toxic substances released by human activities into the environment is still not considered in current procedures of ecological risk assessment. However, strong arguments for mainstreaming such impacts have been formulated for a long time [10]–[14]. Beyond mutagenic compounds affecting the germline, pollutants have proved to be potential sources of evolutionary impact, as supported empirically by an ever growing number of studies [15]–[23].
Agriculture is probably one of the main causes of human-induced microevolution by selection, as a direct consequence of genetic resource management, e.g., domestication, selection programs on productive species, or side effects of intensive agricultural practices [24]–[26]. Furthermore, in the case of pesticides, as substances rarely reach their target species exclusively, non-intentional evolutionary effects are to be expected. Arguably, a substantial loss of biodiversity has resulted from the intensification of arable agriculture over the last five decades [27]. Agriculture, combined with industrial and domestic activities, uses more than one-third of the Earth's accessible renewable freshwater (i.e., approximately 4,430 km3/year in 2006) [28] and often leads to water contamination. About 140 million tons of fertilizers and several million tons of pesticides are applied each year [29]. Natural populations are thus exposed to a diversity of pressures that vary both spatially and temporally, and a succession of wildlife mortality events has marked the evolution of industry and agrichemical practices since the early 20th century [30].
With respect to human-induced stress, some conditions are expected to increase the risk of genetic change in natural populations. For instance, freshwater lentic habitats located within agricultural landscapes are repeatedly contaminated by complex pesticide mixtures, through various modes of transfer from the treated parcels (including aerial drift, run-off, and drainage; see [31]). Although a given pesticide or its metabolites may be only transiently present in water, the succession and combination of different molecules are expected to lead to high environmental variation and recurrent stress. At the population level, if demographic stochasticity is triggered by stressful episodes, random genetic drift and inbreeding will increase, and impede local adaptive processes [32]. Indeed, selection efficiency is positively correlated to effective population size [33], and population response to selection is weaker under high inbreeding, due to linkage disequilibrium and selection interference among loci [34]–[36]. Also, random drift load will accumulate locally and increase the risk of population extinction [33],[37]. Furthermore, population adaptive potential is also predicted to decrease in stressful and changing environments, especially due to genetic erosion [38]. These processes will be exacerbated under limited gene flow, which may result from low dispersal abilities (fully aquatic organisms, e.g., molluscs, crustaceans) or opportunities (weak connectivity among occupied sites, e.g., ponds, temporary ditches) [39]. Also, population tolerance to environmental stress and change will decrease as inbreeding increases, given that inbreeding depression generally worsens under stress [40]. Interestingly, environmental heterogeneity in selection also increases inbreeding depression [41].
Alternatively, faced with stress, populations may adapt in two ways. First, they may adjust through phenotypic plasticity, which should in principle have no evolutionary consequences, although plasticity may itself be under selection [42],[43]. Second, genetic adaptation may occur, provided that adapted alleles are present and selection intensity is strong enough to overcome stochastic interference [44].
To sum up, human-induced environmental stress and heterogeneity may alter population evolutionary trajectory in varied and complex ways, the study of which is anything but an easy task. Processes described above will also have consequences on population genetic divergence: both random genetic drift and local adaptation will cause population divergence, whereas environmental stress and heterogeneity may either act in the same way (random drift or varying selection regimes) or on the contrary, trigger similar adaptive responses among populations (uniform selection acting on general stress responsive pathways or traits).
One method commonly used to decipher population genetic divergence in terms of selective and neutral processes, is the Q ST-F ST approach [45],[46]. The general principle is to partition the total genetic variance at neutral markers and in quantitative traits into within- and between-population components, using Wright's F ST index, and its quantitative analogue Q ST, respectively. Under a purely additive determinism, if quantitative traits evolve neutrally, they should lead to similar levels of differentiation as those inferred from neutral markers (Q ST = F ST). Basically, Q ST and F ST are compared to test the null hypothesis of neutrality. Differences between the two indices indicate selection on the traits, with two possibilities: homogenizing (uniform stabilizing) selection towards a unique fitness optimum is revealed by F ST>Q ST, whereas divergent selection (occurrence of different fitness optima among populations) is reflected by the reverse relationship. This principle has been widely used in the last decade to address population adaptive divergence (see [46] and references therein). It has notably revealed population adaptive divergence as a function of habitat [47], environmental acidification [48], or soil contamination by zinc [49].
In the present study, we applied the Q ST-F ST method to test the hypothesis that anthropogenic pressure may act as a selective force and lead to population adaptive divergence, using a set of 14 natural populations of the freshwater snail Lymnaea stagnalis (Mollusca, Gastropoda, Panpulmonata, Hygrophila; previously classified in the Sub-Order Basommatophora). Due to its habitat characteristics (lentic systems, often close to agricultural zones) and its limited dispersal ability, this species is a good model to test non-intentional evolutionary effects of pesticides. Moreover, L. stagnalis belongs to an ecologically important group, which plays a major role in the transfer of energy across food webs, and can represent up to 20–60% of the total biomass of macro-invertebrates in some freshwater ecosystems [50]. Pesticides and other identified anthropogenic pressures were described qualitatively and considered as contributing to a global and potentially toxic pressure on non-target organisms. In this context, directional selection associated with a particular pesticide was considered irrelevant, and thus not specifically focused. Conversely, we hypothesized that global chronic exposure to various cocktails of pesticides may induce selective response to temporary but recurrent stress. The study aimed at addressing the following questions: (i) is agricultural (pesticide) pressure able to trigger adaptive processes in non-target organisms? (ii) what is the relative strength of such a pressure, compared to other environmental components, including natural and other anthropogenic factors? Indeed, as natural populations in a given level of pesticide pressure are not eco-evolutionary replicates of each other, any other relevant environmental factor should be also considered. Under this rationale, we characterized environments in terms of “pesticide pressure”, “other anthropogenic pressures” (roads, urbanized zones), “global pressure” (combination of agricultural and non agricultural anthropogenic pressures), and “habitat” (pond, ditch, channel, as physical features expected to affect population size and isolation). Population divergence patterns were investigated for a set of life-history traits and compared to neutral genetic differentiation.
Materials and Methods
Population sampling and characterization
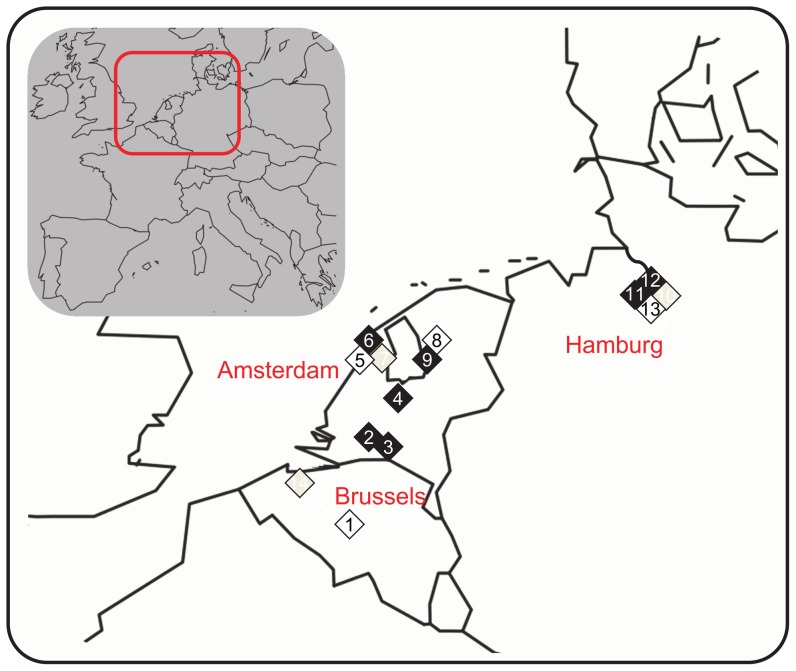
Adult individuals were collected from 14 natural populations during reproduction, in the course of a single campaign (5–12 July 2011) (Fig. 1). Sampling was conducted in public locations that did not require specific authorization, and did not involve endangered or protected species. Locality information and sampling characteristics are summarized in Table 1. Pesticide and other anthropogenic pressures were estimated from land-use patterns observed in the immediate surroundings of sampled sites, and using Google Earth. In the present study, we were not interested in an instant environmental concentration of pesticides at the time of sampling. On the contrary, under the tested hypothesis, we assumed historical contamination (repeated or chronic) as hypothetical selective force driving phenotypic evolution. Our strategy built on the use of landscape geographic information and agricultural land-use data. These have proved to be relevant tools to predict water body contamination by pesticides [51] and to assess pesticide exposure in the field (ditches [52]; ponds [18],[53],[54]). Moreover, in such aquatic systems, most variation may be explained by close land-use patterns (within a 100 m radius area), as demonstrated for water quality and vegetation complexity [55]. Consistently, we characterized each population's environment in terms of percent coverage by forest or moorland, pasture, crop (including potatoes, corn, orchards, bulb plants), and urban zone (Table 1). By crossing field observations and Google Earth satellite views with informations obtained from farmers on their practices, we estimated coverage proportions using the software ImageJ (http://rsbweb.nih.gov/ij/). When available, pesticide concentration data in surface water near the sampled sites were found to be globally consistent with our classification (http://81.93.58.66/bma_nieuw/begin.html; http://www.milieurapport.be/). Finally, populations were also classified according to four criteria: habitat (H: pond, channel, ditch), pesticide pressure (PP: two levels, low vs high), other anthropogenic pressure (OAP: two levels, low vs high), and global environmental pressure, as a combination of pesticide and other anthropogenic pressures (GEP: three levels).
Figure 1. Location of 14 L. stagnalis populations involved in Q ST-F ST comparisons.
Populations are coded from 1 to 14. Colours indicate environmental categories (white, grey, and black, for GEP, GEP1, and GEP2, respectively). See Table 1 for details.
Table 1. Geographical, environmental, and genetic characteristics of 14 L. stagnalis populations involved in FST-QST comparisons.
| Localization | Code | Geographic coordinates | Land use | Genetic/Environmental factor | Population genetics | ||||||||||
| GC | H | PP | OAP | GEP | n | N | A R | H E | F IS | Selfing rate (%) | Ne [CI 95%] | ||||
| CASTRICUM | CAS | 52°33.17N | 100% moor | West | Pond | 0 | 0 | 0 | 32 | 3.3 | 2.60 | 0.341 | 0.327* | 0.0 | 31 |
| (Alkmaar, NL) | (5) | 4°37.22E | [19]; [58] | ||||||||||||
| KUINRE | KUI | 52°47.67N | 98% forest | West | Ditch | 0 | 0 | 0 | 25 | 3.8 | 3.33 | 0.428 | 0.207* | 0.0 | 25 |
| (Nordoostpolder, NL) | (9) | 5°47.69E | 2% urban | [14]; [52] | |||||||||||
| HEDENDORF | HED | 53°29.47N | 90% forest | East | Pond | 0 | 0 | 0 | 26 | 2.8 | 2.39 | 0.315 | 0.258* | 0.0 | 32 |
| (Hamburg, DE) | (13) | 9°36.55E | 10% urban | [18]; [66] | |||||||||||
| OUD-HEVERLEE | OUD | 50°49.39N | 55% forest | West | Pond | 0 | 0 | 0 | 13 | 3.3 | 3.28 | 0.416 | 0.177 | 40.6* | 26 |
| (Brussels, BE) | (1) | 4°39.29E | 45% urban | [13]; [66] | |||||||||||
| SCHOORLDAM | SCH | 52°43.13N | 95% pasture | West | Channel | 0 | 1 | 1 | 26 | 3.3 | 2.89 | 0.324 | 0.225* | 18.5 | 24 |
| (Alkmaar, NL) | (7) | 4°42.2E | 5% urban | [13]; [47] | |||||||||||
| DETSELBERGEN | DET | 51°03.29N | 30% forest | West | Pond | 0 | 1 | 1 | 14 | 2.5 | 2.45 | 0.325 | 0.353* | 48.5* | 33 |
| (Gent, BE) | (14) | 3°49.35E | 30% fallow | [16; 139] | |||||||||||
| 40% urban | |||||||||||||||
| BUXTEHUDE | BUX | 53°29.63N | 80% fallow | East | Channel | 0 | 1 | 1 | 27 | 3.2 | 2.84 | 0.355 | 0.094 | 0.0 | 22 |
| (Hamburg, DE) | (10) | 9°42.78E | 15% crop | [12]; [43] | |||||||||||
| 5% urban | |||||||||||||||
| BAARN | BAA | 52°13.38N | 25% pasture | West | Ditch | 1 | 1 | 2 | 22 | 3.9 | 3.54 | 0.448 | 0.308* | 12.0 | 33 |
| (Utrecht, NL) | (4) | 5°18.53E | 25% crop | [18]; [72] | |||||||||||
| 50% urban | |||||||||||||||
| OOSTEIND | OOS | 51°38.87 | 30% pasture | West | Channel | 1 | 1 | 2 | 32 | 4.8 | 4.18 | 0.590 | 0.175* | 0.0 | 198 |
| (Breda, NL) | (2) | 4°55.82E | 40% crop | [105; 698] | |||||||||||
| 30% urban | |||||||||||||||
| BIEZENMORTEL | BIE | 51°38.03N | 20% forest | West | Channel | 1 | 1 | 2 | 30 | 4.4 | 3.67 | 0.452 | 0.168* | 9.1 | 109 |
| (Breda, NL) | (3) | 5°9.53E | 5% pasture | [64; 259] | |||||||||||
| 75% crop | |||||||||||||||
| PUTTEN | PUT | 52°45.81N | 30% crop | West | Ditch | 1 | 1 | 2 | 32 | 2.9 | 2.62 | 0.370 | 0.216* | 18.6 | 37 |
| (Alkmaar, NL) | (6) | 4°39.82E | 70% urban | [22]; [67] | |||||||||||
| EMMELOORD | EMM | 52°46.27N | 80% crop | West | Ditch | 1 | 1 | 2 | 24 | 3.7 | 3.32 | 0.456 | 0.252* | 0.0 | 36 |
| (Nordoostpolder, NL) | (8) | 5°48.26E | 20% urban | [20]; [80] | |||||||||||
| KOENIGSREICH | KOE | 53°31.6N | 85% crop | East | Channel | 1 | 1 | 2 | 27 | 5.3 | 4.17 | 0.444 | 0.176* | 9.5 | 48 |
| (Hamburg, DE) | (11) | 9°46.29E | 15% urban | [29]; [91] | |||||||||||
| AGATHENBURG | AGA | 53°34.54N | 100% crop | East | Ditch | 1 | 1 | 2 | 25 | 4.3 | 3.69 | 0.491 | 0.266* | 30.7* | 29 |
| (Hamburg, DE) | (12) | 9°32.82E | [17]; [53] | ||||||||||||
n indicates sample size, N mean number of alleles, AR allelic richness, HE expected heterozygosity, FIS inbreeding coefficient, and Ne effective population size. Values significantly different from zero are indicated with an asterisk. GC holds for Genetic cluster and H for Habitat. Environmental pressure is described qualitatively under three alternative factors (PP: Pesticide pressure, OAP: Other Anthropogenic pressure, GEP: Global Environmental Pressure), the levels of which are defined as a function of land use within a radius of 100 m around sample sites (see text for details).
Molecular analyses
DNA was chelex extracted from haemolymph or foot tissue from 399 wild-caught adults (14 to 33 snails per population). Neutral genetic variation was assessed at 12 microsatellite loci, i.e., A2, A112, B117 [56], 2k11 and 2k27 (Genbank accession: EF208747-EF208748 [57]), and EMLS04, EMLS13, EMLS21, EMLS26, EMLS29, EMLS41, EMLS45 [58], following the protocol described in Besnard et al. [58]. Only individuals with less than three missing genotypes were retained for population genetics analysis.
Population neutral genetic structure
Mean allele number (N), allelic richness (AR), expected heterozygosity H E, and observed heterozygosity H O, were calculated with Genetix 4.05.2 [59]. The distribution of neutral genetic diversity within and among populations was estimated from Weir and Cockerham's estimators of Wright's F indices [60] using Fstat 2.9.3.2 [61]. Departures from HWE (heterozygote excess or deficiency) and linkage disequilibria were tested using Genepop 4.0.10 [62]. Population differentiation was tested with a permutation test, in which genotypes were permuted among samples (not assuming HWE within samples; see [61]). As L. stagnalis is a self-fertile hermaphroditic organism, the selfing rate was estimated per population and statistically compared to zero using Rmes [63]. Effective population size was estimated using the sibship assignment method, as implemented in the software Colony 2.0.3.0 [64] and assuming inbreeding, male and female polygamic mating systems, and monoecy.
To estimate the number of genetic clusters in our dataset without taking into account any predefined population, we used Structure 2.2 [65]. Analyses were performed assuming an admixture model and a number of genetic clusters (k) from 1 to 15. Each run started with a burn-in period of 50 000 steps followed by 300 000 Markov Chain Monte Carlo (MCMC) replicates. The most likely number of clusters was determined using the Δk statistic [66] using Structure Harvester [67]. We used Distruct to plot Structure output data [68].
The effect of environmental factors PP, OAP, GEP, H, and genetic cluster was assessed on population genetics parameters (AR, H E, F IS, F ST) with a permutation test using Fstat.
Common garden experiment
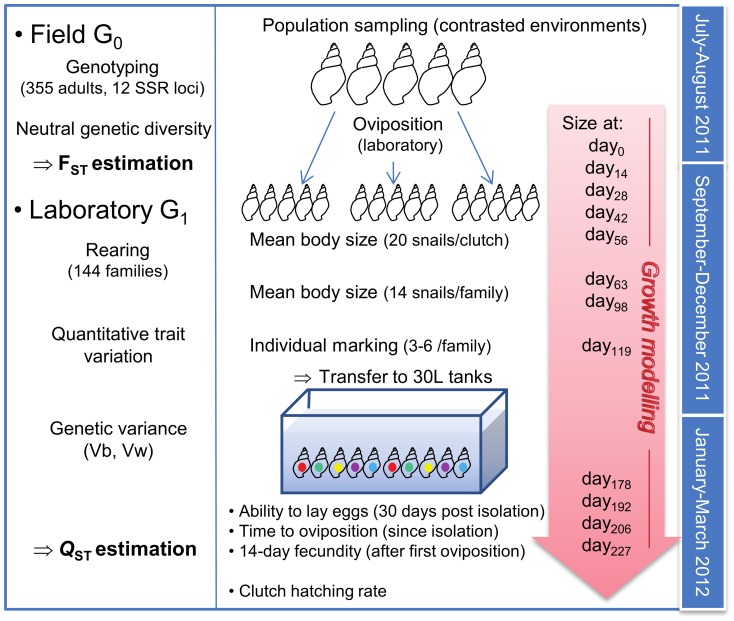
Wild-caught adults were brought to the laboratory and reared under standard conditions at the INRA Experimental Unit U3E (Rennes, France), as previously described [69]. Snails were isolated in plastic vessels filled with 1 L de-chlorinated and charcoal filtered tapwater. They were fed weekly with 1.5 g of organic salad, at each water renewal. Room temperature was maintained at 20±1°C and the photoperiod was 16L/8D. Like other basommatophorans, L. stagnalis lays eggs embedded in mucous enveloppes which are deposited and fixed on available substrates (sediment, vegetation, tank walls, etc.). For a given snail, reproduction was followed during 14 days after the first clutch laid in the laboratory. A total of 228 snails over 399 reproduced after 3 months (57%). Families were characterized at various life-history traits (individual growth, female reproduction, hatching success), measured on the laboratory-born progeny (G1) as illustrated on Fig. 2. G1 snails were reared at 20±1°C, under a 14L/10D photoperiod.
Figure 2. Schematic overview of the common garden experiment used to investigate population divergence in L. stagnalis.
G1 rearing conditions
From hatching to the age of 119 days (roughly corresponding to the end of juvenile growth), G1 snails were reared as groups reflecting clutch origin (n = 427; 1 to 2 clutches per maternal parent). As individuals grew, rearing conditions were adjusted in terms of water volume, individual density, and food supply (see Table S1). Vessels were regularly moved in a randomized way. From the age of 119 days, 7 individuals per clutch were randomly chosen and marked with a honey bee mark (model FC075; diameter: 2.3 mm, weight: 1.8 mg; Ickowicz Apiculture, Bollene, France), and further reared in 30 L tanks (80 individuals per tank), in which they were fed twice a week with organic salad (0.5 g per snail), and from which 2/3 of water was renewed weekly.
Life history traits: individual growth
From hatching to the age of 56 days, individual size (as inferred from shell height) was measured every two weeks on four randomly chosen juveniles per group. From the age of 63 days, all reared snails were individually measured. Measurements were performed with a stereomicroscope fitted with an ocular micrometer until day63, and a digital calliper afterwards. From the age of 119 days, three G1 snails per family (n = 492) were individually followed for growth and reproduction. For these 492 snails, size was measured at the age of 177.71±5.41 days, 191.71±5.41 days, 205.7±5.40 days, and 226.51±5.56 days (owing to variation in clutch age, and because measurements were performed at fixed dates after the age of 119 days), leading to a total of 12 values per individual. Growth was modelled using the Gompertz's model:
where Lt is the shell height at time t, A the asymptotic shell length, b a scaling factor related to shell height at t = 0, and k reflects the growth rate. Using a preliminary subset of 20 individuals, this model performed better than the von Bertalanffy model (lower AIC), and was thus preferred for further growth analysis. Growth parameters were compared among families and populations, as well as size at hatching and size at 119 days.
Life history traits: reproduction
Once clutches started to be observed in all aquaria, the 492 snails followed for growth were isolated in 200 mL plastic vessels and fed weekly with 1 g of organic salad immediately after water renewal. Several female reproductive traits were measured: time to first oviposition under isolation (with a censoring limit of 30 days), ability to lay eggs (as the proportion of reproductive snails per family), number of clutches and eggs laid during two weeks, clutch size (number of eggs per clutch), and clutch hatching rate.
Statistical analyses
All traits were analyzed with generalized linear mixed effect models (GLMM, R-package lme4) [70], with appropriate error distribution (Gaussian for normal data, Poisson for count data, and binomial for proportions). When necessary, data were log- or BoxCox-transformed, and covariates were included in the model (see Table S2). The model structure was: Y ∼ factor1 + factor2 + factor3 + covariate1 + covariate2 +…+ (1|population/family).
Fixed effects were tested by model comparison using a log-likelihood ratio test. Fixed factors were: genetic cluster (see Structure analysis), habitat, and environmental pressure (either pesticide, other anthropogenic, or global pressure). Family was nested within population, and both were treated as random factors. Global tests were followed by post-hoc pairwise comparison tests (Tukey and non parametric equivalent). All statistical analyses were performed using R 2.14.0 (R Core Team 2012).
QST -FST analysis
Q ST was computed for each trait using the equation [71]; [72]:
Where f is the inbreeding coefficient, Vb the between population genetic variance and Vw the within population genetic variance. Vb was obtained directly from the population variance component, while for Vw, as only the maternal origin could be assessed, family variance was used under the full-sib design (2Vf) [73]. Within- and between-population variance components were estimated for each of the 11 studied traits using restricted maximum likelihood (REML) under a simplified mixed model (Y∼1+ (1| pop/fam)).
Population quantitative divergence was tested using the method of Whitlock and Guillaume [74], which accounts for variation among neutral loci used to estimate F ST, and for sampling error and variation in evolutionary history in Q ST estimation. The method has lower type 1 error rate and better statistical power than previous tests, and seems particularly suited when neutral differentiation is high [46], as in L. stagnalis [58],[75]. Briefly, a bootstrapping procedure is applied to compare observed values of the difference Q ST - F ST to the Q ST - F ST distribution expected under the neutral hypothesis, as derived from F ST distribution and variance components [60] and Q ST neutral distribution predicted from the Lewontin-Krakauer distribution. Observed values were considered significant when they fell outside the 95% confidence interval of the neutral distribution [74].
Results
Population genetics
On 399 individual genotypes, 44 were discarded because of missing or unreadable data at more than three loci. Overall, the observed number of alleles per locus varied from three to 29, mean allelic richness ranged from 1.1 to 9.6 (Table S3), and genetic diversity per locus and sample varied from 0 to 0.86. Significant genotypic linkage disequilibrium was observed in 40 out of 656 comparisons. However, after Bonferroni correction, none of these remained significant. F IS-values varied widely across loci and samples, ranging from −0.238 (EMSL41 in BUX) to 1.000 (2k27 and B117 in DET; EMLS13 in HED; EMLS21 in BAA) (Table S3). Genetic parameters estimated per sample over loci reflected discrepancies between populations (Table 1). All populations except BUX and OUD were found significantly inbred (mean F IS-value = 0.219, 95% CI [0.147; 0.287]). The selfing rate was significantly different from zero in three populations (OUD, DET, and AGA). Effective population size Ne was significantly larger in sites exposed to high pesticide pressure (PP1> PP0; Kruskal-Wallis test, P = 0.007).
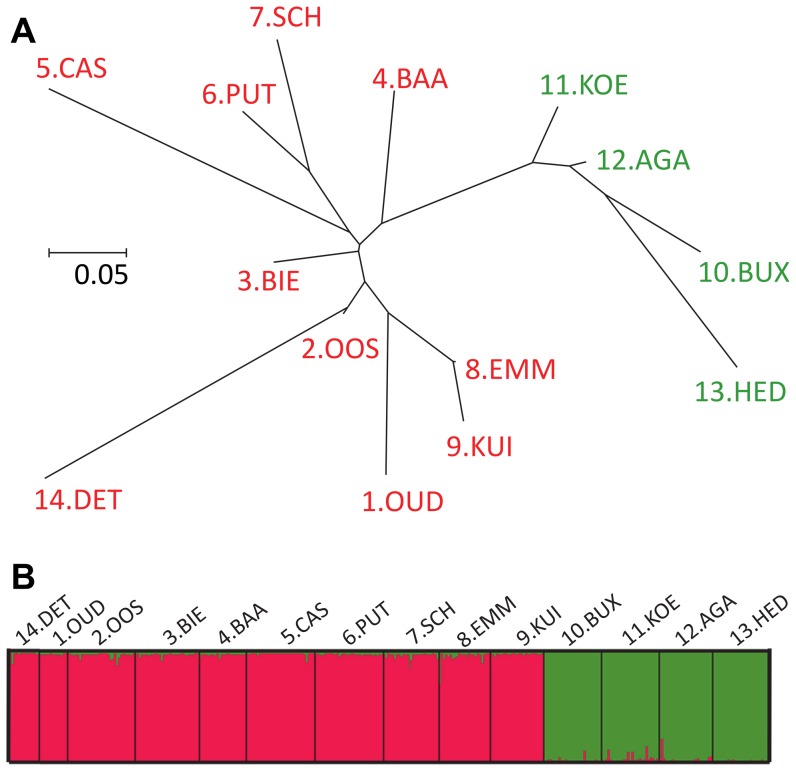
Population differentiation was generally high (mean FST-value = 0.291, 95% CI [0.248; 0.334]), as also reflected by pairwise estimates, which were all significant (Table S4), except between the two geographically close populations KUI and EMM. Interestingly, the four pond populations had the longest branches on the NJ-tree, indicating stronger differentiation associated with this habitat (Fig. 3A). From Structure analysis, individual genotypes clustered into two groups with the highest likelihood, and this was confirmed by the Δk statistic (Fig. 3B). Clusters corresponded to two geographic regions (10 western vs. four eastern populations). Therefore, the effect of “genetic cluster” was tested on life-history traits. No difference in level of genetic diversity, population inbreeding or population differentiation was found between the two clusters (Table 2).
Figure 3. Population genetic differentiation in L. stagnalis.
(A) Bayesian assignment probabilities in Structure analysis, for k = 2 clusters. Each bar represents an individual. Bar colour indicates the posterior probability that the individual belongs to the cluster of that color. (B) Unrooted Neighbour-Joining tree based on population pairwise F ST values (see Table S4).
Table 2. Comparison of L. stagnalis population genetic parameters as a function of different grouping factors.
| Population grouping | A R | H E | F IS | F ST |
| GEP0 | 2.898 | 0.367 | 0.249 | 0.431 |
| GEP1 | 2.725 | 0.353 | 0.190 | 0.450 |
| GEP2 | 3.599 | 0.468 | 0.214 | 0.197 |
| P (GEP) | 0.073 | 0.094 | 0.487 | 0.016 |
| PP0 | 2.824 | 0.362 | 0.226 | 0.413 |
| PP1 | 3.599 | 0.468 | 0.214 | 0.197 |
| P(PP) | 0.007 | 0.008 | 0.773 | 0.001 |
| OAP0 | 2.898 | 0.367 | 0.249 | 0.431 |
| OAP1 | 3.337 | 0.438 | 0.210 | 0.248 |
| P(OAP) | 0.219 | 0.178 | 0.329 | 0.031 |
| H Pond | 2.678 | 0.343 | 0.282 | 0.502 |
| H Ditch | 3.300 | 0.436 | 0.246 | 0.221 |
| H Channel | 3.550 | 0.449 | 0.166 | 0.246 |
| P(H) | 0.062 | 0.095 | 0.016 | 0.001 |
| H Pond | 2.678 | 0.343 | 0.282 | 0.502 |
| H Channel+Ditch | 3.425 | 0.443 | 0.203 | 0.227 |
| P (P vs D+C) | 0.025 | 0.038 | 0.034 | 0.002 |
| H Pond+Ditch | 3.024 | 0.398 | 0.258 | 0.336 |
| H Channel | 3.550 | 0.449 | 0.166 | 0.246 |
| P (P+D vs C) | 0.116 | 0.306 | 0.009 | 0.271 |
| H Pond+Channel | 3.162 | 0.409 | 0.203 | 0.330 |
| H Ditch | 3.300 | 0.436 | 0.246 | 0.221 |
| P (P+C vs D) | 0.686 | 0.641 | 0.285 | 0.178 |
| GC West | 3.187 | 0.422 | 0.229 | 0.244 |
| GC East | 3.272 | 0.410 | 0.196 | 0.162 |
| P (GC) | 0.827 | 0.837 | 0.426 | 0.393 |
A R is allelic richness, H E expected heterozygosity, F IS inbreeding coefficient, and F ST differentiation index.
GEP holds for Global environmental pressure, PP for Pesticide pressure, OAP for Other anthropogenic pressure, H for Habitat (P = Pond, D = Ditch, C = Channel), GC for Genetic cluster. Permutation-based statistical test (P value, 1000 permutations).
Population genetic parameters were affected by most environmental factors (Table 2). Genetic diversity tended to be greater under higher environmental pressure (GEP2, PP1, OAP1), although the difference was significant under pesticide pressure only (A R, P = 0.007; H E, P = 0.008). Population differentiation was significantly affected by global environmental pressure (GEP, P = 0.016) with a lower value among populations exposed to the highest level of pressure. With regard to specific pressures, lower differentiation was associated with pesticide as well as with other anthropogenic pressures (PP, P = 0.001; OAP, P = 0.031). Habitat affected all tested parameters, when ponds were compared to ditch and channel populations: pond populations were significantly less variable (A R, P = 0.025; H E, P = 0.038), more inbred (FIS, P = 0.034) and much more differentiated than were other populations (FST(pond) = 0.502 vs. FST(ditch + channel) = 0.227, P = 0.002).
Life history variation
Most G1 traits showed significant heterogeneity at both population and family levels (Tables S2 and S5). No effect of genetic cluster was detected, excepted on early size, which was larger in eastern populations (size at hatching, P = 0.019; growth parameter b, P = 0.005; Table 3). Among environmental factors, habitat was the most effective, with six traits or parameters significantly different between systems. Growth parameters indicated that body size at t0 increased significantly from ponds to channels and from channels to ditches, whereas asymptotic size was larger in ponds than in channels and ditches (Fig. S1). With respect to reproduction, snails from ponds had a significantly lower reproductive activity than those from ditches and channels: lower ability to lay eggs, and in reproductive snails, longer time to oviposition. Clutch size decreased significantly from ponds to ditches and from ditches to channels, and clutch hatching rate was lower in pond and ditch populations than in channel ones.
Table 3. Summary statistics of fixed effects tested on L. stagnalis life history traits.
| Trait/parameter | Genetic cluster (W/E) | Habitat (P/C/D) | Pesticide pressure (PP0/1) | Other anthropogenic pressure (OAP0/1) | Global environmental pressure (GP0/1/2) | |||||
| χ2 df = 1 | P | χ2 df = 2 | P | χ2 df = 1 | P | χ2 df = 1 | P | χ2 df = 2 | P | |
| Growth | ||||||||||
| Size at hatching | 6.95 | 0.008 | 3.67 | 0.159 | 0.12 | 0.724 | 0.36 | 0.549 | 0.91 | 0.636 |
| W<E | ||||||||||
| Growth parameter b | 9.23 | 0.002 | 17.91 | <0.001 | 0.44 | 0.506 | 9.60 | 0.002 | 10.20 | 0.006 |
| W>E | P>C>D | OAP0>OAP1 | GP0 = GP1>GP2 | |||||||
| Growth parameter k | 1.76 | 0.185 | 0.43 | 0.805 | 0.66 | 0.415 | 0.04 | 0.835 | 1.10 | 0.576 |
| Growth parameter A | 0.25 | 0.614 | 8.21 | 0.016 | 0.010 | 0.919 | 3.01 | 0.083 | 3.68 | 0.159 |
| P>C = D | OAP0>OAP1 | |||||||||
| Size at 119 days | 1.83 | 0.176 | 2.06 | 0.358 | 0.00 | 0.955 | 0.11 | 0.746 | 0.12 | 0.944 |
| Reproduction | ||||||||||
| Ability to lay eggs | 0.11 | 0.745 | 9.45 | 0.009 | 3.30 | 0.069 | 1.75 | 0.186 | 3.96 | 0.138 |
| P<D = C | PP0<PP1 | |||||||||
| Time to oviposition | 2.42 | 0.200 | 4.92 | 0.085 | 0.15 | 0.699 | 1.08 | 0.300 | 1.08 | 0.582 |
| P>D = C | ||||||||||
| Number of clutches | 1.40 | 0.237 | 0.39 | 0.823 | 0.14 | 0.708 | 0.10 | 0.756 | 0.44 | 0.802 |
| Number of eggs | 0.55 | 0.457 | 4.96 | 0.084 | 0.21 | 0.646 | 0.69 | 0.405 | 1.65 | 0.439 |
| P = D<C | ||||||||||
| Clutch size | 2.55 | 0.110 | 11.18 | 0.004 | 0.06 | 0.807 | 1.93 | 0.165 | 3.11 | 0.212 |
| P<D<C | ||||||||||
| Hatching rate | 0.59 | 0.441 | 6.41 | 0.041 | 0.98 | 0.322 | 6.25 | 0.012 | 6.25 | 0.044 |
| P = D<C | OAP0>OAP1 | GP0>GP1 = GP2 | ||||||||
General model: glmer = Y ∼ covar + Genetic cluster + Habitat + Environmental pressure + (1 | pop/fam).
Growth parameters: A is asymptotic size, b is related to size at birth, k to growth rate. Bold values are statistically significant.
Compared to aquatic system, human pressure appeared much less effective. First, pesticide pressure had no effect, except a marginal one on ability to lay eggs, found greater in exposed populations (P = 0.069). Second, other anthropogenic pressures affected growth parameter b (significantly larger size at t0 under OAP1) and hatching rate (significantly lower under OAP1) (Fig. S2). Interestingly, lower hatching rates were already observed on the clutches laid by G0 snails inhabiting OAP1 sites (720 clutches, GLMM test: χ2 = 11.861, P<0.001). Third, global pressure affected b (significantly larger snails at t0 under GEP2), and hatching rate, which was significantly impaired in exposed populations (GEP1 and GEP2) relative to reference ones (GEP0).
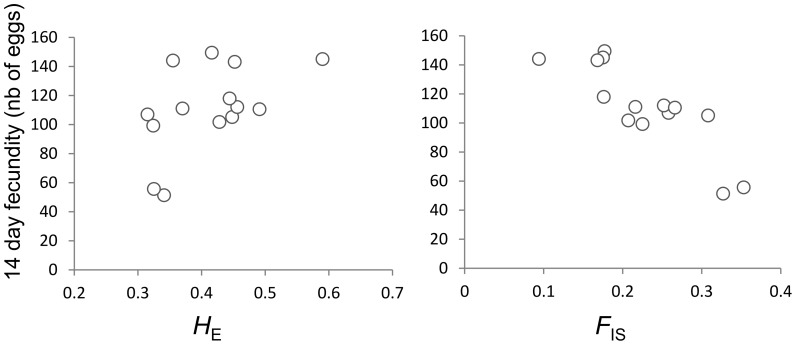
Several life history traits correlated with population genetic characteristics. First, fecundity was positively correlated with H E (r = 0.524, P = 0.031; Fig. 4A), and with A R (r = 0. 489, P = 0.044). Second, a negative correlation was found between population inbreeding and G1 fecundity (r = −0.845, P<0.001; Fig. 4B), ability to reproduce (r = −0.664, P = 0.006), and clutch hatching rate (r = −0.634, P = 0.009) (see Table 1 and Table S5).
Figure 4. Correlation plot between 14-days fecundity and (A) population expected heterozygosity, (B) population inbreeding, as based on 14 L. stagnalis populations.
QST-FST analysis
Results are summarized on Table 4. Over the set of 11 studied traits or parameters, four traits were found to evolve consistently with neutral expectations (hatching size, growth parameter k, number of eggs, clutch size), whereas homogenizing selection was indicated for two traits (growth parameter b, clutch hatching rate), and divergent selection for five traits (size at 119 days, asymptotic size A, ability to lay eggs, time to oviposition, number of clutches laid).
Table 4. Summary of Q ST -F ST analyses on laboratory G1s of 14 L. stagnalis populations.
| Trait/Parameter | N | Nfam | Vb | Vf | Observed Q ST - F ST | Neutral Q ST-F ST 95% CI | P-value left | P-value right | Divergence pattern |
| Growth | |||||||||
| Hatching size | 427 | 211 | 1.46E-03 | 1.86E-03 | −0.098 | [−0.160; 0.150] | 0.141 | 0.859 | Neutrality |
| Parameter b | 492 | 144 | 4.52E-04 | 1.19E-03 | −0.187 | [−0.160; 0.154] | 0.009 | 0.991 | Homogenizing selection |
| Parameter k | 492 | 144 | 2.48E-07 | 2.99E-07 | −0.089 | [−0.155; 0.156] | 0.169 | 0.831 | Neutrality |
| Parameter A | 492 | 144 | 6.02E-03 | 6.56E-04 | 0.446 | [−0.153; 0.155] | 1.000 | 0.000 | Divergent selection |
| Size at 119 days | 2855 | 183 | 2.26E+01 | 5.07E+00 | 0.285 | [−0.151; 0.155] | 1.000 | 0.000 | Divergent selection |
| Reproduction | |||||||||
| Ability to lay eggs | 492 | 144 | 2.70E+00 | 5.39E-08 | 0.709 | [−0.153; 0.157] | 1.000 | 0.000 | Divergent selection |
| Time to oviposition | 492 | 144 | 2.86E-02 | 8.36E-03 | 0.219 | [−0.141; 0.151] | 0.996 | 0.004 | Divergent selection |
| Number of clutches | 492 | 144 | 1.10E-01 | 1.02E-10 | 0.709 | [−0.160; 0.151] | 1.000 | 0.000 | Divergent selection |
| Number of eggs | 492 | 144 | 8.22E+02 | 5.64E+02 | 0.017 | [−0.162; 0.163] | 0.605 | 0.395 | Neutrality |
| Clutch size | 1537 | 144 | 2.95E+00 | 1.47E+00 | 0.088 | [−0.158; 0.168] | 0.885 | 0.115 | Neutrality |
| Hatching rate | 1537 | 144 | 3.27E-01 | 6.29E-01 | −0.154 | [−0.157; 0.160] | 0.025 | 0.975 | Homogenizing selection |
Analyses were performed using a full-sib design and accounting for population inbreeding.
N = sample size, Nfam = number of families, Vb and Vf indicate between population and family variance, respectively. Gompertz growth parameters: A is asymptotic size, b is related to size at birth, k is related to growth rate. Left and right P-values indicate the probability for QST-FST observed value to lie within the neutral distribution (unilateral test based on 1000 bootstraps, α = 0.025).
Discussion
The hypothesis that human activities may induce population adaptive divergence in L. stagnalis was addressed under a common garden experiment involving life history traits and a set of natural populations from contrasted environments. Five traits or parameters over 11 were found under divergent selection, despite a strong neutral genetic structure. The occurrence of two genetic clusters (Eastern and Western populations) had only a weak influence on quantitative genetic divergence, suggesting that population level was the adequate scale to draw selection inferences [76]. Only size at hatching and its corresponding growth parameter (b) were affected by this factor (larger hatchlings to the East). Given that early size was found under uniform selection, it was assumed that the genetic divergence associated to geography did not interfere with selection putatively associated with local environmental pressures.
Selection versus random genetic drift
All traits found to diverge adaptively were adult traits, related to late expressed growth (sub-adult size, asymptotic size A) and to reproduction (ability to reproduce, time to oviposition after isolation, number of clutches). Conversely, uniform selection was the most likely evolutionary hypothesis for early survival and size (hatching success and growth parameter b, which is related to size at t0). Globally, inferred patterns, i.e., early (late) traits under homogenizing (divergent) selection, are in lines with those found in another freshwater snail [47],[77].
Traditional statistical analyses performed on the G1 revealed that, to the exception of a marginal effect of pesticide pressure on ability to lay eggs (P = 0.069), habitat was the only factor affecting most traits found under divergent selection. Indeed, pond populations were characterized by larger ultimate size (growth parameter A), lower ability to lay eggs, and longer time to oviposition after isolation, compared to ditch and channel ones.
Congruent with L. stagnalis low dispersion and weak habitat connectivity, pond populations were particularly differentiated (especially CAS and DET, pairwise F ST) and presented generally lower genetic diversity and higher inbreeding than channel and ditch populations. Moreover, compared to channel populations, pond and ditch populations were also more inbred (P = 0.009). These aquatic systems are likely to imply stronger population isolation, even among ditches (due to frequent drought). A similar influence of habitat was emphasized in another freshwater snail, Physa acuta [78]. Thus, in L. stagnalis, although divergent selection was detected for traits also affected by habitat type, genetic drift is likely to highly contribute to divergence among habitats. This result is in line with the detection of significant drift load in experimental populations occupying outdoor close mesocosms [69].
In the common frog Rana temporaria, habitat fragmentation correlates with low genetic diversity and high differentiation, and negatively impacts tadpole body size [79], which is a critical trait determining individual fitness [80]. In L. stagnalis, although ultimate body size was greater in ponds, the relation between size and fitness is not necessarily positive, because of possible energetic trade-offs between growth and reproduction [81]. Indeed, two pond populations (CAS and DET) exhibited both highest mean asymptotic size and lowest fecundity (see Table S5).
The Q ST-F ST based test concluded to neutral divergence for size at hatching, growth rate (parameter k), and two reproductive traits, clutch size and early fecundity (number of eggs laid during 14 days following first clutch since isolation). On the latter, it seems surprising to find no hint of selection, either uniform or divergent, although this may simply reflect low heritability of fitness traits [82]. In support of this, it is worth noting that fecundity is extremely high in L. stagnalis (from adulthood to death, i.e., about one year under standard conditions, a snail produces more than 10 progeny per day), suggesting the possibility of high variation without drastic fitness consequences. Consistently, population growth rate was found to be insensitive to fecundity under experimental conditions (Leslie matrix modelling [83]). Alternatively, apparent neutral evolution might result from a bias in Q ST estimation. Under genetic drift, dominance and epistasis bias Q ST estimates downward [84]–[86]. As Q ST is traditionally estimated without accounting for these non-additive sources of variation, and given the strong observed global F ST value, divergent selection might have been underestimated for fecundity. On the other side, a hypothetical upward bias in Q ST estimation might result from too high a mutation rate at the markers used to infer F ST, as compared to the mutation rate of loci encoding the studied quantitative trait [87],[88]. This hypothesis is invalidated by observed F ST values, which were rather high and significant, although they may also suffer from a downward bias. Finally, gene flow may well lead to an apparent pattern of adaptive divergence at a trait under uniform selection, if this trait is correlated to a trait under divergent selection [89]. Again, high population neutral differentiation makes this hypothesis unlikely. Besides methodological causes of bias, it might be also mentioned that (1) true lifespan fecundity was not measured, and (2) as a source of uncontrolled variation, no distinction could be made between outcrossing and potential selfing in the G1.
As reported above, homogenizing selection was indicated for early traits, which is expected to reflect the occurrence of a uniform fitness optimum between the studied populations. Q ST estimation was based on the G1 of wild-caught adult snails, with the exception of survival at hatching (measured on G2). G1 families correspond hence to the hermaphroditic equivalent of isofemale lines, as used for broad-sense heritability estimation [73]. Therefore, these “broad-sense” Q ST estimates are potentially biased by genetic non additive sources of variation (dominance, epistasis), and by maternal effects. The latter are likely to operate early in life and tend to recede with age [90]. Hence, in the present study, traits found under uniform selection (early expressed traits) might be more prone to such a bias than traits found under divergent selection (late expressed traits). However, as complex traits seem to vary mostly additively [91],[92], we hypothesized, as done in most Q ST-F ST comparisons [46], that neglecting these sources of variation would have little consequences.
Alternatively to the uniform selection hypothesis, the pattern exhibited by early traits would result from trait canalization, as a phylogenetically inherited characteristics [86]. Since the pattern was similar to that observed in Galba truncatula [77], the criterion of cross-species consistency (supporting the canalization hypothesis) may be met in basommatophorans, but this would need to be confirmed in other species. Further investigation is clearly needed to disentangle both causes of L. stagnalis population phenotypic convergence.
Genetic diversity and fitness
The positive correlation observed between population genetic variability and G1 fecundity, is consistent with previous findings in L. stagnalis [93]. Under the assumption that G1 performances reflect the fitness of natural populations, the present result also supports the hypothesis that local drift load can be strong in this species, as previously suggested [69]. Therefore, under high random genetic drift, as indicated here, it might also be asked to what extent Q ST is modified by the expected stochastic evolution of slightly deleterious alleles as compared to pure selection-based predictions (random drift load [37]).
Furthermore, the negative correlation observed between population inbreeding and several traits related to fecundity (ability to reproduce, time to oviposition, early fecundity, and hatching rate) is in line with the occurrence of reduced fitness in inbred populations, and thus of inbreeding depression in these populations. However, as inbreeding was found significant in most populations and because effective population size was usually small, it is suggested that random genetic drift is actually the main cause of these apparent correlations, through reduced genetic diversity and drift load accumulation caused by population isolation and small size [32]. This hypothesis is still strengthened by the fact that population inbreeding correlated negatively with G2 survival at hatching (i.e., irrespective of individual inbreeding level). Inbreeding depression estimated under selfing relative to random outcrossing (ID) is particularly low in L. stagnalis [69],[93]–[95], despite a clear preference for outcrossing [96],[97]. Thus, although ID was not estimated, it seems unlikely that selfing could be responsible for the observed pattern. In support of this, no correlation was observed between population selfing rate and G1 fecundity.
Evolutionary impact of anthropogenic pressures
Neutral genetic variation was inflated in populations exposed to anthropogenic pressures, including pesticides. This was reflected in terms of genetic diversity, allelic richness, and effective population size, and through lower genetic differentiation. These results are opposed to those found in experimental populations of L. stagnalis exposed to cocktails of pesticides [98]. More generally, they are not consistent with the hypothesis of increased local stochasticity due to anthropogenic pressure (see introduction). High genetic diversity may result either from the maintenance of large population size despite stressful conditions, or from significant gene flow (immigration). Alternatively, undetected population subdivision may also be responsible for the observed patterns, as subdivision is expected to maintain diversity at the global scale. Furthermore, at the same scale, heterozygote deficiencies are also to be expected (Wahlund effect). Therefore, population subdivision into local demes might be responsible for part of the within population fixation observed in the dataset. As a possible impact of anthropogenic pressure, this potential effect should deserve specific attention, e.g., through the scoring of individual location at the time of sampling, and using assignment tests. As already mentioned, the effect of anthropogenic pressures on life history traits was much lower than that of habitat. This was particularly marked for pesticide pressure, which was the primary focus of the study. Three possible causes for this may be discussed: 1) true lack of pesticide effect, 2) effects underestimated due to confounding environmental factors, and 3) experimental conditions too benign to detect adaptive divergence.
Besides the hypothesis of pesticide innocuousness to gastropod populations, one explanation may be that sampling did not reflect the effective magnitude of pesticide pressure. However, as the design included areas of intensive agriculture, and due to the close proximity of water bodies and treated agricultural parcels, this seems unrealistic. Alternatively, evolutionary patterns associated with pesticide exposure may have been masked by other environmental characteristics, an idea supported by the significant habitat effect observed on most studied traits. Thus, although the number of populations and families was close to the recommended values [72], and despite the ability of mixed effect models to treat unbalanced designs [70], our experiment may lack sufficient statistical power, probably due to the lack of ponds within agricultural zones and of channels in reference sites. Meanwhile, the present study emphasizes the need to take habitat into account in Q ST-F ST comparisons [79].
Third, the apparent lack of effect of pesticide pressure on the studied traits might also result from experimental conditions, which were too benign. For example, the trend for enhanced ability to reproduce observed in the G1 from polluted areas, might reflect a specific response of exposed populations to overcome early mortality induced by chemicals (hatching rate: OAP0>OAP1 in G1 and G2), which was unexpected to express under common garden conditions. In the context of evolution under stressful environments, trait expression would be best studied under a gradient of stress conditions, as also recommended for molecular traits and pathways [15]. Stressful conditions may indeed facilitate the detection of divergent selection patterns with respect to pesticide historical exposure, since Q ST has been shown to depend on the experimental environment [48] [99].
Finally, the present study also emphasized the occurrence of high genetic variation both at neutral markers and fitness-related traits in a species used as model in ecotoxicology. This finding provides empirical support for the need to account for genetic variation in toxicity testing and, as a perspective, in future procedures of ecological risk assessment [12].
Supporting Information
L. stagnalis individual growth (G1 snails) according to population habitat. Gompertz growth curves based on 12 dates of measurement, performed on 492 individuals.
(TIFF)
L. stagnalis hatching rate as a function of anthropogenic pressure (OAP, n = 1537 clutches).
(TIFF)
Evolution of L. stagnalis G1 rearing conditions as a function of age.
(DOCX)
Summary of statistical analyses performed on L. stagnalis life history traits.
(DOCX)
Population genetic parameters calculated per locus and population on 14 L. stagnalis population samples (see materials and methods for explanation of population codes).
(DOCX)
Population genetic differentiation estimated among 14 L. stagnalis populations, on the basis of 12 SSR loci.
(DOCX)
Life-history trait/parameter mean-values (SE) estimated in the laboratory-born G1 from 14 L. stagnalis populations used to test patterns of adaptive divergence (Q ST-F ST analysis).
(DOCX)
Acknowledgments
The authors thank the U3E staff (INRA, Rennes), and more particularly Maïra Coke for her technical assistance. Our thanks are also due to Udo Hommen for indicating sites in Germany, to Thierry Caquet and Sabrina Le Cam for their help and advices, and to Patrice David and Nico Van Straalen for constructive comments on the manuscript. The authors finally thank Benoît Heurtault, Gaëlle Méheut, Elise Petitpas, and Arnaud Giusti for hospitality and logistic support during sampling.
Contributions
Conceived and designed the experiments: AB MAC. Performed the experiments: AB ALB MC MAC. Analyzed the data: AB JC MAC. Contributed reagents/materials/analysis tools: AB ALB. Contributed to the writing of the manuscript: AB JC MAC.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Raw data are available from the Dryad database using the DOI doi: 10.5061/dryad.g4d30.
Funding Statement
AB had a PhD fellowship from the French National Institute for Agricultural Research (INRA: CJS Contrat Jeune Scientifique). Experiment and data acquisition were funded by the French National Institute for Agricultural research (INRA: AIP Bioressources and EFPA Department PI). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lankau R, Jørgensen PS, Harris DJ, Sih A (2011) Incorporating evolutionary principles into environmental management and policy. Evol Appl 4: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith TB, Bernatchez L (2008) Evolutionary change in human-altered environments. Mol Ecol 17: 1–8. [DOI] [PubMed] [Google Scholar]
- 3. Hendry AP, Kinnison MT, Heino M, Day T, Smith TB, et al. (2011) Evolutionary principles and their practical application. Evol Appl 4: 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guillemaud T, Lenormand T, Bourguet D, Chevillon C, Pasteur N, et al. (1998) Evolution of resistance in Culex pipiens: allele replacement and changing environment. Evolution 52: 443–453. [DOI] [PubMed] [Google Scholar]
- 5. Hoffmann AA, Hercus MJ (2000) Environmental stress as an evolutionary force. Bioscience 50: 217–226. [Google Scholar]
- 6. Reznick DN, Ghalambor CK (2001) The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica 112: 183–198. [PubMed] [Google Scholar]
- 7. Sänen KR, Laurila A, Merilä J (2003) Geographic variation in acid stress tolerance of the moor frog, Rana arvalis. I. Local adaptation. Evolution 57: 352–362. [PubMed] [Google Scholar]
- 8. Hendry AP, Farrugia TJ, Kinnison MT (2008) Human influences on rates of phenotypic change in wild animal populations. Mol Ecol 17: 20–29. [DOI] [PubMed] [Google Scholar]
- 9. Pomati F, Nizzetto L (2013) Assessing triclosan-induced ecological and trans-generational effects in natural phytoplankton communities: a trait-based field method. Ecotoxicology 22: 779–794. [DOI] [PubMed] [Google Scholar]
- 10. Depledge MH (1994) Genotypic toxicity - Implications for individuals and populations. Environ Health Perspect 102: 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leblanc GA (1994) Assessing deleterious ecosystem-level effects of environmental pollutants as a means of avoiding evolutionary consequences. Environ Health Perspect 102: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breitholtz M, Rudén C, Ove Hansson S, Bengtsson B-E (2006) Ten challenges for improved ecotoxicological testing in environmental risk assessment. Ecotoxicol Environ Saf 63: 324–335. [DOI] [PubMed] [Google Scholar]
- 13. Bickham JW, Sandhu S, Hebert PDN, Chikhi L, Athwal R (2000) Effects of chemical contaminants on genetic diversity in natural populations: implications for biomonitoring and ecotoxicology. Mutat Res 463: 33–51. [DOI] [PubMed] [Google Scholar]
- 14. Klerks PL (2002) Adaptation, ecological impacts, and risk assessment: insights from research at Foundry Cove, Bayou Trepagnier, and Pass Fourchon. Hum Ecol Risk Assess 8: 971–982. [Google Scholar]
- 15. Whitehead A, Roach JL, Zhang S, Galvez F (2011) Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc Natl Acad Sci USA 108: 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varian-Ramos CW, Swaddle JP, Cristol DA (2013) Familial differences in the effects of mercury on reproduction in zebra finches. Environ Pollut 182: 316–323. [DOI] [PubMed] [Google Scholar]
- 17. Belanger-Deschênes S, Couture P, Campbell PGC, Bernatchez L (2013) Evolutionary change driven by metal exposure as revealed by coding SNP genome scan in wild yellow perch (Perca flavescens). Ecotoxicology 22: 938–957. [DOI] [PubMed] [Google Scholar]
- 18. Orsini L, Spanier KI, De Meester L (2012) Genomic signature of natural and anthropogenic stress in wild populations of the waterflea Daphnia magna: validation in space, time and experimental evolution. Mol Ecol 21: 2160–2175. [DOI] [PubMed] [Google Scholar]
- 19. Oleksiak MF, Karchner SI, Jenny MJ, Welch DBM, Hahn ME (2011) Transcriptomic assessment of resistance to effects of an aryl hydrocarbon receptor (AHR) agonist in embryos of Atlantic killifish (Fundulus heteroclitus) from a marine Superfund site. BMC Genomics 12: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bourret V, Couture P, Campbell PGC, Bernatchez L (2008) Evolutionary ecotoxicology of wild yellow perch (Perca flavescens) populations chronically exposed to a polymetallic gradient. Aquat Toxicol 86: 76–90. [DOI] [PubMed] [Google Scholar]
- 21. Morgan AJ, Kille P, Sturzenbaum SR (2007) Microevolution and ecotoxicology of metals in invertebrates. Environ Sci Technol 41: 1085–1096. [DOI] [PubMed] [Google Scholar]
- 22. Matson CW, Lambert MM, McDonald TJ, Autenrieth RL, Donnelly KC, et al. (2006) Evolutionary toxicology: population-level effects of chronic contaminant exposure on the marsh frogs (Rana ridibunda) of Azerbaijan. Env Health Perspect 114: 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Medina MH, Correa JA, Barata C (2007) Micro-evolution due to pollution: Possible consequences for ecosystem responses to toxic stress. Chemosphere 67: 2105–2114. [DOI] [PubMed] [Google Scholar]
- 24. Yuan JS, Tranel PJ, Stewart CN (2006) Non-target-site herbicide resistance: a family business. Trends Plant Sci 12: 6–13. [DOI] [PubMed] [Google Scholar]
- 25. Via S (1990) Ecological genetics and host adaptation in herbivorous species: the experimental study of evolution in natural and agricultural systems. Annu Rev Entomol 35: 421–446. [DOI] [PubMed] [Google Scholar]
- 26. Jones BA, Grace D, Kock R, Alonso S, Rushton J, et al. (2013) Zoonosis emergence linked to agricultural intensification and environmental change. Proc Natl Acad Sci USA 110: 8399–8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Memmott J (2009) Food webs: a ladder for picking strawberries or a practical tool for practical problems? Philos Trans R Soc B Biol Sci 364: 1693–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwarzenbach RP, Escher BI, Fenner K, Hofstetter TB, Johnson CA, et al. (2006) The challenge of micropollutants in aquatic systems. Science 313: 1072–1077. [DOI] [PubMed] [Google Scholar]
- 29. Schwarzenbach RP, Egli T, Hofstetter TB, von Gunten U, Wehrli B (2010) Global water pollution and human health. Annu Rev Environ Resour 35: 109–136. [Google Scholar]
- 30. Rattner BA (2009) History of wildlife toxicology. Ecotoxicology 18: 773–783. [DOI] [PubMed] [Google Scholar]
- 31. Brown CD, van Beinum W (2009) Pesticide transport via sub-surface drains in Europe. Environ Pollut 157: 3314–3324. [DOI] [PubMed] [Google Scholar]
- 32. Willi Y, Van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37: 433–458. [Google Scholar]
- 33. Lynch M (1995) Mutation accumulation and the extinction of small populations. Am Nat 146: 489–518. [Google Scholar]
- 34. Haddrill PR, Halligan DL, Tomaras D, Charlesworth B (2007) Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. B, Morgan MT, Charlesworth D (1993) The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gordo I, Charlesworth B (2001) Genetic linkage and molecular evolution. Curr Biol 11: 684–686. [DOI] [PubMed] [Google Scholar]
- 37. Whitlock MC, Ingvarsson PK, Hatfield T (2000) Local drift load and the heterosis of interconnected populations. Heredity 84: 452–457. [DOI] [PubMed] [Google Scholar]
- 38. Bijlsma R, Loeschcke V (2012) Genetic erosion impedes adaptive responses to stressful environments. Evol Appl 5: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coutellec M-A, Collinet M, Caquet T (2011) Parental exposure to pesticides and progeny reaction norm to a biotic stress gradient in the freshwater snail Lymnaea stagnalis . Ecotoxicology 20: 524–534. [DOI] [PubMed] [Google Scholar]
- 40. Fox CW, Reed DH (2011) Inbreeding depression increases with environmental stress: an experimental study and meta-analysis. Evolution 65: 246–258. [DOI] [PubMed] [Google Scholar]
- 41. Long TAF, Rowe L, Agrawal AF (2013) The effects of selective history and environmental heterogeneity on inbreeding depression in experimental populations of Drosophila melanogaster . Am Nat 181: 532–544. [DOI] [PubMed] [Google Scholar]
- 42. Pelletier F, Réale D, Watters J, Boakes EH, Garant D (2009) Value of captive populations for quantitative genetics research. Trends Ecol Evol 24: 263–270. [DOI] [PubMed] [Google Scholar]
- 43. Via S, Lande R (1985) Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39: 505–522. [DOI] [PubMed] [Google Scholar]
- 44. Hoffmann AA, Willi Y (2008) Detecting genetic responses to environmental change. Nat Rev Genet 9: 421–432. [DOI] [PubMed] [Google Scholar]
- 45. Spitze K (1993) Population Structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135: 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leinonen T, McCairns RS, O'Hara RB, Merilä J (2013) QST–FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat Rev Genet 14: 179–190. [DOI] [PubMed] [Google Scholar]
- 47. Chapuis E, Trouvé S, Facon B, Degen L, Goudet J (2007) High quantitative and no molecular differentiation of a freshwater snail (Galba truncatula) between temporary and permanent water habitats. Mol Ecol 16: 3484–3496. [DOI] [PubMed] [Google Scholar]
- 48. Hangartner S, Laurila A, Räsänen K (2012) Adaptive divergence in moor frog (Rana arvalis) populations along an acidification gradient: inferences from QST-FST correlations. Evolution 66: 867–881. [DOI] [PubMed] [Google Scholar]
- 49. Jiménez-Ambriz G, Petit C, Bourrié I, Dubois S, Olivieri I, et al. (2007) Life history variation in the heavy metal tolerant plant Thlaspi caerulescens growing in a network of contaminated and noncontaminated sites in southern France: role of gene flow, selection and phenotypic plasticity. New Phytol 173: 199–215. [DOI] [PubMed] [Google Scholar]
- 50. Habdija I, Lajtner J, Belinić I (1995) The contribution of gastropod biomass in macrobenthic communities of a karstic river. Int Rev Gesamten Hydrobiol Hydrogr 80: 103–110. [Google Scholar]
- 51. Brown CD, Holmes C, Williams R, Beulke S, van Beinum W, et al. (2007) How does crop type influence risk from pesticides to the aquatic environment? Environ Toxicol Chem 26: 1818–1826. [DOI] [PubMed] [Google Scholar]
- 52. De Zwart D (2005) Ecological effects of pesticide use in The Netherlands: Modeled and observed effects in the field ditch. Integr Environ Assess Manag 1: 123–134. [DOI] [PubMed] [Google Scholar]
- 53. Coors A, Vanoverbeke J, De Bie T, De Meester L (2009) Land use, genetic diversity and toxicant tolerance in natural populations of Daphnia magna . Aquat Toxicol 95: 71–79. [DOI] [PubMed] [Google Scholar]
- 54. Cothran RD, Brown JM, Relyea RA (2013) Proximity to agriculture is correlated with pesticide tolerance: evidence for the evolution of amphibian resistance to modern pesticides. Evol Appl 6: 832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Declerck S, De Bie T, Ercken D, Hampel H, Schrijvers S, et al. (2006) Ecological characteristics of small farmland ponds: associations with land use practices at multiple spatial scales. Biol Conserv 131: 523–532. [Google Scholar]
- 56. Knott E, Puurtinen M, Kaitala V (2003) Primers for nine microsatellites loci in the hermaphroditic snail Lymnaea stagnalis . Mol Ecol Notes 3: 333–335. [Google Scholar]
- 57.Kopp KC, Wolf K (2007) Lymnaea stagnalis microsatellite sequences. Genbank Access EF208747–EF208748.
- 58.Besnard A-L, Bouétard A, Azam D, Coutellec M-A (2013) Isolation and characterization of three new multiplex sets of microsatellite markers in the hermaphroditic freshwater snail Lymnaea stagnalis (Mollusca, Gastropoda, Heterobranchia, Panpulmonata, Hygrophila) using 454-pyrosequencing technology. Mol Ecol Resour 13. doi: http://tomato.bio.trinity.edu/manuscripts/13-1/mer-12-0292.pdf.
- 59.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5171, Université de Montpellier II, Montpellier (France).
- 60. Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 61.Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3).
- 62. Raymond M, Rousset F (1995) GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- 63. David P, Pujol B, Viard F, Castella V, Goudet J (2007) Reliable selfing rate estimates from imperfect population genetic data. Mol Ecol 16: 2474–2487. [DOI] [PubMed] [Google Scholar]
- 64. Jones OR, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10: 551–555. [DOI] [PubMed] [Google Scholar]
- 65. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 67. Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361. [Google Scholar]
- 68. Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138. [Google Scholar]
- 69. Coutellec M-A, Caquet T (2011) Heterosis and inbreeding depression in bottlenecked populations: a test in the hermaphroditic freshwater snail Lymnaea stagnalis . J Evol Biol 24: 2248–2257. [DOI] [PubMed] [Google Scholar]
- 70.Pinheiro JC, Bates DM (2002) Mixed Effects Models in S and S-Plus. New York: Springer Verlag New York, LLC.
- 71. Bonnin I, Prosperi M, Olivieri I (1996) Genetic markers and quantitative genetic variation in Medicago truncatula (Leguminosa): a comparative analysis of population structure. Genetics 143: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goudet J, Büchi L (2006) The effects of dominance, regular inbreeding and sampling design on QST, an estimator of population differentiation for quantitative traits. Genetics 172: 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates.
- 74. Whitlock MC, Guillaume F (2009) Testing for spatially divergent selection: comparing QST to FST . Genetics 183: 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kopp K, Wolff K, Jokela J (2012) Natural range expansion and human-assisted introduction leave different genetic signatures in a hermaphroditic freshwater snail. Evol Ecol 26: 483–498. [Google Scholar]
- 76. Volis S, Yakubov B, Shulgina I, Ward D, Mendlinger S (2005) Distinguishing adaptive from nonadaptive genetic differentiation: comparison of QST and FST at two spatial scales. Heredity 95: 466–475. [DOI] [PubMed] [Google Scholar]
- 77. Chapuis E, Martin G, Goudet J (2008) Effects of selection and drift on G matrix evolution in a heterogeneous environment: a multivariate QST-FST test with the freshwater snail Galba truncatula . Genetics 180: 2151–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Escobar JS, Nicot A, David P (2008) The different sources of variation in inbreeding depression, heterosis and outbreeding depression in a metapopulation of Physa acuta . Genetics 180: 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Johansson M, Primmer CR, Merilä J (2007) Does habitat fragmentation reduce fitness and adaptability? A case study of the common frog Rana temporaria . Mol Ecol 16: 2693–2700. [DOI] [PubMed] [Google Scholar]
- 80. Bridges CM, Semlitsch RD (2000) Variation in pesticide tolerance of tadpoles among and within species of Ranidae and patterns of amphibian decline. Conserv Biol 14: 1490–1499. [Google Scholar]
- 81. Koene JM, Ter Maat A (2004) Energy budgets in the simultaneously hermaphroditic pond snail, Lymnaea stagnalis: A trade-off between growth and reproduction during development. Belg J Zool 134: 41–45. [Google Scholar]
- 82. Mousseau TA, Roff DA (1987) Natural selection and the heritability of fitness components. Heredity 59: 181–197. [DOI] [PubMed] [Google Scholar]
- 83.Coutellec M-A, Caquet T, Lagadic L (2008) Lymnaea stagnalis: the effects of experimental demographic reduction on population dynamics. in: Akçakaya HR, Stark JD, Bridges TS, editors. Demographic Toxicity: Methods in Ecological Risk Assessment. Oxford, UK. pp.152–167. [Google Scholar]
- 84. Goudet J, Martin G (2007) Under neutrality, QST<FST when there is dominance in an island model. Genetics 176: 1371–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. López-Fanjul C, Fernandez A, Toro MA (2003) The effect of neutral nonadditive gene action on the quantitative index of population divergence. Genetics 164: 1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lamy J-B, Plomion C, Kremer A, Delzon S (2012) QST < FST as a signature of canalization. Mol Ecol 21: 5646–5655. [DOI] [PubMed] [Google Scholar]
- 87. Le Corre V, Kremer A (2012) The genetic differentiation at quantitative trait loci under local adaptation. Mol Ecol 21: 1548–1566. [DOI] [PubMed] [Google Scholar]
- 88. Edelaar M, Björklund M (2011) If FST does not measure neutral genetic differentiation, then comparing it with QST is misleading. Or is it? Mol Ecol 20: 1805–1812. [DOI] [PubMed] [Google Scholar]
- 89. Guillaume F (2011) Migration-induced phenotypic divergence: the migration-selection balance of correlated traits. Evolution 65: 1723–1738. [DOI] [PubMed] [Google Scholar]
- 90. Wolf JB (2000) Gene interactions from maternal effects. Evolution 54: 1882–1898. [DOI] [PubMed] [Google Scholar]
- 91. Hill WG, Goddard ME, Visscher PM (2008) Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pujol B, Wilson AJ, Ross RIC, Pannell JR (2008) Are QST-FST comparisons for natural populations meaningful? Mol Ecol 17: 4782–4785. [DOI] [PubMed] [Google Scholar]
- 93. Puurtinen M, Knott KE, Suonpää S, Ooik T van, Kaitala V (2004) Genetic variability and drift load in populations of an aquatic snail. Evolution 58: 749–756. [DOI] [PubMed] [Google Scholar]
- 94. Coutellec M-A, Lagadic L (2006) Effects of self-fertilization, environmental stress and exposure to xenobiotics on fitness-related traits of the freshwater snail Lymnaea stagnalis . Ecotoxicology 15: 199–213. [DOI] [PubMed] [Google Scholar]
- 95. Puurtinen M, Knott KE, Suonpää S, Nissinen K, Kaitala V (2007) Predominance of outcrossing in Lymnaea stagnalis despite low apparent fitness costs of self-fertilization. J Evol Biol 20: 901–912. [DOI] [PubMed] [Google Scholar]
- 96. Koene JM, Montagne-Wajer K, Roelofs D, Ter Maat A (2009) The fate of received sperm in the reproductive tract of a hermaphroditic snail and its implications for fertilisation. Evol Ecol 23: 533–543. [Google Scholar]
- 97. Escobar JS, Auld JR, Correa AC, Alonso JM, Bony YK, et al. (2011) Patterns of mating-system evolution in hermaphroditic animals: correlations among selfing rate, inbreeding depression, and the timing of reproduction. Evolution 65: 1233–1253. [DOI] [PubMed] [Google Scholar]
- 98. Coutellec M-A, Besnard A-L, Caquet T (2013) Population genetics of Lymnaea stagnalis experimentally exposed to cocktails of pesticides. Ecotoxicology 22: 879–888. [DOI] [PubMed] [Google Scholar]
- 99. Palo JU, O'Hara RB, Laugen AT, Laurila A, Primmer CR, et al. (2003) Latitudinal divergence of common frog (Rana temporaria) life history traits by natural selection: evidence from a comparison of molecular and quantitative genetic data. Mol Ecol 12: 1963–1978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L. stagnalis individual growth (G1 snails) according to population habitat. Gompertz growth curves based on 12 dates of measurement, performed on 492 individuals.
(TIFF)
L. stagnalis hatching rate as a function of anthropogenic pressure (OAP, n = 1537 clutches).
(TIFF)
Evolution of L. stagnalis G1 rearing conditions as a function of age.
(DOCX)
Summary of statistical analyses performed on L. stagnalis life history traits.
(DOCX)
Population genetic parameters calculated per locus and population on 14 L. stagnalis population samples (see materials and methods for explanation of population codes).
(DOCX)
Population genetic differentiation estimated among 14 L. stagnalis populations, on the basis of 12 SSR loci.
(DOCX)
Life-history trait/parameter mean-values (SE) estimated in the laboratory-born G1 from 14 L. stagnalis populations used to test patterns of adaptive divergence (Q ST-F ST analysis).
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Raw data are available from the Dryad database using the DOI doi: 10.5061/dryad.g4d30.