Abstract
Poly(ethylene glycol) (PEG) hydrogels represent a versatile material scaffold for culturing cells in two or three dimensions with the advantages of limited protein fouling and cytocompatible polymerization to enable cell encapsulation. By using light-based chemistries for gelation and for incorporating biomolecules into the network, dynamic niches can be created that facilitate the study of how cells respond to user-dictated or cell-dictated changes in environmental signals. Specifically, we demonstrate integration of a photo-cleavable molecule into network cross-links and into pendant functional groups to construct gels with biophysical and biochemical properties that are spatiotemporally tunable with light. Complementary to this approach, an enzymatically cleavable peptide sequence can be introduced within hydrogel networks, in this case through photoinitiated addition reactions between thiol-containing biomacromolecules and ene-containing synthetic polymers, to enable cellular remodeling of their surrounding hydrogel microenvironment. With such tunable material platforms, researchers can employ a systematic approach for 3D cell culture experiments, spatially and temporally modulating physical properties (e.g., stiffness) as well as biological signals (e.g., adhesive ligands) to study cell behavior in response to environmental stimuli. Collectively, these material systems suggest routes for new experimentation to study and manipulate cellular functions in four dimensions.
Introduction
Hydrogels represent a robust material system for answering fundamental biological questions relating to three-dimensional cell culture and have been especially effective in investigating the question: How do cells receive and exchange information with their extracellular environment? In living tissues, cells are surrounded by an extracellular matrix (ECM), a network of various protein fibers (e.g., collagen and fibrin) interlaced with glycosaminoglycan chains that provide support and signaling essential for proper development and maintenance of the tissue. The native ECM is actively involved in providing cues that influence cellular processes; for example, adhesion proteins bind to cell-surface receptors that prevent cell death,1 facilitate attachment,2 and influence motility3 in a manner that depends on their concentration and composition (Figure 1).4 In addition, the ECM serves as a reservoir for important growth factors, often sequestering them in both latent and active forms.5 Rapid proteolytic release and activation of these factors enables localized signaling that is important in promoting wound healing, dictating development, or even furthering cancer progression.5–7 From a general perspective, the ECM is enzymatically degradable to allow for local cellular remodeling.8 These proteinaceous matrices have a high water content and resist mechanical stresses, both of which are properties thought to influence chemical signaling between and within cells through mechanotransduction and diffusion of paracrine, autocrine, and hormone signaling molecules.9
Figure 1.
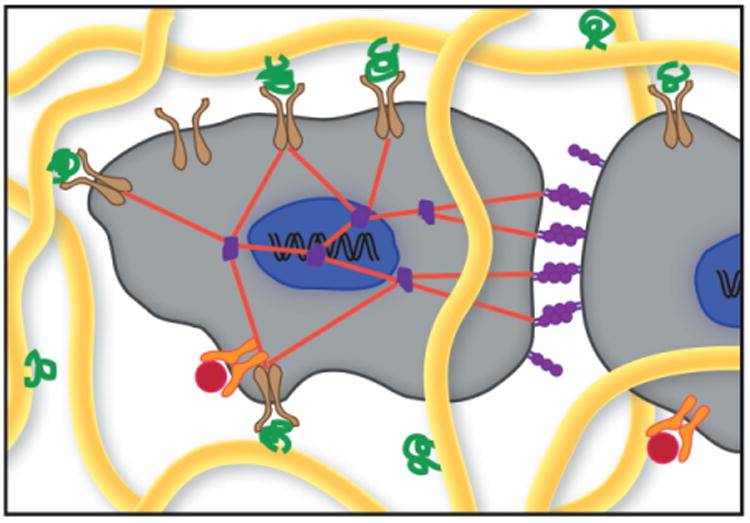
Schematic of cells within a native extracellular matrix (ECM). Cells (gray) bind specific ECM proteins (green) with cell-surface receptors, such as integrins (brown), to form adhesions important in cell viability, migration, and mechanotransduction. Cell-cell junctions (purple) in addition to cell-ECM adhesions allow the cell to feel mechanical forces in its environment through cytoskeletal stress fibers (red). The ECM is also a reservoir for important soluble cytokines and chemokines (red), which bind to specific cell-surface receptors (orange). The structural fibers of the ECM (yellow) can be cleaved by proteinases secreted by the cells, allowing for localized matrix remodeling. Adapted from Reference 4.
More recent work has focused on de-convoluting the complex roles that mechanical and chemical signals play in dictating cellular development. Diverse tissues in the body have an ECM environment with different compositions and different stiffnesses. For example, the elasticity of various mammalian tissues ranges from about 10 Pa for soft tissues, such as the brain, up to hundreds of kPa for cartilage.10 The mechanical properties of the surrounding cellular environment have been shown to greatly influence cell fate. For example, matrix elasticity is known to affect cytoskeletal tension, which can translate to intracellular signals and has been shown to direct stem cell differentiation.11 In addition, tumor microenvironments are often described as much stiffer than healthy tissue, a fact supported by the familiar cancer screening technique of inspection for a hard mass within a compliant tissue. In a striking example of mechanical signaling, mammary epithelial cells cultured on substrates with an elastic modulus comparable to that of malignant breast tissue took on cancer-like traits, whereas soft substrates promoted normal tissue growth.12 Clearly, there is much still to be learned about mechanotransduction processes and how mechanical and chemical cues work in concert to regulate cell function and fate, and the design of tunable material culture systems will be important in advancing the field.
Animal models have provided an important approach to test some of these complex signals and can illuminate the effects of the introduction of small molecules or macromolecular signals in the context of the entire body. This approach can be particularly useful, for example, in the drug development process when potential damage to off-target organs needs to be discovered. For fundamental research, however, animal models have the obvious drawback of being complicated systems with countless signaling pathways, redundancies, and environmental influences that can confound experimental variables. Therefore, many studies in vitro have aimed to mimic aspects of the native ECM, creating more biologically relevant models, through the use of natural and synthetic hydrogels.13,14 At the most basic definition, hydrogels are three-dimensional networks of hydrophilic polymers that are rendered water swellable, but insoluble, because of chemical or physical interactions between the polymer chains. The high water content imparts tissue-like elastic properties and facilitates rapid transport of small molecules, as well as gradients in cell-secreted growth factors and matrix molecules.15
From the applied perspective, gels used for three-dimensional cell culture in vitro should allow for cell migration, matrix degradation, and distribution of evolving tissue. However, numerous challenges with engineering the properties of these hydrogels remain; state-of-the-art tissue-engineered cartilage is still weak and inferior compared to native cartilage, and regenerating complex tissues from multiple cell sources continues to be elusive. Beyond basic structural and mechanical function, it is often challenging to define a minimum functionality for the encapsulating matrix (i.e., decide what chemical signals to include at relevant doses) and to work with cells that must perform metabolic functions. As a result, the field has much to learn, starting with the basic understanding of how cells receive signals from their ECM and progressing toward strategies to dynamically alter this environment in a cellularly appropriate manner. Thus, many research groups are revisiting advances in material science and engineering to develop highly controlled cellular microenvironments to improve our understanding and apply this knowledge toward the manipulation and engineering of critical cellular functions.
Ideally, an in vitro scaffold should provide the user with the ability to modulate the presentation of biochemical and physical signals in space and time in order to elucidate the effect of each individual cue, as well as their synergies, on desired cell fates. The following sections illustrate some of our perspectives on critical advances and recent progress toward the development of cytocompatible chemistries and their applications in synthesizing cell-laden material matrices. In a simplified sense, this might be described as understanding biology in four dimensions, where experimenters control the cellular niche in three-dimensional space and the fourth dimension of time.
PEG hydrogels as ECM mimics for cell culture
From a fundamental perspective, synthetic materials provide a basic platform to engineer cell culture matrices with defined mechanical properties, transport properties, and even degradation rates. In such systems, poly(ethylene glycol) or PEG is a frequent choice as the base material for synthetic hydrogel matrices because of its low level of protein adsorption, which prevents nonspecific interactions between cells and the network, providing a “blank slate” that can be easily modified to present chemical and physical cues in a defined manner. For example, the photoinitiated solution polymerization of PEG-diacrylate (PEGDA) in the presence of cells creates a cell-laden network of polyacrylate kinetic chains cross-linked by elongated PEG chains (Figure 2a–b).19 Cytocompatible reaction conditions and monomer formulations have been identified for PEGDA hydrogels, which enable both 2D culture on preformed materials as well as 3D encapsulation and cell culture within a minimal ECM.16,17
Figure 2.
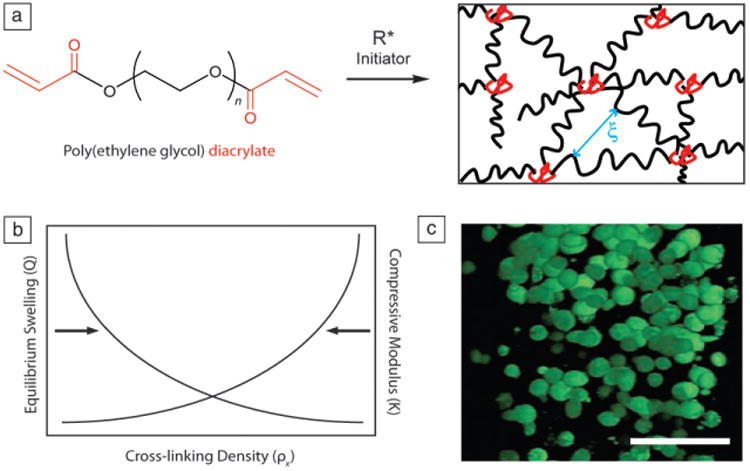
(a) Polyethylene glycol) diacrylate (PEGDA), of varying number of repeat units (n), can be chain polymerized in the presence of a radical initiator (R*) to form a network of polyacrylate chains (red) cross-linked by PEG (black). Mesh size (ξ) is a measure of the distance between cross-links and inversely related to cross-linking density. (b) Generic plot showing the relationship between equilibrium swelling and compressive modulus as a function of cross-linking density. As cross-linking density increases, compressive modulus of the hydrogel increases while swelling decreases. (c) Chondrocytes encapsulated within a PEGDA hydrogel, stained with LIVE/DEAD. Live cells fluoresce green. In this “blank slate” hydrogel, cells do not form attachments to the matrix and maintain a rounded morphology. Scale bar = 50 μm. Adapted from Reference 19.
While synthetic systems, such as cross-linked PEG, have numerous advantages, a major limitation of this material chemistry is the lack of biochemical signals that cells recognize. Thus, encapsulated cells generally adopt a rounded morphology (Figure 2c), and tissue engineering approaches rely on the ability of cells to rapidly and appropriately remodel their local matrix environment. Such minimal systems are permissive to cell function and can be suitable for the long-term culture of cells with low metabolic activities or a natural rounded morphology (e.g., cartilage cells, a.k.a. chondrocytes)18,19 or alternatively when one wishes to promote cell-cell interactions (e.g., islets,20 neurospheres21). However, a significant drawback of purely synthetic and nondegradable extracellular matrix analogs is the inability of cells to spread or migrate, which can severely limit the broad utility of such a system.
To enable cell motility through these materials, network cross-links must be cleaved. As a result, researchers have explored methods to integrate and control degradation in cell-laden matrices. These approaches range from pre-engineered hydrolytic degradation22–24 to cell-dictated degradation through secretion of enzymes25,26 to emerging paradigms in user-dictated degradation.27 Systems that depend on hydrolytic degradation often employ the cleavage of ester bonds, and the rate at which these materials degrade can be tuned to allow for cell proliferation and ECM deposition. The degradation kinetics of ester hydrolysis can be controlled by changing either the monomer chemistry or the number of cleavable links in the macromolecular backbone.28 Because of the high water content of hydrogels, the degradation kinetics and mass loss can be engineered to proceed in a predictable and homogeneous manner (i.e., bulk degradation).29 However, correctly timing the exchange of newly synthesized ECM with the degrading synthetic scaffold requires a precise understanding of both the material degradation kinetics and the rate of tissue deposition, time scales that are not always well-characterized. Therefore, many researchers have moved to enzymatically degradable hydrogels for 3D cell culture, where scaffold degradation is caused by cellular activity (i.e., the production of active proteinases). This localized degradation allows cells to modify and migrate through their immediate surroundings, which limits the need for a priori knowledge of the appropriate degradation kinetics. However, in these materials, the researcher is a passive observer of cell function and is often unaware of how the material is being altered locally.
While scaffolds with cell-dictated and pre-engineered degradation have certainly provided robust solutions to numerous tissue engineering applications, there has also been an evolution in thought toward the development of material platforms that respond to user-dictated inputs, which enable systematic and external modification of the cellular environment. Such systems allow the experimenter to test how cells react to these dynamic and controlled changes in matrix material properties. These user-controlled techniques are often combined with cell-dictated degradation mechanisms to both observe and manipulate cell function in a more native-like culture environment. Through appropriate chemical modifications, researchers can spatially and temporally vary both biophysical properties (stiffness, shape) and biochemical signals (adhesive ligands, growth factors) incorporated in cellular matrices, progressing the field toward a dynamic niche for culturing encapsulated cells.
User control over environmental signals presented to cells
To provide insight as to how cells respond to specific matricellular cues, researchers have recently pursued ways to actively manipulate the cellular microenvironment. One convenient and versatile method is to initiate chemical reactions with light. A hallmark advantage of photochemical reactions is that they provide the user with precise spatial and temporal control over bond formation and cleavage, allowing for directed network formation and degradation as well as pendant ligand tethering and release. In photoinitiated processes, reactions only occur where light is present. Light can be controlled in space by using standard techniques such as photomasks and focused lasers, and controlled in time by turning the switch on and off. One specific example of a photolabile (photo-cleavable) functionality employed to create cell culture matrices relies on a nitrobenzyl ether moiety (Figure 3a) that exhibits high photolytic efficiency.27
Figure 3.
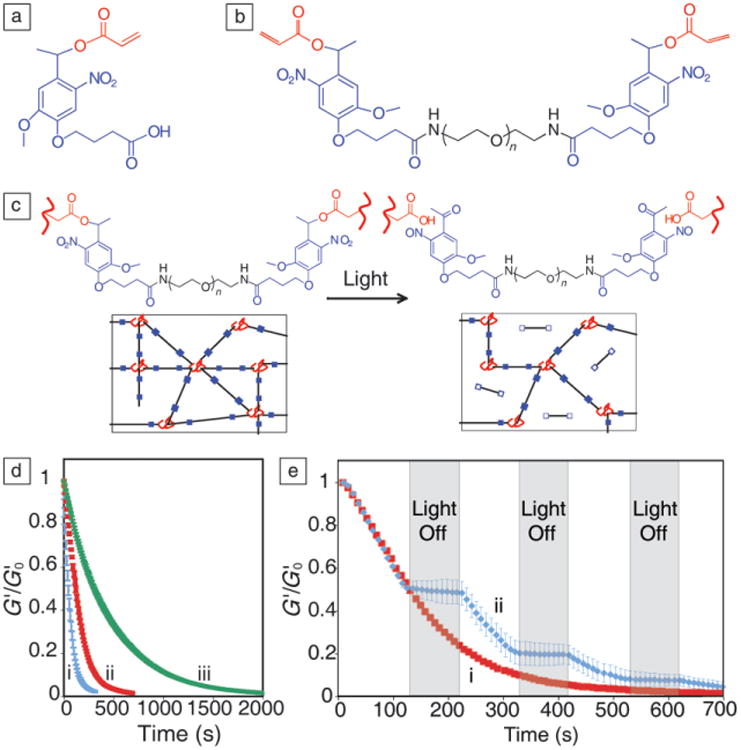
(a) Photolabile nitrobenzyl ether acrylic monomer. (b) Polyethylene glycol) di-photodegradable acrylate (PEGdiPDA) macromer, composed of PEG (black), photolabile moieties (blue), and acrylate end groups (red), polymerizes to form a gel. (c) Irradiation with specific wavelengths of light cleaves the nitrobenzyl ether moiety, degrading network cross-links. (d) Wavelength and intensity of the light controls degradation time, as determined by in situ rheometry to measure elastic modulus (G′) normalized to initial modulus (G′0), which is directly proportional to cross-linking density. (i) 365 nm at 20 mW/cm2, (ii) 365 nm at 10 mW/cm2, and (iii) 405 nm at 25 mW/cm2. (e) The degradation reaction only proceeds during exposure to light. (i) Continuous or (ii) periodic irradiation with 365 nm at 10 mW/cm2. Adapted from Reference 27. Note: n, number of repeat units.
Acrylated nitrobenzyl ether has been attached to both ends of PEG-bis-amine through a pendant carboxylic acid, creating a photocleavable cross-linking molecule, PEGdiPDA (PEG di-photodegradable acrylate, Figure 3b). This macromer is readily copolymerized with PEG monoacrylates or other monomers to create photodegradable hydrogels. Upon irradiation with single photon light between 365–420 nm or multiphoton light centered at 740 nm, the ester bond connecting the nitrobenzyl ether to the rest of the network is cleaved, forming a carboxylic acid and a ketone (Figure 3c). As the PEGdiPDA segments are detached from the network, the overall cross-linking density decreases until complete degradation is achieved. This process is fully controlled by the intensity and wavelength of light dictated by the photophysical properties of the linker, giving the user spatio-temporal control over gel modulus and network geometry. Figure 3d illustrates that irradiating with either a lower intensity of the same wavelength of light or with a different wavelength of light that corresponds to a lower molar absorptivity for nitrobenzyl ether will result in a longer degradation time.27,30 Network degradation only occurs during exposure to light, so the gel modulus can be softened to a specific value, remaining constant while the light source is shuttered, and then it can be decreased again at a later point in time (Figure 3e).
This controlled degradation technique has been used to study the differentiation of the resident cell population found in heart valves, valvular interstitial cells (VICs), into activated myofibroblasts, a process that has been shown to be correlated with substrate stiffness.31,32 VICs become activated from a quiescent fibroblast phenotype to an active myofibroblast phenotype in response to injury, and these activated myofibroblasts assist with the process of wound healing and valve homeostasis. Little is known about the fate of myofibroblasts after healing, but their persistence is often linked to the progression of fibrotic diseases. Since elasticity of the culture platform has been shown to influence VIC activation, matrices that allow fine-tuning of the modulus during culture are uniquely qualified to investigate thresholds and reversibility of activation in vitro.
Thus, the light-tunable properties of these PEG materials were harnessed to investigate questions related to the fibroblast-myofibroblast transition in VICs.33,34 First, it was asked whether there was a critical threshold in matrix elasticity above which VIC activation would be promoted and below which VICs would remain quiescent. By passing a mask over a photodegradable hydrogel during irradiation, a modulus gradient was achieved, in this case ranging from ∼7–32 kPa for Young's modulus, which is a physiologically relevant range for heart valves (Figure 4a).33 VICs were seeded on top of these gradient gels, and after three days, the number of myofibroblasts at each position on the gel was counted, as determined by the presence of α-smooth muscle actin (αSMA) organized into stress fibers (Figure 4b). Significantly more cells were activated on the stiff side of the gel than on the soft side, and the activation threshold was determined to be approximately 15 kPa.
Figure 4.
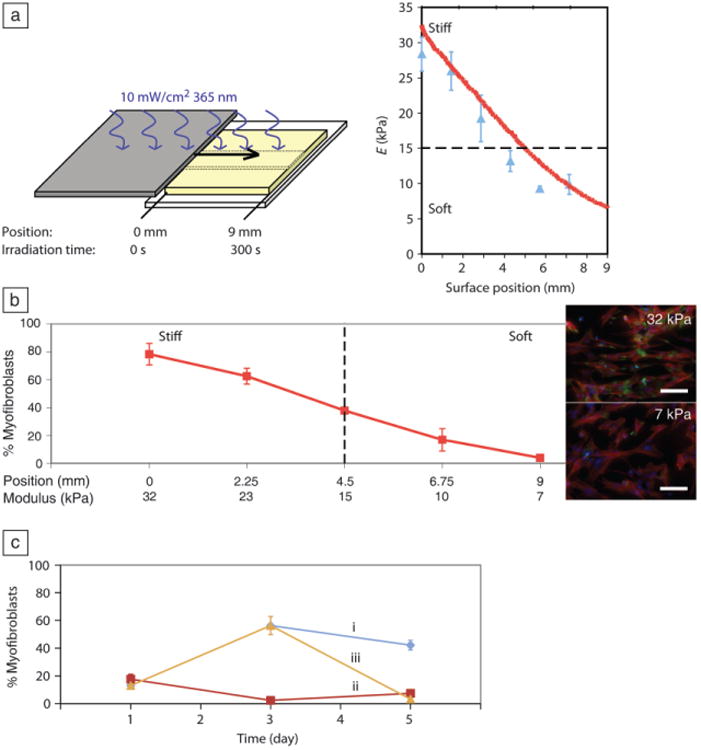
Valvular interstitial cells (VICs) activation on gradient and stiff-to-soft gels. (a) Modulus gradient across a gel created by moving a mask over the surface during irradiation (left). Modulus (E) measured by atomic force microscopy (blue triangles) and rheometric measurements (red line) as a function of position. (b) The stiff side of the gel promoted VIC activation, whereas cells grown on the soft side remained quiescent on day 3. Percentage of myofibroblasts determined by counting the fraction of cells with α-smooth muscle actin (αSMA) stress fibers, a classic marker for myofibroblasts. Inset: fluorescent images of VICs on stiff (32 kPa) and soft (7 kPa) sides of the gel on day 3, immunostained for αSMA (green), f-actin (red), and nuclei (blue). Scale bar = 100 μm. (c) By softening stiff gels during culture, VICs can be de-activated. (i) 32 kPa “stiff gels,” (ii) 7 kPa “soft gels,” and (iii) 32 kPa-7 kPa “stiff-to-soft gels.” Adapted from Reference 33.
While there are numerous materials and approaches to create hydrogels of varying elasticity, either through the preparation of discrete gels or the formation of gradient systems,35–37 photodegradable gels allow unique experiments to be performed where the gels can be softened in situ, and the influence of this softening on cell fate can be observed. Specifically, the cytocompatible nature of the photodegradation reaction was exploited to soften gels that were initially stiff at later time points during culture of VICs to determine if de-activation of the myofibroblast state is possible. VICs cultured on ∼32 kPa gels became activated over three days, then were exposed to 365 nm light to soften the underlying gel to ∼7 kPa and cultured for another two days (Figure 4c). This temporal change in substrate modulus caused de-activation of the myofibroblasts, without cell death,34 highlighting the importance of substrate stiffness in tissue engineering applications and informing strategies for potential therapies to treat fibrosis.
The biomechanics of material culture systems are not the only environmental signals that can be modulated with this user-controlled photodegradable functionality; one can also dynamically present biochemical ligands to cells. Typically, the amino acid sequence derived from fibronectin, arginine-glycine-aspartic acid-serine (RGDS), is pendantly incorporated into synthetic matrices to promote cell adhesion and survival.38–40 However, biochemical signal presentation is not static in vivo, and sometimes a ligand used for one purpose can inhibit other important functions. In general, there is a complex exchange of signals between cells and the ECM, and materials-based strategies can be exploited to better understand the influence of key signals on cellular fates.
In vivo, differentiating human mesenchymal stem cells (hMSCs) initially secrete high levels of fibronectin, which is later down regulated between days 7–10 followed by up-regulation of glycosaminoglycans, an early marker for chondrogenesis (i.e., differentiation toward cartilage-forming cells, chondrocytes).41,42 While RGDS is necessary for cell viability in these PEG hydrogels during the first week of culture, the question is whether its persistence is necessary or detrimental after a certain period of time. To provide an illustrative example of biochemical control of matricellular signaling, pendant peptides (Figure 5a, black) were tethered to the hydrogel network through the acrylated nitrobenzyl ether group (Figure 5a, blue and red), allowing for removal of the signal at a defined place and time. In this instance, RGDS was attached to a PEGDA hydrogel and photoreleased to study the influence of dynamic RGDS presentation on chondrogenesis of encapsulated hMSCs.27 When RGDS was photoreleased from the gels on day 10 and diffused out of the network, GAG production was found to increase four-fold over gels persistently presenting RGDS for the full three weeks of culture, indicating a significant increase in an important chondrogenic marker in the photoreleasable gels (Figure 5b). This example provides one simple demonstration where the dynamic nature of biochemical signal presentation can be mimicked to influence cellular functions and hints at the potential need for multiple aspects of control over the cell microenvironment for in vitro culture systems.
Figure 5.
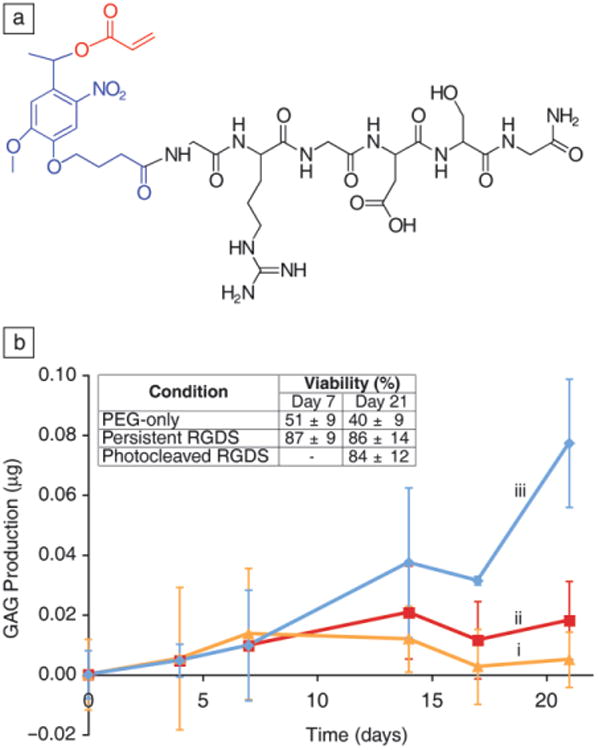
(a) Structure of the photoreleasable adhesion peptide monomer, arginine-glycine-aspartic acid-serine (RGDS) in black, photolabile moiety in blue, acrylate in red. (b) RGDS presentation maintains human mesenchymal stem cells viability within PEG-based gels (inset table). A chondrogenic marker (glycosaminoglycan, GAG, production) is increased four-fold by day 21 when the adhesive ligand RGD is photoreleased on day 10. (i) PEG-only gels. (ii) Persistently presented RGDS. (iii) Photolytic removal of RGDS on day 10. Adapted from Reference 27. Note: PEG, polyethylene glycol).
Cellular remodeling of their environment
While materials with user-controlled properties are beneficial for performing certain types of experiments, there are many other instances when one may wish to be a passive observer, watching cells in a more physiologically relevant environment. In this case, materials researchers have sought to develop methods to formulate hydrogel environments that promote specific cellular interactions, such as survival cues, degradable sequences, and growth factor binding functionalities. In the context of PEG hydrogels, one way in which a more advanced synthetic ECM can be realized is to apply the cytocompatible, photoinitiated, step-growth, thiol-ene polymerization.43,44 In this example, a multi-arm PEG-norbornene is reacted with dicysteine-peptide cross-linkers with or without monocysteine-peptide pendant groups (Figure 6a–c). This approach exploits certain aspects of the biochemical properties of peptides, as mimics of their full protein counterparts, and builds from the pioneering early work of Hubbell et al. to create peptide functionalized PEG hydrogels using a complementary type of step-growth polymerization based on Michael-type addition reactions.45–48
Figure 6.
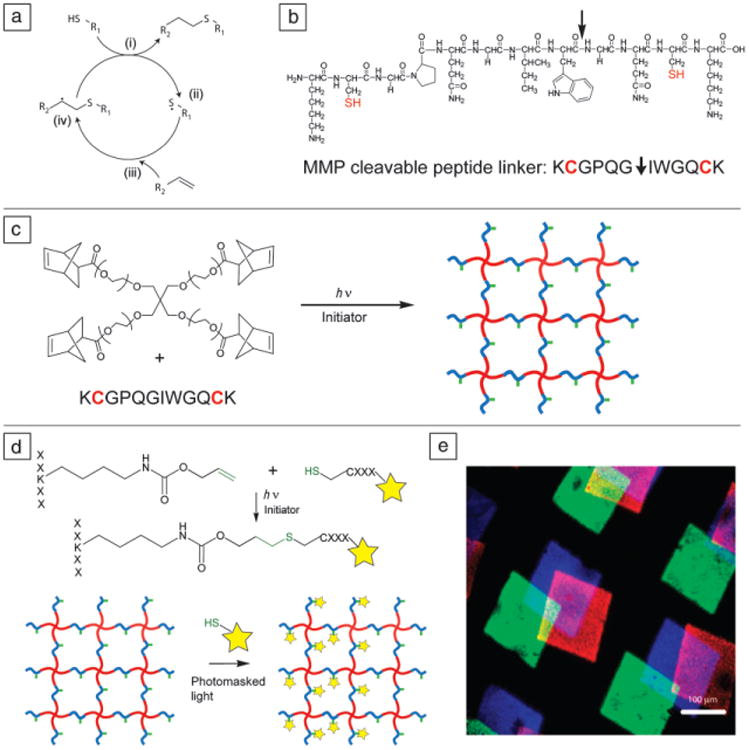
(a) Thiol-ene reaction cycle. An initiator abstracts a proton from a thiol (i), forming a thiyl radical (ii), which then reacts with a carbon-carbon double bond (iii). The resulting carbon radical (iv) abstracts a proton from another thiol, completing the thio-ether bond and regenerating a thiyl radical. (b) Di-cysteine polypeptide monomer, chemical structure (top) and amino acid abbreviation (bottom). This sequence, derived from collagen I, is cleavable by cell-secreted matrix metalloproteinases (MMPs) (cleavage site indicated by arrow) and is often used as a cellularly degradable cross-linker. (c) PEG-tetranorbornene and di-cysteine peptides react via the thiol-ene reaction cycle to form a step-growth network. Note: PEG (red), peptide cross-linker (blue), pendant peptide (green). (d) Peptide tethering with the thiol-ene reaction. Using photomasked light or focused laser light, cysteine-containing peptides can be covalently attached to pendant ene groups on the polymer network exclusively in user-defined regions. Fluorescent label on pendant peptide is indicated by a star. (e) Three different fluorescently labeled peptides (blue, green, and red) are sequentially attached at user-defined locations and times using photomasks and radical-initiated thiol-ene coupling reactions. Adapted from References 50, 54, and 55. Note: PEG, polyethylene glycol); h, Planck constant; ν, frequency of light; R1 and R2, side chains; X, any amino acid residue.
As with the PEGDA system, the photo-initiation technique provides both spatial and temporal control over network formation. However, the thiol-ene reaction has the extra advantages of rapid gelation and ideal network formation, maintaining uniform physical properties throughout the gel. Moreover, biochemical signals can easily be tethered to the network through the same thiol-ene reaction between excess PEG-norbornene or other ene functionalities pendant to the PEG backbone and cysteine-containing proteins or peptides either during the initial network formation for uniform signal distribution or in a post-gelation step allowing for spatial patterning within the gel (Figure 6d).43,49 For example, the commercially available protective group allyloxycarbonyl (alloc) used during peptide synthesis contains a terminal vinyl group that can be employed as a handle for covalently attaching cysteine-containing peptides to an existing peptide network cross-link post-gelation. Gels are swollen with the desired peptide and a photoinitiator, and then exposed to light. Since radicals are only formed in irradiated areas of the gel, these ligands can be patterned into the gel using a photomask. Typically, low concentrations of pendant peptides are used, leaving plenty of vinyl handles available for the addition of other peptides at later times (Figure 6e). In this way the permissive “blank slate” PEG gel becomes a promoting environment, presenting cells with biologically relevant cues from the native ECM and promoting specific cell functions through cell-matrix interactions.
These peptide-functionalized PEG hydrogels are readily tuned to permit cellular remodeling of their surroundings. Specifically, enzymes naturally produced by cultured cells can be exploited to create a flexible milieu in which network cross-links are locally cleaved. As part of many physiologic and pathologic processes, numerous cell types produce matrix metalloproteinases (MMPs), a family of enzymes that cleave proteins found in the ECM, causing degradation of the matrix and allowing for cell migration and remodeling of the matrix composition or geometry. An oligopeptide sequence derived from collagen I (Figure 6b) has been shown to be efficiently cleaved by several MMPs25 and has been incorporated into hydrogels to facilitate migration of many cell types during 3D cell culture.50–52 This designer peptide contains a cysteine on one or both ends to enable thiol-ene reactions with ene-functionalized PEG monomers and create enzymatically cleavable pendant groups or cross-links. A noteworthy advantage of this thiol-ene reaction is that any cysteine-containing peptide can be used and requires no post-synthetic modifications.
Cells encapsulated within these promoting scaffolds are able to form attachments to the hydrogel network, as integrins found on the cell surface will bind to many peptide sequences. These cell-material interactions allow the cell to adhere and spread (Figure 7a), which is very different from cells encapsulated in the PEGDA “blank slate” gels. In addition, enzymatically cleavable cross-links allow for local remodeling of the hydrogel matrix, which enables cell migration through the material. Individual cell paths can be tracked using real-time microscopy, and such quantitative data allow researchers to better understand how matricellular cues influence parameters, including cell speed and persistence, both of which are important to fields such as cancer biology and wound healing (Figure 7b). Peptide functionalized gels can provide a minimal mimic of the extracellular matrix, and thiol-ene chemistry is one easy avenue to synthesize materials decorated with these biologically functional molecules and represents a significant advance toward more native-like material platforms for culturing encapsulated cells.
Figure 7.
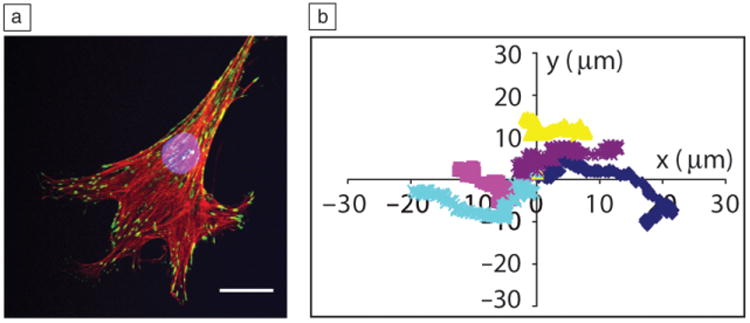
Cells attach, proliferate, and migrate in MMP-degradable thiol-ene hydrogels. (a) Human mesenchymal stem cells exhibit a spread morphology that depends on the degradability and integrin binding motifs in thiol-ene gels. Single cell immunostained for nucleus (blue), actin (red), and vinculin (green, focal adhesions). Scale bar = 25 μm. Image courtesy of Chelsea Kirschner. (b) Plot of five different cell migration paths of valvular interstitial cells encapsulated within the same MMP-degradable gel. When the enzymatically cleavable cross-link is used for gel formation, cells are able to degrade the surrounding isotropic material and migrate freely in a random walk. Cells were imaged and tracked in a real-time microscope. Adapted from Reference 51. Note: MMP, matrix metalloproteinase.
Sequential and orthogonal reactions create dynamic material niches for 3D cell culture
When combined into one system, the techniques and reactions described previously empower the user to design a highly tunable scaffold for 3D cell cultures that can evolve in real time through both cell- and user-dictated processes (Figure 8a–e). Namely, one can envision strategies where one reaction is used to form a cell-laden gel, while bio-orthogonal reactions are used to introduce or remove biochemical signals or to erode the matrix. Bio-orthogonal reactions, which are chemical reactions that do not interfere with native biochemical processes, are of growing interest in numerous applications. Here, we show one recent example where a strain-promoted azide-alkyne cycloaddition (SPAAC) between cyclooctyne-functionalized PEGs and azide-functionalized peptides is used to encapsulate live cells.53–56 Next, an orthogonal photoinitiated thiol-ene reaction between a cysteine-containing peptide or protein and a pendant ene functionality incorporated in the hydrogel backbone is used to pattern cues in a spatially defined manner.54–56 Third, enzymatically cleavable peptides are incorporated to enable cell migration or serve as reporters for local cell behavior.54 Lastly, the photolabile nitrobenzyl ether moiety described earlier is included to allow user-directed softening or complete degradation of the material upon irradiation.56
Figure 8.
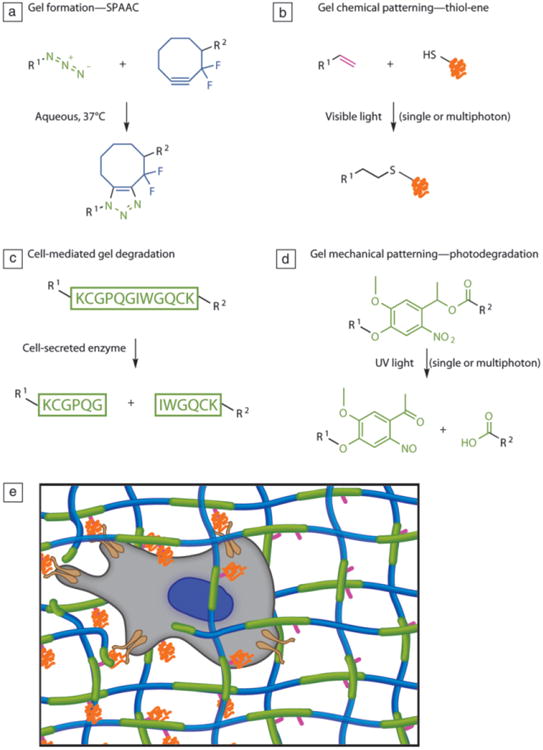
Sequential and orthogonal reactions. (a) Gels can be formed via a strain-promoted azide-alkyne cycloaddition (SPAAC) between cyclooctyne-functionalized polyethylene glycols) (PEGs, blue) and azide-functionalized peptides (green). (b) Peptide ligands can be patterned into the gel through the photoinitiated thiol-ene reaction between a cysteine-containing peptide (orange) and a free carbon-carbon double bond (pink) from the alloc protecting group located on the peptide cross-linker. (c) By choosing an enzymatically cleavable peptide sequence for the di-azide cross-linker (green), the gel can be locally degraded by cells. (d) User-dictated degradation can be accomplished by incorporating a photolabile moiety (green) within the cross-linker. (e) In this synthetic scaffold, the cell (gray) can attach to the PEG matrix (blue) through integrins (brown) binding to adhesive peptides (orange), which are covalently linked to the network through a pendant ene functionality (pink) during a post-gelation photoinitiated reaction, enabling spatial patterning of the peptide. Enzymatically degradable peptide sequences and/or photolabile groups form the cross-links (green), allowing for cellular and/or user remodeling of the microenvironment. Adapted from Reference 56. Note: R1 and R2, side chains.
To demonstrate the potential for this type of multifunctional and responsive material system, two different biochemical cues were sequentially patterned within a gel containing hMSCs to create a three-dimensional culture environment with spatially distinct biochemical regions (Figure 9a–b).56 Later, columns of user-defined cross-sections were eroded through the gel with two-photon microscopy to remove cells from selected niches (Figure 9c). The recovered cells were then plated, expanded, and further processed to provide more detailed molecular characterization, all processes that are difficult to do in a typical three-dimensional culture.56 Alternatively, using the same techniques, channels were degraded within SPAAC gels and used to direct fibroblasts and their migration down user-directed paths by patterning the adhesive peptide RGD only in the desired channels (Figure 9d).56 Such advanced patterning in three-dimensions may eventually address technical challenges in guiding formation of complex tissue structures involving multiple cell types.
Figure 9.
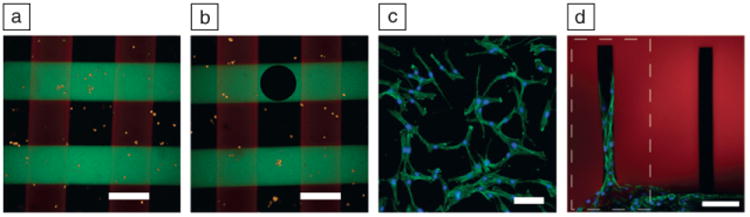
(a) Fluorescently labeled arginine-glycine-aspartic acid-serine-cysteine (RGDSC, green) was swollen into cell-laden gels along with a photoinitiator and exposed to UV light through a photolithography mask, causing a radical-mediated addition of a thiol across an olefin and creating stripes of the adhesive ligand in the gel. After swelling in fresh media to remove unreacted RGDSC, fluorescently labeled proline-histidine-serine-arginine-asparagine-cysteine (PHSRNC, red) was similarly patterned in perpendicular stripes, creating an array of four unique environments in a single cell-laden gel: blank, RGD-only, PHSRN-only, and RGD plus PHSRN. Cells encapsulated within the gel are labeled orange. Scale bar = 200 μm. (b) Circle-shaped columns were degraded to remove cells from a certain environmental niche. Scale bar = 200 μm. (c) Those cells were then plated on tissue culture polystyrene and grown to show continued viability. Immunostained for F-actin (green) and cell nuclei (blue). Scale bar = 50 μm. (d) Fibroblast outgrowth is directed by degrading channels in the strain-promoted azide-alkyne cycloaddition gel and tethering RGD only in the desired path by using focused laser light only in the channel enclosed by the dotted line. Scale bar = 100 μm. Adapted with permission from Reference 56.
Conclusions and challenges
By designing poly(ethylene glycol) hydrogel systems with the functionalities described, researchers can easily control and observe cell behavior with precise spatial and temporal resolution. Cytocompatible photochemistries allow the synthesis of versatile 3D cell culture material systems in which users can dictate material stiffness, matrix geometry, and biochemical signal presentation. These dynamic environments have been used to investigate complex biological processes, such as the de-activation of valvular interstitial cell myofibroblasts in response to decreases in microenvironmental elasticity. Employing multiple orthogonal chemistries within a single scaffold greatly improves our ability to mimic the biophysical and biochemical cues present in the native ECM environment and to study and direct cell function.
Despite these significant advances, engineering of 3D cell culture remains a complex problem with considerable challenges to overcome in the pursuit of complete understanding of cell behavior. For one, biological assays that are trivial when cells are plated in 2D on a surface become much more difficult when cells are entrapped within a matrix. Co-cultures of multiple cell types within hydrogels have yet to be fully explored, especially when spatial patterning of different cell populations is desired. At the individual cell level, little is known about what cells are doing to remodel their local environment. What extracellular matrix proteins are being laid down, and how is the local modulus changing as the cells go about altering their microenvironment? These and other issues will continue to be at the forefront of research in the tissue engineering field. And as material platforms evolve, greater advances in cell biology will follow.
Acknowledgments
K.L. would like to thank Ryan Lewis and Mark Tibbitt for helpful discussions while drafting the manuscript. The authors would like to acknowledge funding from the Howard Hughes Medical Institute and NSF (DMR 1006711).
Biographies

Katherine J.R. Lewis received her BA degree in chemistry from Whitman College (2008) and is currently working on her PhD degree in chemical and biological engineering at the University of Colorado at Boulder in the laboratory of Kristi Anseth. Lewis is developing a method to direct alveoli formation in PEG hydrogels to use as a model for studying lung development and disease. Lewis can be contacted by email at katherinejrlewis@gmail.com.

Kristi S. Anseth is presently a Howard Hughes Medical Institute Investigator and Distinguished Professor of Chemical and Biological Engineering at the University of Colorado at Boulder. She is an elected a member of the National Academy of Engineering and the Institute of Medicine. Anseth has also been active in MRS, serving on the Board of Directors, chair of the Planning Committee, co-chair of the Fall MRS Meeting in 2009, and elected MRS Fellow in 2009. Anseth can be contacted by email at Kristi.anseth@colorado.edu.
Footnotes
This article is based on the Mid-Career Researcher Award lecture, presented by Kristi S. Anseth on April 11, 2012, at the 2012 Materials Research Society Spring Meeting in San Francisco. Anseth is recognized for “exceptional achievement at the interface of materials and biology enabling new, functional biomaterials that answer fundamental questions in biology and yield advances in regenerative medicine, stem-cell differentiation, and cancer treatment.”
Contributor Information
Katherine J.R. Lewis, Email: katherinejrlewis@gmail.com, University of Colorado at Boulder.
Kristi S. Anseth, Email: Kristi.anseth@colorado.edu, University of Colorado at Boulder.
References
- 1.Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK, Brugge JS. Nat Cell Biol. 2003;5:733. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 2.Magnusson MK, Mosher DF. Arterioscler Thromb Vasc Biol. 1998;18:1363. doi: 10.1161/01.atv.18.9.1363. [DOI] [PubMed] [Google Scholar]
- 3.Parsons JT, Martin KH, Slack JK, Taylor JM, Weed SA. Oncogene. 2000;19:5606. doi: 10.1038/sj.onc.1203877. [DOI] [PubMed] [Google Scholar]
- 4.Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taipale J, Keski-Oja J. FASEB J. 1997;11:51. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
- 6.Kim SH, Turnbull J, Guimond S. J Endocrinol. 2011;209:139. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 7.Bacac M, Stamenkovic I. Annu Rev Pathol. 2008;3:221. doi: 10.1146/annurev.pathmechdis.3.121806.151523. [DOI] [PubMed] [Google Scholar]
- 8.Daley WP, Peters SB, Larsen M. J Cell Sci. 2008;121:255. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 9.Ingber D. Annu Rev Physiol. 1997;59:575. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- 10.Levental I, Georges PC, Janmey PA. Soft Matter. 2007;3:299. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 11.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King DA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Cancer Cell. 2005;8:241. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Rosso F, Marino G, Giordano A, Barbarisi M, Parmeggiani D, Barbarisi A. J Cell Physiol. 2005;203:465. doi: 10.1002/jcp.20270. [DOI] [PubMed] [Google Scholar]
- 14.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 15.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv Mater. 2006;18:1345. [Google Scholar]
- 16.Sawhney A, Pathak C, Hubbell J. Macromolecules. 1993;26:581. [Google Scholar]
- 17.Bryant SJ, Nuttelman CR, Anseth KS. J Biomater Sci, Polym Ed. 2000;11:439. doi: 10.1163/156856200743805. [DOI] [PubMed] [Google Scholar]
- 18.Bryant SJ, Anseth KS. J Biomed Mater Res. 2002;59:63. doi: 10.1002/jbm.1217. [DOI] [PubMed] [Google Scholar]
- 19.Cushing MC, Anseth KS. Science. 2007;316:1133. doi: 10.1126/science.1140171. [DOI] [PubMed] [Google Scholar]
- 20.Weber LM, He J, Bradley B, Haskins K, Anseth KS. Acta Biomater. 2006;2:1. doi: 10.1016/j.actbio.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Cordey M, Limacher M, Kobel S, Taylor V, Lutolf MP. Stem Cells. 2008;26:2586. doi: 10.1634/stemcells.2008-0498. [DOI] [PubMed] [Google Scholar]
- 22.Rice MA, Anseth KS. J Biomed Mater Res Part A. 2004;70:560. doi: 10.1002/jbm.a.30106. [DOI] [PubMed] [Google Scholar]
- 23.Nuttelman CR, Henry SM, Anseth KS. Biomaterials. 2002;23:3617. doi: 10.1016/s0142-9612(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 24.Bryant SJ, Anseth KS. J Biomed Mater Res Part A. 2003;64:70. doi: 10.1002/jbm.a.10319. [DOI] [PubMed] [Google Scholar]
- 25.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proc Natl Acad Sci USA. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salinas CN, Anseth KS. Biomaterials. 2008;29:2370. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant SJ, Anseth KS. In: Scaffolding in Tissue Engineering. Ma PX, Elisseeff J, editors. chap. 6. CRC Press; Boca Raton, FL: 2006. p. 71. [Google Scholar]
- 29.Metters A, Hubbell J. Biomacromolecules. 2005;6:290. doi: 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- 30.Kloxin AM, Tibbitt MW, Kasko AM, Fairbairn JA, Anseth KS. Adv Mater. 2010;22:61. doi: 10.1002/adma.200900917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benton JA, Kern HB, Anseth KS. J Heart Valve Dis. 2008;17:689. [PMC free article] [PubMed] [Google Scholar]
- 32.Yip CYY, Chen JH, Zhao R, Simmons CA. Arterioscler Thromb Vasc Biol. 2009;29:936. doi: 10.1161/ATVBAHA.108.182394. [DOI] [PubMed] [Google Scholar]
- 33.Kloxin AM, Benton JA, Anseth KS. Biomaterials. 2010;31:1. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Haeger SM, Kloxin AM, Leinwand LA, Anseth KS. PloS One. 2012;7:e39969. doi: 10.1371/journal.pone.0039969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Biophys J. 2004;86:617. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tse JR, Engler AJ. PloS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Adv Mater. 2004;16:2133. [Google Scholar]
- 38.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 39.Hubbell J. Nat Biotechnol. 1995;13:565. [Google Scholar]
- 40.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 41.Tavella S, Bellese G, Castagnola P, Martin I, Piccini D, Doliana R, Colombatti A, Cancedda R, Tacchetti C. J Cell Sci. 1997;110:2261. doi: 10.1242/jcs.110.18.2261. [DOI] [PubMed] [Google Scholar]
- 42.DeLise AM, Fischer L, Tuan RS. Osteoarth Cartil. 2000;8:309. doi: 10.1053/joca.1999.0306. [DOI] [PubMed] [Google Scholar]
- 43.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv Mater. 2009;21:5005. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoyle CE, Bowman CN. Angew Chem Int Ed. 2010;49:1540. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 45.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Bioconjugate Chem. 2001;12:1051. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 46.Elbert DL, Hubbell JA. Biomacromolecules. 2001;2:430. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 47.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 48.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Adv Mater. 2003;15:888. [Google Scholar]
- 49.Polizzotti BD, Fairbanks BD, Anseth KS. Biomacromolecules. 2008;9:1084. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 50.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. Biomaterials. 2011;32:3564. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benton JA, Fairbanks BD, Anseth KS. Biomaterials. 2009;30:6593. doi: 10.1016/j.biomaterials.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Biomaterials. 2001;22:3045. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 53.Codelli JA, Baskin JM, Agard NJ, Bertozzi CR. J Am Chem Soc. 2008;130:11486. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeForest CA, Sims EA, Anseth KS. Chem Mater. 2010;22:4783. doi: 10.1021/cm101391y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeForest CA, Anseth KS. Nat Chem. 2011;3:925. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]


