Abstract
MicroRNAs (miRNAs) are involved in the post-transcriptional regulation of gene expression during development. This study was performed to determine gestational age-dependent changes in miRNA expression in the chorioamniotic membranes and to assess the significance of miRNAs in human pregnancy and parturition. The expression profile of 455 miRNAs was compared between patients at term without labor (TNL: n=10), in labor (TL: n=10), and preterm labor (PTL: n=10) using microarrays. A total of 39 miRNAs were differentially expressed between term and preterm cases, of which 31 (79.5%) were down-regulated at term. Expression of 10 miRNAs, including miR-338, differentially expressed between PTL and TL groups was decreased at term. Computational analyses using miRBase Targets have identified PLA2G4B, a phospholipase implicated in parturition, as a putative target of miR-338. Inhibition of endogenous miR-338 with anti-miR-338 increased the mRNA and protein expression of PLA2G4B in decidual cells. Luciferase assay with reporter constructs confirmed that the suppression of PLA2G4B occurs through binding of miR-338 to the 3’UTR of PLA2G4B. Interestingly, the expression of Dicer, a key miRNA-processing enzyme, was markedly decreased at term, particularly with labor in the chorioamniotic membranes. Collectively, the novel findings reported herein strongly suggest that post-transcriptional regulation of genes by miRNAs, coupled with the changes of miRNA processing machinery in the chorioamniotic membranes, plays a role in pregnancy and parturition. Furthermore, the expression level of Dicer in the chorioamniotic membranes dichotomizes pathological preterm labor and physiological spontaneous labor at term.
Keywords: miR-338, group IVB phospholipase A2, Dicer, development, decidua, labor, preterm birth
INTRODUCTION
MicroRNAs (miRNAs) are a class of single-stranded, non-coding small RNA molecules that play a central role in the post-transcriptional regulation of gene expression [1]. Primary miRNAs, transcribed in the nucleus, are sequentially cleaved by the processing enzymes Drosha and Dicer to form mature ~22 nt miRNAs. Mature miRNAs within an RNA-induced silencing complex negatively modulate protein expression [2–4]. In animals, repression by miRNAs occurs through base pairing between miRNA and messenger RNA (mRNA) targets leading to mRNA degradation or inhibition of translation. [5,6]. More than 800 miRNAs are predicted to exist in humans, and bioinformatic analyses estimate that over 30% of protein coding genes are regulated by miRNAs [7,8]. Since their discovery in Caenorhabditis elegans, miRNAs have been extensively studied within the context of eukaryotic development [9]. In mice, deletion of miR-1–2 family members results in aberrant cardiac development due to defective conduction, cell cycle, and morphogenesis [10]. During zebrafish embryogenesis, miR-430 promotes deadenylation and clearance of maternal mRNA transcripts, thus playing a critical role in the transition from maternal to zygotic gene expression [11].
The biological significance of miRNAs has mainly been analyzed in the placenta. Dysregulated miRNA expression in the placentas of women with preeclampsia and small-for-gestational-age neonates has been reported [12,13]. Hypoxia also altered the expression of miR-93 and miR-424 in human trophoblasts, suggesting an association of these miRNAs with the pathophysiology of preeclampsia and intrauterine growth restriction [14]. The chorioamniotic membranes encasing the amniotic cavity are physically and functionally critical to pregnancy and parturition, and are involved in physiological and pathological responses [15,16]. It has been shown that the transcriptases of the chorioamniotic membranes before and after spontaneous labor at term are different [15]. However, the detailed expression patterns and any role for miRNAs in the human chorioamniotic membranes are yet to be determined. Previous screening of 157 miRNAs in the chorioamniotic membranes by real-time PCR revealed changes in the expression of a limited number of miRNAs with progression of gestation in preterm cases and acute inflammation (chorioamnionitis) [17]. The present study was conducted to examine miRNA expression using a more comprehensive screening method in the chorioamniotic membranes, and to assess the functional significance of miRNAs in human pregnancy and parturition.
MATERIALS AND METHODS
Tissue Collection
Tissues samples were retrieved from the Bank of Biological Materials of the Perinatology Research Branch, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). Chorioamniotic membranes were obtained from patients in the following groups: (1) normal pregnancy without labor at term (TNL; n=10); (2) normal pregnancy with spontaneous labor at term (TL; n=10); and (3) preterm labor (PTL; n=10). Preterm labor was defined by the presence of regular uterine contractions (at a frequency of at least 2 every 10 minutes) associated with cervical changes leading to preterm delivery before 37 completed weeks of gestation. Normal pregnancy was defined by the absence of any medical, surgical, or obstetrical complications. All of the snap-frozen chorioamniotic membranes used were free of histopathological lesions such as histological chorioamnionitis. All women provided written informed consent for the collection of clinical data and tissue samples under protocols approved by the Institutional Review Boards of the participating institutions.
MicroRNA Microarray Analysis
The expression profiles of 455 miRNAs were determined using miRCURY™ locked nucleic acid (LNA) microarrays (Exiqon, Denmark). Total RNA isolated using TRIzol® (Invitrogen, Carlsbad, CA, USA) was labeled using the miRCURY™ Hy3™/Hy5™ labeling kit and hybridized on the miRCURY™ LNA Array (v.8.1). All study samples were labeled with Hy3 and compared with a common total RNA reference pool (Ambion, Foster City, CA, USA) labeled with Hy5.
Cell Culture
Decidual cells from the chorioamniotic membranes were obtained by collagenase digestion from a TNL patient who delivered a male newborn. Cells were maintained in McCoy’s 5A media (Mediatech, Herndon, VA, USA) supplemented with 10% fetal bovine serum and antibiotics. Cells were incubated in a humidified atmosphere of 5% CO2 at 37°C. The cells had an intact female karyotype (46, XX).
Anti-miR-338 Transfection
Anti-miR-338 molecules (Ambion) were used to inhibit the function of endogenous miR-338 in decidual cells. Using siPORT NeoFX Transfection Agent (Ambion) and optiMEM (Invitrogen), 2 × 105 decidual cells were transfected with anti-miR-338 at the concentration of 100 nM. Scrambled-sequence anti-miR negative controls were added in separate wells at equimolar concentrations. The cells were harvested for 24 hours and 48 hours, respectively, after transfection for total RNA and protein isolation.
Generation of PLA2G4B 3’UTR Reporter Construct
The 3’UTR of PLA2G4B, whose sequence was obtained from ENSEMBL [18], was PCR-amplified and cloned into a SacI and HindIII site of the pMIR-REPORT™ miRNA Expression Reporter Vector (Ambion). This procedure put the firefly luciferase gene under the control of PLA2G4B 3’UTR. The upstream primer (5’-GAGCTCTGGAGCACTGGTGAACACCAC-3’) bearing a SacI site and the downstream primer (5’-AAGCTTCACAGCTACCTTTTATTCAAC-3’) bearing a HindIII site were used for PCR reactions. PCR conditions consisted of an initial denaturation at 94°C for 5 minutes; 40 cycles of 94°C for 1 minute, 50°C for 1 minute, 72°C for 1 minute; and a final extension at 72°C for 1 minute. The PCR product was purified from a gel using a QiaQuick gel extraction kit (Qiagen, Valencia, CA, USA), and cloned into the vector pCRII-TOPO (Invitrogen). The identity of inserts was confirmed by restriction digests and DNA sequencing using an ABI 3100 sequencer (Applied Biosystems, Foster City, CA, USA).
Reporter Gene Assay
For luciferase analysis of the PLA2G4B 3’UTR region activity, 2×105 decidual cells were transfected with 80 ng of the firefly luciferase plasmid containing the PLA2G4B 3’UTR or a mock plasmid containing no PLA2G4B sequence using Lipofectamine 2000. Alternatively, decidual cells were co-transfected with reporter plasmids and 10 nM anti-miR-338 as described above. Renilla luciferase reporter pSV40-RL (Promega, Madison, WI, USA) was used for the control of transfection efficiency. Luciferase activity was measured using Dual-Luciferase Reporter Assay kits according to the manufacturer’s instructions (Promega).
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Reverse transcription of total RNA was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and the SuperScript® III First-Strand Synthesis System (Invitrogen) for miRNA and mRNA analyses, respectively. All assays were performed using TaqMan assays (Applied Biosystems). RPLP0 was used for the normalization of PLA2G4B mRNA expression (Hs00192661_m1) and a custom-designed TaqMan assay for 5S ribosomal RNA (4332078) was used to normalize miR-338 (4373043), miR-449 (4373207), miR-136 (4373173), and miR-199a* (4378068) expression. Reactions were carried using a 7500 Fast Real-Time PCR System (Applied Biosystems).
Immunofluorescent Staining
Chorioamniotic membranes, villous placenta, and umbilical cord obtained from a normal term delivery were probed with rabbit polyclonal anti-Dicer (Atlas Antibodies, Sweden) and mouse monoclonal anti-cytokeratin (Dako, Denmark). Slides were incubated with Alexa Fluor 594-labeled donkey anti-rabbit and Alexa Fluor 488-labeled goat anti-mouse (Invitrogen). Nuclear staining was performed with DAPI (Invitrogen).
Immunoblot Analysis
Proteins were isolated using RIPA buffer, and 40 μg of protein were subjected to SDS-polyacrylamide gel electrophoresis and electro-transferred onto PVDF membranes. Transferred proteins were probed with mouse monoclonal anti-cytosolic phospholipase A2 (Abnova, Taiwan) or mouse monoclonal anti-Dicer (Genetex, Inc., San Antonio, TX, USA). Loading was assessed by detecting β-actin.
Statistical Analysis
Statistical significance of the data obtained was inferred using a two-tailed Student’s t-test and a paired t-test using SPSS version 12.0 (SPSS Inc., Chicago, IL, USA). MicroRNA microarray data were background-corrected to account for non-specific hybridization and local artefacts. The corrected signal intensities of the two channels were transformed into a ratio (sample/common reference). The ratios were log-transformed (base 2) and then normalized using a global loess normalization procedure [19]. A moderated t-test [20] was used to compute p values for miRNA differential expression between the study groups, based on the normalized log ratios. The resulting p values were corrected for multiple testing using the False Discovery Rate algorithm [21]. The R-statistical environment (www.r-project.org) was used for all microarray analyses using specialized packages from the bioconductor project (www.bioconductor.org).
RESULTS
microRNA Transcriptome of Term and Preterm Chorioamniotic Membranes
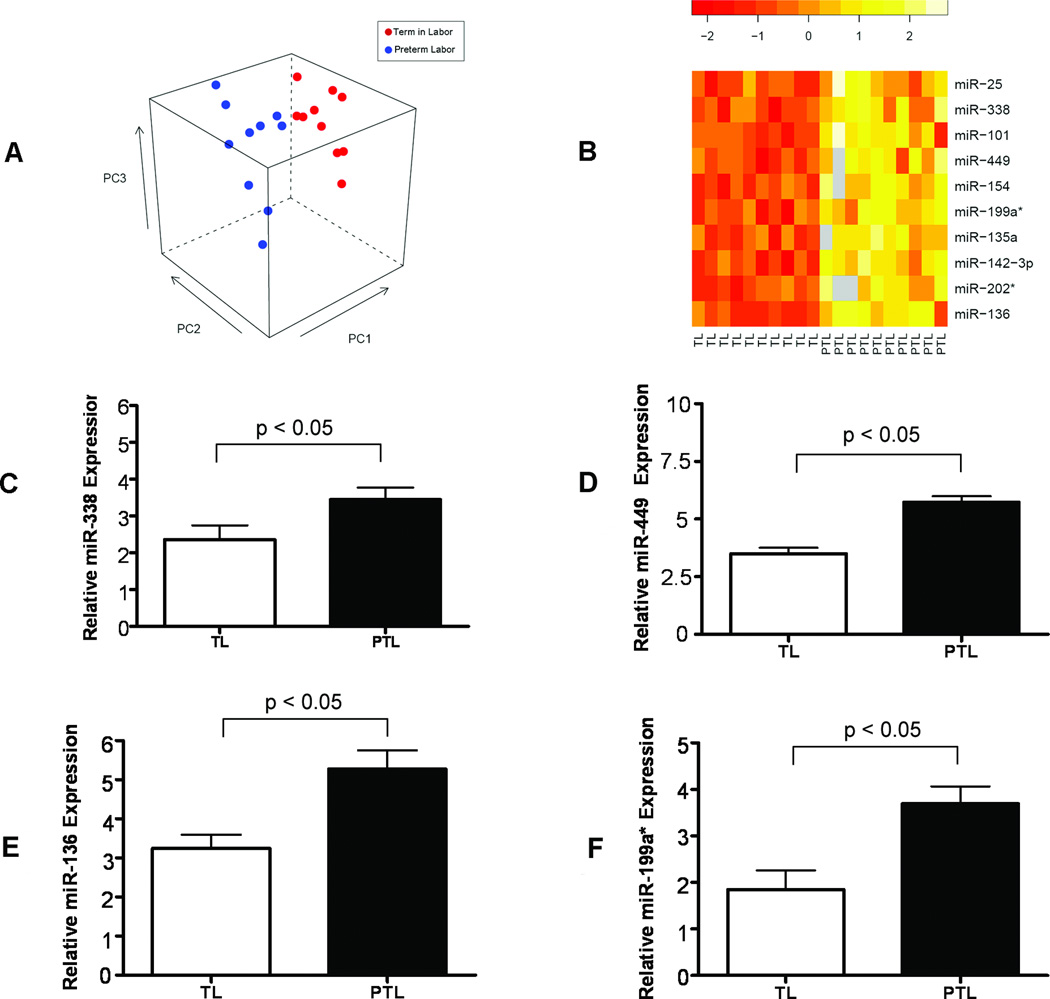
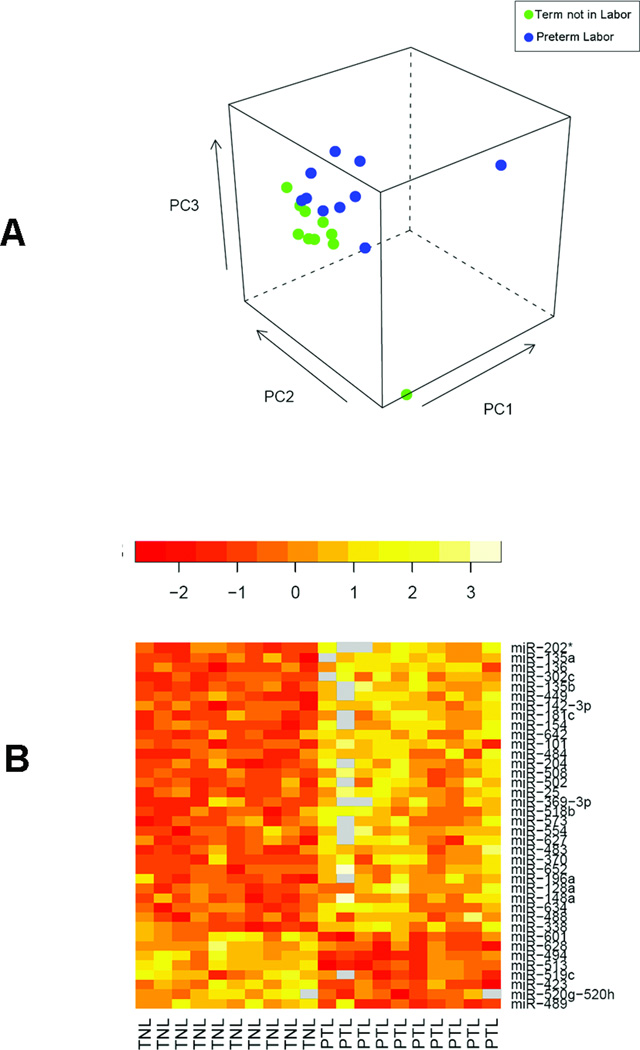
Demographic characteristics of the study population are summarized in Table 1. The expression profiles of 455 miRNAs were compared between the following groups: 1) PTL versus TL; 2) PTL versus TNL; and 3) TNL versus TL (ArrayExpress accession: E-TABM-469). An unsupervised visualization of the microarray data intended for the observation of within/between group similarity/dissimilarity, where each sample is shown in the space of the first three principal components computed from miRNA expression levels, is provided in Figures 1A and 2A. More details about this representation have been provided elsewhere [22]. A total of 39 miRNAs were differentially expressed between term and preterm groups. A comparison between PTL and TL groups revealed differential expression of 10 miRNAs, all of which were down-regulated at term (Figure 1B). A comparison between the PTL and TNL cases also showed decreased expression of 28 miRNAs and increased expression of 10 miRNAs at term (Figure 2B). The down-regulation of nine miRNAs at term (miR-25, 338, 101, 449, 154, 135a, 142-3p, 202*, and 136) was shared by the two groups compared, PTL vs. TL and PTL vs. TNL. Overall, the majority (79.5%) of all differentially expressed miRNAs had decreased expression at term. Decreased expression of miR-338, miR-449, miR-136, and miR-199a* at term was confirmed by qRT-PCR (p<0.05; Figures 1C, 1D, 1E, and 1F). The effect of labor was also assessed in the chorioamniotic membranes from TNL and TL cases. However, after adjustment for multiple comparisons, no significant differences were observed in the miRNA expression profiles between the two groups (data not shown).
Table I.
Patient demographics and clinical characteristics
| Characteristics | TNL (n=10) |
TL (n=10) |
PTL (n=10) |
|---|---|---|---|
| Maternal age, years | 27 (20–38) | 23 (19–32) | 21 (18–31) |
| Parity | 1 (0–5) | 1 (0–7) | 1 (0–5) |
| Gestational age at delivery, weeks | 39.3 (37.1–40.9) | 39.3 (37.1–41.9) | 33.3 (24.1–34.7) |
| Birth weight, g | 3495 (2950–4020) | 3373 (2695–3880) | 2275 (710–3175) |
Values expressed as median (range).
TNL = term no labor; TL = term in labor; PTL = preterm labor
Figure 1.
Comparison of miRNA expression levels in the chorioamniotic membranes between preterm labor and term in labor cases. (A) Principal component analysis shows a segregation of the two groups of samples in the space of the first three principal components computed from normalized log ratios (sample/reference). (B) A heat map showing the log ratios of the 10 differentially expressed miRNAs between TL (term in labor) and PTL (preterm labor) cases, all of which were down-regulated at term (p < 0.05). Grey boxes represent missing values. (C) Confirmation of miR-338 down-regulation at term by real-time qRT-PCR (fold-change = 2.12; p < 0.05). (D) Confirmation of miR-449 down-regulation at term by qRT-PCR. (fold-change = 4.86; p < 0.05). (E) Confirmation of miR-136 down-regulation at term by qRT-PCR. (fold-change = 4.11; p < 0.05). (F) Confirmation of miR-199a* down-regulation at term by qRT-PCR (fold-change = 3.59; p < 0.05). Data are presented as mean ± SEM.
Figure 2.
Microarray analysis of miRNA expression in the chorioamniotic membranes between preterm labor (PTL) and term no labor (TNL) cases. (A) An unsupervised analysis based on principal component analysis shows segregation of two a priori known groups of samples. (B) A heat map showing the log ratios of the 38 differentially expressed miRNAs between TNL and PTL cases. The chorioamniotic membranes at term without labor have decreased expression of 28 miRNAs and increased expression of 10 miRNAs. (p < 0.05). Grey boxes represent missing values.
miR-338 targets PLA2G4B mRNA
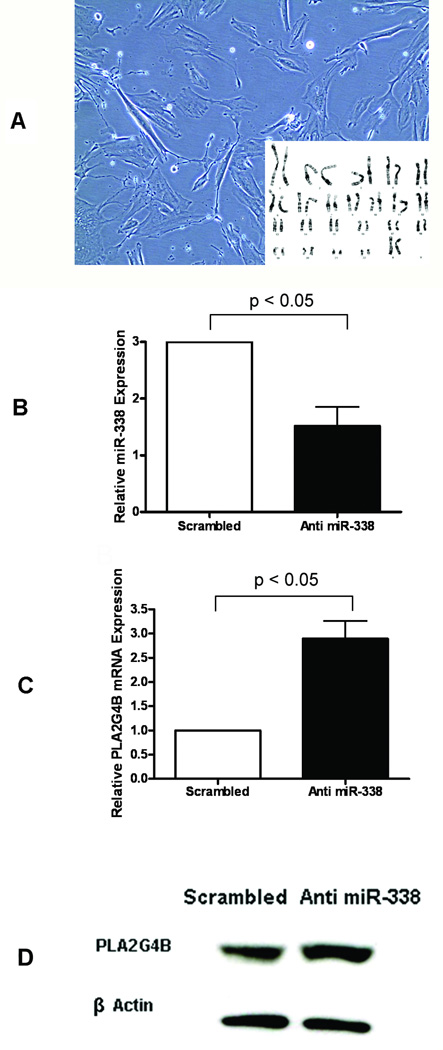
In our previous analysis, the expression of miR-338 was found to decrease with the progression of gestation in preterm cases, and Gene Ontology analysis of its potential targets revealed CRHR1 and PLA2G4B under the “biological process” category of parturition [17]. Decreased expression of miR-338 at term in both comparisons led us to explore whether miR-338 is involved in the translational inhibition of PLA2G4B, since CRHR1 mRNA was rarely detected in decidual cells (data not shown). Transfection of decidual cells with anti-miR-338 induced a 2.8-fold reduction in miR-338 expression (Figures 3A and 3B), while increasing PLA2G4B mRNA 3.7-fold (Figure 3C). Immunoblotting also demonstrated an increase in PLA2G4B protein expression in anti-miR-338 transfected cells (Figure 3D).
Figure 3.
Inhibition of PLA2G4B mRNA by miR-338 in decidual cells. (A) The cells were harvested by collagenase digestion of the decidua from a TNL patient who delivered a male baby. The cells have a normal female karyotype. (B) Transfection of the cells with anti-miR-338 induced a decrease in miR-338 expression (n=4; fold-change = 2.79, p < 0.05). (C) Anti-miR-338 transfection inversely increased PLA2G4B mRNA expression (n=5; fold-change = 3.72; p < 0.05). Data are presented as mean ± SEM and relative to the expression of a scrambled-sequence negative control. (D) PLA2G4B protein expression was also increased following anti-miR-338 transfection.
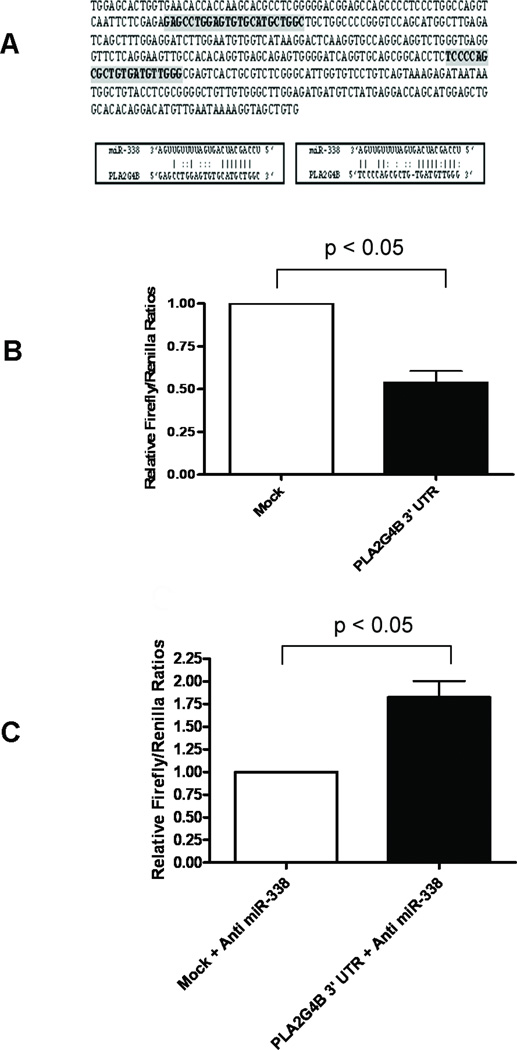
The post-transcriptional regulation of gene expression by miRNAs is the result of their complementary binding to the 3’UTR of mRNA targets [23]. Analysis of the PLA2G4B 3’UTR using miRBase [8] revealed two miR-338 binding sites (Figure 4A). To determine whether endogenous miR-338 suppresses PLA2G4B by targeting 3’UTR, decidual cells were transiently transfected with a reporter luciferase vector containing the full-length PLA2G4B 3’UTR sequence. The vector pMIR-REPORT Luciferase contains a firefly luciferase reporter gene under the control of a cytomegalovirus promoter/termination system. Cloning of the PLA2G4B 3’UTR sequence into pMIR-REPORT subjects the luciferase reporter to regulation that mimics the miRNA target. The cells transfected with the vector containing PLA2G4B 3’UTR showed a significantly reduced luciferase activity (46%) compared to mock transfected cells (Figure 4B). The cells co-transfected with anti-miR-338 and the luciferase vector with PLA2G4B 3’UTR showed a 1.82-fold increase in luciferase activity compared to cells co-transfected with anti-miR-338 and a mock reporter vector (Figure 4C).
Figure 4.
Suppression of PLA2G4B in decidual cells occurs through binding of miR-338 to PLA2G4B 3’UTR. (A) Target scanning of the PLA2G4B 3’UTR identified two putative miR-338 binding sites. The full-length PLA2G4B 3’UTR sequence was cloned in the downstream of the firefly luciferase gene in the reporter plasmid pMIR-REPORT™. (B) Transfection with the reporter plasmid containing the PLA2G4B 3’UTR decreased luciferase activity by 46% compared to a mock reporter plasmid. Renilla luciferase plasmid was used as a transfection control (n=3; p < 0.05). (C) Co-transfection of anti-miR-338 and a reporter plasmid with the PLA2G4B 3’UTR caused a 1.82 fold-increase in luciferase activity compared to cells co-transfected with anti-miR-338 and a mock reporter plasmid (n=3; p < 0.05).
Dicer Expression in Term and Preterm Chorioamniotic Membranes and Placenta
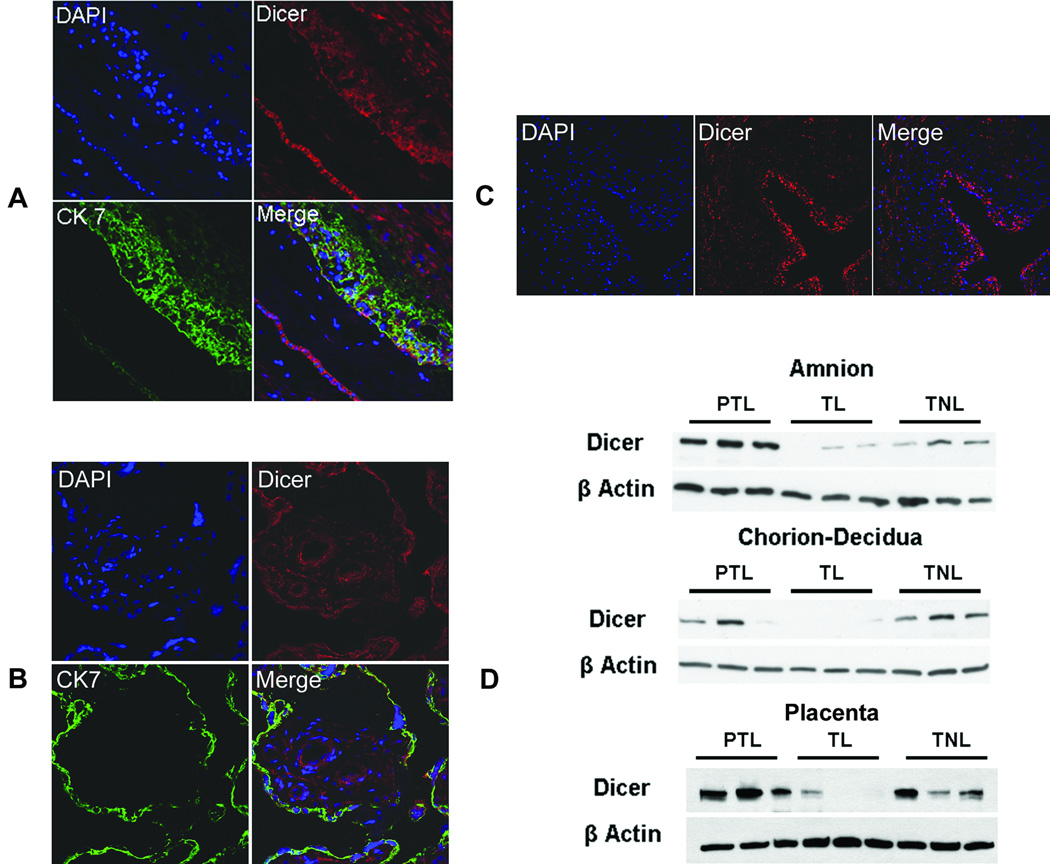
The overrepresentation of miRNAs down-regulated at term compared to preterm cases led us to evaluate the expression of the miRNA processing enzyme Dicer, which is involved in the generation of mature miRNAs in the chorioamniotic membranes and the placenta. Immunofluorescence staining showed the ubiquitous expression of Dicer in the cells comprising the chorioamniotic membranes such as amnion epithelial cells, chorionic trophoblasts, and decidual cells (Figure 5A). Dicer expression was also ubiquitous in the chorionic villi (Figure 5B) and umbilical cord (Figure 5C), with more distinct immunoreactivity in the vascular endothelial and smooth muscle cells in both structures. In the amnion, immunoblotting revealed a profound decrease in Dicer expression in TNL and TL cases compared to PTL cases. In the chorion-decidua, TL cases showed drastically reduced expression of Dicer compared to PTL and TNL cases (Figure 5D). Furthermore, immunoblotting of the villous placenta revealed a strikingly similar pattern of Dicer expression. Of special note is that the molecular weights of Dicer in the chorioamniotic membranes and the placenta were different. The chorioamniotic membranes predominantly expressed 219 kD (1,922 aa) full-length protein, while the placenta largely expressed 113 kD (997 aa) and 93 kD (820 aa) variants (Figure 5D). Therefore, preterm labor was distinct from spontaneous labor at term in terms of Dicer content in the chorioamniotic membranes and placenta.
Figure 5.
Dicer expression in term and preterm chorioamniotic membranes. (A) Immunofluorescent staining showing ubiquitous expression in the cells comprising the chorioamniotic membranes. (B, C) The expression is also ubiquitous in the chorionic villi and umbilical cord, with more distinct immunoreactivity in the vascular endothelial cells and smooth muscle cells in both structures. (D) In the amnion, immunoblotting demonstrates decreased Dicer expression in TNL and TL cases compared to PTL cases. In the chorion-decidua, Dicer expression was substantially reduced in TL cases compared to PTL and TNL cases. The expression pattern of Dicer in the placenta is strikingly similar to those in the amnion and chorion. Of special note is that the molecular weights of Dicer in the chorioamniotic membranes and the placenta are different. The chorioamniotic membranes predominantly expressed a 219 kD (1,922 aa) full-length protein, while the placenta largely expressed 113 kD (997 aa) and 93 kD (820 aa) variants.
DISCUSSION
The principal findings from extended profiling of miRNAs in the human chorioamniotic membranes are, firstly, subsets of miRNAs differentially expressed between term and preterm chorioamniotic membranes and the majority are down-regulated at term; secondly, one of these miRNAs, miR-338, is involved in the post-transcriptional regulation of group IVB phospholipase A2 (PLA2G4B) in decidual cells; and, thirdly, the expression of Dicer, a key component of the miRNA processing machinery, is markedly reduced in the chorioamniotic membranes at term after spontaneous labor, which is in stark contrast to preterm labor.
MicroRNA expression profiles have been widely used for the identification of miRNAs important in the development of organisms and for the clinicopathological characterization of human cancers [24,25]. A comprehensive miRNA signature of the human chorioamniotic membranes at term and preterm gestation revealed an over-representation of miRNAs down-regulated at term, including miR-338. Although computer algorithms are helpful in the identification of a large number of putative miRNA target genes, the experimental validation of authentic targets is a challenge, as less than 600 miRNA targets have been experimentally confirmed so far [26]. MicroRNA loss-of-function is an effective approach for the validation of predicted miRNA targets [27,28]. PLA2G4B is a member of the cytosolic phospholipase A2 protein family, whose function in the chorioamniotic membranes is crucial for prostaglandin biosynthesis during parturition [29–31]. In decidual cells, miR-338 was readily detected and its inhibition increased the expression of PLA2G4B mRNA, a predicted target of miR-338, and thereby PLA2G4B protein. Furthermore, luciferase reporter constructs confirmed that miR-338 represses PLA2G4B expression by binding to the 3’UTR. Collectively, these novel observations provide the first biological evidence that miR-338, a miRNA differentially expressed with advancing gestation and inflammation in the chorioamniotic membranes, regulates post-transcriptionally the expression of PLA2G4B, whose expression is closely linked to human parturition.
The tendency for down-regulation of differentially expressed miRNAs between preterm and term cases led us to examine potential changes in miRNA processing by Dicer in the chorioamniotic membranes. The expression level of Dicer in the amnion, which is composed purely of fetal cells, and in the chorion-decidua, which is composed of a variety of both maternal and fetal cells, strongly suggests that Dicer expression changes with the progression of gestation, especially with labor at term. In a mouse model, the conditional knockout of Dicer induced comparable reductions in miRNA expression [32]. Therefore, it is very likely that the changes in miRNA processing by Dicer are responsible for the differential expression of miRNAs between term and preterm cases. However, despite a notable difference in the expression of Dicer, significant differences in miRNA expression were not observed between TNL and TL cases, and this needs further clarification. It is noteworthy that preterm labor and labor at term are different in terms of Dicer expression in the chorioamniotic membranes and the placenta. The former is Dicer-rich while the latter is Dicer-deficient. Therefore, Dicer expression represents a novel aspect of the biochemical dichotomy between pathological preterm labor and physiological spontaneous labor at term. In contrast to preterm labor, spontaneous labor at term seems to be associated with the release of a negative post-transcriptional modulation of target genes by miRNAs. As there is no ethically acceptable way to obtain placentas from normal women in the second and third trimesters of pregnancy that could serve as controls for women with preterm labor, comparisons between these groups could not be performed. Mouse models have been widely used in reproductive biology and could have been an option, but there is a considerable difference in the anatomical constitution of the fetal membranes between humans and mice [33].
Our data show unique patterns of miRNA regulation in the human chorioamniotic membranes, although we could not evaluate all miRNAs identified in humans, as new miRNAs are continuously identified. The observations presented herein strongly support a role for miRNAs in human pregnancy and parturition.
Acknowledgements
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U. S. Department of Health and Human Services.
Footnotes
Conflict of Interest Statement:
The authors of this study do not have conflicts of interest to declare.
Supplemental Data:
Micro Array data available through ArrayExpress, accession: E-TABM-469.
Reference List
- 1.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 6.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.miRBase. Cambridge, UK: 2005. [12-16-2005]. Sanger Wellcome Trust. Ref Type: Data File. [Google Scholar]
- 9.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 12.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261–266. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Donker RB, Mouillet JF, Nelson DM, Sadovsky Y. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod. 2007;13:273–279. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- 15.Haddad R, Tromp G, Kuivaniemi H, Chaiworapongsa T, Kim YM, Mazor M, et al. Human spontaneous labor without histologic chorioamnionitis is characterized by an acute inflammation gene expression signature. Am J Obstet Gynecol. 2006;195:394.e1–394.e24. doi: 10.1016/j.ajog.2005.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YM, Romero R, Chaiworapongsa T, Kim GJ, Kim MR, Kuivaniemi H, et al. Toll-like receptor-2 and-4 in the chorioamniotic membranes in spontaneous labor at term and in preterm parturition that are associated with chorioamnionitis. Am J Obstet Gynecol. 2004;191:1346–1355. doi: 10.1016/j.ajog.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Montenegro D, Romero R, Pineles BL, Tarca AL, Kim YM, Draghici S, et al. Differential expression of microRNAs with progression of gestation and inflammation in the human chorioamniotic membranes. Am J Obstet Gynecol. 2007;197:289.e1–289.e6. doi: 10.1016/j.ajog.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth GK. Limma: linear models for microarray data. New York, NY: Springer; 2005. [Google Scholar]
- 21.Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 22.Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3:e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 25.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 29.Okazaki T, Sagawa N, Bleasdale JE, Okita JR, MacDonald PC, Johnston JM. Initiation of human parturition: XIII. Phospholipase C, phospholipase A2, and diacylglycerol lipase activities in fetal membranes and decidua vera tissues from early and late gestation. Biol Reprod. 1981;25:103–109. doi: 10.1095/biolreprod25.1.103. [DOI] [PubMed] [Google Scholar]
- 30.Skannal DG, Brockman DE, Eis AL, Xue S, Siddiqi TA, Myatt L. Changes in activity of cytosolic phospholipase A2 in human amnion at parturition. Am J Obstet Gynecol. 1997;177:179–184. doi: 10.1016/s0002-9378(97)70459-9. [DOI] [PubMed] [Google Scholar]
- 31.Terakawa K, Itoh H, Sagawa N, Yura S, Yoshida M, Korita D, et al. Site-specific augmentation of amnion cyclooxygenase-2 and decidua vera phospholipase-A2 expression in labor: possible contribution of mechanical stretch and interleukin-1 to amnion prostaglandin synthesis. J Soc Gynecol Investig. 2002;9:68–74. doi: 10.1016/s1071-5576(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 32.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross JC. Formation of the placenta and extraembryonic membranes. Ann N Y Acad Sci. 1998;857:23–32. doi: 10.1111/j.1749-6632.1998.tb10104.x. [DOI] [PubMed] [Google Scholar]