Abstract
The identification of effective polypeptide ligands for magnetic iron oxide nanoparticles (IONPs) could considerably accelerate the high-throughput analysis of IONP-based reagents for imaging and cell labeling. We developed a procedure for screening IONP ligands and applied it to compare candidate peptides that incorporated carboxylic acid side chains, catechols, and sequences derived from phage display selection. We found that only l-3,4-dihydroxyphenylalanine (DOPA)-containing peptides were sufficient to maintain particles in solution. We used a DOPA-containing sequence motif as the starting point for generation of a further library of over 30 peptides, each of which was complexed with IONPs and evaluated for colloidal stability and magnetic resonance imaging (MRI) contrast properties. Optimal properties were conferred by sequences within a narrow range of biophysical parameters, suggesting that these sequences could serve as generalizable anchors for formation of polypeptide–IONP complexes. Differences in the amino acid sequence affected T1- and T2-weighted MRI contrast without substantially altering particle size, indicating that the microstructure of peptide-based IONP coatings exerts a substantial influence and could be manipulated to tune properties of targeted or responsive contrast agents. A representative peptide–IONP complex displayed stability in biological buffer and induced persistent MRI contrast in mice, indicating suitability of these species for in vivo molecular imaging applications.
Magnetic iron oxide nanoparticles (IONPs) have become popular tools for biotechnology and medical imaging,1 and techniques for rapid molecular engineering of IONP species could be enormously valuable for design and optimization of magnetic reagents. In most applications, IONP cores are coated with a passivating layer of hydrophilic material, such as polyethylene glycol (PEG) or dextran, and then conjugated to functional moieties that promote target binding or detection of analytes.2 The complexity of multistep synthesis and conjugation chemistry is an impediment to production of large collections of IONPs. In addition, traditional passivation and functionalization of IONPs adds substantial bulk to IONPs, limiting steric access to the mineral cores where magnetic fields are highest; this in turn limits the effectiveness and manipulability of IONPs for molecular-scale applications.
In an effort to discover simple and versatile IONP modification strategies suitable for high-throughput analysis and identification of desirable IONP reagents, we explored the ability of short polypeptides to act as chemically tunable direct iron oxide ligands. Peptides that bind iron oxide cores could form an ideal basis for large-scale investigation of the determinants of IONP properties in applications such as magnetic resonance imaging (MRI) contrast manipulation. Effective stabilization of IONP cores by individual peptide sequences has not previously been demonstrated, but both functional groups and peptide sequences with iron oxide binding characteristics have been identified. We constructed a small initial set of 6–15 residue sequences using some of these moieties, including carboxylate3 and catechol groups,4,5 as well as peptide motifs derived from phage display screens for binding to magnetite6 (DSPHRHS) and hematite7 (LSTVQTISPSNH). Catechol-containing sequences were formulated with variations in net charge and with polyserine moieties to ensure hydrophilicity.
In order to screen the peptide library for IONP binding and MRI contrast properties, we developed a procedure for parallel analysis of candidate peptide–IONP complexes. Iron oxide cores were prepared for complexation by exchanging oleate-stabilized iron oxide nanocrystals into aqueous solutions of tetramethylammonium hydroxide (TMA) at basic pH (Figure 1a).8,9 Candidate peptides from the initial collection were added to the resulting TMA-associated cores and neutralized by dilution into phosphate-buffered saline (PBS). Peptides were assessed for their ability to stabilize IONPs in the resulting solutions by evaluating relative optical density at 450 nm (OD450) following formulation (Figure 1b). Lower stability results in precipitation and lower OD450 (Figure 1b images). Examination of results from the initial library showed that only an anionic l-3,4-dihydroxyphenylalanine (DOPA)-terminated peptide, SSSSSSDDZ, where Z = DOPA, provided significant enhancement of stability with respect to the negative control of no peptide addition (p = 0.02, n = 3). The stabilization afforded by SSSSSSDDZ was statistically indistinguishable from that provided by citrate, an established IONP ligand (p = 0.4, n = 3).10 Among peptides that did not appear to stabilize IONPs in PBS were the two phage display motifs, two polyglutamate-containing peptides, and additional sequences that lacked either acidic residues or a DOPA moiety. Some of these sequences, including polyacidic but not phage display peptides, were better able to stabilize IONPs under low salt conditions (Supporting Information), suggesting that poor performance of some of the candidate peptides in the absorbance assay of Figure 1 may result from a combination of low affinity for IONP cores and lack of electrostatic properties conducive to colloidal behavior. The assay conditions applied in our screening procedure are likely therefore to select simultaneously for effective peptide–IONP binding and complexes with favorable colloidal stability.
Figure 1.
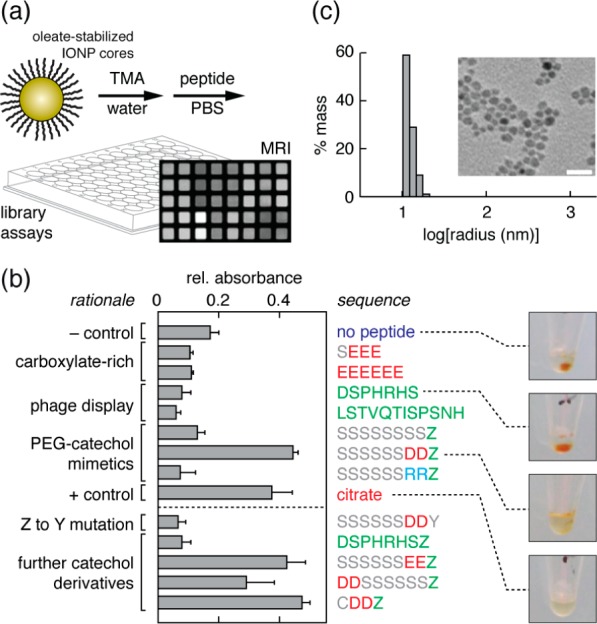
Identification of stable peptide–IONP complexes. (a) Schematic of peptide–IONP library production. Oleate-stabilized iron oxide cores are exchanged into TMA/water mixtures, library peptides (50 μM) are added to IONP aliquots (4.2 mM Fe), and suspensions are neutralized with PBS. Samples are arrayed into microtiter plates for imaging-based assays. (b) A small library of candidate IONP ligands was screened for ability to stabilize cores in PBS. Greater relative absorbance values indicate more effective stabilization. Design rationales for each peptide are noted on the left, and sequences reported to their right, with color coding to denote positively charged residues (cyan), negatively charged residues (red), iron oxide binding groups (green), and hydrophilic residues (gray). Error bars denote s.e.m. of three independent measurements. On the far right-hand side, images are given illustrating formation of pellets (dark spots) or lack thereof, following centrifugation of representative samples. (c) Dynamic light scattering and transmission electron micrographic data (inset) illustrating size parameters of the CDDZ–IONP complex. Scale bar = 20 nm.
To further probe molecular determinants of IONP stabilization by SSSSSSDDZ, a second set of peptides was constructed. OD450 results obtained using this library (Figure 1b and Supporting Information) indicated that replacement of the catechol by phenol (Z to Y substitution) or addition of DOPA to a phage display-selected sequence failed to stabilize colloidal IONPs. Separation of aspartate residues from DOPA, substitution of D by E residues, and shortening of the hydrophilic polyserine tract were tolerated, however. We found that a short, thiol-functionalized peptide, CDDZ, was ∼25% more effective than SSSSSSDDZ at stabilizing IONPs. Dynamic light scattering (DLS) analysis of IONP cores complexed with CDDZ (Figure 1c) indicated a hydrodynamic radius of 5.8 ± 0.7 nm, and transmission electron microscopy (TEM) of the material (inset) showed single, compact, electron-dense cores of 6.1 ± 0.2 nm diameter separated by gaps of 0.89 ± 0.03 nm; this result was also consistent with TEM data obtained in negative stain (Supporting Information). The zeta potential of CDDZ–IONP complexes was determined to be −20.9 mV (Supporting Information). These measurements are consistent with a nanoparticle structure in which individual cores from the oleate-stabilized starting material are coated and stabilized by a thin layer of anionic peptides. The results also suggested that the DDZ motif might be a generalizable anchor from which further libraries of polypeptide–IONP conjugates could be formed and screened for functional characteristics.
We used these findings as a basis for synthesis of a further collection of 39 peptide sequences, leading to a library of 55 peptides in total, and assessed these peptides for their ability yield effective contrast agents for MRI. This approach enabled us to examine the dependence of MRI contrast induction in parallel on a variety of parameters, such as hydrophilicity, charge, peptide length, and the presence of specific functional groups or sequence motifs. Twenty-eight out of the 55 peptide–IONP complexes formed showed OD450 values within 30% of the maximum value, indicating relative stability in PBS. These conjugates were examined by MRI and by DLS (Figure 2a and Supporting Information). MRI data obtained consisted of measurements of the apparent longitudinal and transverse relaxivity, r1 and r2, which reflect the ability of each IONP reagent to produce contrast in T1- and T2-weighted imaging, respectively. Mean values of r1, r2, and DLS radius across the library were 1.8 ± 0.2 (mM Fe)−1 s–1, 37 ± 6 (mM Fe)−1 s–1, and 25 ± 5 nm, respectively.
Figure 2.
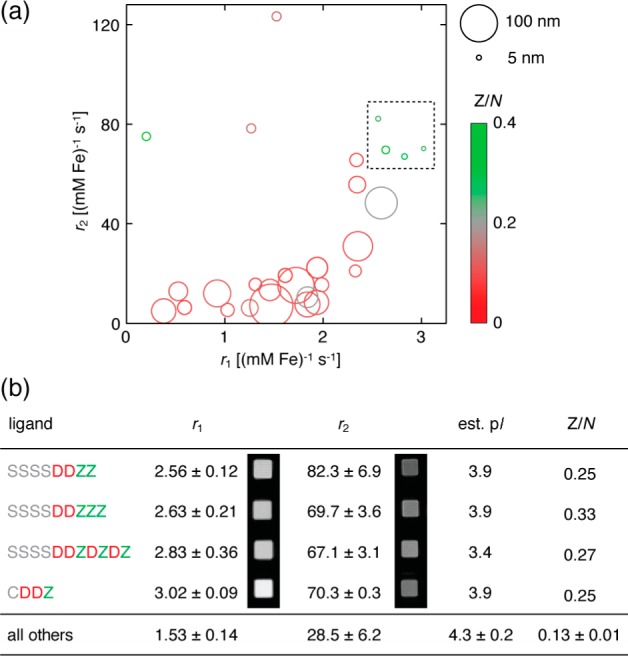
Analysis of peptide–IONP libraries. (a) Characterization of 28 peptide–IONP complexes that passed initial screening for stability from a library of 55 candidates. Each peptide–IONP complex is represented by an open circle centered on coordinates corresponding to its measured relaxivity values (r1 and r2) at 7 T. DLS radius measurements from each species are indicated by symbol sizes (key on right), and fractional contents of DOPA residues (Z/N) are indicated by the color scale. Individual measurements are reported by peptide sequence in Supporting Information. (b) Properties of optimized contrast agents selected for consistent high r2 values [dashed box in (a)]. Relaxivity values, estimated pI, and fractional DOPA content all differed significantly from the rest of the library. T1- and T2-weighted MRI scans illustrating contrast produced by these peptide–IONP complexes (500 μM Fe) are inset at left (TE/TR = 10/477) and right (TE/TR = 40/3125), respectively. Error margins reflect s.e.m. (n = 3).
The data of Figure 2a yield a number of notable results. First, dramatic variation of both r1 and r2, from a maximum r2/r1 ratio of 380 ± 150 to a minimum of 4.4 ± 1.4, is apparent among peptide–IONP complexes. This suggests that determinants of r1 and r2 are separable, perhaps due to differential effects of the peptide ligands on the inner vs outer sphere interactions of water molecules with IONP cores, which underlie the MRI contrast mechanism. Second, some variations in r2 are not explained by differences in DLS radius, despite experimental and theoretical accounts of nanoparticle relaxivity that depend heavily on particle size.11 In particular, the highest r2 value of 123 ± 11 (mM Fe)−1 s–1 is displayed by an IONP complex with sequence SSSCEEZ, which exhibits one of the smallest hydrodynamic radii. Third, the highest r1 values are displayed by IONP complexes with a set of peptides with 20% or higher DOPA content, well above the average of 0.11 ± 0.01%. These complexes also show uniformly compact size (DLS radii <23 nm), which could contribute mechanistically to their relatively strong effects on longitudinal relaxation.12 The most DOPA-rich peptide in the library, SSSSDDZZZZ, displayed a relatively low r1 [0.2 ± 0.1 (mM Fe)−1 s–1] and large size (71 nm), however.
The presence of a number of very large IONP complexes with r2 < 30 (mM Fe)−1 s–1 suggested that some of the peptides promote agglomeration to sizes above the so-called “static dephasing limit”, at which r2 decreases with apparent size.13 Agglomeration is a dynamic process and can lead to undesirable changes in MRI contrast over time. To screen against such effects, we compared r2 values obtained before and after an incubation period and discovered that many conjugates displayed instability (Supporting Information). Four peptide–IONP complexes showed optimal properties, however, with substantial r2 values with variation of less than 25%. The corresponding ligand sequences were all DOPA-rich peptides that also produced high r1 values in Figure 2a (dashed box). The properties of these sequences were examined in greater detail (Figure 2b). With respect to the rest of the library, the four peptides showed significantly lower pI (p < 10–6), in addition to higher DOPA content (p = 0.025) and r1 (p < 10–6). Within the set, some variation of r1 and r2 was also observed; the r1 difference between CDDZ and SSSSDDZZ was significant (p = 0.05), suggesting the possibility that microstructural differences even among this narrow set of sequences can influence relaxivity.
To test the ability of optimized peptide–IONP complexes to function as MRI contrast agents in vivo, we examined the behavior of CDDZ–IONP in biological buffer and in mice. Incubation of the particles (4.2 mM Fe, 50 μM peptide) in 10% fetal bovine serum (FBS) resulted in ∼10% change in optical absorbance over a 5 h period, and stability could be further prolonged by adding excess CDDZ (Figure 3a). Intravenous injection of 150 μL of CDDZ–IONP aliquots containing 4 mM Fe and 500 μM peptide into mice (Figure 3b,c) resulted in T2-weighted MRI changes that persisted for up to 12 h and corresponded to T2 decreases of close to 20% in muscle tissue (significant at 1 and 12 h with t-test p = 0.016 and 0.013, respectively).
Figure 3.
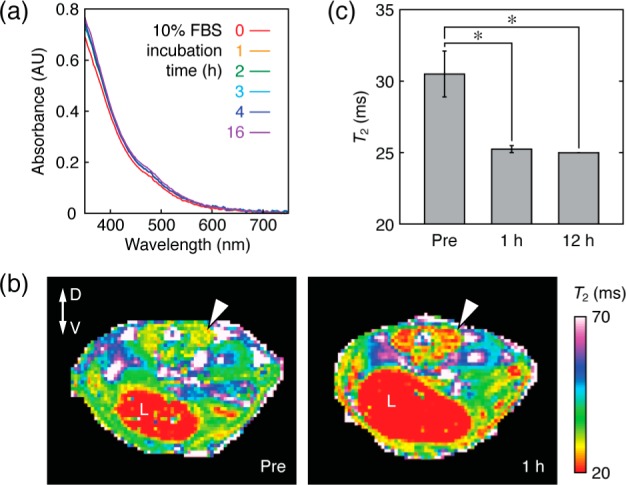
Biostability and in vivo MRI contrast induced by a peptide–IONP complex. (a) UV–visible spectra of CDDZ–IONP (3.6 mM Fe, 450 μM peptide) incubated for varying lengths of time in a 10% FBS biomimetic solution. Near invariance of the spectrum indicates relative stability of the particles in a biomimetic milieu. (b) T2 maps obtained from a representative mouse injected with 150 μL of CDDZ–IONP suspension (4 mM Fe, 500 μM peptide) and imaged by MRI at 7 T both before (left) and 1 h after (right) tail vein infusion. T2 shortening is visible in the musculature surrounding the spinal column (arrowheads), and the dorsoventral (D-V) axis and liver (L) are identified to facilitate orientation. (c) Group T2 measurements from the perispinal musculature of animals injected with contrast agent and imaged pre-injection and 1 and 12 h following injection. Error bars denote s.e.m. over multiple animals (n = 3).
Both the screening approach and specific results we report on here could have general applicability to the design of molecular imaging reagents. Discovery of optimized peptide–IONP MRI T2 contrast agents was enabled by our ability to screen a large collection of sequences for MRI and colloidal properties; this was in turn enabled by identification of the DDZ motif as a basic building block for forming IONP complexes, and rejection of alternative motifs. Results of the study show that the combination of catechol and carboxylate groups in peptide ligands promotes, but is not sufficient for stabilizing colloidal iron oxide cores. By screening peptide complexes for MRI properties, we found considerable diversity of r1 and r2 values, which is likely to reflect fundamental mechanisms of contrast generation by IONP agents. In the future, these findings might become bases for synthesis of isomorphous peptide–IONP complexes with different r2/r1 ratios for dual-probe molecular imaging applications, or for design of responsive MRI contrast agents in which peptide–IONP interactions are explicitly modulated. The peptide ligand family we demonstrate here incorporates a non-genetically encodable amino acid, DOPA; this limits possible application of genetic techniques for generating IONP ligand libraries, but fusion of DOPA moieties into genetically encoded libraries might still be possible using various chemical approaches.14,15 Screening of new reagents for MRI and other applications will be most straightforward if based on synthetic peptides, where analysis of libraries of over 103 molecules is feasible.16
Acknowledgments
Support was provided by NIH grants R01-DA28299, R01-NS76462, and R21-MH102470 to A.J. S.O. was supported by a JSPS Postdoctoral Fellowship for Research Abroad. He Wei and Moungi Bawendi are thanked for assistance with zeta potential measurement. Angela Belcher is thanked for helpful discussions.
Supporting Information Available
Detailed methods and further characterization of nanoparticle complexes. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Niemirowicz K.; Markiewicz K. H.; Wilczewska A. Z.; Car H. Adv. Med. Sci. 2012, 57, 196. [DOI] [PubMed] [Google Scholar]
- Na H. B.; Song I. C.; Hyeon T. Adv. Mater. 2009, 21, 2133. [Google Scholar]
- Sahoo Y.; Pizem H.; Fried T.; Golodnitsky D.; Burstein L.; Sukenik C. N.; Markovich G. Langmuir 2001, 17, 7907. [Google Scholar]
- Amstad E.; Gillich T.; Bilecka I.; Textor M.; Reimhult E. Nano Lett. 2009, 9, 4042. [DOI] [PubMed] [Google Scholar]
- Xu C.; Xu K.; Gu H.; Zheng R.; Liu H.; Zhang X.; Guo Z.; Xu B. J. Am. Chem. Soc. 2004, 126, 9938. [DOI] [PubMed] [Google Scholar]
- Whaley S. R. Ph.D. Dissertation, The University of Texas at Austin, 2001. [Google Scholar]
- Lower B. H.; Lins R. D.; Oestreicher Z.; Straatsma T. P.; Hochella M. F. Jr.; Shi L.; Lower S. K. Environ. Sci. Technol. 2008, 42, 3821. [DOI] [PubMed] [Google Scholar]
- Massart R. IEEE Trans. Magn. 1981, 17, 1247. [Google Scholar]
- Taboada E.; Rodriguez E.; Roig A.; Oro J.; Roch A.; Muller R. N. Langmuir 2007, 23, 4583. [DOI] [PubMed] [Google Scholar]
- Sahoo Y.; Goodarzi A.; Swihart M. T.; Ohulchanskyy T. Y.; Kaur N.; Furlani E. P.; Prasad P. N. J. Phys. Chem. B 2005, 109, 3879. [DOI] [PubMed] [Google Scholar]
- Roch A.; Gossuin Y.; Muller R. N.; Gillis P. J. Magn. Magn. Mater. 2005, 293, 532. [Google Scholar]
- Kim B. H.; Lee N.; Kim H.; An K.; Park Y. I.; Choi Y.; Shin K.; Lee Y.; Kwon S. G.; Na H. B.; Park J. G.; Ahn T. Y.; Kim Y. W.; Moon W. K.; Choi S. H.; Hyeon T. J. Am. Chem. Soc. 2011, 133, 12624. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y.; Jasanoff A. Magn. Reson. Imaging 2008, 26, 994. [DOI] [PubMed] [Google Scholar]
- Noren C. J.; Anthony-Cahill S. J.; Griffith M. C.; Schultz P. G. Science 1989, 244, 182. [DOI] [PubMed] [Google Scholar]
- Muir T. W. Annu. Rev. Biochem. 2003, 72, 249. [DOI] [PubMed] [Google Scholar]
- Winkler D. F. H.; Campbell W. D. Methods Mol. Biol. 2008, 494, 47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


