Abstract
Myxococcus xanthus , a predatory soil bacterium, has long been used as a model organism to study bacterial gliding motility. Research has revealed that two fundamentally distinct motor systems power gliding in this bacterium: repeated extensions and retractions of pili mediate social or (S-) motility, whereas the motor powering adventurous or (A-) motility has not yet been identified with certainty. Several different hypotheses to explain A-motility have been suggested and differ with respect to the involved motor structures as well as the mechanics of motility. As some of the more recent models invoke helically arranged structures and processes that require rotations of the cell, we decided to re-examine myxobacterial motility using microcinematographic techniques. This re-examination was also prompted by the lack of direct experimental data on the rotation of M. xanthus during gliding. Microcinematographic observations of deformed cells and cells containing large stationary intracellular structures reveal clearly that M. xanthus gliding does not require cell rotation.
Keywords: Cytoskeletal proteins, Bactofilin, Cell morphology, Gliding motility, Myxococcus xanthus
Introduction
The soil-dwelling social bacterium Myxococcus xanthus is a swarm-forming predator. Upon starvation, it aggregates into multicellular mounds that eventually develop into spore-filled fruiting bodies. For all aspects of this life style – swarming, hunting and aggregation – motility is of paramount importance [1, 2]. The cells use two different motors to achieve locomotion under these diverse circumstances [3, 4]. Coordinated movements of cell groups during swarming and aggregation appear to rely predominantly upon social or (S-) motility. In contrast, substrate exploration by isolated individual cells utilizes a completely different type of motility termed adventurous or (A-) motility.
Genetic and behavioral studies have shown that in order to perform S-motility, the cells repeatedly extend and retract polarly arranged type IV pili [5, 6]. The identification of the molecular motor of A-motility has proven more challenging. Despite intense studies, no unambiguous candidate for a cellular structure comprising an A-motility motor has been identified so far, although accumulating evidence including cytological evidence strongly supports the idea that motors distributed along the cell body are involved in gliding motility [3, 4]. Ultrastructural studies have linked two different structures to A-motility: linear periodic chain-like structures [7, 8], and nozzle-like slime secretion organelles [9]. However, the precise role of these two structures in A-motility is difficult to assess because their protein components have not been identified yet, and consequently no mutant strains lacking these structures have been described. Genetic studies have identified about 40 genes relevant for A-motility [10, 11]. Gene deletion studies together with light microscopic localizations of fluorescent protein fusions suggest that some of these A-motility-related proteins (Agl/Glt) may form large envelope-spanning complexes that have been proposed to represent the A-motor [12]. It has been suggested that components of this Agl/Glt-motor harness the proton motive force across the cytoplasmic membrane, while using the cell envelope-associated MreB filaments as tracks to generate force and to guide their own locomotion during gliding [13–15]. Another yet unsolved question is the function of the slime that is secreted during gliding [16]. It has variously been proposed to propel the cells, to facilitate adhesion, or to decrease friction through lubrication [3]. A recent examination of slime secretion using wet-surface-enhanced ellipsometric contrast microscopy revealed that the polymer is distributed all over the cell body and predominantly deposited underneath the cells where it associates with proteins of the Agl/Glt complex potentially providing adhesion sites against which the complexes can push to generate propulsion [17].
Based on these data, three models have been developed [3]. The slime nozzle model suggests that an expandable gel is secreted from many polarly and a few laterally arranged nozzles. Upon gel adherence to the substrate and further secretion, the cells are propelled in the opposite direction [9]. According to the focal adhesion model, clusters of A-motility-related Agl/Glt proteins connect the MreB cytoskeleton with hypothetical extra-cellular adhesion sites. During locomotion, motor proteins push against the slime-attached focal adhesion sites using the MreB helix as support thereby moving these sites from the front to the rear pole and, as a consequence, the cell in the opposite direction [17, 18]. In contrast, the helical rotor model suggests that the A-motor complexes track along helical MreB filaments thereby generating waves of surface deformations that push against the substrate resulting in forward motion of the cells [14].
The currently available evidence is inconclusive with respect to the nature of the A-motor, as well as the model to describe A-motility. On the other hand, predicted differences between these models can be assessed experimentally. In particular, the aspect whether cellular motility requires rotation is of central importance to our understanding of the process because it is linked to the arrangement and potential movement of A-motor complexes. While the slime nozzle model does not predict cellular rotation during gliding, both other models envisage some form of rotation due to A-motor complexes interacting with and tracking along helically arranged MreB filaments. Surprisingly, given the importance of rotation, no direct experimental data exist regarding this aspect of M. xanthus motility. We therefore decided to re-examine gliding motility in this bacterium in order to solve the question whether cellular rotation is an essential characteristic of A-motility.
Detecting rotations in gliding M. xanthus cells is challenging due to the small size of the cells, the absence of an easily detectable irregular morphological feature, and the slow speed of cellular movements. Several approaches have been used with other bacterial species to successfully address these challenges. One such approach relies on the light microscopic observation of intracellular inclusions. Since bacteria lack plasma streaming [19], these inclusions should not change their relative cellular positions and can therefore be used as fiduciary markers for rotations. An obvious drawback is that most bacteria lack inclusions that are sufficiently refractile to be microscopically observable, or they contain only large centrally arranged globules that do not allow detection of rotation.
Another commonly used approach is to attach latex micro beads or India ink particles to the surface of an organism to create easily traceable surface markers and to monitor their movements during locomotion [15, 20, 21]. Although this method has been used successfully to gain insight into gliding for a number of bacteria, it is not always clear how bead and cell movements correlate and, more importantly, whether these two processes are necessarily linked to each other. For example, while Lapidus and Berg [20] reported that for Cytophaga bead movements are invariably linked to cell movements, Ridgway and Lewin [21] observed just the opposite, namely that bead movements occurred even on non-motile cells indicating that the two processes in this organism were not obligatorily linked. Since the nature of the binding sites in these two organisms is not known, it could be that some beads are bound to stationary sites while others are bound to sites that can freely diffuse along the cell body. It is furthermore not implausible that these two scenarios might occur in the same cell simultaneously, making interpretations difficult. Additional ambiguity results from the fact that all gliding bacteria secrete slime while moving [16] and that some beads may attach to the secreted slime. Since the slime is hyaline and invisible by light microscopy, it is virtually impossible to tell whether the beads are attached to the surface of the organism or to the secreted slime.
In contrast, the most unambiguous results have been obtained using cells that show morphological irregularities like lateral buds or terminal branches [21]. Since these irregularities are permanent and easily observable, they make detection of rotation during motility straightforward. However, under normal culture conditions, most bacteria including M. xanthus do not develop morphological irregularities that could be used for this purpose.
In this study, we re-examined gliding motility in M. xanthus in order to solve the question whether the cells rotate or not, by taking advantage of our recent observation that the absence of the cytoskeleton protein BacM results in morphologically irregular cells [22]. We used cells derived from a parent strain incapable of pilus-driven S-motility so that the irregularly shaped cells could only move using A-motility. To rule out the possibility that bacM deletion might cause the disappearance of potentially existing rotation during cellular gliding, we also investigated wild-type cells that contained a prominent polarly arranged fluorescent intra-cellular screw-like structure composed of a BacM-mCherry fusion protein.
Materials and methods
All Myxococcus xanthus strains used in this study were either derived from wild-type strain DK1622, or from S-motility defective strain DK8615 (ΔpilQ). Strains EH302 (ΔpilQ, ΔbacM) and EH362 (Poar-bacM-mCherry) have been described in detail elsewhere [22]; strain EH367 (ΔpilQ, ΔbacM, Poar-bacM-mCherry) was generated from EH302 in the same way EH362 was generated from DK1622.
To analyze motility, all strains were grown for 9 d on 1.5% agar containing CTT (1% casitone, 10 mM Tris pH 8.0, 8 mM MgSO4, 1 mM KH2PO4) harvested and suspended in TPM buffer (10 mM Tris pH 8.0, 8 mM MgSO4, 1 mM KH2PO4) to an OD600 of 0.1. One microliter of each individual cell suspension was then spotted onto the surface of a small 0.7 mm thick 1.5% agar patch containing TPM buffer, and covered with a glass cover slip leaving a 3 mm air gap on all sides of the patch. A molten mixture of paraffin/vaseline (2:1 w/w) was used to seal the edges of the cover slip onto the microscopic slide carrying the patch to protect the cells from desiccation while at the same time allowing oxygen diffusion. As described earlier [22], light and fluorescence images were acquired 24 to 60 h after spotting using a Nikon Eclipse 90i microscope with a 100x oil immersion phase contrast objective to produce videos at a rate of 60 exposures per minute (epm) for rotational analysis and at 30 epm for cell tracking.
To generate tracking data for comparison of gliding speeds in the different strains, cell movements were recorded 48 h after spotting, and the generated videos were analyzed with Volocity software (Improvision) as described in detail in the Supporting Information (see Fig. S1 and text).
Results and discussion
We had previously established that Myxococcus xanthus cells lacking bactofilin BacM exhibit wild-type S-motility [22]. To further analyze whether deletion of bacM, or expression of bacM-mCherry in cis from the oar promoter, would affect A-motility of the respective strains, we investigated their gliding motility by tracking individual isolated cells. Table 1 summarizes the results of these measurements. Cells of DK1622 (wild type) and EH362 (Poar-bacM-mCherry) move at similar speeds. Cells of the A+S− strain DK8615 (ΔpilQ) are capable of A- but not S-motility, and these cells as well as those of daughter strains EH302 (ΔpilQ, ΔbacM) and EH367 (ΔpilQ, ΔbacM, Poar-bacM-mCherry) exhibit a lower average speed. This speed difference between A+S+ wild-type cells and A+S− cells has previously been reported, and attributed to the loss of the S-motor that normally operates synergistically with the A-motor to allow for full wild-type speed [23, 24]. The observation that strain EH302 is slightly slower than the other two A+S− strains might be explained by increased friction during gliding caused by their cellular deformations. Single cells of all strains examined retained the ability to glide on agar. Based on the observed similarity in gliding speeds, we conclude that there is no fundamental difference in A-motility gliding between the parental strains (wild type and ΔpilQ) and their respective daughter strains, which were used to analyze cellular movements.
Table 1.
Tracking of individual isolated Myxococcus xanthus cells.
| Strain (motility phenotype) | Relevant genotype | Average speed and standard deviation (μm/min) | Median speed (μm/min) | Maximum speed (μm/min) | Minimum speed (μm/min) | Number of tracked cells |
|---|---|---|---|---|---|---|
| DK1622 (A+S+) | wild type | 6.6 +/− 1.5 | 6.8 | 8.8 | 2.1 | 36 |
| 6.2 +/− 1.0 | 6.1 | 8.4 | 2.7 | 30 | ||
| EH362 (A+S+) | Poar-bacM-mCherry | 5.6 +/− 0.8 | 5.6 | 7.3 | 3.7 | 69 |
| 5.6 +/− 1.3 | 5.7 | 9.5 | 1.9 | 60 | ||
| DK8615 (A+S−) | ΔpilQ | 4.6 +/− 1.1 | 4.6 | 7.9 | 2.3 | 41 |
| EH302 (A+S−) | ΔpilQ, ΔbacM | 3.7 +/− 0.9 | 3.6 | 6.0 | 2.0 | 33 |
| 3.5 +/− 1.0 | 3.6 | 5.4 | 1.3 | 47 | ||
| EH367 (A+S−) | ΔpilQ, ΔbacM, Poar-bacM-mCherry | 4.1 +/− 0.8 | 4.2 | 6.2 | 2.3 | 36 |
| 4.4 +/− 1.0 | 4.3 | 7.5 | 3.0 | 18 |
For all strains, with the exception of DK8615, tracking was performed using videos recorded at two different positions on the agar patch, and the results are presented separately.
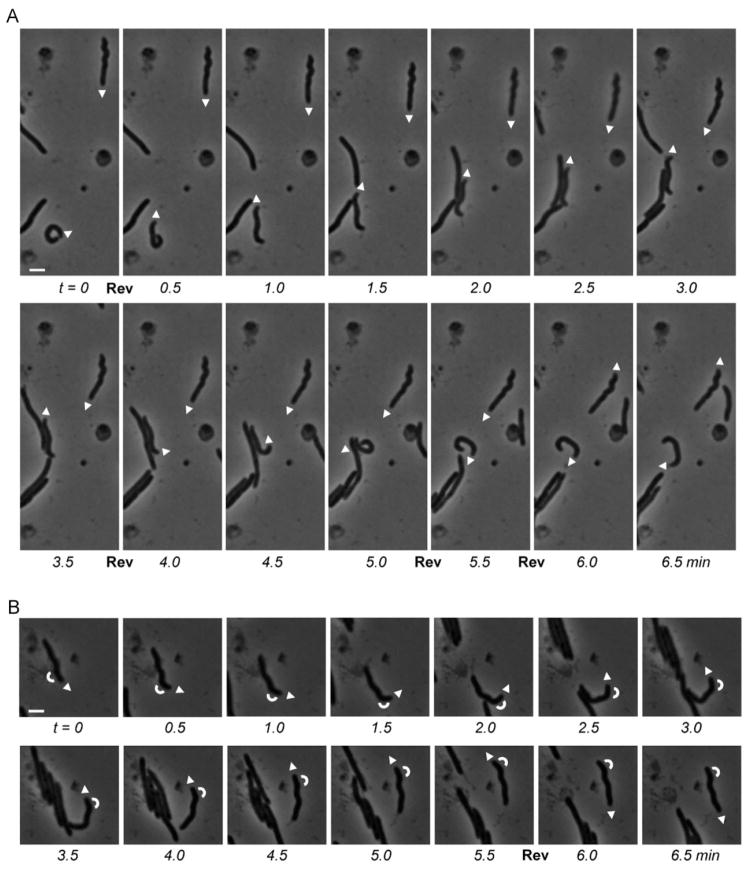
To explore whether isolated M. xanthus cells rotate while gliding, we first analyzed deformed A+S− cells of strain EH302 which lacked the genes pilQ as well as bacM. These cells do not produce the outer membrane secretin channel protein PilQ, consequently do not have type IV pili, and can no longer move using S-motility. In addition, they have irregular cell morphologies due to the absence of BacM [22]. Since these irregular morphologies result from perturbations of the cell’s peptidoglycan layer, they are stable and ideally suited to detect rotations. In the case of cellular rotations, bent or crooked tips of these cells would have to show oscillations from one side to the other. Our analysis showed, however, that the tips of isolated gliding cells never oscillated, and that these cells consequently did not rotate during gliding (see Fig. 1A, 1B and corresponding Movies 1A and 1B). Even during complex motility maneuvers, like performing a U-turn as seen for the cell shown in Fig. 1B, we observed no signs of rotation. Only when cells were gliding in groups, which generated cell-cell contacts, rotations could occasionally be seen (Fig. S2; Movie S2). These rotations are most likely the consequence of lateral cell-cell interactions that are known to occur in M. xanthus [25]. The fact that rotations were occasionally observed for a very minor fraction of cells gliding in groups but not for gliding isolated cells shows that our setup was indeed capable of detecting rotations when they occur, and that isolated gliding cells did not rotate.
Figure 1.
S-motility-deficient cells of Myxococcus xanthus EH302 (ΔpilQ, ΔbacM) gliding over a TPM agar patch 24 h after spotting. Two (A) and one (B) individual crooked cells were marked at the leading pole by arrowheads that indicate the direction of gliding. White bars represent 2 μm; reversals of gliding direction are marked by “Rev”. Arcs in (B) indicate the orientation of the bend near the end of the tracked cell. Note, that the zigzag-shaped cells maintain their orientation during gliding rather than changing into their mirror image (see also corresponding Movies 1A and 1B).
In addition to morphologically defective cells, we also investigated wild type-derived cells that contained a bacM-mCherry fusion gene under control of the oar promoter at their chromosomal attB site. These cells also express their genomic untagged bacM and, as a consequence, possess wild-type morphology. During A-motility their gliding speeds were similar to those seen for wild-type cells (Table 1). Although the average gliding speed of 5.6 μm/min observed for strain EH362 (Poar-bacM-mCherry) was slightly slower than that of DK1622 (wild type), it was faster than the 4.6 μm/min seen for DK8615 (ΔpilQ), which is generally considered an A+S− strain capable of normal A-motility.
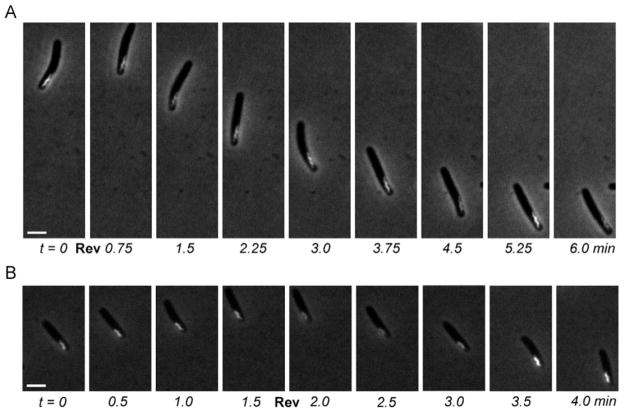
By fluorescence microscopy, the BacM-mCherry fusion protein can be visualized in the cell as a unique and easily detectable fluorescent screw-like structure that is generally located close to one of the poles (Fig. 2A, 2B). While the center of the cell is occupied by the nucleoid, this screw structure is usually positioned laterally, which makes it an ideal marker structure to detect rotations. As can be seen in the pictures and the movies showing the gliding of these cells (Fig. 2A, 2B and corresponding Movies 2A, 2B as well as Fig. S3A and S3B and Movies S3A and S3B), the screw structure always stayed on the same side of the cells. This is in contrast to a change of sides that would be expected for rotating cells. The same results were observed for both normal forward motion, as well as changes of direction during reversals. To exclude pilus-driven S-motility as the driving force for the gliding of the cells, we additionally analyzed A+S− strain EH367 (ΔpilQ, ΔbacM, Poar-bacM-mCherry). Like in strain EH362 (Poar-bacM-mCherry), the BacM-mCherry screws always stayed at the same side of these cells confirming that these cells also glide without rotating (Fig. 2B and Movie 2B as well as Fig. S3B and Movie S3B).
Figure 2.
Gliding cells of Myxococcus xanthus producing a BacM-mCherry fusion protein, which forms a fluorescent screw-like structure near one of the cell poles. (A) Cell of A+S+ strain EH362, which expresses bacM-mCherry; 48 h after spotting on TPM agar. (B) Cell of A+S− strain EH367, which expresses bacM-mCherry in the absence of bacM and pilQ; 60 h after spotting on TPM agar. Gliding was observed under phase contrast while BacM-mCherry fluorescence was recorded concomitantly. White bars represent 2 μm; reversals of gliding direction are marked by “Rev”. Note, that during gliding none of the screw structures changes its relative orientation within any of the cells (see corresponding Movies 2A and 2B).
Concluding remarks
Based on these data, we conclude that Myxococcus xanthus cells glide without rotation during A-motility, regardless whether both types of motility or only A-motility are functional in the cells. The data presented here are inconsistent with certain aspects of the focal adhesion and the helical rotor models. For the cellular motor proteins to follow the track of the MreB helix, the cell would have to rotate several times around its own axis when gliding the distance of just one cell length. The finding that the cells exhibit A-motility in the absence of rotation, questions the suggested A-motility motor track function of the MreB filaments. This is in line with the recent observation that, in a number of bacteria, MreB does not form continuous helical structures but instead is arranged in short filaments [26, 27]. However, if the Agl/Glt complexes would instead track along an unknown linear cytoskeleton, the focal adhesion model should result in movements that would be in agreement with the observed lack of rotation during A-motility.
On the other hand, although the absence of cellular rotation during gliding would be consistent with the slime nozzle model, our findings do not prove this model. Nevertheless, our data provide useful mechanistic insights into this type of motility and can serve as a framework for the development of novel A-motility models in M. xanthus. A true understanding of A-motility in this organism will require the identification and characterization of the A-motor.
Supplementary Material
Acknowledgments
We would like to thank the members of the Hoiczyk laboratory for helpful discussions and comments on the manuscript. The research in our laboratory is funded by a grant from the National Institutes of Health (GM85024) and a fellowship from the Jane Russell Fund for Innovative Research in Infectious Diseases to MKK.
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- 1.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 2.Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu Rev Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 3.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Ducret A, Shaevitz JW, Mignot T. From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol Rev. 2011;36:149–164. doi: 10.1111/j.1574-6976.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- 5.Wu SS, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 6.Sun H, Zusman DR, Shi W. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 7.Freese A, Reichenbach H, Lünsdorf H. Further characterization and in situ localization of chain-like aggregates of the gliding bacteria Myxococcus fulvus and Myxococcus xanthus. J Bacteriol. 1997;179:1246–1252. doi: 10.1128/jb.179.4.1246-1252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lünsdorf H, Reichenbach H. Ultastructural details of the apparatus of gliding motility of Myxococcus fulvus (Myxobacterales) J Gen Microbiol. 1989;135:1633–1641. [Google Scholar]
- 9.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 10.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 11.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 12.Luciano J, Agrebi R, Le Gall AV, Wartel M, et al. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;9:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauriello EMF, Mouhamar F, Nan B, Ducret A, et al. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan B, Chen J, Neu JC, Berry RM, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reichenbach H, Dworkin M. Introduction to the gliding bacteria. In: Starr MP, Stolp H, Trüper HG, Balows A, Schlegel HG, editors. The Prokaryotes. Vol. 1. Springer Verlag; Berlin, Heidelberg, New York: 1981. pp. 315–327. [Google Scholar]
- 17.Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci USA. 2012;109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mignot T, Shaevitz JW, Hartzell P, Zusman DR. Evidence that focal adhesions complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewin RA. Flexibacter polymorphus, a new marine species. J Gen Microbiol. 1974;82:393–403. [Google Scholar]
- 20.Lapidus IR, Berg HC. Gliding motility of Cytophaga sp strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridgway HF, Lewin RA. Characterization of gliding motility in Flexibacter polymorphus. Cell Motil Cytoskeleton. 1988;11:46–63. doi: 10.1002/cm.970110106. [DOI] [PubMed] [Google Scholar]
- 22.Koch MK, McHugh CA, Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for cell shape. Mol Microbiol. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser D, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motility. 1983;3:227–245. [Google Scholar]
- 24.Spormann AM, Kaiser AD. Gliding movements in Myxococcus xanthus. J Bacteriol. 1995;177:5846–5852. doi: 10.1128/jb.177.20.5846-5852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc Natl Acad Sci USA. 2009;106:4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garner EC, Bernard R, Wang W, Zhuang X, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, et al. Processive movements of MreB-associated biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.