Abstract
Since discovery of the centromere-specific histone H3 variant CENP-A, centromeres have come to be defined as chromatin structures that establish the assembly site for the complex kinetochore machinery. In most organisms, centromere activity is defined epigenetically, rather than by specific DNA sequences. In this review, we describe selected classic work and recent progress in studies of centromeric chromatin with a focus on vertebrates. We consider possible roles for repetitive DNA sequences found at most centromeres, chromatin factors and modifications that assemble and activate CENP-A chromatin for kinetochore assembly, plus the use of artificial chromosomes and kinetochores to study centromere function.
Main Text
Introduction
In the 1930s, the site where chromosomes associate with the spindle during cell division was independently given two names: the “centromere” (Darlington, 1936) and the “kinetochore” (Schrader, 1939). The terms were long thought redundant, but it has recently proven useful to differentiate between them.
Centromeres, classically defined in genetics as regions of suppressed meiotic recombination (Beadle, 1932), were later recognized as the primary constriction of mitotic chromosomes. Centromeres are enriched in satellite repeats (Pardue and Gall, 1970) that stain dark by C-banding (McKay, 1973). When electron microscopy revealed a multilayered structure that binds to microtubules at the surface of centromeres (Luykx, 1965, Brinkley and Stubblefield, 1966, Jokelainen, 1967), that structure was termed the “kinetochore.” The centromere is now generally accepted to be a chromatin structure that specifies where the kinetochore will form. Kinetochores are complex protein structures lacking DNA (Cooke et al., 1993).
Molecular studies of the centromere/kinetochore began with the discovery that some patients with scleroderma spectrum disease (e.g., CREST syndrome) have anti-centromere autoantibodies (ACA) (Moroi et al., 1980). Three antigens, CENP-A, CENP-B, and CENP-C, are recognized by those sera (Earnshaw and Rothfield, 1985). CENP-A is a centromere-specific histone H3 variant (Palmer et al., 1987, Earnshaw et al., 2013). CENP-A and CENP-C localize to the inner kinetochore (Saitoh et al., 1992, Vafa and Sullivan, 1997, Warburton et al., 1997). CENP-B is an α-satellite (human centromeric DNA)-binding protein (Earnshaw et al., 1987). To date, over 100 kinetochore components have been identified. Aspects of kinetochore function that are beginning to be well understood include microtubule binding, chromosome movement, and checkpoint signaling. (For reviews, see Rieder, 1982, Maiato et al., 2004, Cheeseman and Desai, 2008, Santaguida and Musacchio, 2009, Perpelescu and Fukagawa, 2011, Vleugel et al., 2012).
The organization and functions of centromeric chromatin remain less understood subjects of active study. In this review, we discuss recent progress in understanding the organization, composition, and assembly of centromeric chromatin and DNA.
Point and Regional Centromeres of Model Organisms
Budding yeast Saccharomyces cerevisiae centromeres occupy a ∼125 bp DNA sequence (Hegemann and Fleig, 1993, Clarke, 1998), now termed a “point centromere” (Pluta et al., 1995). They include three conserved DNA elements (CDE), CDEI, CDEII, and CDEIII that form a single nucleosome containing CENP-A (Cse4 in S. cerevisiae) (Stoler et al., 1995). The CDEI-III sequences are necessary and sufficient for active centromere formation, and a single base mutation in CDEIII can abolish centromere function (Clarke, 1998).
Centromeres of the fission yeast Schizosaccharomyces pombe encompass 40–100 kb containing a central core (cnt) of 4–7 kb, where the kinetochore forms, flanked by repeated sequences (otr) that form heterochromatin (Takahashi et al., 1992). This configuration is known as a “regional centromere” (Pluta et al., 1995). Both cnt and otr sequences are required for centromere function (Baum et al., 1994). Heterochromatin is required during de novo centromere formation (Folco et al., 2008). Transcription of otr and subsequent transcript processing by the RNAi machinery direct formation of pericentromeric heterochromatin (Volpe et al., 2002, Chen et al., 2008).
Regional centromeres of the pathogenic yeast Candida albicans map to unique sequences of ∼3 kb on each of the 8 chromosomes (Sanyal et al., 2004). Although a truncated minichromosome containing a fragment from the centromere retained centromere activity, centromeric activity was not reconstituted when naked minichromosome DNA was introduced back into yeast (Baum et al., 2006). It was suggested that an unusual chromatin structure detected at endogenous centromeres might be involved in epigenetic specification of the centromere in this yeast.
The regional centromere is the most common organization for centromeres in humans and most model organisms characterized to date, including Neurospora crassa (Centola and Carbon, 1994), Arabidopsis thaliana (Copenhaver et al., 1999), Drosophila melanogaster (Sun et al., 2003), and Oryza sativa (rice) (Nagaki et al., 2004). Common features of regional centromeres include the lack of a “magic” DNA sequence, the presence of complex repeated DNAs (satellite repeats together with centromeric retrotransposons), and an involvement of some sort of epigenetic mechanism in specification of the site for kinetochore assembly.
Nematodes, some insects, and plants assemble a “holocentromere” (originally, a holokinetochore) that extends along the entire length of the chromosome (Hughes-Schrader and Ris, 1941). Recent chromatin immunoprecipitation-on-chip (ChIP-chip) analyses in Caenorhabditis elegans (Gassmann et al., 2012) revealed that CENP-A occupies nonrepeated regions of 10–12 kb dispersed across about half of the genome and is excluded from loci that are transcribed in the germline and early embryo (for a contrasting view, see Steiner and Henikoff, 2014). Holocentromeres may consist of numerous CENP-A “seeds” dispersed along the chromosome that somehow cooperate to direct assembly of a functional kinetochore.
Kinetochores Assemble on Repetitive DNA Sequences in Most Organisms
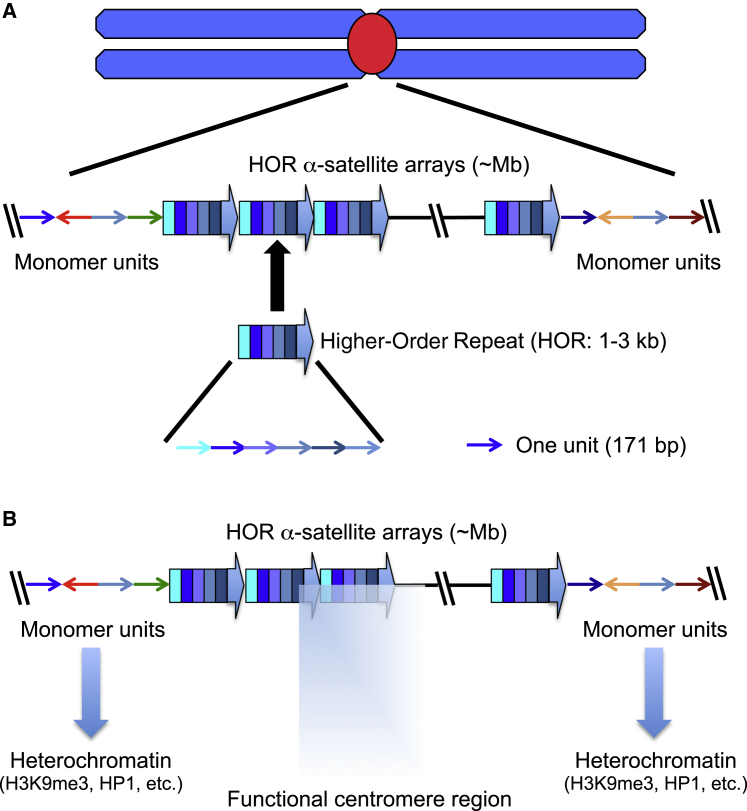
In situ hybridization first revealed that satellite repeats were located at centromeres of mouse chromosomes (Pardue and Gall, 1970). Subsequent studies identified human centromeric DNA as α-satellite (Manuelidis, 1978). This DNA, with its 171 bp consensus sequence (Vissel and Choo, 1987), exhibits a complex higher-order repeat (HOR) pattern, in which adjacent monomers may share no more than 50% sequence identity, but corresponding monomers among the HORs share >90% identity (Waye and Willard, 1989, Aldrup-Macdonald and Sullivan, 2014). Different-sized HORs are observed on differing human chromosomes, where they typically create a chromosome-specific array that spans 0.3–5 Mb within the centromere (Figure 1).
Figure 1.
Genomic Organization of Human Centromeres
(A) Human centromeres contain α-satellite sequences. In the inner core of the centromere, α-satellite monomers 171 bp long are organized into higher-order repeat (HOR) units, which are amplified, spanning 0.3–5 Mb. Unordered monomer units flank the HOR region.
(B) Functional centromeres form on a portion of the HOR, which might be newly evolved and homogenized. Disordered monomer unit sequences in pericentromeres are derived from an ancestral primate centromere. Pericentromere regions are highly heterochromatinized.
Sequence assembly across α-satellite arrays at human centromeres is difficult due to high sequence similarities within the arrays and variability between individuals. To date, complete maps across the HOR and into pericentromeric heterochromatin are available only for chromosomes 8, X, and Y (Schueler et al., 2001, Nusbaum et al., 2006, Miga et al., 2014). On the X, a repeating HOR structure is flanked by divergent α-satellite monomers not ordered into HOR (Figure 1) (Schueler et al., 2001). The functional kinetochore assembles on the HOR core, with monomeric α-satellite sequences comprising pericentromeric heterochromatin (Schueler et al., 2001).
The conservation of α-satellite sequences at all natural human centromeres suggested that these HORs are required for centromere identity or function (Schueler et al., 2001). However, this was ruled out by the discovery of human dicentric chromosomes containing α-satellite DNA arrays that do not nucleate kinetochore formation (Earnshaw and Migeon, 1985, Earnshaw et al., 1989) and functional neocentromeres lacking α-satellite sequences (Voullaire et al., 1993).
Thus, although there are exceptions (see below), centromere regions from most organisms contain repetitive sequences, suggesting that those sequences contribute to important aspects of centromere function.
Repetitive Sequences in Centromeres May Allow Kinetochore Plasticity
If kinetochores can form on nonrepetitive sequences, why do most centromeres contain repetitive sequences? One possibility is that repetitive sequences direct the formation of pericentromeric heterochromatin (Ekwall, 2007), with its molecular signature of heterochromatin protein 1 (HP1) bound to histone H3 trimethylated on lysine 9 (H3K9me3) (Minc et al., 1999, Nakayama et al., 2001, Yamagishi et al., 2008) (Figure 1). Although neocentromeres assembled on nonrepetitive DNA lack heterochromatin and yet function perfectly well in mitosis and meiosis (Alonso et al., 2010, Shang et al., 2013), pericentromeric heterochromatin, which consists of unordered monomer units of α-satellite in human, appears to provide a boundary between the kinetochore and flanking euchromatin regions in natural centromeres and might act as a barrier to centromere migration (Figure 1).
Pericentromeric heterochromatin recruits cohesin (Nonaka et al., 2002, Yamagishi et al., 2008, Gartenberg, 2009), and centromeric cohesion is particularly important in meiosis I where sister chromatids must remain paired. A requirement for strong centromeric cohesion in meiosis I could select for the accumulation of repetitive sequences in centromeres over evolutionary time scales.
In humans, the CENP-A associated domain at natural centromeres varies between 200 and 2,000 kb on different chromosomes and individuals (Sullivan et al., 2011). Thus, kinetochores form only over a portion of the α-satellite arrays (Figure 1). Due to the repetitive nature of the underlying DNA at natural centromeres, the kinetochore region cannot be mapped unambiguously by modern ChIP methods. This can be done at human neocentromeres, where the CENP-A domain spans just 80–100 kb (Alonso et al., 2010, Hasson et al., 2013). Thus, neocentromeres are much smaller than native centromeres.
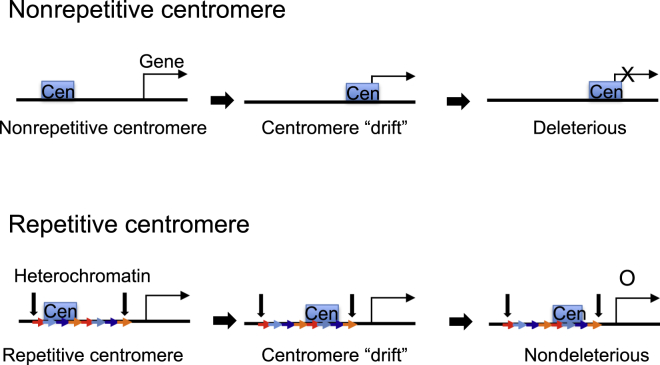
Although active genes are present within rice centromeres (Nagaki et al., 2004), kinetochore formation across a transcribed gene strongly suppressed transcription of that gene on the chicken Z chromosome (Shang et al., 2013). Thus, kinetochore formation on essential genes would be expected to be deleterious. Because regional centromere position is not strictly specified by DNA sequence, it is possible that the kinetochore position on the underlying DNA might drift slightly (Figure 2). In this case, repetitive arrays could provide a safety buffer within which such drift would be harmless (Figure 2). It will be interesting in future studies of unique sequence centromeres to test whether kinetochore position is fixed or plastic.
Figure 2.
Proposed Role of Repetitive Sequences in Centromeres
We postulate that the kinetochore location might be able to “drift” on the DNA. If a kinetochore moved over a gene, then it would suppress its expression. This would be deleterious for essential genes. At nonrepetitive centromeres, the probability of this drift affecting a flanking gene is higher than at repetitive centromeres, where the probability of affecting a gene is relatively low. We suggest that repetitive sequences might provide a safety area for centromere “drift.” Pericentromere regions at edge of centromeres might also function as a heterochromatin barrier between centromere and euchromatin regions.
Neocentromeres Reveal Insights into Centromere Specification
Very rarely, disruption or inactivation of a natural centromere is followed by formation of a neocentromere at a new locus on a chromosome arm. Over 100 neocentromeres have been described in human clinical samples (Marshall et al., 2008). They form on diverse DNA sequences and are not associated with α-satellite arrays, thus providing strong evidence that human centromeres are specified by sequence-independent epigenetic mechanisms.
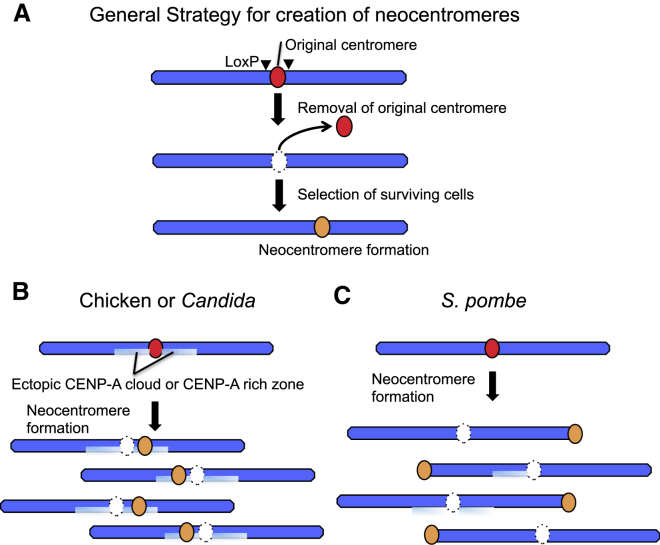
Experimental generation of neocentromeres in model organisms has yielded insights into the process of neocentromere formation. In D. melanogaster, neocentromeres obtained following γ-irradiation-based chromosome breakage formed near the pericentromeric region of the X chromosome (Murphy and Karpen, 1995). Subsequently, neocentromeres were generated in S. pombe (Ishii et al., 2008) and C. albicans (Ketel et al., 2009) following targeted deletion of the original centromere and genetic selection for retention of the chromosome (Figure 3). C. albicans neocentromeres formed either in transcriptionally active or intergenic regions near the natural centromeres (Ketel et al., 2009, Thakur and Sanyal, 2013). In contrast, S. pombe neocentromeres preferentially formed near telomeric heterochromatin and efficient neocentromere formation required heterochromatin proteins (Ishii et al., 2008).
Figure 3.
Neocentromere Generation in Various Experimental Systems
(A) Isolation of surviving cells after removal of the endogenous centromere on a chromosome by homologous recombination between two LoxP loci flanking the centromere. Surviving cells usually have segregation of the chromosome directed by a neocentromere.
(B) Ectopic CENP-A is assembled into a “CENP-A cloud” flanking the endogenous centromere in chicken cells. A CENP-A-rich region might be created by specific 3D packaging of the genome in Candida nuclei. The ectopic CENP-A functions as a site of preferential neocentromere formation in chicken or Candida cells when the endogenous centromere is disrupted. In contrast, neocentromeres are preferentially formed on telomeres in S. pombe.
Recently, chromosome engineering has allowed the efficient isolation of neocentromeres in chicken DT40 cells. Conditional deletion of the centromeres of chromosome Z or 5 led to neocentromere formation at a frequency of ∼3 × 10−6 on a wide range of both transcriptionally active and inactive sequences (Figure 3) (Shang et al., 2013). Thus, neocentromeres can be “seeded” on either transcriptionally active or inactive regions of the genome, and heterochromatin is not required for centromere formation. However, neocentromere formation caused a significant drop in gene transcription when a neocentromere formed on an actively transcribed gene on the Z chromosome.
Chicken neocentromeres have a remarkably constant size of ∼40 ± 6 kb, and CENP-A domains did not expand even in cells overexpressing CENP-A (Shang et al., 2013). In contrast, increased levels of CENP-A expression human cells resulted in increased CENP-A incorporation at centromeres (Bodor et al., 2014). CENP-A levels are regulated by a ubiquitin-dependent pathway in yeasts (Ranjitkar et al., 2010, Kitagawa et al., 2014), and the mechanisms governing centromere size may vary.
Drosophila, barley, Candida, and chicken neocentromeres all formed close to the natural centromeres (Figure 3) (Williams et al., 1998, Nasuda et al., 2005, Shang et al., 2013, Thakur and Sanyal, 2013). Possibly, patterns of neocentromere formation are determined by noncentromeric incorporation of CENP-A (Figure 3) in regions with high histone turnover (Lacoste et al., 2014). Remarkably, significant levels of CENP-A in vertebrates are incorporated into chromatin at noncentromeric sites (30% in chicken and 74% in human RPE1 cells [Shang et al., 2013, Bodor et al., 2014]). Importantly, given the size of human genome, this still corresponds to a ∼50× concentration increase at centromeres (Bodor et al., 2014). In chicken, the ectopic CENP-A is enriched in chromatin flanking the natural centromeres (the “CENP-A cloud”). Noncentromeric Cse4 (CENP-A) is also observed in budding yeast (Camahort et al., 2009, Lefrançois et al., 2009, Lefrançois et al., 2013) and might form a CENP-A “cloud” (Kerry Bloom, personal communication). The “cloud” was not detected in Candida (Thakur and Sanyal, 2013), where it was proposed that neocentromeres might form in a CENP-A-rich zone created by specific three-dimensional packaging of the genome in nuclei.
We suggest that the ectopic “CENP-A cloud” functions to seed neocentromere formation when natural centromeres are disrupted (Figure 3). How centromere formation is suppressed in the ectopic “CENP-A cloud” and how this suppression is lifted once natural centromeres are compromised remain interesting questions for further study.
Nonrepetitive Centromeres Found in Horses, Orangutans, and Chickens
Sequencing the horse genome revealed that the centromere of chromosome 11 lacked repetitive sequences (Wade et al., 2009). The CENP-A domain of horse centromere 11 is ∼90 kb (Wade et al., 2009), similar to a typical human neocentromere (80–100 kb) (Alonso et al., 2010). Chickens also have nonrepetitive centromeres on chromosomes 5, 27, and Z (the others are repetitive) (Shang et al., 2010). These nonrepetitive centromeres are only ∼40kb long based on CENP-A ChIP-seq analysis (Shang et al., 2010). Interestingly, orangutans also have one nonrepetitive centromere (Locke et al., 2011).
These nonrepeated centromeres might be evolutionally new centromeres (ENCs) that formed initially as neocentromeres, then become fixed in the population. It appears that over time, ENCs acquire repetitive DNA elements—presumably to stabilize them as proposed above.
Human Artificial Chromosomes and Epigenetic Engineering of Centromeric Chromatin
Given the powerful insights obtained by formation of artificial chromosomes in yeasts (Murray and Szostak, 1983), it was thought that formation of artificial chromosomes in human cells might lead to important insights into centromere structure and function. The first human artificial chromosomes (HACs, also known as MACs or mammalian artificial chromosomes) were formed in cells transfected with a DNA cocktail including an α-satellite array, genomic DNA, and telomeric sequences (Harrington et al., 1997). Subsequently, it was reported that yeast artificial chromosomes containing α-satellite DNA and retrofitted with telomeres could also form HACs (Ikeno et al., 1998). Circular BACs lacking telomeric sequences also give rise to stable HACs and are easier to construct (Ebersole et al., 2000).
A number of general principles emerged from these studies. Only α-satellite DNA with a regular HOR repeat structure and with CENP-B binding sites is functional for HAC formation (Ohzeki et al., 2002). This was paradoxical as CENP-B knockout mice are viable and fertile (Hudson et al., 1998, Kapoor et al., 1998, Perez-Castro et al., 1998). Subsequent studies revealed that CENP-B appears to manage heterochromatin formation during de novo centromere formation (Okada et al., 2007), and it also appears to be required for efficient recruitment of CENP-C to kinetochores (Daniele Fachinetti and Don Cleveland, personal communication). Interestingly, either excessive heterochromatin or excessive transcription flanking the α-satellite DNA sequences interferes with de novo centromere formation (Nakashima et al., 2005, Ohzeki et al., 2012).
HACs have thus far proved to be of only limited utility in understanding the mechanisms of human centromere formation. HACs are observed only after multiple generations in cell culture during which an unknown sequence of events has occurred. HACs identified to date are considerably larger than the input DNA, and physical characterization of one HAC revealed that it had undergone a complex set of rearrangements and acquired sequences from the arm of chromosome 13 (Kouprina et al., 2013).
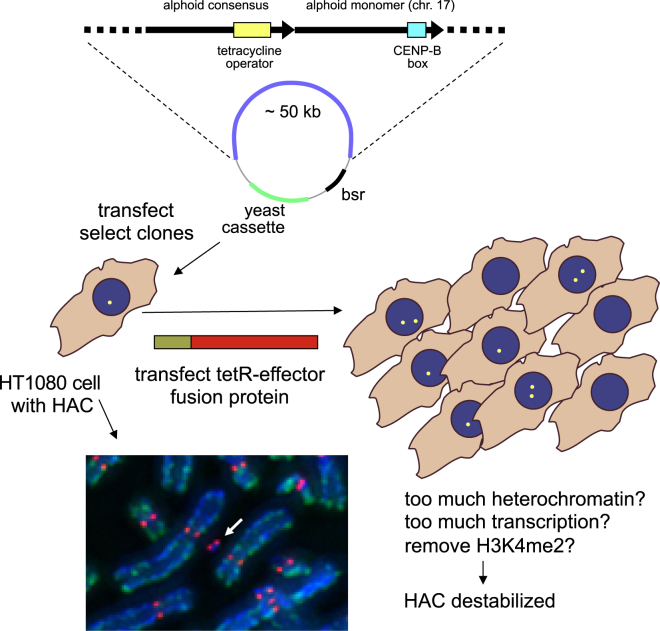
Despite these limitations, the alphoidtetO HAC has recently begun to yield insights about the chromatin environment required within centromeric DNA in and around the kinetochore (Nakano et al., 2008) (Figure 4). When tetracycline repressor fusion proteins were used to direct various chromatin modifiers into the centromeric chromatin, this revealed that excessive heterochromatin or excessive transcriptional activity within the centromere are incompatible with kinetochore assembly and propagation (Nakano et al., 2008, Cardinale et al., 2009, Bergmann et al., 2012). Interestingly, a moderate (10×) activation of transcription within the centromere is tolerated (Bergmann et al., 2012) (Figure 4). Consistent with these results, transcription and RNA polymerase have been detected in centromeric sequences, even in mitotic cells (Bergmann et al., 2011, Chan et al., 2012). It thus appears that kinetochore stability at human centromeres requires finely balanced RNA transcription within an otherwise silent chromatin environment.
Figure 4.
Use of Human Artificial Chromosomes to Study Centromeric Chromatin
A synthetic α-satellite DNA construct was used to generate human artificial chromosomes (HACs) in HT1080 fibrosarcoma cells. An example of the HAC (white arrow) stained for kinetochore protein CENP-C (red) and DNA (blue) is shown. The presence of tetracycline operator sequences in the synthetic array allows targeting of chimeric proteins containing a range of chromatin modification activities into the HAC centromere. These studies reveal that centromere activity requires finely balanced transcription in a repressive environment. Micrograph by Jan Bergmann.
Are CENP-A-Containing Nucleosomes Different from Bulk Nucleosomes?
Centromeric chromatin forms a specialized structure in budding (Bloom and Carbon, 1982) and fission yeasts (Takahashi et al., 1992). CENP-A sequences from model organisms have highly variable N-terminal tails (Black and Cleveland, 2011), but in human cells, centromere targeting is directed by the 22 amino acid (aa) CENP-A targeting domain (CATD) located in the histone-fold region (Black et al., 2004). The CATD binds the CENP-A-specific chaperone HJURP (Dunleavy et al., 2009, Foltz et al., 2009). The CENP-A nucleosome core is rigid (Black et al., 2004), but overall the DNA wraps less tightly in both natural and neocentromeres than in conventional nucleosomes (Hasson et al., 2013). These differences suggest that CENP-A nucleosomes are distinct from canonical H3 nucleosomes. Indeed, conflicting models have been proposed for the structure of CENP-A-containing nucleosomes (Black and Cleveland, 2011), and a spirited ongoing controversy concerns whether they are octameric or tetrameric.
Octameric CENP-A nucleosomes can be reconstituted with recombinant histones (Yoda et al., 2000, Sekulic et al., 2010) and CENP-A-containing nucleosomes purified from human cells contained stoichiometric CENP-A, H4, H2A, and H2B, with two CENP-A molecules per nucleosome (Shelby et al., 1997, Foltz et al., 2006). X-ray crystallography revealed that the structure of reconstituted CENP-A nucleosomes resembles canonical nucleosomes with subtle differences (Tachiwana et al., 2011).
In contrast, Henikoff and Dalal argued that CENP-A nucleosomes form a tetrameric hemisome containing a single copy of CENP-A, H4, H2A, and H2B in Drosophila cells (Dalal et al., 2007). They also proposed that DNA wraps around CENP-A hemisomes with a handedness opposite to that found in canonical nucleosomes (Furuyama and Henikoff, 2009). Using atomic force microscopy (AFM), they suggested that CENP-A nucleosomes are half the height of canonical nucleosomes (Dimitriadis et al., 2010). More recently, studies in human cells (Bui et al., 2012) and budding yeast (Shivaraju et al., 2012) proposed that the CENP-A-containing nucleosomes are dynamic, oscillating between octameric and tetrameric forms during cell-cycle progression.
Although this debate is still active, recent crosslinking (Zhang et al., 2012), photobleaching (Padeganeh et al., 2013), and AFM experiments (Miell et al., 2013) suggest that most CENP-A-containing nucleosomes are octameric, with a more rigid core than canonical nucleosomes (Black et al., 2007). A recent study using fully functional Cse4 with an internal tdEos tag after Leu81 in the N-terminal region showed clearly that budding yeast kinetochores have two copies of Cse4 in a single nucleosome (Wisniewski et al., 2014). The authors also demonstrated that studies in which Cse4 copy number appeared to vary across the cell cycle can be explained by delays in the activation of fluorescent proteins.
This controversy appears to have a life of its own, and studies using isolated CENP-A nucleosomes could be confounded by the fact that many CENP-A nucleosomes are noncentromeric. However, although there is still no absolute consensus, most emerging data appear to support the existence of octameric CENP-A nucleosomes in vivo.
Even if CENP-A-containing nucleosomes are octameric, some of these could be heterotypic, with both CENP-A and H3. CENP-A/H3.3 heterotypic nucleosomes can indeed form following CENP-A overexpression, but this is mostly in noncentromere regions (Lacoste et al., 2014). Homotypic CENP-A/CENP-A nucleosomes predominate at centromeres (Shelby et al., 1997, Hori et al., 2014, Lacoste et al., 2014).
How Are CENP-A Nucleosomes Distributed in Centromeres?
A critical question is how many CENP-A nucleosomes are required to define a kinetochore. Immunofluorescence on extended chromatin fibers (Blower et al., 2002), superresolution microscopy (Ribeiro et al., 2010), and biochemical analyses (Blower et al., 2002, Hori et al., 2008) all suggest that centromeres consist of islets of CENP-A nucleosomes interspersed between regions containing H3 nucleosomes (Figure 5). In S. cerevisiae, elegant biochemical and quantitative imaging experiments detected a single Cse4/CENP-A nucleosome in the point centromere (Furuyama and Biggins, 2007, Joglekar et al., 2008). Other microscopy experiments suggest that other budding yeast strains might have up to three CENP-A nucleosomes per centromere (Lawrimore et al., 2011). However, these are technically deceptively complex experiments (Wisniewski et al., 2014).
Figure 5.
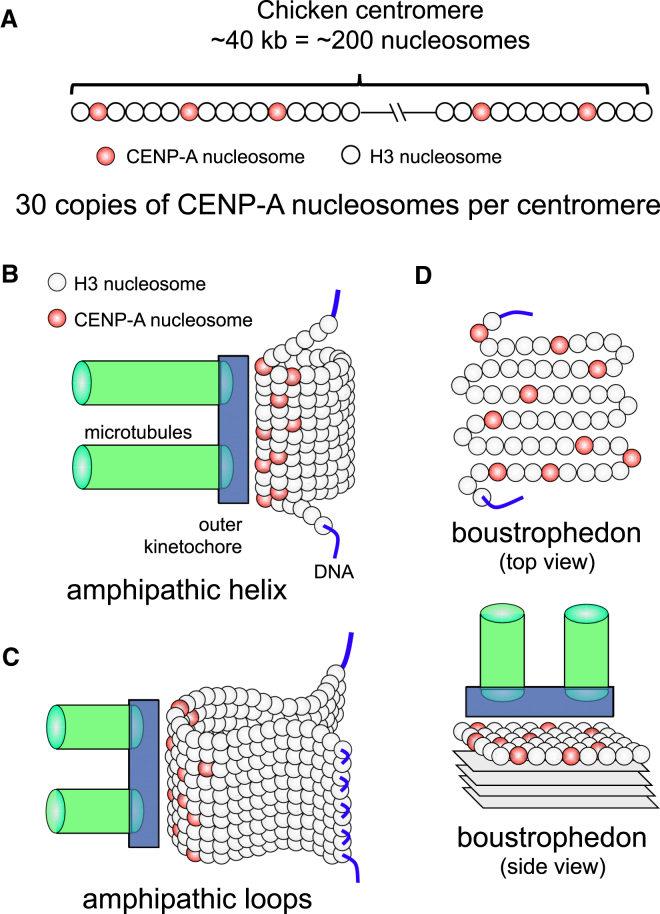
Organization of CENP-A and H3 Nucleosomes in Centromeres
(A) Based on ChIP-seq analysis centromeres are ∼40 kb long in chicken, corresponding to 200 nucleosomes per centromere. Of these, 30 are predicted to contain CENP-A (roughly 1 in 6–8 centromeric nucleosomes). Thus, centromeric chromatin is largely composed of nucleosomes containing histone H3.
(B and C) The CENP-A chromatin was originally suggested to form an amphipathic organization, with CENP-A on the exterior facing the kinetochore, and H3 largely on the interior. This chromatin was proposed to form either a helix or loop structure. The diagram in (B and C) is based on Blower et al. (2002), but modified to show the lower occupancy of CENP-A nucleosomes in the centromeric chromatin.
(D) The boustrophedon model of centromeric CENP-A-containing chromatin was proposed based on super-resolution microscopy (Ribeiro et al., 2010).
Using S. cerevisiae as a standard (assuming a single CENP-A nucleosome per centromere), S. pombe centromeres were estimated to have 3 CENP-A nucleosomes (Joglekar et al., 2008) and chicken DT40 cells to have 30 (Johnston et al., 2010). Because CENP-A associated regions are ∼5 kb (∼25 nucleosomes) in S. pombe and ∼40 kb (∼200 nucleosomes) in chicken, and assuming 200 bp per nucleosome, this suggests that one in six to eight centromeric nucleosomes contains CENP-A (Figure 5). Recent studies report larger numbers of CENP-A nucleosomes in S. pombe (Coffman et al., 2011, Lando et al., 2012). A typical human neocentromere of 80–100 kb (400–500 nucleosomes) should contain ∼100 CENP-A nucleosomes (200 copies of CENP-A) per centromere (Figure 5). This is in remarkable agreement with the recent conclusion that human RPE1 cells have ∼100 CENP-A nucleosomes per mitotic kinetochore (Bodor et al., 2014). Considering copy-number estimates for other kinetochore proteins and numbers of microtubules (10–20 per kinetochore), this value of 30–100 CENP-A nucleosomes per vertebrate kinetochore appears reasonable (Figure 5).
Folding of the CENP-A Chromatin Fiber at Centromeres
Analysis of extended centromeric chromatin fibers revealed that CENP-A forms clusters that alternate with chromatin containing canonical histone H3 (Blower et al., 2002). The H3 nucleosomes carry the transcription-associated modification H3K4me2 and define a specialized class of chromatin termed “centrochromatin” (Sullivan and Karpen, 2004). It was proposed that CENP-A was packaged either into an amphipathic-like solenoidal superhelix or as radial loops, with CENP-A nucleosomes clustered on the outer surface of the chromatin and H3 internal (Blower et al., 2002) (Figure 5).
Two types of super-resolution microscopy combined to challenge this interpretation. High-resolution dual-label microscopy showed that CENP-T, an important linker between the chromatin and the outer kinetochore (Hori et al., 2008, Suzuki et al., 2011), was located significantly outside of CENP-A (Joglekar et al., 2009, Wan et al., 2009, Varma et al., 2013). PALM microscopy of unfolded fibers derived from chicken kinetochores found CENP-T in regions of H3 nucleosomes (Ribeiro et al., 2010). In the amphipathic helix-loop model, this would place CENP-T on the inside, in direct contradiction with the dual-label microscopy mapping measurements.
To account for the superresolution mapping data, it was suggested that CENP-A chromatin might be organized as a sinusoidally folded patch, or boustrophedon, at the surface of the centromeric chromatin (Ribeiro et al., 2010). The boustrophedon was proposed to be 4–5 layers deep (Figure 5), consistent with the 10 nm diameter of the nucleosome and the ∼60 nm thickness of the kinetochore plate typically observed in electron micrographs (Rieder, 1982). Thus, the architecture of CENP-A chromatin and the boustrophedon model remains questions in need of further experimentation.
The Paradoxical Timing of CENP-A Incorporation into Centromeres
New CENP-A is incorporated into the centromere during early G1 phase in vertebrate cells, following the drop in mitosis-associated CDK activity (Jansen et al., 2007, Silva et al., 2012). This was unexpected, because most canonical H3 is incorporated into chromatin during DNA replication. Furthermore, it means that cells traverse mitosis with only half of their maximal complement of CENP-A. This is not what one might have predicted given that centromeres perform their most important functions during mitosis.
Budding yeast Cse4 (CENP-A) is incorporated into centromeres during S-phase coupled with DNA replication and following the complete removal of preexisting Cse4 (Pearson et al., 2004, Wisniewski et al., 2014). Thus CENP-A incorporation timing differs between yeast and vertebrates. In the latter, histone H3.3 incorporated into centromeres during S-phase might function as a placeholder for new CENP-A deposition in the next G1 (Dunleavy et al., 2011).
Genetic analysis in S. pombe identified Mis16 and Mis18 as factors involved in CENP-A localization (Hayashi et al., 2004). Mis16 is the S. pombe homolog of vertebrate pRab46/48, which is involved in histone H3 incorporation into chromatin. In vertebrates, the two isoforms Mis18α/β form a complex with M18BP1 (Fujita et al., 2007), also known as KNL-2 (Maddox et al., 2007). Depletion of pRab46/48 or Mis18α/β in human cells reduces CENP-A incorporation into centromeres (Hayashi et al., 2004, Fujita et al., 2007), suggesting a conserved role for these proteins.
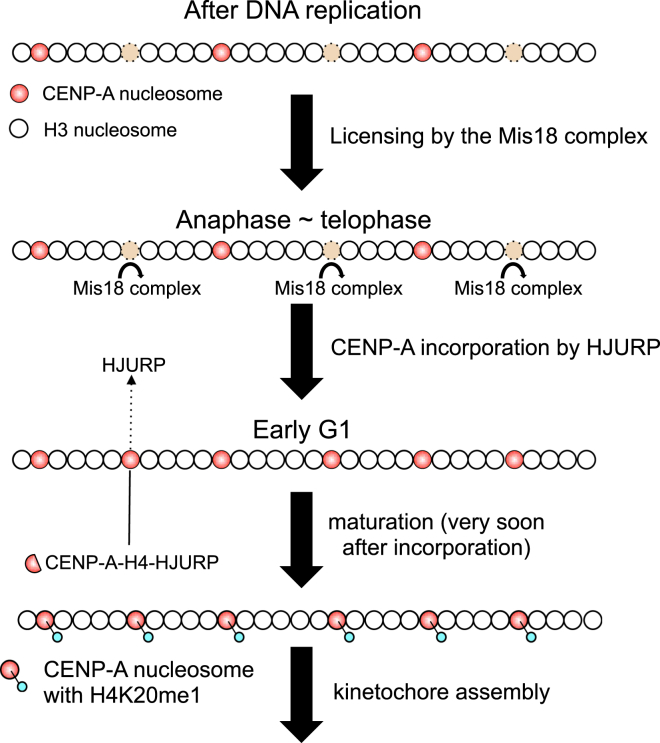
The timing of CENP-A incorporation is regulated by Mis18α/β, which is recruited to centromeres at least in part via Mis18BP1 binding to CENP-C (Moree et al., 2011, Dambacher et al., 2012, McKinley and Cheeseman, 2014). Mis18α/β starts to localize to centromeres during anaphase and remains there during telophase. Its levels fall dramatically in G1 phase (Hayashi et al., 2004, Fujita et al., 2007). Mis18α/β thus appears to function as a licensing factor for CENP-A incorporation (Figure 6).
Figure 6.
The Mis18 Complex Licenses Chromatin for CENP-A Incorporation
During replication, CENP-A nucleosomes are divided between the two daughter DNA molecules and are either replaced with H3 (H3.1/H3.3) nucleosomes or leave nucleosome free gaps. During anaphase to telophase, the Mis18 complex localizes to centromeres. Soluble CENP-A-H4 binds to the chaperone HJURP before incorporation, and during early G1 the CENP-A-H4-HJUPRP complex is recruited into the licensed chromatin by binding to the Mis18 complex. Following CENP-A incorporation, H4K20 in CENP-A nucleosomes is monomethylated. This maturation step is required for certain of the constitutive centromere associated network (CCAN) proteins to assemble and subsequently direct assembly of a functional kinetochore.
The Mis18 complex must be phosphorylated by Polo-like kinase 1 (Plk1) during G1 to facilitate CENP-A incorporation (McKinley and Cheeseman, 2014). Because CDK1 activity negatively regulates Mis18 complex-derived CENP-A incorporation (Silva et al., 2012), CENP-A incorporation might be controlled by a two-step regulatory mechanism governed by Plk1 and CDK1.
The Mis18 complex does not directly bind to soluble CENP-A. Instead a CENP-A-specific chaperone, HJURP, binds soluble CENP-A-H4 complex before its chromatin incorporation during G1 phase (Dunleavy et al., 2009, Foltz et al., 2009) (Figure 6). The yeast and Drosophila homologs of HJURP are apparently Scm3 (Mizuguchi et al., 2007, Pidoux et al., 2009, Sanchez-Pulido et al., 2009, Williams et al., 2009) and Cal1 (Erhardt et al., 2008), respectively. Scm3 is found in hexameric Cse4 nucleosomes (Mizuguchi et al., 2007), which appear to be assembly intermediates (Camahort et al., 2007). The mechanism of CENP-A incorporation is thus widely conserved. Current data suggest that the Mis18 complex licenses centromeric chromatin by binding and recruiting a complex of HJURP-CENP-A-H4 (Figure 6).
The timing of CENP-A incorporation is critical for kinetochore assembly and function. If CENP-A incorporation is artificially deregulated by constitutive targeting of Mis18α to centromeres (this causes constitutive insertion of CENP-A across the cell cycle), then mitotic kinetochore function is strongly disrupted (McKinley and Cheeseman, 2014). It is not known why unscheduled incorporation of CENP-A has such a strong disruptive effect.
How Does CENP-A Chromatin Become Competent for Kinetochore Assembly?
With the exception of Trypanosomids (Akiyoshi and Gull, 2014), CENP-A containing chromatin is a near universal feature of centromere specification. Yet with few examples (Guse et al., 2011, Mendiburo et al., 2011), targeting of ectopic CENP-A is not sufficient to trigger centromere formation (Van Hooser et al., 2001, Gascoigne et al., 2011). Indeed, because most chromosomal CENP-A is incorporated at ectopic sites that lack centromere activity (Shang et al., 2013, Bodor et al., 2014), this begs the question of how cells distinguish centromeric CENP-A from ectopic CENP-A. It is possible that additional modifications might “license” CENP-A containing chromatin, making it competent for kinetochore assembly. A recent study revealed that histone H4 Lys20 monomethylation (H4K20me1) specifically occurs at centromeric CENP-A chromatin (Hori et al., 2014). Because reducing H4K20me1 levels at centromeres causes mislocalization of CENP-H and CENP-T, this modification might help render CENP-A chromatin competent for kinetochore assembly (Figure 6). Other histone modifications of CENP-A nucleosomes or of centromeric nucleosomes containing canonical H3 might also be involved in formation of functional centromeric chromatin, possibly by influencing interactions with other kinetochore proteins such as CENP-C or CENP-N, which bind to CENP-A nucleosomes concentrated in centromeres (Carroll et al., 2009, Guse et al., 2011, Kato et al., 2013).
Understanding the mechanisms by which CENP-A chromatin induces subsequent kinetochore assembly remains an important area for future studies.
Centromeric Chromatin Contributes to Sister Chromatid Cohesion
In addition to serving as a mark for kinetochore assembly, centromeric chromatin also contributes to chromosome segregation by binding cohesin and the chromosomal passenger complex (CPC) of Aurora B kinase, INCENP, Survivin, and Borealin (Carmena et al., 2012).
Cohesin regulates the cohesion of sister chromatids so that kinetochores can orient to opposite spindle poles and chromosomes can segregate equally during mitosis (Nasmyth and Haering, 2009). Pericentromeric heterochromatin in S. pombe recruits cohesin (Nonaka et al., 2002), and the link between heterochromatin and cohesin is conserved in other organisms (Gartenberg, 2009). Cohesin is also concentrated in the pericentromere in budding yeast where there is no canonical heterochromatin (Kiburz et al., 2005). Cohesion at centromeres is protected by proteins of the shugoshin family (Watanabe, 2005), which have recently been observed to also function in recruitment of the CPC (Gutiérrez-Caballero et al., 2012).
Shugoshin (Sgo1) forms part of a two-part mechanism for recruiting the CPC to centromeric heterochromatin. First, the checkpoint kinase Bub1 phosphorylates histone H2A at threonine 120 (H2AT120ph) in inner centromeres. Sgo1 binds to this histone mark and then recruits the CPC via an interaction with Borealin (Kawashima et al., 2007). Second, Haspin kinase phosphorylation of histone H3 threonine 3 (H3T3ph). Survivin binding to this histone mark is required for CPC targeting to centromeres (Kelly et al., 2010, Wang et al., 2010, Yamagishi et al., 2010). At centromeres, the CPC regulates kinetochore-microtubule interactions by phosphorylation of several kinetochore proteins and regulates spindle checkpoint signaling to delay mitotic progression if there are incorrect kinetochore-microtubule attachments (Carmena et al., 2012).
Histone-fold Proteins that Bridge between the Centromere and Kinetochore
CENP-A and H3 nucleosomes are not the only proteins that directly bind to centromeric DNA and contribute to the formation of centromere-specific chromatin. Experiments aimed at isolating unknown centromere proteins identified CENP-T, CENP-W, CENP-S, and CENP-X, all of which directly bind to centromeric DNA (Okada et al., 2006, Hori et al., 2008, Amano et al., 2009). These proteins have histone folds and make a tetrameric CENP-T-W-S-X complex (Nishino et al., 2012). Because this complex can induce supercoils into DNA and its DNA binding surface resembles that of canonical nucleosomes, it might form a nucleosome-like structure at centromeres (Nishino et al., 2012).
Interestingly, the CENP-T-W-S-X complex induces positive supercoils into DNA (Takeuchi et al., 2014), whereas canonical histones induce negative supercoils. Analysis of yeast mini-chromosomes suggested that there are positive supercoils in centromere chromatin (Furuyama and Henikoff, 2009). Thus, in addition to CENP-A nucleosomes, the CENP-T-W-S-X nucleosome-like complex might contribute to formation of centromere-specific chromatin (Figure 7) (Takeuchi et al., 2014).
Figure 7.
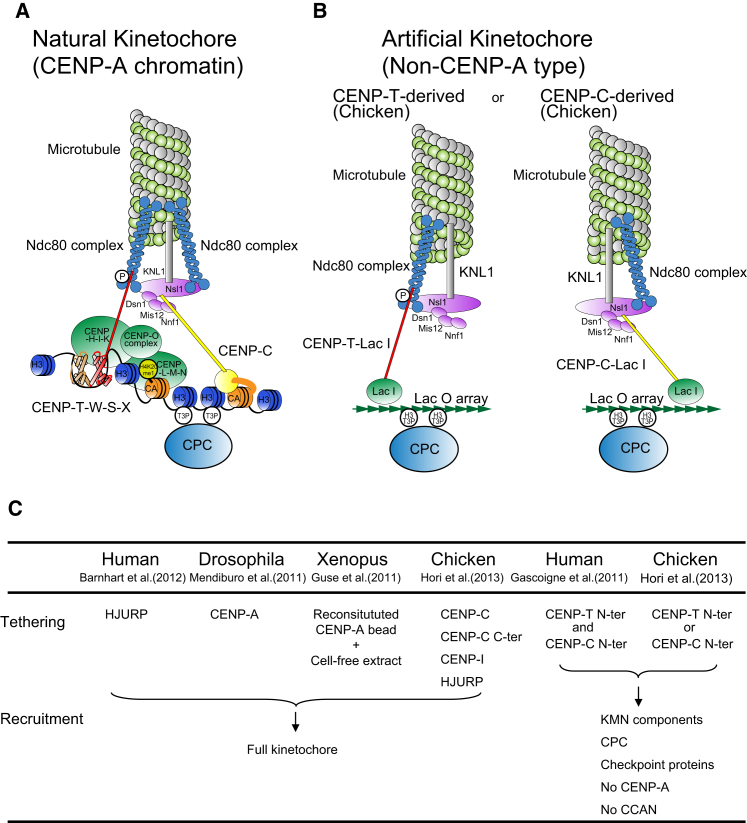
Molecular Architecture of Kinetochores
(A) Molecular architecture of natural kinetochores. Centromere chromatin is crucial for centromere specification and kinetochore assembly at natural centromeres. At the base of the structure are CENP-A containing nucleosomes, centromere specific H3 nucleosomes, and a CENP-T-W-S-X nucleosome-like structure in centromere chromatin. Centromere-specific chromatin structure is established by coordination of these components. CCAN proteins assemble on the centromeric chromatin and the microtubule-binding complex is subsequently recruited to assemble the functional kinetochore.
(B) When the CENP-T N terminus or CENP-C N terminus is tethered at a noncentromere locus using the LacI-LacO system, an artificial kinetochore forms on the noncentromeric LacO site. Most centromeric chromatin proteins, including CENP-A, are not detected in the artificial kinetochores. However, the chromosome passenger complex (CPC) and Ndc80 complex are recruited and the artificial kinetochores are fully functional.
(C) Summary of studies on creation of artificial kinetochores. Tethering of CENP-T N terminus and/or CENP-C-N terminus can bypass the need for centromere-specific chromatin including CENP-A. CENP-A-mediated artificial kinetochores also have been created in human, chicken, and Drosophila cells and in Xenopus egg extracts.
Although CENP-T-W-S-X tetramers are critical for centromere function, chromosome segregation still occurs in CENP-S- or CENP-X-deficient cells (Amano et al., 2009). This suggests that CENP-T-W and CENP-S-X can function independently. Indeed, CENP-S-X binds FANCM proteins at DNA damage sites (Singh et al., 2010, Yan et al., 2010). Further studies are needed to clarify how CENP-T-W, CENP-S-X, and CENP-T-W-S-X bind to DNA and how the tetramer contributes to formation of centromeric chromatin.
Artificial Kinetochores Bypass the Need for Centromere-Specific Chromatin
One key function of the centromere is to regulate chromosomal interactions with microtubules. This interaction requires the Ndc80 complex in the kinetochore (Cheeseman et al., 2006, DeLuca et al., 2006). This suggested that if the Ndc80 complex could be artificially localized to an ectopic chromosomal locus, then that locus might function as an artificial kinetochore and direct chromosome segregation.
The CENP-T N-terminal region directly binds to the Ndc80 complex (Gascoigne et al., 2011, Nishino et al., 2013), while its C terminus contacts the centromeric DNA (Nishino et al., 2012). Remarkably, tethering the CENP-T N terminus at a noncentromeric locus using the LacI-LacO system resulted in creation of an artificial kinetochore that efficiently directed chromosome segregation following deletion of the natural centromere of the corresponding chromosome in chicken DT40 cells (Gascoigne et al., 2011, Hori et al., 2013). A second artificial kinetochore was also constructed by similar tethering of CENP-C to a Lac operator array (Hori et al., 2013). CENP-C recruits the Mis12 complex, which binds the Ndc80 complex (Gascoigne et al., 2011, Przewloka et al., 2011, Screpanti et al., 2011; Figure 7). Surprisingly, many centromere proteins, including CENP-A, were not detected in either artificial kinetochore (Hori et al., 2013). Together, these experiments reveal that recruiting the Ndc80 complex to centromeres is a major function of centromere chromatin. Thus, artificial recruitment of the complex can bypass the need for centromere-specific chromatin structure during kinetochore formation.
CENP-A-mediated artificial kinetochores also have been created in human, chicken, and Drosophila cells (Figure 7) (Barnhart et al., 2011, Mendiburo et al., 2011, Hori et al., 2013). In another approach, several aspects of kinetochore function were reconstituted in vitro on CENP-A nucleosome-coated beads using Xenopus egg extracts (Guse et al., 2011).
Artificial kinetochores are promising tools for kinetochore studies and genetic engineering. It will therefore be important to determine the efficiency with which they are able to direct chromosome segregation in animals and their ability to cope with attachment errors, which are a natural hazard of mitotic chromosome segregation.
Perspectives
Even though centromeric chromatin can form both on specialized (often repetitive) DNA sequences and on other sequences that are not normally centromeric, its structure and composition are distinct from that of other chromatin regions. Centromeric chromatin consists of a relatively small number of CENP-A-containing nucleosomes distributed among centromere-specific H3 nucleosomes together with additional specialized DNA-binding proteins including the CENP-T-W-S-X complex (Figure 7). The centromere-specific chromatin structure is established by coordination of these factors with modification of the CENP-A nucleosomes, and together this lays the essential foundation for functional kinetochore assembly. Throughout this review, we have stressed that kinetochore function is distinct from centromeric chromatin. However, because kinetochores assemble on the surface of centromeres, it can be difficult to separate the functions of the two. By bypassing centromere function, artificial kinetochores might provide an excellent tool to tackle this issue.
Acknowledgments
The authors thank members of their labs for useful discussions and Iain Cheeseman and Beth Sullivan for critical reading of the manuscript. Work in the Fukagawa Laboratory was supported by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to T.F. Work in the Earnshaw Laboratory was supported by the Wellcome Trust (grant number 073915).
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/).
Contributor Information
Tatsuo Fukagawa, Email: tfukagaw@lab.nig.ac.jp.
William C. Earnshaw, Email: bill.earnshaw@ed.ac.uk.
References
- Akiyoshi B., Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156:1247–1258. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrup-Macdonald M.E., Sullivan B.A. The past, present, and future of human centromere genomics. Genes (Basel) 2014;5:33–50. doi: 10.3390/genes5010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Hasson D., Cheung F., Warburton P.E. A paucity of heterochromatin at functional human neocentromeres. Epigenetics & Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M., Suzuki A., Hori T., Backer C., Okawa K., Cheeseman I.M., Fukagawa T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M., Ngan V.K., Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol. Biol. Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M., Sanyal K., Mishra P.K., Thaler N., Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. USA. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G.W. A Possible Influence of the Spindle Fibre on Crossing-Over in Drosophila. Proc. Natl. Acad. Sci. USA. 1932;18:160–165. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J.H., Rodríguez M.G., Martins N.M., Kimura H., Kelly D.A., Masumoto H., Larionov V., Jansen L.E., Earnshaw W.C. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann J.H., Jakubsche J.N., Martins N.M., Kagansky A., Nakano M., Kimura H., Kelly D.A., Turner B.M., Masumoto H., Larionov V., Earnshaw W.C. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Cleveland D.W. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Foltz D.R., Chakravarthy S., Luger K., Woods V.L., Jr., Cleveland D.W. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Black B.E., Brock M.A., Bédard S., Woods V.L., Jr., Cleveland D.W. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K.S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Blower M.D., Sullivan B.A., Karpen G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., Jansen L.E. The quantitative architecture of centromeric chromatin. eLife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley B.R., Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- Bui M., Dimitriadis E.K., Hoischen C., An E., Quénet D., Giebe S., Nita-Lazar A., Diekmann S., Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J.L., Gerton J.L. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale S., Bergmann J.H., Kelly D., Nakano M., Valdivia M.M., Kimura H., Masumoto H., Larionov V., Earnshaw W.C. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol. Biol. Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Wheelock M., Funabiki H., Earnshaw W.C. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., Straight A.F. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centola M., Carbon J. Cloning and characterization of centromeric DNA from Neurospora crassa. Mol. Cell. Biol. 1994;14:1510–1519. doi: 10.1128/mcb.14.2.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F.L., Marshall O.J., Saffery R., Kim B.W., Earle E., Choo K.H., Wong L.H. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. USA. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chen E.S., Zhang K., Nicolas E., Cam H.P., Zofall M., Grewal S.I. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- Coffman V.C., Wu P., Parthun M.R., Wu J.Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C.A., Bazett-Jones D.P., Earnshaw W.C., Rattner J.B. Mapping DNA within the mammalian kinetochore. J. Cell Biol. 1993;120:1083–1091. doi: 10.1083/jcb.120.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G.P., Nickel K., Kuromori T., Benito M.I., Kaul S., Lin X., Bevan M., Murphy G., Harris B., Parnell L.D. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- Dalal Y., Furuyama T., Vermaak D., Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S., Deng W., Hahn M., Sadic D., Fröhlich J., Nuber A., Hoischen C., Diekmann S., Leonhardt H., Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington C.D. The external mechanics of chromosomes. Proc. R. Soc. Lond. B Biol. Sci. 1936;121:264–319. [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Dimitriadis E.K., Weber C., Gill R.K., Diekmann S., Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Almouzni G., Karpen G.H. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Migeon B.R. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Sullivan K.F., Machlin P.S., Cooke C.A., Kaiser D.A., Pollard T.D., Rothfield N.F., Cleveland D.W. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Ratrie H., 3rd, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Allshire R.C., Black B.E., Bloom K., Brinkley B.R., Brown W., Cheeseman I.M., Choo K.H., Copenhaver G.P., Deluca J.G. Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome Res. 2013;21:101–106. doi: 10.1007/s10577-013-9347-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole T.A., Ross A., Clark E., McGill N., Schindelhauer D., Cooke H., Grimes B. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- Ekwall K. Epigenetic control of centromere behavior. Annu. Rev. Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H.D., Pidoux A.L., Urano T., Allshire R.C. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., 3rd, Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., 3rd, Bassett E.A., Wood S., Black B.E., Cleveland D.W. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Furuyama S., Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T., Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. Heterochromatin and the cohesion of sister chromatids. Chromosome Res. 2009;17:229–238. doi: 10.1007/s10577-008-9012-z. [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Rechtsteiner A., Yuen K.W., Muroyama A., Egelhofer T., Gaydos L., Barron F., Maddox P., Essex A., Monen J. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484:534–537. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A., Carroll C.W., Moree B., Fuller C.J., Straight A.F. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Caballero C., Cebollero L.R., Pendás A.M. Shugoshins: from protectors of cohesion to versatile adaptors at the centromere. Trends Genet. 2012;28:351–360. doi: 10.1016/j.tig.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Hasson D., Panchenko T., Salimian K.J., Salman M.U., Sekulic N., Alonso A., Warburton P.E., Black B.E. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hegemann J.H., Fleig U.N. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Toyoda A., Misu S., Monma N., Ikeo K., Molina O., Vargiu G., Fujiyama A., Kimura H. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev. Cell. 2014;29:740–749. doi: 10.1016/j.devcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D.F., Fowler K.J., Earle E., Saffery R., Kalitsis P., Trowell H., Hill J., Wreford N.G., de Kretser D.M., Cancilla M.R. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes-Schrader S., Ris H. The diffuse spindle attachment of coccids, verified by the mitotic behavior of induced chromosome fragments. J. Exp. Zool. 1941;87:429–456. [Google Scholar]
- Ikeno M., Grimes B., Okazaki T., Nakano M., Saitoh K., Hoshino H., McGill N.I., Cooke H., Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., Kakuma T., Hiraoka Y., Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K., Joglekar A., Hori T., Suzuki A., Fukagawa T., Salmon E.D. Vertebrate kinetochore protein architecture: protein copy number. J. Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokelainen P.T. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J. Ultrastruct. Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- Kapoor M., Montes de Oca Luna R., Liu G., Lozano G., Cummings C., Mancini M., Ouspenski I., Brinkley B.R., May G.S. The cenpB gene is not essential in mice. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S.A., Tsukahara T., Langegger M., Hauf S., Kitajima T.S., Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C., Wang H.S., McClellan M., Bouchonville K., Selmecki A., Lahav T., Gerami-Nejad M., Berman J. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiburz B.M., Reynolds D.B., Megee P.C., Marston A.L., Lee B.H., Lee T.I., Levine S.S., Young R.A., Amon A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 2005;19:3017–3030. doi: 10.1101/gad.1373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa T., Ishii K., Takeda K., Matsumoto T. The 19S proteasome subunit Rpt3 regulates distribution of CENP-A by associating with centromeric chromatin. Nature Communications. 2014;5:3597. doi: 10.1038/ncomms4597. [DOI] [PubMed] [Google Scholar]
- Kouprina N., Earnshaw W.C., Masumoto H., Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell. Mol. Life Sci. 2013;70:1135–1148. doi: 10.1007/s00018-012-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., Almouzni G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Lando D., Endesfelder U., Berger H., Subramanian L., Dunne P.D., McColl J., Klenerman D., Carr A.M., Sauer M., Allshire R.C. Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast. Open Biology. 2012;2:120078. doi: 10.1098/rsob.120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J., Bloom K.S., Salmon E.D. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois P., Euskirchen G.M., Auerbach R.K., Rozowsky J., Gibson T., Yellman C.M., Gerstein M., Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois P., Auerbach R.K., Yellman C.M., Roeder G.S., Snyder M. Centromere-like regions in the budding yeast genome. PLoS Genet. 2013;9:e1003209. doi: 10.1371/journal.pgen.1003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke D.P., Hillier L.W., Warren W.C., Worley K.C., Nazareth L.V., Muzny D.M., Yang S.P., Wang Z., Chinwalla A.T., Minx P. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luykx P. The structure of the kinetochore in meiosis and mitosis in Urechis eggs. Exp. Cell Res. 1965;39:643–657. doi: 10.1016/0014-4827(65)90068-6. [DOI] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., DeLuca J., Salmon E.D., Earnshaw W.C. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma. 1978;66:23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R.D. The mechanism of G and C banding in mammalian metaphase chromosomes. Chromosoma. 1973;44:1–14. doi: 10.1007/BF00372569. [DOI] [PubMed] [Google Scholar]
- McKinley K.L., Cheeseman I.M. Polo-like Kinase 1 Licenses CENP-A Deposition at Centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo M.J., Padeken J., Fülöp S., Schepers A., Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Miell M.D., Fuller C.J., Guse A., Barysz H.M., Downes A., Owen-Hughes T., Rappsilber J., Straight A.F., Allshire R.C. CENP-A confers a reduction in height on octameric nucleosomes. Nat. Struct. Mol. Biol. 2013;20:763–765. doi: 10.1038/nsmb.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., Kent W.J. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E., Allory Y., Worman H.J., Courvalin J.C., Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moree B., Meyer C.B., Fuller C.J., Straight A.F. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi Y., Peebles C., Fritzler M.J., Steigerwald J., Tan E.M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.D., Karpen G.H. Localization of centromere function in a Drosophila minichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W., Szostak J.W. Construction of artificial chromosomes in yeast. Nature. 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Nakano M., Cardinale S., Noskov V.N., Gassmann R., Vagnarelli P., Kandels-Lewis S., Larionov V., Earnshaw W.C., Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima H., Nakano M., Ohnishi R., Hiraoka Y., Kaneda Y., Sugino A., Masumoto H. Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. J. Cell Sci. 2005;118:5885–5898. doi: 10.1242/jcs.02702. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Nasuda S., Hudakova S., Schubert I., Houben A., Endo T.R. Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Nusbaum C., Mikkelsen T.S., Zody M.C., Asakawa S., Taudien S., Garber M., Kodira C.D., Schueler M.G., Shimizu A., Whittaker C.A. DNA sequence and analysis of human chromosome 8. Nature. 2006;439:331–335. doi: 10.1038/nature04406. [DOI] [PubMed] [Google Scholar]
- Ohzeki J., Nakano M., Okada T., Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J., Bergmann J.H., Kouprina N., Noskov V.N., Nakano M., Kimura H., Earnshaw W.C., Larionov V., Masumoto H. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., 3rd, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Okada T., Ohzeki J., Nakano M., Yoda K., Brinkley W.R., Larionov V., Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Padeganeh A., Ryan J., Boisvert J., Ladouceur A.M., Dorn J.F., Maddox P.S. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr. Biol. 2013;23:764–769. doi: 10.1016/j.cub.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Palmer D.K., O’Day K., Wener M.H., Andrews B.S., Margolis R.L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M.L., Gall J.G. Chromosomal localization of mouse satellite DNA. Science. 1970;168:1356–1358. doi: 10.1126/science.168.3937.1356. [DOI] [PubMed] [Google Scholar]
- Pearson C.G., Yeh E., Gardner M., Odde D., Salmon E.D., Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Perez-Castro A.V., Shamanski F.L., Meneses J.J., Lovato T.L., Vogel K.G., Moyzis R.K., Pedersen R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 1998;201:135–143. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X., Allshire R.C. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A.F., Mackay A.M., Ainsztein A.M., Goldberg I.G., Earnshaw W.C. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., Glover D.M. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P., Press M.O., Yi X., Baker R., MacCoss M.J., Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Vagnarelli P., Dong Y., Hori T., McEwen B.F., Fukagawa T., Flors C., Earnshaw W.C. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H., 3rd, Maurer M., Rothfield N.F., Earnshaw W.C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Pidoux A.L., Ponting C.P., Allshire R.C. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K., Baum M., Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader F. Kinetic Regions in Chromosomes. Nature. 1939;143:122. [Google Scholar]
- Schueler M.G., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulic N., Bassett E.A., Rogers D.J., Black B.E. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Toyoda A., Kato J., Popendorf K., Sakakibara Y., Fujiyama A., Fukagawa T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010;20:1219–1228. doi: 10.1101/gr.106245.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Martins N.M., Toyoda A., Misu S., Monma N., Hiratani I., Maeshima K., Ikeo K., Fujiyama A. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell. 2013;24:635–648. doi: 10.1016/j.devcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R.D., Vafa O., Sullivan K.F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M., Unruh J.R., Slaughter B.D., Mattingly M., Berman J., Gerton J.L. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.C., Bodor D.L., Stellfox M.E., Martins N.M., Hochegger H., Foltz D.R., Jansen L.E. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Singh T.R., Saro D., Ali A.M., Zheng X.F., Du C.H., Killen M.W., Sachpatzidis A., Wahengbam K., Pierce A.J., Xiong Y. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner F.A., Henikoff S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife. 2014;3:e02025. doi: 10.7554/eLife.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K.C., Curnick K.E., Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Sullivan B.A., Karpen G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L.L., Boivin C.D., Mravinac B., Song I.Y., Sullivan B.A. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Le H.D., Wahlstrom J.M., Karpen G.H. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Hori T., Nishino T., Usukura J., Miyagi A., Morikawa K., Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J. Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., Tachiwana H., Hori T., Kurumizaka H., Fukagawa T. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 2014;42:1644–1655. doi: 10.1093/nar/gkt1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J., Sanyal K. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 2013;23:638–652. doi: 10.1101/gr.141614.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O., Sullivan K.F. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Van Hooser A.A., Ouspenski I.I., Gregson H.C., Starr D.A., Yen T.J., Goldberg M.L., Yokomori K., Earnshaw W.C., Sullivan K.F., Brinkley B.R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Varma D., Wan X., Cheerambathur D., Gassmann R., Suzuki A., Lawrimore J., Desai A., Salmon E.D. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 2013;202:735–746. doi: 10.1083/jcb.201304197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B., Choo K.H. Human alpha satellite DNA—consensus sequence and conserved regions. Nucleic Acids Res. 1987;15:6751–6752. doi: 10.1093/nar/15.16.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleugel M., Hoogendoorn E., Snel B., Kops G.J. Evolution and function of the mitotic checkpoint. Dev. Cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Voullaire L.E., Slater H.R., Petrovic V., Choo K.H. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am. J. Hum. Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wade C.M., Giulotto E., Sigurdsson S., Zoli M., Gnerre S., Imsland F., Lear T.L., Adelson D.L., Bailey E., Bellone R.R., Broad Institute Genome Sequencing Platform. Broad Institute Whole Genome Assembly Team Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Dai J., Daum J.R., Niedzialkowska E., Banerjee B., Stukenberg P.T., Gorbsky G.J., Higgins J.M. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton P.E., Cooke C.A., Bourassa S., Vafa O., Sullivan B.A., Stetten G., Gimelli G., Warburton D., Tyler-Smith C., Sullivan K.F. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Waye J.S., Willard H.F. Human beta satellite DNA: genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc. Natl. Acad. Sci. USA. 1989;86:6250–6254. doi: 10.1073/pnas.86.16.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.C., Murphy T.D., Goldberg M.L., Karpen G.H. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- Williams J.S., Hayashi T., Yanagida M., Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J., Hajj B., Chen J., Mizuguchi G., Xiao H., Wei D., Dahan M., Wu C. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife. 2014;3:e02203. doi: 10.7554/eLife.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Sakuno T., Shimura M., Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Colmenares S.U., Karpen G.H. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol. Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]