Abstract
Background
In a genome-wide association study of autism, zinc finger protein 804A (ZNF804A) single nucleotide polymorphisms (SNPs) were found to be nominally associated in verbally deficient individuals with autism. Zinc finger protein 804A copy number variations (CNVs) have also been observed in individuals with autism. In addition, ZNF804A is known to be involved in theory of mind (ToM) tasks, and ToM deficits are deemed responsible for the communication and social challenges faced by individuals with autism. We hypothesized that ZNF804A could be a risk gene for autism.
Methods
We examined the genetic association and CNVs of ZNF804A in 841 families in which 1 or more members had autism. We compared the expression of ZNF804A in the postmortem brains of individuals with autism (n = 8) and controls (n = 13). We also assessed in vitro the effect of ZNF804A silencing on the expression of several genes known to be involved in verbal efficiency and social cognition.
Results
We found that rs7603001 was nominally associated with autism (p = 0.018). The association was stronger (p = 0.008) in the families of individuals with autism who were verbally deficient (n = 761 families). We observed ZNF804A CNVs in 7 verbally deficient boys with autism. In ZNF804A knockdown cells, the expression of synaptosomal-associated protein, 25kDa (SNAP25) was reduced compared with controls (p = 0.009). The expression of ZNF804A (p = 0.009) and SNAP25 (p = 0.009) were reduced in the anterior cingulate gyrus (ACG) of individuals with autism. There was a strong positive correlation between the expression of ZNF804A and SNAP25 in the ACG (p < 0.001).
Limitations
Study limitations include our small sample size of postmortem brains.
Conclusion
Our results suggest that ZNF804A could be a potential candidate gene mediating the intermediate phenotypes associated with verbal traits in individuals with autism.
Introduction
Autism is a complex neurodevelopmental disorder characterized by deficiencies in social interaction and communication, and by repetitive and stereotyped behaviours. The abnormalities are usually identified in the early years of childhood. Autism is one of the most heritable neurodevelopmental disorders. According to a recent report, the prevalence of this pervasive developmental disorder has risen to 1 in 88. Owing to the genetic heterogeneity and phenotypic variability of autism, classic genetic studies in search of risk genes have not yielded consistent results.
Although autism has been recognized as a distinct diagnostic entity from schizophrenia, several clinical, biological and genetic overlaps have been observed between these 2 neurodevelopmental disorders. Several psychopathological traits, such as deficits in social interaction and cognition, disruption of emotional processing and sensorimotor gating, and impairments in executive functions, are shared between schizophrenia and autism.1 Other shared features include abnormalities in brain morphology, neurochemical anomalies and epigenetic risk factors.1 Whole-genome studies have provided ample evidence for a genetic overlap between these 2 disorders, suggesting common biological pathways in their pathogenesis.2
A genome-wide association study (GWAS)3 and several other independent studies4–6 have identified zinc finger protein 804A (ZNF804A) as the most compelling candidate gene for schizophrenia. Interestingly, in a GWAS of autism, 5 single nucleotide polymorphisms (SNPs) at the ZNF804A locus were found to be associated (p < 0.001) in verbally deficient individuals with autism (supplementary data of Anney and colleagues, 2010).7 In addition to the GWAS evidence, copy number variation (CNV) and gene disruption have also been observed at the ZNF804A locus (2q32.1) of individuals with autism.8,9
ZNF804A has been found to affect neural activation during theory of mind (ToM; also called mentalizing) tasks.10 Theory of mind is a higher-order form of social cognition representing the ability to infer the mental state of others.11 It is reported to be impaired in individuals with autism12 and schizophrenia13 and is therefore considered as a promising intermediate phenotype for these neurodevelopmental disorders. It is a crucial factor for efficient social interaction.14 The development of linguistic/verbal abilities and ToM are closely intertwined from infancy.15 Linguistic abilities have been reported to influence the development of ToM through children’s exposure to conversing with people about mental states.16 Children with linguistic/verbal impairments have been found to perform poorly in verbally dependent ToM tasks.17 Owing to the presence of a zinc finger domain at its N-terminal end, ZNF804A is deemed to be involved in DNA binding and transcriptional regulation.18
On the basis of the previous GWAS7 linking ZNF804A with verbal deficits in individuals with autism and on the role of ZNF804A in ToM that, in turn, relates to social cognition and verbal skills, we hypothesized that ZNF804A could play a role in predisposing individuals to autism by mediating the intermediate phenotypes associated with verbal traits. We evaluated our hypothesis by conducting a genetic association study of ZNF804A with autism, performing a CNV analysis at the ZNF804A locus, comparing the expression of ZNF804A in the postmortem brains of individuals with autism and healthy controls, and assessing the effect of ZNF804A silencing on the expression of genes previously reported to be involved in verbal efficiency and social cognition.
Methods
This study was approved by the Ethics Committee of Hamamatsu University School of Medicine, Hamamatsu, Japan.
Genetic association study
Samples
We obtained DNA samples from the Autism Genetic Resource Exchange (AGRE; www.agre.org).19 The AGRE has obtained informed consent for the distribution of biological samples to approved researchers. We used DNA samples from 841 families (3211 individuals in total), most of whom were white.
The AGRE website provides pedigree information on each individual along with a diagnosis based on the Autism Diagnostic Interview—Revised (ADI-R).20 In all, 1467 individuals (1178 male; 289 female) had autism diagnosed based on the ADI-R. Families with a nonidiopathic autism flag (e.g., fragile-X, abnormal brain imaging results, dysmorphic features, birth trauma) recorded for any of its members were not included in the study. Based on the ADI-R score on overall level of language (scores of 0–2), which is an indicator of verbal abilities, individuals with autism were grouped into low verbal (Lvrb; score of 0 or 1) and healthy (Hvrb; score of 2) categories. Verbal deficits were recorded for 1222 individuals with autism belonging to 761 families (Lvrb category).
SNP selection
The genomic structure of ZNF804A (positions 185, 171, 338–185, 512, 457 in chromosome 2) is based on the National Center for Biotechnology Innovation B36 human genome assembly (dbSNP b126).
We selected SNPs (MAF > 0.1) from white populations in the International HapMap Project (www.hapmap.org) database. We selected 16 SNPs by aggressive tagging (r2 thresh-old = 0.8) using Haploview version 4.1 (www.broad.mit.edu/mpg/haploview). All the SNPs except rs3731834 (missense mutation in exon 4) were located in the introns (see the Appendix, Fig. S1A, available at jpn.ca).
Genotyping
We genotyped the SNPs using the TaqMan method. We purchased Assay-on-Demand TaqMan SNP genotyping assays from Applied Biosystems (ABI). Genotyping polymerase chain reaction (PCR) was carried out in ABI PRISM 7900HT SDS software (ABI) and analyzed using SDS software version 2.0 (ABI).
Statistical analysis
We performed a power analysis using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/dtdt.html). We used FBAT version 2.0.3 (http://biosun1.harvard.edu/~fbat/fbat.htm) to examine the genetic association of ZNF804A SNPs with autism in a family-based association test under an additive model. We used the FBAT–MM option for the multimarker test. Statistical analyses were carried out separately for the whole set of 841 families (hereafter referred to as “all families”) and for the 761 families with Lvrb children with autism (hereafter referred to as “Lvrb families”).
We estimated pairwise linkage disequilibrium (LD) between SNPs, based on the r2 correlation coefficient, using Haploview. Linkage disequilibrium blocks were defined by the confidence interval algorithm. We examined haplotype association, and the significance was evaluated by permutation testing (100 000 permutations).
Copy number variation at the ZNF804A locus
Copy number variation was examined in the DNA samples of 841 families obtained from AGRE. We analyzed CNV using the TaqMan method in ABI PRISM 7900HT SDS software. The TaqMan CNV assays for ZNF804A (Assay ID: Hs00815147_cn; target CNV ID based on the Database of Genomic Variants: Variation_50357) and for the reference gene (telomerase reverse transcriptase [TERT]) were purchased from ABI. The CNV analysis of ZNF804A and TERT were run simultaneously in a duplex real-time PCR. We analyzed 5 ng of each sample in triplicate according to the manufacturer’s protocol.
We determined the copy number at the ZNF804A locus using CopyCaller software version 2.0 (ABI). The number of copies of the target sequence in each sample was determined by relative quantification using the comparative Ct (ΔΔCt) method, which measures the Ct difference (ΔCt) between target and reference sequences and then compares the ΔCt values of samples to a calibrator sample known to have 2 copies of the target sequence. The copy number of the target is estimated to be 2 times the relative quantity.
ZNF804A silencing
The expression of ZNF804A was found to be low in the commonly used cell lines, such as HEK 293 and SK-N-SH, whereas a robust expression was observed in SH-SY5Y human neuroblastoma cell line (data not shown). We therefore examined the effect of ZNF804A silencing in SH-SY5Y cell lines.
The expression of ZNF804A was knocked down in SH-SY5Y cells by RNA interference (RNAi) using gene-specific small interfering RNAs (siRNAs). Sufficient gene silencing could not be achieved using the routine methods of transfection (Lipofectamine 2000, FuGENE HD, Accell SMARTpool siRNA). Efficient silencing of ZNF804A was achieved by electroporation using the Neon Transfection System (Invitrogen). Electroporation was performed according to the manufacturer’s instructions. Briefly, 2 × 105 cells (5 replicates each for ZNF804A RNAi and negative control RNAi) were suspended in 10 μL electroporation buffer containing either 100 nM ZNF804A siRNA (ID: s40770; Ambion) or 100 nM negative control siRNA (Negative Control #1 siRNA; Ambion) and electroporated (1500 V, 20 ms, 1 pulse) in 10 μL tips. The cells (10 μL electroporated cells in 2 mL medium [Ham’s F12 and Eagle’s minimum essential medium in 1:1 ratio, supplemented with 2 mM glutamine, 1% nonessential amino acids and 15% fetal bovine serum]) were grown (37°C; 5% CO2) in 6-well plates for 72 hours.
Extraction of RNA
We extracted total RNA from SH-SY5Y cells using TRIzol Reagent (Invitrogen) in accordance with the manufacturer’s protocol. The RNA samples were further purified using RNeasy Micro Kit (QIAGEN GmbH); this protocol includes a DNase treatment step. The quantity (absorbance at 260 nm) and quality (ratio of absorbance at 260 nm and 280 nm) of RNA were estimated with a NanoDrop ND-1000 Spectrophotometer (Scrum).
Real-time quantitative PCR
We synthesized complementary DNA (cDNA) from total RNA using the ImProm-II Reverse Transcription System (Promega) following the manufacturer’s protocol for oligo (dT) primer.
We performed quantitative PCR (qPCR) analysis using the TaqMan method in ABI PRISM 7900HT SDS software. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous reference. TaqMan assays for ZNF804A (Hs00290118_s1) and GAPDH (Pre-developed Taq-Man Assay Reagent) were purchased from ABI. Each assay was performed in triplicate. Cycle threshold (Ct) values of the target gene were normalized (ΔCt) to that of GAPDH (ΔCt = target gene Ct – GAPDH Ct). Any alteration in gene expression in the ZNF804A-silenced cells was analyzed by relative quantification (ΔΔCt) against the negative control cells (ΔΔCt = ΔCt of ZNF804A RNAi – ΔCt of negative control). We determined the fold-change in gene expression between the 2 groups of cells by calculating 2−ΔΔCt. Any difference in ZNF804A expression between the 2 groups of cells was evaluated using the t test.
Further, the expression of the following genes, previously reported to be associated with verbal/linguistic abilities and social cognition, was compared between ZNF804A-silenced cells and negative control cells by SYBR Green qPCR: BDNF,21 CNTNAP2,22 DISC1,23 DRD2,24 FOXP2,25 NRG1,26 OXTR,27 SHANK3,28 SNAP25,29 SRPX230 and TCF4.31 We designed qPCR primers (see the Appendix, Table S1) using Primer Express version 2.0 (ABI). The efficiency of these primers ranged between 0.93 and 1.03. The specificity of amplicons was demonstrated by melting curve analysis (single peak at 83–86°C).
We used the QuantiTect SYBR Green PCR kit (QIAGEN) for qPCR assays; each assay was carried out in triplicate. We used GAPDH as the reference gene. The qPCR analysis was performed in ABI PRISM 7900HT SDS software. Any alteration in gene expression between the 2 groups of cells was estimated by the relative quantification method described earlier. We evaluated the difference in gene expression between ZNF804A silenced cells and negative control cells using a t test, and any correlation between the expression of ZNF804A and other genes was examined using the Pearson correlation coefficient.
Western blot confirmation of ZNF804A silencing
The protein expression of ZNF804A and SNAP25 in ZNF804A-silenced SH-SY5Y cells and negative control siRNA-transfected cells were compared using Western blot. The cells were homogenized in radioimmunoprecipitation assay buffer. The total protein in the lysate was quantified using Pierce bicinchoninic acid assay kit (Thermo Scientific). We separated 10 μg of each sample on 10% SDS/ polyacrylamide gel electrophoresis. The separated proteins were electroblotted onto a polyvinylidene fluoride membrane (Millipore), blocked and incubated with the primary antibody at 4°C overnight. The following primary antibodies were used: anti-ZNF804A (Santa Cruz Biotechnology) at 1:200 dilution for the detection of ZNF804A, anti-SNAP25 (Abcam) at 1:500 dilution for the detection of SNAP25 and anti-GAPDH (Abcam) at 1:5000 dilution for the detection of GAPDH, which was used as the loading control. The blots were then washed, incubated with 1:15 000 diluted IRDye-conjugated secondary antibody (Rockland) for 1 hour and washed again. The blots were scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Gene expression in postmortem brain samples
Postmortem brain tissues
Postmortem brain samples from individuals with autism and healthy controls were provided by the Autism Tissue Program (ATP; www.autismtissueprogram.org), National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders (NICHD BTB; http://medschool.umaryland.edu/btbank/) and the Harvard Brain Tissue Resource Center (www.brainbank.mclean.org/). Frozen tissue samples from the anterior cingulate gyrus (ACG), motor cortex (MC) and thalamus were used in the study.
Extraction of RNA
The brain tissues (~75 mg obtained by macrodissection) were homogenized by ultrasonication, and total RNA was extracted using TRIzol Reagent (Invitrogen). We performed RNA purification and quantification as described previously.
Quantitative PCR
We performed cDNA synthesis as described previously. The expression of ZNF804A and synaptosomal-associated protein, 25kDa (SNAP25) were compared in the postmortem brains of individuals with autism and healthy controls. We performed qPCR analysis using the TaqMan method in ABI PRISM 7900HT SDS software. We used GAPDH as the endogenous reference. The Ct values of the target gene were normalized (ΔCt) to that of GAPDH. Any alteration in gene expression in the autism group was analyzed by relative quantification (ΔΔCt) against the control group. We determined the fold change in gene expression between the autism and control groups by calculating 2−ΔΔCt.
Statistical analysis
We examined the difference in age, postmortem interval (PMI) and gene expression between the autism and control groups using a t test, and the χ2 test was used to examine the difference in sex distribution between the 2 groups. Any correlation between the expression of ZNF804A and SNAP25 was examined using the Pearson correlation coefficient.
Results
Genetic association study
Power analysis showed that the overall sample size of 841 families provides 91% power to detect an odds ratio of 1.5 for an allele frequency of 0.1 at an α of 0.05.
In the family-based association test (Table 1), rs7603001 located in intron 2 of ZNF804A was nominally associated with autism (z score for risk allele A = 2.362, p = 0.018). When individuals with autism were categorized based on verbal abilities, a stronger association of this SNP was found in the Lvrb families (z score for risk allele A = 2.657, p = 0.008), whereas no association was observed in the Hvrb families (z score = 0, p > 0.99; data not shown). The A allele of rs7603001 was over-transmitted to the individuals with autism (transmission 53% in all families v. 54% in Lvrb families). The genetic association, however, did not withstand multiple testing correction. None of the other SNPs showed any significant association with autism. Genotypic distribution of SNPs were in Hardy–Weinberg equilibrium.
Table 1.
Family-based association test analysis of ZNF804A with autism
| Families† | Frequency | p value‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| SNP | Physical position | Allele* | Location | All§ | Lvrb§ | All§ | Lvrb§ | All§ | Lvrb§ |
| rs13393273 | 185185922 | A | Intron 1 | 591 | 532 | 0.619 | 0.618 | 0.24 | 0.08 |
| G | 0.381 | 0.382 | |||||||
| rs12613195 | 185197466 | C | Intron 1 | 551 | 499 | 0.682 | 0.679 | 0.57 | 0.59 |
| G | 0.318 | 0.321 | |||||||
| rs12693385 | 185215474 | T | Intron 1 | 604 | 548 | 0.520 | 0.518 | 0.60 | 0.40 |
| C | 0.480 | 0.482 | |||||||
| rs990844 | 185227330 | T | Intron 1 | 323 | 287 | 0.867 | 0.869 | 0.24 | 0.10 |
| G | 0.133 | 0.131 | |||||||
| rs7597593 | 185241825 | C | Intron 1 | 617 | 549 | 0.600 | 0.602 | 0.50 | 0.22 |
| T | 0.400 | 0.398 | |||||||
| rs1038197 | 185265516 | A | Intron 1 | 480 | 429 | 0.760 | 0.764 | 0.11 | 0.08 |
| G | 0.240 | 0.236 | |||||||
| rs13026742 | 185313227 | C | Intron 1 | 597 | 536 | 0.579 | 0.579 | 0.18 | 0.25 |
| T | 0.421 | 0.421 | |||||||
| rs1987025 | 185355840 | T | Intron 1 | 479 | 425 | 0.750 | 0.746 | 0.12 | 0.09 |
| A | 0.250 | 0.254 | |||||||
| rs17509608 | 185440823 | C | Intron 2 | 295 | 270 | 0.892 | 0.892 | 0.74 | > 0.99 |
| T | 0.108 | 0.108 | |||||||
| rs7603001 | 185475061 | G | Intron 2 | 596 | 539 | 0.510 | 0.506 | 0.018 | 0.008 |
| A | 0.490 | 0.494 | |||||||
| rs1344706 | 185486673 | T | Intron 2 | 584 | 524 | 0.637 | 0.635 | 0.16 | 0.13 |
| G | 0.363 | 0.365 | |||||||
| rs7593816 | 185490557 | C | Intron 2 | 412 | 375 | 0.809 | 0.806 | 0.59 | 0.45 |
| T | 0.191 | 0.194 | |||||||
| rs3731834 | 185511609 | C | Exon 4 (L/V) | 388 | 349 | 0.830 | 0.833 | 0.29 | 0.48 |
| G | 0.170 | 0.167 | |||||||
| rs10931157 | 185513698 | A | 3′ | 542 | 484 | 0.704 | 0.702 | 0.21 | 0.06 |
| G | 0.296 | 0.298 | |||||||
| rs12693402 | 185516324 | C | 3′ | 396 | 351 | 0.822 | 0.826 | 0.20 | 0.07 |
| T | 0.178 | 0.174 | |||||||
| rs4380187 | 185520185 | A | 3′ | 616 | 554 | 0.570 | 0.567 | 0.50 | 0.46 |
| C | 0.430 | 0.433 | |||||||
L/V = leucine/valine; Lvrb = autistic, low verbal; SNP = single nucleotide polymorphism; ZNF804A = zinc finger protein 804A.
Major allele is listed first.
No. of informative families used.
p < 0.05, additive model.
Whole set of 841 pedigrees; Lvrb: 761 pedigrees.
Three LD blocks were identified in ZNF804A (Table 2; Appendix, Fig. S1B). The haplotype ACTCATC in the second LD block (rs1038197, rs13026742, rs1987025, rs17509608, rs7603001, rs1344706, rs7593816) showed a significant association with autism in the Lvrb families (z score = 3.103, p = 0.004). This haplotype includes the risk allele A of rs7603001. The association remained significant (p = 0.047) following multiple testing correction by permutation analysis (100 000 permutations). Interestingly, the haplotype ACTC-GTC that includes the protective G allele of rs7603001 showed a tendency toward association with autism in the Lvrb families (z score = −1.907, p = 0.05).
Table 2.
Haplotype association analysis of ZNF804A with autism in the low verbal subgroup
| Block; haplotype | Frequency | p value |
|---|---|---|
| Block 1 (SNPs 01–04) | ||
| GCTT | 0.377 | 0.09 |
| AGCT | 0.317 | 0.57 |
| ACCT | 0.16 | 0.06 |
| ACTG | 0.135 | 0.09 |
| Block 2 (SNPs 06–12) | ||
| GTACATC | 0.234 | 0.08 |
| ACTCGGT | 0.193 | 0.69 |
| ACTCGGC | 0.178 | 0.13 |
| ACTCGTC | 0.143 | 0.05 |
| ATTTATC | 0.104 | 0.57 |
| ATTCATC | 0.073 | 0.54 |
| ACTCATC | 0.057 | 0.004 |
| Block 3 (SNPs 14,15) | ||
| AC | 0.531 | 0.73 |
| GC | 0.292 | 0.07 |
| AT | 0.177 | 0.08 |
SNP = single nucleotide polymorphism; ZNF804A = zinc finger protein 804A.
Taken together, the A allele of rs7603001 may be considered as a risk allele and the G allele as a protective allele of autism in individuals with verbal defects.
Copy number variation at the ZNF804A locus
We observed CNV at the ZNF804A locus in the same DNA samples that we used in our genetic association study (Table 3): copy number gain (3 copies) in 6 samples and copy number loss (1 copy) in 2 samples. One of the CNVs (gain) was inherited from the mother, whereas the other CNVs were caused by de novo events. All the CNVs were observed in boys with autism (age 7–16 yr); all but 1 of them belonged to the Lvrb category. We also observed CNVs in 7 maternal samples (gain in 6 and loss in 1 sample) and in 2 paternal samples (gain in 1 and loss in 1 sample).
Table 3.
Copy number variation at ZNF804A locus
| Sample ID* | Sex | Age, yr | Affection status | CNV | Gain/loss | De novo/inherited | Lvrb/Hvrb |
|---|---|---|---|---|---|---|---|
| AU0154302 | Male | 14 | Autism | 3 | Gain | De novo | Lvrb |
| AU023803 | Male | 8 | Autism | 3 | Gain | De novo | Lvrb |
| AU077304 | Male | 16 | Autism | 3 | Gain | De novo | Lvrb |
| AU0871302 | Male | 7 | Autism | 1 | Loss | De novo | Hvrb |
| AU1092302 | Male | 3 | Autism | 3 | Gain | Inherited | Lvrb |
| AU1466302 | Male | 10 | Autism | 1 | Loss | De novo | Lvrb |
| AU1650305 | Male | 7 | Autism | 3 | Gain | De novo | Lvrb |
| AU1655301 | Male | 16 | Autism | 3 | Gain | De novo | Lvrb |
CNV = copy number variation; Hvrb = autistic, healthy; Lvrb: autistic, low verbal; ZNF804A = zinc finger protein 804A.
Autism Genetic Resource Exchange (AGRE) identifier.
ZNF804A silencing
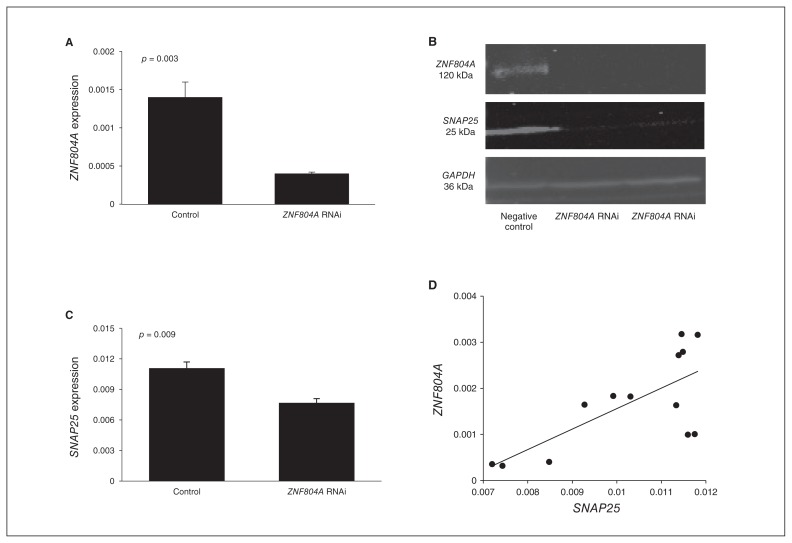
Figure 1A shows a significant difference in the expression of ZNF804A between the cells electroporated with ZNF804A-specific siRNA and the negative control (p = 0.003). In qPCR, the expression of ZNF804A was knocked down by 77%. ZNF804A silencing was confirmed by Western blot (Fig. 1B).
Fig. 1.
Zinc finger protein 804A (ZNF804A) silencing in SH-SY5Y cells. (A) ZNF804A expression was knocked down by 77% (p = 0.003) in the SH-SY5Y cells electroporated with ZNF804A-specific small interfering RNA (siRNA) compared with the negative controls. (B) Comparison of the expression of ZNF804A and SNAP25 between ZNF804A-silenced SH-SY5Y cells and negative control siRNA-transfected SH-SY5Y cells in Western blot. The expression of SNAP25 was downregulated in ZNF804A-silenced cells. GAPDH was used as the loading control. (C) SNAP25 expression was significantly lower in the ZNF804A-silenced cells compared with the negative controls (p = 0.009). (D) Positive correlation between the expression of ZNF804A and SNAP25 in SH-SY5Y cells (Pearson r = 0.713; p = 0.006).
In the ZNF804A-knockdown SH-SY5Y cells, the expression of SNAP25 was significantly reduced compared with the negative controls (p = 0.009; Fig. 1C). This was confirmed by Western blot (Fig. 1B). We also found a significant positive correlation between the expression of ZNF804A and SNAP25 (Pearson r = 0.713, p = 0.006; Fig. 1D). There was no significant alteration in the expression of other genes (data not shown).
Gene expression in postmortem brain
We obtained postmortem brain samples from the ACG (8 autism, 13 control), MC (7 autism, 8 control) and thalamus (8 autism, 9 control). Demographic characteristics of the individuals from whom the samples were obtained are described in Table 4.
Table 4.
Postmortem brain tissue information
| Sample ID* | Diagnosis | Age, yr | Sex | PMI, h | Race | Cause of death | Brain region† |
|---|---|---|---|---|---|---|---|
| 818 | Control | 27 | M | 10 | White | Multiple injuries | ACG |
| 1065 | Control | 15 | M | 12 | White | Multiple injuries | ACG, THL |
| 1297 | Control | 15 | M | 16 | African American | Multiple injuries | ACG, MC, THL |
| 1407 | Control | 9 | F | 20 | African American | Asthma | ACG, MC, THL |
| 1541 | Control | 20 | F | 19 | White | Head injuries | ACG, MC, THL |
| 1649 | Control | 20 | M | 22 | Hispanic | Multiple injuries | ACG, MC, THL |
| 1708 | Control | 8 | F | 20 | African American | Asphyxia, multiple injuries | ACG, MC, THL |
| 1790 | Control | 13 | M | 18 | White | Multiple injuries | ACG |
| 1793 | Control | 11 | M | 19 | African American | Drowning | ACG, MC, THL |
| 1860 | Control | 8 | M | 5 | White | Cardiac arrhythmia | ACG |
| 4543 | Control | 28 | M | 13 | White | Multiple injuries | ACG, MC, THL |
| 4638 | Control | 15 | F | 5 | White | Chest injuries | ACG |
| 4722 | Control | 14 | M | 16 | White | Multiple injuries | ACG, MC, THL |
| 797 | Autism | 9 | M | 13 | White | Drowning | ACG, THL |
| 1638 | Autism | 20 | F | 50 | White | Seizure | ACG, MC, THL |
| 4231 | Autism | 8 | M | 12 | African American | Drowning | ACG, MC, THL |
| 4721 | Autism | 8 | M | 16 | African American | Drowning | ACG, MC, THL |
| 4899 | Autism | 14 | M | 9 | White | Drowning | ACG, MC, THL |
| 5000 | Autism | 27 | M | 8.3 | NA | NA | ACG, MC, THL |
| 6294 | Autism | 16 | M | NA | NA | NA | ACG, MC, THL |
| 6640 | Autism | 29 | F | 17.83 | NA | NA | ACG, MC, THL |
ACG = anterior cingulate gyrus; F = female; M = male; MC = motor cortex; NA = not available; PMI = postmortem interval; THL = thalamus.
Autism Tissue Program (ATP) identifier.
Brain regions for which each sample was available.
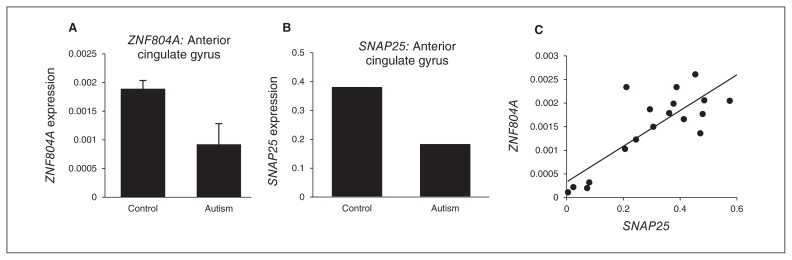
There was no significant difference in age, postmortem interval and sex distribution between the control and autism groups (see the Appendix, Table S2). The expression of ZNF804A (fold-change 2−ΔΔCt = 0.277, p = 0.009) and SNAP25 (2−ΔΔCt = 0.258, p = 0.009) were significantly reduced in the ACG of individuals with autism compared with controls (Fig. 2A and B). We also found a strong positive correlation between the expression of ZNF804A and SNAP25 in the ACG (Pearson r = 0.837, p < 0.001; Fig. 2C). In the MC and thalamus, the expression of ZNF804A or SNAP25 did not differ significantly between the control and autism groups (data not shown).
Fig. 2.
Gene expression in postmortem brain. The expression of (A) zinc finger protein 804A (ZNF804A; p = 0.009) and (B) SNAP25 (p = 0.009) were significantly reduced in the anterior cingulate gyrus (ACG) of individuals with autism compared with healthy controls. (C) Positive correlation between the expression of ZNF804A and SNAP25 in the ACG (Pearson r = 0.837; p < 0.001).
Discussion
We suggest that ZNF804A could be a risk gene mediating the intermediate phenotypes related to verbal skills in individuals with autism. In a GWAS of autism, Anney and colleagues (supplementary data)7 reported nominal association of several ZNF804A SNPs (rs17508877, rs1038197, rs7585738, rs6730122, rs10199843) with the Lvrb subset of individuals with autism. To our knowledge, the present study is the first to confirm the association of ZNF804A with a subgroup of individuals with autism characterized by verbal deficits.
The SNP rs7603001, which showed nominal association with autism in all families and in the subset of Lvrb families, is located in intron 2 of ZNF804A. Even though this SNP may not have a functional significance, putative regulatory regions have been predicted (FastSNP; http://fastsnp.ibms.sinica.edu.tw/pages/inputSNPListAnalysis.jsp) for the SNPs included in the LD bin of rs7603001. The r2 LD value between rs7603001, the SNP that was associated with autism in our study, and the SNPs that were associated with autism in the GWAS7 ranged between 0.25 and 0.28. The GWAS finding was thus replicated at the gene level, not at the level of specific SNPs.
In addition to genetic association, CNVs (gain and loss), mostly de novo, were observed at the ZNF804A locus of boys with autism who had a verbal deficit. Griswold and colleagues8 and Talkowski and colleagues9 have also reported CNVs at the ZNF804A locus in individuals with autism. Since the penetrance of CNVs is variable, it is not possible to predict the effect of these CNVs in the pathogenesis of autism. Copy number gain and loss were observed in autistic individuals, and similar CNVs were observed in unaffected parents. Furthermore, similar CNVs have also been observed in patients with other neuropsychiatric disorders,32 suggesting pleiotropic effects. Future studies to correlate specific CNVs with detailed clinical characteristics and to assess their effects on neurodevelopment are warranted.
Impaired linguistic/verbal ability is a key cognitive defect in individuals with autism.33,34 Based on our results, we suggest that ZNF804A could be a modulator of verbal traits in individuals with autism. There is ample evidence of the involvement of ZNF804A in the development of ToM,10 which in turn, is closely intertwined with the development of linguistic/verbal abilities from infancy.15–17
Genetic, neuropsychological and neuroimaging studies have suggested that ZNF804A is involved in higher-order cognitive processes such as ToM,10 working memory35 and executive control of attention.36 It has been found to play a pivotal role in the maintenance of functional connectivity in the brain.37,38 We observed a reduced expression of ZNF804A in the ACG of individuals with autism compared with controls. The ACG, a brain region vital for cognitive and behavioural abilities, is involved in emotion formation and processing, learning and memory.39,40 Downregulated expression of ZNF804A could lead to adverse effects on the cognitive processes associated with this gene.
Even though the previous studies on ZNF804A were focused on schizophrenia, overwhelming evidence suggests that the risk variants of this gene may be involved in the modulation of intermediate cognitive phenotypes associated with the disorder rather than the disorder itself.10,35,36,38 Adult- onset schizophrenia and early-onset autism, despite being 2 clinically distinct, complex neurodevelopmental disorders, share several deficits in cognitive functioning.41–43 A deficient ToM has been identified as a potential contributor to the social cognitive dysfunction in individuals with schizophrenia and autism,44,45 and it could be a common factor mediating ToM-related key intermediate phenotypes in people with these disorders. Several studies have shown the association of ZNF804A variants with cognitive dysfunction in individuals with schizophrenia.46–48 Interestingly, we observed a stronger association of ZNF804A in individuals with an autism subtype characterized by verbal deficits.
The protein sequence of ZNF804A shows a C2H2-type zinc-finger domain at its N-terminal end, suggesting that it may bind DNA and have a role in regulating gene expression.18 ZNF804A has been found to modulate the expression of several genes implicated in the pathogenesis of schizophrenia.18,49
We examined the possible role of ZNF80A as a regulator of the expression of genes previously reported to be associated with verbal/linguistic abilities and/or social cognition. The expression of SNAP25 was downregulated in ZNF804A-silenced cells compared with control cells. Furthermore, the expression of SNAP25 was significantly reduced in the ACG of individuals with autism, and a strong positive correlation was observed between the expression of ZNF804A and SNAP25 in the ACG.
SNAP25 is a presynaptic plasma membrane protein that is specifically and abundantly expressed in nerve cells. It participates in synaptic vesicle exocytosis through the formation of a soluble NSF attachment protein receptor complex50 and plays a pivotal role in modulating calcium homeostasis.51 SNAP25 is important for axonal growth and synaptic plasticity, 2 essential steps in the wiring of the central nervous system.50,52 SNAP25 variants have been found to modulate cognitive performances.29,53,54 SNAP25 is located in a chromosomal region (20p12–p11.2) with a previously suggested linkage to intelligence.55 Moreover, polymorphisms in SNAP25 have been associated with hyperactivity in individuals with autism.56 However, at present, there is no literature linking ZNF804A and SNAP25.
Limitations
A replication study in a larger cohort of verbally deficient individuals with autism from different racial backgrounds would have been more informative. Further studies on the functional implications of ZNF804A CNVs and on the nature of the interaction between ZNF804A and SNAP25 in the pathogenesis of autism are warranted. The small number of postmortem brain samples used is another limitation of our study.
Conclusion
We suggest that ZNF804A could have a pivotal role in mediating the intermediate phenotypes associated with verbal traits in individuals with autism. It could be a common factor modulating the ToM-related intermediate phenotypes in individuals with schizophrenia and autism.
Acknowledgements
We gratefully acknowledge the resources provided by the AGRE Consortium and the participating AGRE families. The AGRE is a program of Autism Speaks and is supported in part by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (P.I.). We thank Dr. Jane Pickett, Director of Brain Resources and Data, ATP, for facilitating brain tissue collection. Human tissue was obtained from the NICHD BTB for Developmental Disorders at the University of Maryland. Tissue samples were also provided by the Harvard Brain Tissue Resource Center, which is supported in part by PHS grant number R24 MH 068855. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (A.A., K.N.), the Takeda Science Foundation, Japan (K.S.), and partly by the Strategic Research Program for Brain Sciences (Integrated research on neuropsychiatric disorders). We thank Tae Takahashi and Mika Oyaizu for technical assistance.
Footnotes
Competing interests: None declared.
Contributors: A. Anitha, I. Thanseem and K. Nakamura designed the study, acquired and analyzed the data and wrote the article. M.M. Vasu, K. Yamada, T. Ueki, Y. Iwayama and T. Toyota acquired and analyzed the data and reviewed the article. K.J. Tsuchiya, Y. Iwata and K. Suzuki analyzed the data and reviewed the article. T. Sugiyama, M. Tsujii, T. Yoshikawa and N. Mori designed the study and reviewed the article. All authors approved the final version for publication.
References
- 1.Meyer U, Feldon J, Dammann O. Schizophrenia and autism: Both shared and disorder-specific pathogenesis via perinatal inflammation? Pediatr Res. 2011;69:26R–33R. doi: 10.1203/PDR.0b013e318212c196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll LS, Owen MJ. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009;1:102. doi: 10.1186/gm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–5. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 4.Riley B, Thiselton D, Maher BS, et al. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Luo XJ, Xiao X, et al. Allelic differences between Han Chinese and Europeans for functional variants in ZNF804A and their association with schizophrenia. Am J Psychiatry. 2011;168:1318–25. doi: 10.1176/appi.ajp.2011.11030381. [DOI] [PubMed] [Google Scholar]
- 6.Williams HJ, Norton N, Dwyer S, et al. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16:429–41. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anney R, Klei L, Pinto D, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–82. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griswold AJ, Ma D, Cukier HN, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012;21:3513–23. doi: 10.1093/hmg/dds164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talkowski ME, Rosenfeld JA, Blumenthal I, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–37. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter H, Schnell K, Erk S, et al. Effects of a genome-wide supported psychosis risk variant on neural activation during a theory-of-mind task. Mol Psychiatry. 2011;16:462–70. doi: 10.1038/mp.2010.18. [DOI] [PubMed] [Google Scholar]
- 11.Baron-Cohen S. The autistic child’s theory of mind: a case of specific developmental delay. J Child Psychol Psychiatry. 1989;30:285–97. doi: 10.1111/j.1469-7610.1989.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 12.Yirmiya N, Erel O, Shaked M, et al. Meta-analyses comparing theory of mind abilities of individuals with autism, individuals with mental retardation, and normally developing individuals. Psychol Bull. 1998;124:283–307. doi: 10.1037/0033-2909.124.3.283. [DOI] [PubMed] [Google Scholar]
- 13.Bora E, Yucel M, Pantelis C. Theory of mind impairment in schizophrenia: meta-analysis. Schizophr Res. 2009;109:1–9. doi: 10.1016/j.schres.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Frith CD, Frith U. Interacting minds — a biological basis. Science. 1999;286:1692–5. doi: 10.1126/science.286.5445.1692. [DOI] [PubMed] [Google Scholar]
- 15.Miller CA. Developmental relationships between language and theory of mind. Am J Speech Lang Pathol. 2006;15:142–54. doi: 10.1044/1058-0360(2006/014). [DOI] [PubMed] [Google Scholar]
- 16.Ruffman T, Slade L, Crowe E. The relation between children’s and mothers’ mental state language and theory-of-mind understanding. Child Dev. 2002;73:734–51. doi: 10.1111/1467-8624.00435. [DOI] [PubMed] [Google Scholar]
- 17.Dahlgren S, Dahlgren Sandberg A, Larsson M. Theory of mind in children with severe speech and physical impairments. Res Dev Disabil. 2010;31:617–24. doi: 10.1016/j.ridd.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Girgenti MJ, Loturco JJ, Maher BJ. ZNF804a regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PLoS ONE. 2012;7:e32404. doi: 10.1371/journal.pone.0032404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geschwind DH, Sowinski J, Lord C, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–6. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–85. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg TE, Iudicello J, Russo C, et al. BDNF Val66Met polymorphism significantly affects d’ in verbal recognition memory at short and long delays. Biol Psychol. 2008;77:20–4. doi: 10.1016/j.biopsycho.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Vernes SC, Newbury DF, Abrahams BS, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–45. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palo OM, Antila M, Silander K, et al. Association of distinct allelic haplotypes of Drosoph Inf ServC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet. 2007;16:2517–28. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- 24.Beaver KM, Delisi M, Vaughn MG, et al. Association between the A1 allele of the DRD2 gene and reduced verbal abilities in adolescence and early adulthood. J Neural Transm. 2010;117:827–30. doi: 10.1007/s00702-010-0421-8. [DOI] [PubMed] [Google Scholar]
- 25.Balter M. Genetics. First gene linked to speech identified. Science. 2001;294:32. doi: 10.1126/science.294.5540.32a. [DOI] [PubMed] [Google Scholar]
- 26.Kircher T, Krug A, Markov V, et al. Genetic variation in the schizophrenia-risk gene neuregulin 1 correlates with brain activation and impaired speech production in a verbal fluency task in healthy individuals. Hum Brain Mapp. 2009;30:3406–16. doi: 10.1002/hbm.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J, Willmott M, Vetuz G, et al. Evidence that genetic variation in the oxytocin receptor (OXTR) gene influences social cognition in ADHD. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:697–702. doi: 10.1016/j.pnpbp.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 28.Waga C, Okamoto N, Ondo Y, et al. Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr Genet. 2011;21:208–11. doi: 10.1097/YPG.0b013e328341e069. [DOI] [PubMed] [Google Scholar]
- 29.Cagliani R, Riva S, Marino C, et al. Variants in SNAP25 are targets of natural selection and influence verbal performances in women. Cell Mol Life Sci. 2012;69:1705–15. doi: 10.1007/s00018-011-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roll P, Vernes SC, Bruneau N, et al. Molecular networks implicated in speech-related disorders: FOXP2 regulates the SRPX2/ uPAR complex. Hum Mol Genet. 2010;19:4848–60. doi: 10.1093/hmg/ddq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lennertz L, Rujescu D, Wagner M, et al. Novel schizophrenia risk gene TCF4 influences verbal learning and memory functioning in schizophrenia patients. Neuropsychobiology. 2011;63:131–6. doi: 10.1159/000317844. [DOI] [PubMed] [Google Scholar]
- 32.Steinberg S, Mors O, Borglum AD, et al. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry. 2011;16:59–66. doi: 10.1038/mp.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner MA. Generating novel ideas: fluency performance in high-functioning and learning disabled individuals with autism. J Child Psychol Psychiatry. 1999;40:189–201. [PubMed] [Google Scholar]
- 34.Yirmiya N, Gamliel I, Shaked M, et al. Cognitive and verbal abilities of 24- to 36-month-old siblings of children with autism. J Autism Dev Disord. 2007;37:218–29. doi: 10.1007/s10803-006-0163-5. [DOI] [PubMed] [Google Scholar]
- 35.Linden DE, Lancaster TM, Wolf C, et al. ZNF804A genotype modulates neural activity during working memory for faces. Neuropsychobiology. 2013;67:84–92. doi: 10.1159/000344001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balog Z, Kiss I, Keri S. ZNF804A may be associated with executive control of attention. Genes Brain Behav. 2011;10:223–7. doi: 10.1111/j.1601-183X.2010.00657.x. [DOI] [PubMed] [Google Scholar]
- 37.Esslinger C, Kirsch P, Haddad L, et al. Cognitive state and connectivity effects of the genome-wide significant psychosis variant in ZNF804A. Neuroimage. 2011;54:2514–23. doi: 10.1016/j.neuroimage.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Rasetti R, Sambataro F, Chen Q, et al. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry. 2011;68:1207–17. doi: 10.1001/archgenpsychiatry.2011.103. [DOI] [PubMed] [Google Scholar]
- 39.Takenouchi K, Nishijo H, Uwano T, et al. Emotional and behavioral correlates of the anterior cingulate cortex during associative learning in rats. Neuroscience. 1999;93:1271–87. doi: 10.1016/s0306-4522(99)00216-x. [DOI] [PubMed] [Google Scholar]
- 40.Bush G, Luu P, Posner MI. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci. 2000;4:215–22. doi: 10.1016/s1364-6613(00)01483-2. [DOI] [PubMed] [Google Scholar]
- 41.Frith CD, Corcoran R. Exploring ‘theory of mind’ in people with schizophrenia. Psychol Med. 1996;26:521–30. doi: 10.1017/s0033291700035601. [DOI] [PubMed] [Google Scholar]
- 42.Baron-Cohen S, Wheelwright S, Hill J, et al. The “Reading the Mind in the Eyes” Test revised version: a study with normal adults, and adults with Asperger syndrome or high-functioning autism. J Child Psychol Psychiatry. 2001;42:241–51. [PubMed] [Google Scholar]
- 43.Couture SM, Penn DL, Losh M, et al. Comparison of social cognitive functioning in schizophrenia and high functioning autism: more convergence than divergence. Psychol Med. 2010;40:569–79. doi: 10.1017/S003329170999078X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muris P, Steerneman P, Meesters C, et al. The ToM test: a new instrument for assessing theory of mind in normal children and children with pervasive developmental disorders. J Autism Dev Disord. 1999;29:67–80. doi: 10.1023/a:1025922717020. [DOI] [PubMed] [Google Scholar]
- 45.Couture SM, Penn DL, Roberts DL. The functional significance of social cognition in schizophrenia: a review. Schizophr Bull. 2006;32(Suppl 1):S44–63. doi: 10.1093/schbul/sbl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walters JT, Corvin A, Owen MJ, et al. Psychosis susceptibility gene ZNF804A and cognitive performance in schizophrenia. Arch Gen Psychiatry. 2010;67:692–700. doi: 10.1001/archgenpsychiatry.2010.81. [DOI] [PubMed] [Google Scholar]
- 47.Chen M, Xu Z, Zhai J, et al. Evidence of IQ-modulated association between ZNF804A gene polymorphism and cognitive function in schizophrenia patients. Neuropsychopharmacology. 2012;37:1572–8. doi: 10.1038/npp.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto R, Ohi K, Yasuda Y, et al. The impact of a genome-wide supported psychosis variant in the ZNF804A gene on memory function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1459–64. doi: 10.1002/ajmg.b.31123. [DOI] [PubMed] [Google Scholar]
- 49.Umeda-Yano S, Hashimoto R, Yamamori H, et al. The regulation of gene expression involved in TGF-beta signaling by ZNF804A, a risk gene for schizophrenia. Schizophr Res. 2013;146:273–8. doi: 10.1016/j.schres.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 50.Oyler GA, Higgins GA, Hart RA, et al. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol. 1989;109:3039–52. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pozzi D, Condliffe S, Bozzi Y, et al. Activity-dependent phosphorylation of Ser187 is required for SNAP-25-negative modulation of neuronal voltage-gated calcium channels. Proc Natl Acad Sci U S A. 2008;105:323–8. doi: 10.1073/pnas.0706211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osen-Sand A, Catsicas M, Staple JK, et al. Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature. 1993;364:445–8. doi: 10.1038/364445a0. [DOI] [PubMed] [Google Scholar]
- 53.Gosso MF, de Geus EJ, van Belzen MJ, et al. The SNAP-25 gene is associated with cognitive ability: evidence from a family-based study in two independent Dutch cohorts. Mol Psychiatry. 2006;11:878–86. doi: 10.1038/sj.mp.4001868. [DOI] [PubMed] [Google Scholar]
- 54.Söderqvist S, McNab F, Peyrard-Janvid M, et al. The SNAP25 gene is linked to working memory capacity and maturation of the posterior cingulate cortex during childhood. Biol Psychiatry. 2010;68:1120–5. doi: 10.1016/j.biopsych.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 55.Posthuma D, Luciano M, Geus EJ, et al. A genome-wide scan for intelligence identifies quantitative trait loci on 2q and 6p. Am J Hum Genet. 2005;77:318–26. doi: 10.1086/432647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerini FR, Bolognesi E, Chiappedi M, et al. SNAP-25 single nucleotide polymorphisms are associated with hyperactivity in autism spectrum disorders. Pharmacol Res. 2011;64:283–8. doi: 10.1016/j.phrs.2011.03.015. [DOI] [PubMed] [Google Scholar]