Summary
Assembly of cilia and flagella requires intraflagellar transport (IFT), a highly regulated kinesin-based transport system that moves cargo from the basal body to the tip of flagella [1]. The recruitment of IFT components to basal bodies is a function of flagellar length, with increased recruitment in rapidly growing short flagella [2]. The molecular pathways regulating IFT are largely a mystery. Since actin network disruption leads to changes in ciliary length and number, actin has been proposed to have a role in ciliary assembly. However, the mechanisms involved are unknown. In Chlamydomonas reinhardtii, conventional actin is found in both the cell body and the inner dynein arm complexes within flagella [3, 4]. Previous work showed that treating Chlamydomonas cells with the actin-depolymerizing compound cytochalasin D resulted in reversible flagellar shortening [5], but how actin is related to flagellar length or assembly remains unknown. Here, we utilize small-molecule inhibitors and genetic mutants to analyze the role of actin dynamics in flagellar assembly in Chlamydomonas reinhardtii. We demonstrate that actin plays a role in IFT recruitment to basal bodies during flagellar elongation, and that when actin is perturbed, the normal dependence of IFT recruitment on flagellar length is lost. We also find that actin is required for sufficient entry of IFT material into flagella during assembly. These same effects are recapitulated with a myosin inhibitor suggesting actin may act via myosin in a pathway by which flagellar assembly is regulated by flagellar length.
Results and Discussion
Inhibiting Actin Polymerization Shortens Flagella
In Chlamydomonas, Cytochalasin D (CD), which acutely disrupts filamentous actin (F-actin) network-dependent processes, causes reversible flagellar shortening [5]. CD binds multiple actin species and increases the critical concentration needed for polymerization [6].
Because standard F-actin localization methods do not label vegetative cells [7], we expressed Lifeact, an F-actin binding peptide [8], tagged with the YFP variant Venus. Lifeact-Venus localized to perinuclear regions (Cell diagram, Figure 1A; Lifeact localization, Figures 1B, S1A) similar to localization seen with anti-actin antibodies [4]. In addition, some cells showed Lifeact-Venus near the flagellar base (Figure 1B); these dots were variable in number (one to three) and found in less than 10% of cells (Figure 1D), hence we speculate that this localization may be transient.
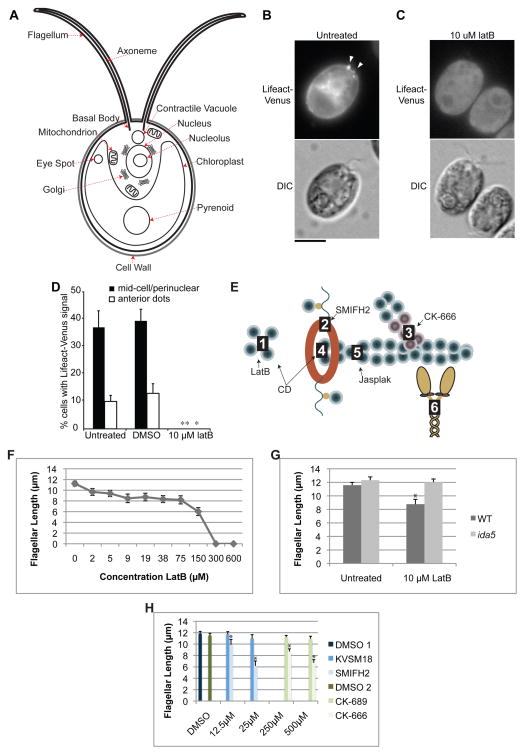
Figure 1.
The actin polymerization inhibitor Latrunculin B causes flagellar shortening. (A) Diagram of a Chlamydomonas cell with relevant or prominent organelles identified. (B) Representative images showing Lifeact-Venus localization to the mid-cell portion as filaments and to the cell-anterior region as patches (arrowheads) and (C) its complete loss upon treatment with latrunculin B. Bar 5μm. (D) Lifeact-Venus expressing cells were cultured to OD = 0.3 and treated with 0.1% DMSO or 10 μM latrunculin B. 100–250 cells were counted by fluorescence microscopy for localization of Lifeact-Venus at the mid-cell portion and as cell-anterior patches. The error bars indicate 95% confidence intervals from three replicates. * and ** indicate p < 0.05 and 0.01, respectively. (E) Diagram of point of action for inhibitors. Latrunculin B (LatB) binds actin monomers to prevent filament assembly (1); SMIFH2 inhibits Formin mediated actin nucleation (2); CK-666 inhibits Arp2/3 mediated branched actin assembly (3); Cytochalasin D (CD) binds barbed ends of actin filaments (4) with high affinity and G-actin (1) with low affinity; jasplakinolide (jasplak) binds F-actin at the interface of 3 actin subunits (5); blebbistatin binds myosin motor (6). (F) Treatment with latrunculin B to impair actin polymerization and ultimately disassemble the actin network results in a dose dependent decrease in flagellar length. Concentrations indicated correspond to the markers on the line graph. (G) Treatment with 10μM latrunculin B shortens wild-type flagella but not ida5 mutant flagella demonstrating that the flagellar shortening effect is caused by targeting actin and not a nonspecific effect of impaired cell health (*:p< .00000005). (H) Inhibiting an actin nucleation factor, formin, with SMIFH2 or inhibiting actin branch promoting Arp2/3 with CK-666, shortens flagellar length in a dose dependent manner. KVSM18 and CK-689 are inactive controls for SMIFH2 and CK-666 respectively and have no effect. 1% DMSO are also included as controls as inhibitors are diluted in DMSO. DMSO 1 refers to the control for the formin experiment and DMSO 2 refers to the control for the Arp2/3 experiment. Error bars are 95% confidence intervals. Asterisks indicate significant difference from controls (p<0.05). See also Figure S1.
To explore the effect of actin disruption on flagellar length, we utilized Latrunculin B (LatB), a compound that inhibits actin polymerization by binding to monomeric (G-actin). While only a small percentage of cells have detectable anterior localized Lifeact, G-actin is prominent near the basal bodies that nucleate flagella (Figure S1B) and is excluded from sub-basal body regions even in the absence of filaments connecting the nucleus to basal bodies (Figure S1C). The point of action for all inhibitors is shown in Figure 1E. Both perinuclear and anterior Lifeact labeling were sensitive to LatB at concentrations of 1 μM and higher (Figures 1C,D and data not shown), indicating Lifeact-Venus indeed represents F-actin and confirming the efficacy of 10 μM LatB used elsewhere in this study. When measuring flagellar length, LatB treatment of vegetative cells resulted in a dose dependent shortening (Figure 1F). Unlike the broad length distribution seen with CD [5], flagella are uniformly shortened in the population under LatB treatment (Figure S1D).
We tested specificity of LatB using a Chlamydomonas actin null mutant, ida5 [9, 10]. ida5 mutants are non-lethal, apparently because the essential actin mediated functions are provided by an actin isoform called Novel Actin-like Protein (NAP) which is upregulated in the ida5 mutant [4]. Although ida5 mutants have a flagellar motility defect due to lack of a subset of inner dynein arms, they have been reported to have normal flagellar length, suggesting NAP may be able to sufficiently compensate for actin in its functions related to acquisition of normal steady-state flagellar length. Because ida5 mutants are completely missing conventional actin protein, they provide a way to test whether the effects of LatB require conventional actin. When ida5 actin mutants were treated with 10μM LatB, flagellar length was unaltered (Figure 1G), demonstrating that the flagellar shortening effect of LatB is actin-dependent and is not a non-specific effect of impaired cell health. At 10μM, LatB did not impair cell division (not shown).
To confirm that altering actin polymerization state can shorten flagellar length, we utilized inhibitors of additional proteins known to mediate actin polymerization [11, 12], formin and Arp2/3. The formin inhibitor, SMIFH2, and the Arp2/3 inhibitor, CK-666, both caused a dose dependent decrease in flagellar length compared to inactive analogs of each drug (Figures 1H), suggesting that parallel actin bundles and branched networks are both important for flagellar maintenance. A similar Arp2/3 inhibitor, CK-869, completely eliminates flagella in ida5 mutants, potentially indicating an interaction of Arp2/3 with both actin isoforms (Figure S1E).
Flagellar Regeneration is Impaired in Actin Mutants
Flagellar length is dynamically maintained by the balance between growth and disassembly. In order to determine if loss of actin reduces flagellar assembly rates, we performed a regeneration experiment by inducing cells to sever and release their flagella (a process termed deflagellation) by transient reduction of pH. Following deflagellation, wild-type cells regrow their flagella to steady state wild-type length within a few hours while ida5 mutants regenerate at a slower rate during the rapid elongation phase within the first 90 minutes and then ultimately reach the same length as wild-type cells (Figure 2A). P-values all less than 0.001 for nine independent experiments at the 90 minute time point are shown in Table S1. We repeated the experiment using ida5 mutant cells that expressed a GFP tagged wild-type version of the gene (referred to here as ida5-GFP). Expression was confirmed by western blots of flagellar preparations using both GFP and actin antibodies (Figure S1F). Regeneration in ida5-GFP was similar to wild-type and did not exhibit the slow flagellar assembly kinetics of the ida5 mutants (Figure S1G). This demonstrates that altered regeneration kinetics is due to loss of the actin gene and not any other mutations in the background. Since the quantity of tagged actin in the ida5-GFP strain is less than the quantity of actin in wild-type cells (Figure S1H, left), expression of a small amount of actin is sufficient to function in flagellar assembly, perhaps with the help of NAP which is elevated in these lines (Figure S1H, right). Flagellar assembly is biphasic in wild-type cells, with a rapid early phase that depends on IFT and requires utilization of cytoplasmic precursor pools of unassembled flagellar protein, followed by a second slower phase of assembly that requires new protein synthesis [13]. The fact that ida5 mutants initially grow more slowly, while ultimately reaching the same final length, suggests loss of actin affects some process important for the rapid phase of assembly. The actin network itself does not appear to be visibly depleted or rearranged during regeneration as perinuclear and anterior Lifeact-Venus localization is unchanged (Figure S1I). In addition to their slow regeneration, ida5 mutants failed to elongate when treated with LiCl which is known to cause flagellar elongation in wild-type cells (Figure S1J). LiCl induced elongation was also impaired in cells treated with LatB and Jasplakinolide (Figures S1K,L). Flagellar regeneration was also impaired in cells treated with latB or Jasplakinolide (Figure S1M).
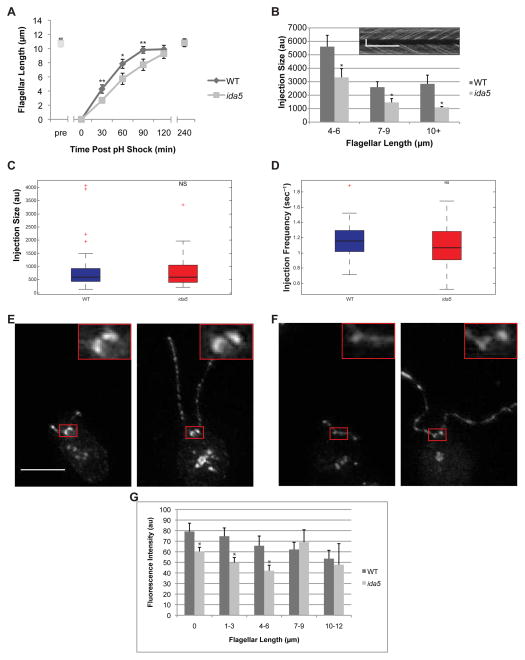
Figure 2.
Flagellar length, IFT train size and basal body accumulation of IFT material are perturbed during flagellar regeneration in a mutant lacking conventional actin. (A) Flagellar regeneration following pH shock mediated flagellar shedding results in a rapid initial phase of growth and slow late phase as flagella near final length in wild type cells. In ida5 actin mutants, initial phase of growth is much slower but ultimately flagellar length equals wild type. Error bars are 95% confidence intervals. (*:p<0.0005, **:p<0.00005)(B) As flagella regenerate, wild-type cells show typical IFT injection behavior with larger trains injected in shorter flagella and smaller trains injected in longer flagella. ida5 mutants retain length-dependent injection of IFT material but the injection sizes are significantly smaller than wild-type (p<0.05 for each bin). Means and standard error are shown. Inset is an example of the type of kymograph data that is analyzed for this graph. Vertical scale bar is 5μm. Horizontal scale bar is 5 sec. (C) Box and whisker plot of injection sizes shows median (black bar), 25th and 75th percentiles (box top and bottom edges), extreme data points not considered outliers (whiskers) and outliers (red crosses). There is no significant difference (p=0.6581) in injection sizes between wild type and ida5 mutants at steady state. (D) Box and whisker plot of injection frequency shows no significant difference between wild type and ida5 mutants (p=0.1214). Injection frequency is the inverse of the measured wait time. (E) Projection of z-stacks taken of a representative image for KAP-GFP expressing cells. 0.2μm step TIFFs were used for image analysis for Figure 3C. Red box shows region surrounding basal bodies. Scale bar is 5μm. Insets are 3x magnifications. Left panel shows short flagella early in regeneration. Right panel shows full length flagella late in regeneration. (F) Projection of z-stacks of a representative image for KAP-GFP expressing cells on ida5 mutant background. Left panel shows short flagella early in regeneration. Right panel shows full length flagella late in regeneration. (G) Intensity of KAP-GFP motor fluorescence at basal bodies is reduced in ida5 mutants compared to wild type initially, mirroring the initial decrease in flagellar regeneration kinetics. Wild type accumulation of KAP-GFP is known to decrease with flagellar length, as less IFT material is required when flagellar growth slows. When growth slows and ida5 flagella reach wild-type lengths (7–12 microns), ida5 mutant accumulation of KAP-GFP at basal bodies is no longer statistically significantly different from wild-type levels. Asterisks indicate significant difference from wild type (p<0.05). See also Figure S2.
When flagella regenerate, new flagellar proteins are synthesized and then transported into the flagellum by IFT. Impairment of either precursor pool regeneration or IFT could cause slow regeneration. As shown in Figure S2E, the ida5 mutant did not impair flagellar precursor pool regeneration. Therefore we looked at IFT by analyzing the sizes of IFT trains injected into flagella. IFT train sizes are inversely correlated with flagellar length [14], suggesting larger trains are injected in short, rapidly growing flagella to support rapid growth, with trains becoming smaller as flagella reach their final length and growth slows. IFT train size also correlates with the time since the previous injection [2]. We used the same methods as the previous study [2] of analyzing Chlamydomonas strains expressing a GFP-tagged motor component, KAP, on a KAP mutant background via real-time TIRF microscopy. Consistent with our previous results, when we examined regenerating flagella we found that the size of IFT trains in wild-type cells initially increased compared to steady-state preshock levels at the beginning of regeneration, when flagella are short and rapidly growing, and then decreased as regeneration progressed (Figure 2B, S2A). In ida5 mutants, train size never attained the large size seen in wild-type flagella early in regeneration, and remained smaller than train sizes at corresponding lengths in wild-type cells throughout the course of regeneration (Figures 2B, S2A). Reduced IFT could explain why regeneration of ida5 mutants is impaired. However, when cells at steady state were analyzed, we saw no clear differences in sizes of IFT material injected into the flagellum with respect to flagellar length (Figures 2C, S2B) or time since last injection (Figures 2D, S2C) between wild-type and ida5 actin mutants. The fact that the ida5 mutation affects IFT train size during rapid flagellar regeneration but not once flagella have reached steady state length matches our finding that actin mutant flagella regenerate slowly but eventually reach lengths equal to wild-type, further suggesting that actin functions predominantly during the rapid early phase of regeneration.
Accumulation of IFT Machinery is Reduced at Basal Bodies in Actin Mutants During Regeneration
IFT machinery is recruited to the base of flagella during regeneration [2], as confirmed with KAP-GFP in Figures 2E–G, which show that IFT recruitment is highest in the early stages of regeneration and decreases as regeneration proceeds. In ida5 mutants, initial KAP-GFP recruitment following deflagellation was lower than wild-type, and then decreased more steeply during the first 6 microns of flagellar regrowth (Figures 2F,G). This is consistent with the results of Figure 2B in which IFT train size shows less of an increase immediately after deflagellation in ida5 mutants compared to wild-type cells. Ultimately IFT recruitment caught up with levels seen in wild-type as the final steady-state length was reached (Figure 2G). We thus obtain consistent results whether we examine regeneration rate, IFT train size, or IFT recruitment to the basal body - ida5 mutants lack the increased recruitment of IFT normally seen early in regeneration when growth is most rapid, but can maintain wild-type recruitment levels when flagella have reached steady state length. Using an experiment to allow flagellar growth and regrowth in the presence of the protein synthesis inhibitor, cycloheximide (Figure S2D), we were able to conclude that impaired basal body IFT recruitment was not due to decreased unassembled flagellar precursor (Figure S2E) or decreased synthesis of flagellar protein during regeneration (Figure S2F).
F-actin in Chlamydomonas appears to be present in vegetative cells
In Chlamydomonas, fluorescent phallacidin labels fertilization tubules in gametes [7], but does not clearly identify additional regions in vegetative cells. However phallotoxin staining can miss short actin filaments [16, 17]. Based on our perinuclear and anterior localization of Lifeact-Venus as well as the dramatic effects of actin polymerization, we believe filamentous actin is present in vegetative cells.
Myosin inhibitor mimics effect of actin mutation
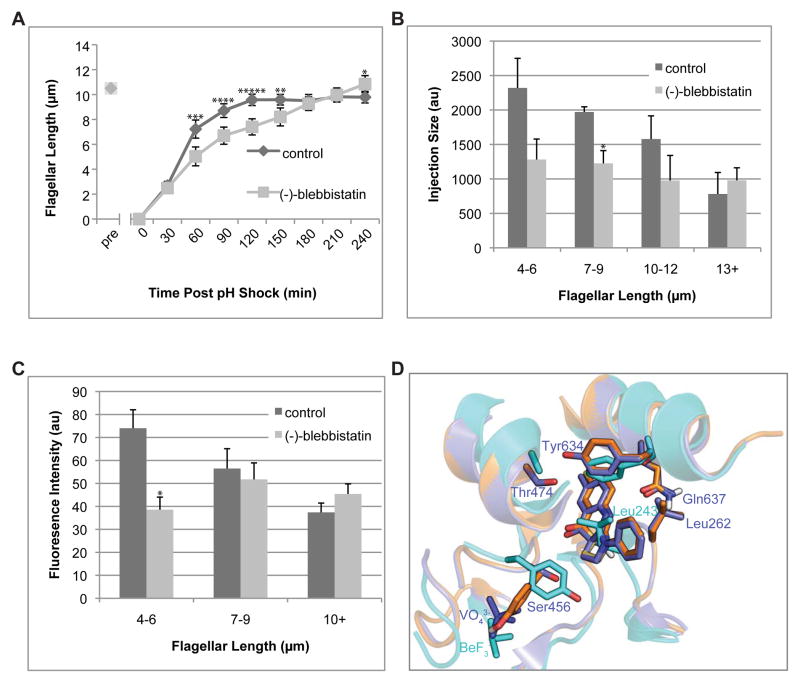
Our results indicate a role for actin in the regulation of IFT recruitment, based on both actin inhibitors and an actin mutant. Are these effects mediated by myosin? We evaluated the effects of blocking myosin activity using the myosin-II inhibitor blebbistatin. Blebbistatin up to 300μM blebbistatin did not alter steady state flagellar length (Figure S3A). However, when cells were treated with blebbistatin during regeneration following pH shock, flagellar growth was significantly impaired (Figure 3A). A myosin V inhibitor [15], did not affect regeneration (Figure S3B). Regeneration in blebbistatin is less severe than in latB (Figure S3C) suggesting a role for actin in IFT regulation beyond its functions requiring myosin. Unlike what was previously reported for Cytochalasin D [5], blebbistatin treated cells do not begin to shorten at longer time points following pH shock (Figures S3D and S3E). Blebbistatin also abrogated the normal length dependence of both IFT injection (Figure 3B) and basal body IFT recruitment (Figure 3C). These effects resemble the ida5 mutant phenotype, suggesting a role for myosin in regulating IFT recruitment.
Figure 3.
Myosin inhibition alters flagellar assembly kinetics. (A) In contrast to the deceleratory kinetics seen in wild-type flagellar regeneration, cells regenerating in the presence of the active (−)-blebbistatin enantiomer regeneration at a roughly constant rate. All error bars are 95% confidence intervals. (*:p<0.05, **:p<0.005, ***:p<0.0005, ****:p<0.00005, *****:p<0.000005) (B) For regeneration in inactive (+)-blebbistatin enantiomer, kymograph analysis of fluorescence intensity of IFT trains injected into flagella shows typical wild type behavior. As flagella elongate, IFT trains injected become smaller. In contrast, IFT injection sizes are reduced (*:p<.005) and do not significantly decrease with flagellar length in active (−)-blebbistatin. (C) Unlike inactive control, accumulation of KAP-GFP at basal bodies is also reduced (*:p<0.005) at shorter lengths and flagellar length-independent in (−)-blebbistatin, suggesting that myosin-II activity is required for length-dependent mobilization of IFT material for flagellar assembly. Error bars are the standard error of the mean. (D) Superimposition of Dictyostelium myosin II (PDB ID: 1YV3) crystallized with bound (−)-blebbistatin (purple), chicken myosin Va (PDB ID: 1W7J, cyan), and the homology model of putative Chlamydomonas myosin VIII (Phytozyme ID Myo2/Cre09.g416250.t1), with (−)-blebbistatin docked (orange). The loop containing Leu262 in myosin II moves away to allow (−)-blebbistatin to bind; in myosin V, the homologous Leu243 completely obstructs (−)-blebbistatin binding. All three putative Chlamydomonas myosins also have a leucine in this position. In myosin II, Ser456 is small enough to not encroach the (−)-blebbistatin binding site. However, the homologous bulky Tyr439 substitution forces not only the entire loop to move away from the binding site, but it also forces ATP, here represented by the BeF3− ion, to bind lower than the VO43− ion in myosin II. All three putative Chlamydomonas myosins also have a tyrosine in this position. See also Figure S3.
The Chlamydomonas genome contains three myosins: Two type-XI myosins (Myo1/Cre16.g658650.t1 and Myo3/Cre13.g563800.t1) and a type-VIII myosin (Myo2/Cre09.g416250.t1). Neither Type-XI nor Type-VIII myosins have been tested in vitro for blebbistatin sensitivity. In myosin II, Ser456, Thr474, Tyr634, and Gln637 were implicated in blebbistatin binding based on crystal structures [19], but sequence alignment of myosins in which blebbistatin has been tested (Figure S4) indicates these residues do not fully determine blebbistatin activity. Acanthamoeba myosin Ic has the same homologous residues but is blebbistatin insensitive, whereas Acanthamoeba myosin II and turkey smooth muscle myosin II do not fit the myosin II-like profile but are blebbistatin sensitive. The Chlamydomonas myosins showed ambiguous myosin II/myosin V profiles for the four residues, along with similar overall sequence identities to both (ranging from 40–45%).
We asked whether it would be structurally possible for (−)-blebbistatin to bind Chlamydomonas myosins. Homology models of all three based on the (−)-blebbistatin-bound myosin II structure (PDB ID: 1YV3) revealed many amino acids in the blebbistatin binding site of myosin II are identical in Chlamydomonas myosins, including the entire loop containing the homologous Leu262 residues. We then docked (−)-blebbistatin using DOCK 3.7 [20]. Figure 3D shows that blebbistatin could be successfully docked onto the predicted structure of Chlamydomonas Myo2/Cre09.g416250.t1 (orange) provided that the Leucine corresponding to Leu262 of myosin II is able to flip out of the binding pocket. In contrast, the homologous residue in myosin V, Leu243, completely occludes blebbistatin binding.
Given the positive docking result with the homology model structure and the fact that we apply blebbistatin at a concentration (150μM) exceeding that previously tested in other myosin types in vitro, we conclude it is possible that blebbistatin targets Chlamydomonas myosins.
Potential roles of actin and myosin in IFT recruitment
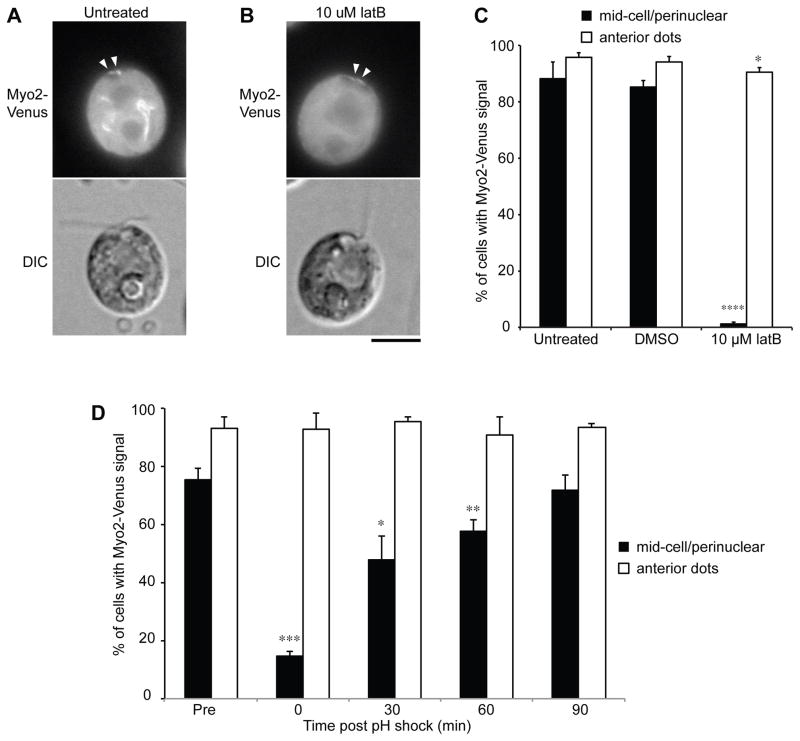
We have successfully tagged one of the three Chlamydomonas myosins, Myo2, with Venus. Myo2-Venus showed a similar localization as Lifeact (Figures 4A,C). The mid-cell/perinuclear localization was LatB-sensitive (Figures 4B,C). Myo2-Venus was lost from perinuclear regions following deflagellation and recovered with time during regeneration (Figure 4D), suggesting myosin is regulated during deflagellation or flagellar regrowth.
Figure 4.
Myo2-Venus shows similar localization to LifeAct-Venus; this localization is sensitive to latrunculin B and is transiently lost during regeneration. (A) Representative images showing Myo2-Venus localization to the mid-cell portion as filaments and to the cell-anterior region as dots (arrowheads). (B) Only the mid-cell localization was lost upon latrunculin B addition. Bar, 5 μm. (C) Myo2-Venus expressing cells were cultured and counted as in (Figure 1D). * and **** indicate p < 0.05 and 0.0001, respectively. (D) Loss of mid-cell Myo2 localization during initial phase of flagellar regeneration. 100 cells were counted at each time point. Error bars indicate 95% confidence intervals from three replicates. *, **, and *** indicates p < 0.05, 0.01, and 0.001, respectively. Bar, 5 μm. See also Figure S4.
Plant myosin XI has similar domain structure and functions to mammalian myosin V [21]. Myosin V transports organelles, vesicles and protein cargo over long distances, including Golgi-derived vesicles which are known to be required for ciliogenesis and ciliary trafficking [22–29]. IFT20 is Golgi-localized and involved in vesicular transport of membrane proteins to the cilium [30–32]. Thus actin might regulate basal body IFT recruitment by regulating transport from the Golgi. Class VIII plant myosins are found at the cell periphery and are implicated in protein delivery to plasmodesmata [33–35]. The loss of perinuclear Myo2-Venus following deflagellation might indicate that myosin is departing the perinuclear region for trafficking towards the plasma membrane at the base of the flagella. Further investigations are required to determine the precise role of Myo2, as well as involvement of the other two myosins, in flagellar regeneration. However, we have provided here the first description of a specific role for actin and myosin in the regulation of well-characterized mechanisms essential for proper ciliogenesis.
Supplementary Material
Highlights.
Compounds affecting actin polymerization and turnover shorten flagellar length.
Actin mutants have impaired flagellar growth.
Actin mutants have impaired IFT recruitment and injection into flagella.
Myosin inhibition recapitulates the actin mutant phenotype.
Acknowledgments
We thank current and former members of the Marshall lab and Lillian Fritz-Laylin for critical reading of the manuscript. Thanks to Will Ludington for his help utilizing software for fluorescence and IFT quantification and to David Kovar for providing Formin inhibitors and many helpful discussions. We also thank Haru-aki Yanagisawa for sharing the pKF18-CrAct vector and Takako Kato-Minoura for sharing the anti-NAP antibody. This work was funded by National Institutes of Health grant R01 GM097017 (WM), GM051173 (WS), a Postdoctoral Fellowship for Research Abroad from Japan Society for the Promotion of Science (RY) and an NRSA postdoctoral fellowship (PA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 2.Ludington WB, Wemmer KA, Lechtreck KF, Witman GB, Marshall WF. Avalanche-like behavior in ciliary import. Proc Natl Acad Sci U S A. 2013;110:3925–3930. doi: 10.1073/pnas.1217354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirono M, Uryu S, Ohara A, Kato-Minoura T, Kamiya R. Expression of conventional and unconventional actins in Chlamydomonas reinhardtii upon deflagellation and sexual adhesion. Eukaryotic Cell. 2003;2:486–493. doi: 10.1128/EC.2.3.486-493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato-Minoura T, Uryu S, Hirono M, Kamiya R. Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem Biophys Res Commun. 1998;251:71–76. doi: 10.1006/bbrc.1998.9373. [DOI] [PubMed] [Google Scholar]
- 5.Dentler WL, Adams C. Flagellar microtubule dynamics in Chlamydomonas: cytochalasin D induces periods of microtubule shortening and elongation; and colchicine induces disassembly of the distal, but not proximal, half of the flagellum. J Cell Biol. 1992;117:1289–1298. doi: 10.1083/jcb.117.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goddette DW, Frieden C. Actin polymerization. The mechanism of action of cytochalasin D. J Biol Chem. 1986;261:15974–15980. [PubMed] [Google Scholar]
- 7.Detmers PA, Carboni JM, Condeelis J. Localization of actin in Chlamydomonas using antiactin and NBD-phallacidin. Cell Motil. 1985;5:415–430. doi: 10.1002/cm.970050505. [DOI] [PubMed] [Google Scholar]
- 8.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato T, Kagami O, Yagi T, Kamiya R. Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct. 1993;18:371–377. doi: 10.1247/csf.18.371. [DOI] [PubMed] [Google Scholar]
- 10.Kato-Minoura T, Hirono M, Kamiya R. Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J Cell Biol. 1997;137:649–656. doi: 10.1083/jcb.137.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol. 2013;20:701–712. doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J Cell Biol. 2009;187:81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam K, Chin HF, Olivares AO, Saunders LP, De La Cruz EM, Kapoor TM. A myosin V inhibitor based on privileged chemical scaffolds. Angew Chem Int Ed Engl. 2010;49:8484–8488. doi: 10.1002/anie.201004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell. 2013;24:982–994. doi: 10.1091/mbc.E12-09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuler H, Mueller AK, Matuschewski K. Unusual properties of Plasmodium falciparum actin: new insights into microfilament dynamics of apicomplexan parasites. FEBS Lett. 2005;579:655–660. doi: 10.1016/j.febslet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 18.Sebe-Pedros A, Grau-Bove X, Richards TA, Ruiz-Trillo I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome biology and evolution. 2014;6:290–305. doi: 10.1093/gbe/evu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allingham JS, Smith R, Rayment I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nature structural & molecular biology. 2005;12:378–379. doi: 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- 20.Coleman RG, Carchia M, Sterling T, Irwin JJ, Shoichet BK. Ligand pose and orientational sampling in molecular docking. PLoS One. 2013;8:e75992. doi: 10.1371/journal.pone.0075992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sparkes I. Recent advances in understanding plant myosin function: life in the fast lane. Molecular plant. 2011;4:805–812. doi: 10.1093/mp/ssr063. [DOI] [PubMed] [Google Scholar]
- 22.Jin Y, Sultana A, Gandhi P, Franklin E, Hamamoto S, Khan AR, Munson M, Schekman R, Weisman LS. Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev Cell. 2011;21:1156–1170. doi: 10.1016/j.devcel.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schott DH, Collins RN, Bretscher A. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deretic D. Crosstalk of Arf and Rab GTPases en route to cilia. Small GTPases. 2013;4:70–77. doi: 10.4161/sgtp.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiao YC, Tuz K, Ferland RJ. Trafficking in and to the primary cilium. Cilia. 2012;1:4. doi: 10.1186/2046-2530-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, Moritz OL, Randazzo PA, Deretic D. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. The EMBO Journal. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen LB, Veland IR, Schroder JM, Christensen ST. Assembly of primary cilia. Dev Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- 29.Reiter JF, Mostov K. Vesicle transport, cilium formation, and membrane specialization: the origins of a sensory organelle. Proc Natl Acad Sci U S A. 2006;103:18383–18384. doi: 10.1073/pnas.0609324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldari CT, Rosenbaum J. Intraflagellar transport: it’s not just for cilia anymore. Curr Opin Cell Biol. 2010;22:75–80. doi: 10.1016/j.ceb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genetics. 2008;4:e1000315. doi: 10.1371/journal.pgen.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichelt S, Knight AE, Hodge TP, Baluska F, Samaj J, Volkmann D, Kendrick-Jones J. Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 1999;19:555–567. doi: 10.1046/j.1365-313x.1999.00553.x. [DOI] [PubMed] [Google Scholar]
- 34.Avisar D, Prokhnevsky AI, Dolja VV. Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. Journal of virology. 2008;82:2836–2843. doi: 10.1128/JVI.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baluska F, Cvrckova F, Kendrick-Jones J, Volkmann D. Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol. 2001;126:39–46. doi: 10.1104/pp.126.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.