Abstract
Genome sequencing studies have shown that human malignancies often bear mutations in four or more driver genes1, but it is difficult to recapitulate this degree of genetic complexity in mouse models using conventional breeding. Here we use the CRISPR-Cas9 system of genome editing2–4 to overcome this limitation. By delivering combinations of small guide RNAs (sgRNAs) and Cas9 with a lentiviral vector, we modified up to five genes in a single mouse hematopoietic stem cell (HSC), leading to clonal outgrowth and myeloid malignancy. We thereby generated models of acute myeloid leukemia (AML) with cooperating mutations in genes encoding epigenetic modifiers, transcription factors, and mediators of cytokine signaling, recapitulating the combinations of mutations observed in the human disease. Our results suggest that lentivirus-delivered sgRNA:Cas9 genome editing should be useful to engineer a broad array of in vivo cancer models that better reflect the complexity of human disease.
Murine models of myeloid malignancies and other cancers have traditionally been generated using genetic engineering of germline alleles, retroviral overexpression of putative oncogenes, retrovirus-based insertional mutagenesis, or RNA interference. Genome sequencing studies have identified a host of recurrently mutated genes,1 so technologies are now required to examine the function of large numbers of cancer genes and to recapitulate the complex combinations of mutations characteristic of human malignancies. While short hairpin RNA screens have been used to screen for functionally important genes in malignancy,5, 6 the use of RNA interference is complicated by incomplete genetic inactivation and substantial off-target effects.
Targeted nucleases, most recently the Cas9-based system,2 have been successfully used to engineer the genomes of multiple model organisms,7–12 but their use in primary hematopoietic stem and progenitor cells (HSPC), the disease-initiating cell populations for myeloid malignancies, is complicated by the difficulty of using common non-viral gene transfer methods in these cells.
To perform genome editing with high efficiency in primary HSPCs, and to track the engineered cells in vivo, we generated a modular lentiviral sgRNA:Cas9 vector for modeling of myeloid malignancies by genome editing in primary HSPCs in vivo (Figure 1A and Supplementary Figure 1). This lentiviral vector simultaneously delivers the Streptococcus pyogenes cas9 gene, a chimeric sgRNA and a fluorescent marker, similar to our recently developed system.2, 3, 13, 14 This enables the targeting of any genomic locus in a broad range of cell types, and consequent NHEJ-mediated gene disruption, by a one-step exchange of the target site (spacer).
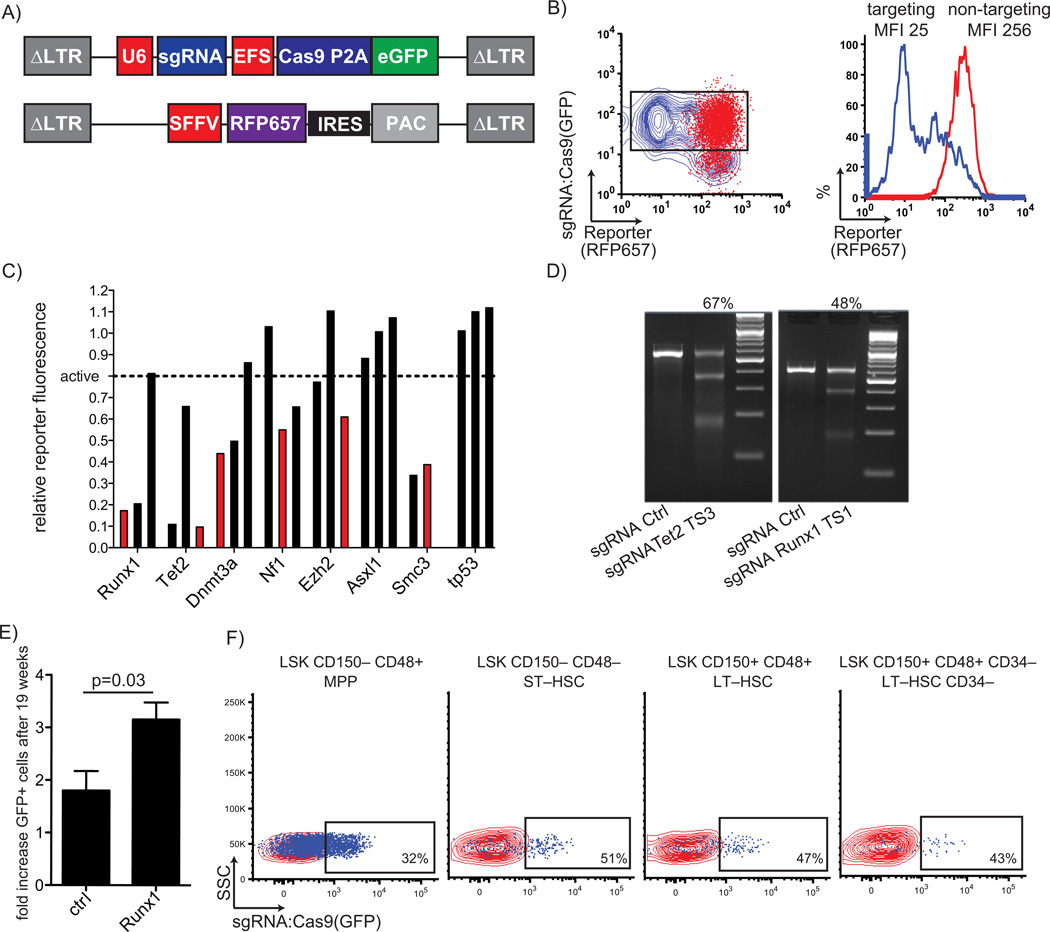
FIGURE 1. Stable modification of hematopoietic stem cells by a lentiviral sgRNA:Cas9 delivery system.
A) Depiction of a lentiviral vector for bi-cistronic expression of the sgRNA from a U6 promoter (U6) and cas9 from a short EF1a promoter (EFS) with a fluorescent protein marker (eGFP) from a picorna virus derived 2A auto-cleavage site (P2A). A lentiviral vector that allows incorporation of target sites into the coding sequence of a fluorescent protein (RFP657) is depicted at the bottom. A Puromycin resistance gene (PAC) is expressed from an internal ribosomal entry site (IRES) B) Super-transduction of a reporter cell line with an eGFP tagged sgRNA:Cas9 vector targeting the respective site (blue) induces loss of reporter fluorescence. A non-targeting sgRNA:Cas9 vector (red) does not affect reporter fluorescence. Quantification by plotting the MFI of the reporter in targeting (blue) vs non-targeting (red) sgRNA transduced cells (black box, left plot). C) Efficacy of spacers for recurrently mutated genes in myeloid malignancy. Efficacy was assessed with the RFP657 reporter system described in A) and B). Results were normalized to non-targeting spacer. Spacer marked in red were used in following experiments. D) Surveyor assay based confirmation of target site cleavage at genomic loci for Tet2 and Runx1 spacers indicated in panel C compared to non-targeting spacer. Percentages above are quantified cleavage efficacies. E) Peripheral blood sgRNA:Cas9 vector marking tracked over a period of 19 weeks. A significant increase of sgRNA:Cas9 expressing cells with a spacer targeting Runx1 (n=4) was observed in comparison to mice expressing a non-targeting spacer (n=4). F) Stable expression of the sgRNA:Cas9 vector after 19 weeks in hematopoietic stem and progenitor cells. Overlay of eGFP expression in sgRNA:Cas9 transduced cells (blue) and non-transduced cells (red) of the respective cell population. MPP=Multipotent progenitors; ST-HSC=short-term HSCs; LT-HSC=long-term HSCs; LT-HSC CD34-=most quiescent long-term-HSCs
We sought to model loss-of-function mutations in 8 genes that are recurrently inactivated in myeloid malignancies (Tet2, Runx1, Dnmt3a, Ezh2, Nf1, Smc3, p53 and Asxl1). We validated the targeting efficacy using a fluorescent reporter, similar to former testing of genome editing,15 harboring up to 20 sgRNA:Cas9 target sites as an alternative approach to commonly used endonuclease assays (Figure 1A/1B, Supplementary Data & Methods). At least one effective sgRNA was identified for 6 genes (Figure 1C, Supplementary Table 1), whereas no functional sgRNAs were found for p53 and Asxl1. Results for two spacers were confirmed by the standard T7 endonuclease assay, validating efficient cutting at genomic loci (Figure 1D). These findings demonstrate that our lentiviral system achieves efficient cleavage of target sites, and that engineered fluorescent reporter cell lines can provide rapid, quantitative evaluation of spacer efficacy
Having demonstrated the efficiency of our lentiviral delivery system in vitro, we examined whether we could engineer the genomes of primary murine HSPCs ex vivo. We isolated Lineage−/Sca1+/cKit+ (LSK) cells, which are enriched for HSPC activity, from mice with knock-in of the Flt3 internal tandem duplication (Flt3-ITD) mutation.16 Although Flt3-ITD mutations frequently occur in acute myeloid leukemia (AML), Flt3-ITD knock-in is insufficient to cause AML in mice, and we hypothesized that additional co-operating oncogenic lesions would induce transformation. Heterozygous Flt3-ITD expressing LSK cells were transduced with eGFP-expressing sgRNA:Cas9 lentiviral vectors targeting the Runx1 gene or a non-targeting sgRNA (Figure 1A) and were then transplanted into lethally irradiated wild-type (WT) congenic recipient mice. In serial analysis of the peripheral blood of recipient mice, we found that the population of cells expressing Cas9 and the Runx1 sgRNA expanded significantly over a period of 19 weeks in comparison to the population of cells expressing Cas9 and the non-targeting sgRNA (p=0.03), thus indicating that loss of Runx1 may induce a growth advantage in heterozygous Flt3-ITD expressing cells in vivo (Figure 1E, Supplementary Figure 2A).
Detection of eGFP-positive cells at 19 weeks post-transplant in all HSPC compartments indicated that we transduced long-term repopulating HSCs (Figure 1E and Supplementary Figure 2B and 2C). Stable expression of Cas9-eGFP over the 19 weeks in HSPCs and normal differentiation capacity indicates that continuous Cas9 expression at the levels provided does not cause significant toxicity in hematopoietic cells (Supplementary Figure 3). As further validation of genome modification of HSCs, we performed deep sequencing of the genomic sequence targeted in Runx1 in peripheral blood at serial time points. We identified at least 16 distinct insertions or deletions at the targeted Runx1 site in peripheral blood harvested 5 weeks post-transplant that were also present in blood 19 weeks post-transplant (Supplementary Figure 4). The majority of genome modifications were deletions ranging from 1–251bp. Overall, these results demonstrate that lentiviral delivery of the sgRNA:Cas9 system in HSCs is feasible and well tolerated, and that this system can be employed to edit the genomes of HSCs to interrogate gene function in hematopoiesis.
Because myeloid malignancies, and cancer more broadly, are caused by multiple cooperating genetic lesions (at least 3–4 oncogenic lesions in AML1), we next sought to perform multiplex targeting of cancer-associated genes to induce myeloid malignancy. To facilitate multiplex genome editing, we generated a two-vector system in which Cas9 and an eGFP are delivered from one lentiviral vector, whereas additional sgRNAs and a RFP657 fluorescent marker are delivered by a second lentiviral vector (Figure 2A). The small size of the sgRNA-only vector enables high viral titers and introduction of multiple sgRNAs into a single LSK cell.
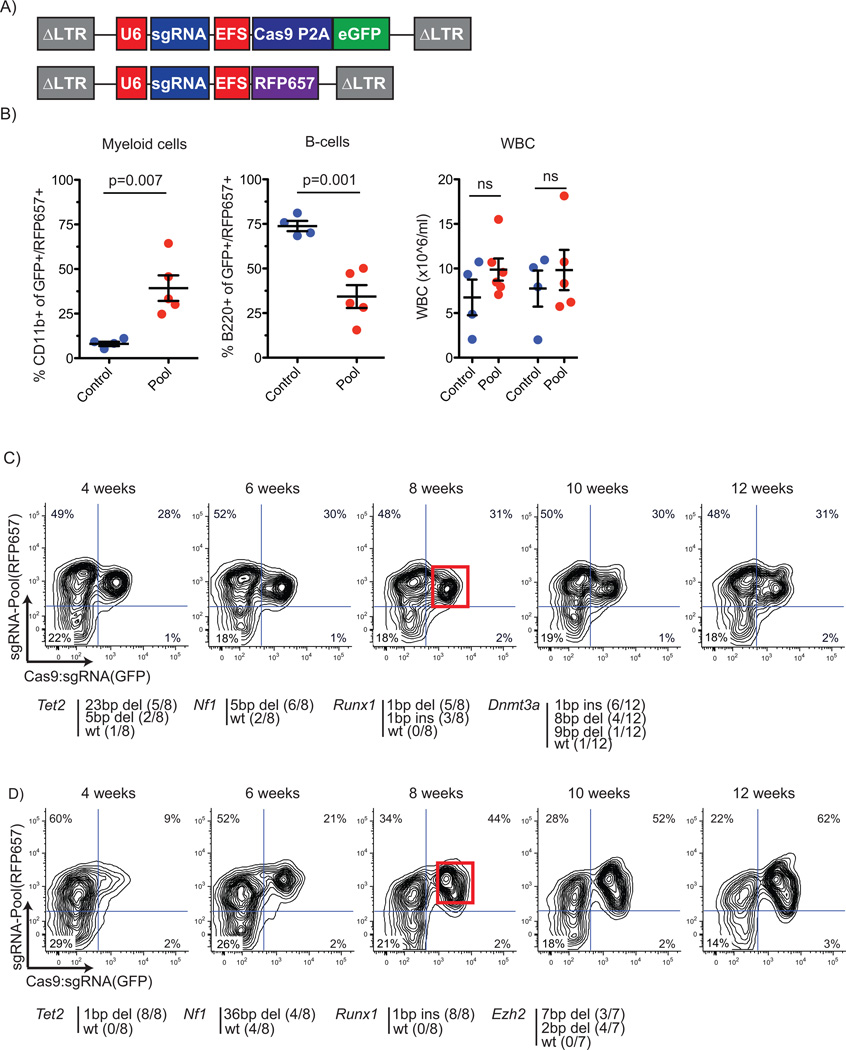
FIGURE 2. Multiplex gene targeting induces clonal development in vivo.
A) Schematic for multiplex gene targeting with two lentiviral vectors: a sgRNA:Cas9-eGFP vector expressing sgRNA, Cas9 and eGFP, and a vector expressing additional sgRNAs with a RFP657 fluorescent reporter gene from a short EF1a promoter.
B) Multiplexed gene targeting in mice (n=5) significantly increases myelomonocytic cells (CD11b+), decreases B-cells (B220+) and increases white blood cell counts at 12 weeks post-transplant in comparison to mice that were transplanted with control sgRNA vector transduced cells (n=4).
C) Peripheral blood sgRNA:Cas-eGFP vector expression (x-axis) and expression of a pool of vectors expressing RFP657 (y-axis) over time starting 4 weeks post-transplant. Clonal populations were detected by the pattern of eGFP/RFP657 expression. Two clones were isolated (red gates indicate clones) and genomic regions of targeted genes were sequenced (displayed in red box). Mono- and bi-allelic deletions in Tet2, Dnmt3a, Nf1 and Runx1 were detected in one case. Detected mutations at 8 weeks are shown below. Sequences of the deletions are shown in Supplementary Figure 8A. D) Another clone was analyzed as in panel C and was found to have mutations in Tet2, Ezh2, Nf1 and Runx1 B-II. Detected mutations at 8 weeks are shown below. Sequences of the deletions are shown in Supplementary Figure 8B.
We generated a pool of vectors with the single most effective sgRNA for the Tet2, Runx1, Dnmt3a, Nf1, Ezh2 and Smc3 genes. Transduction of LSK cells from C57Bl/6 wild-type mice with the pooled virus, followed by transplantation into lethally irradiated recipient mice, caused significant myeloid skewing of hematopoiesis (p=0.007) with reduction of B-cells (p=0.001) within the transduced cell population, and leukocytosis in some mice, as compared to the control group that received LSK cells transduced with control vectors expressing Cas9 and a non-targeting sgRNA (Figure 2B).
To determine the efficacy of multiplex genome editing and to assess the selective pressure for combinations of mutations we examined the genetics of putative clones in the peripheral blood of mice. As we used pooled virus expressing different sgRNAs, transduced cells bear different combinations of mutations, and cells with a genotype that drives a competitive advantage would gain a clonal dominance as occurs naturally during leukemogenesis. Two-dimensional flow cytometry analysis of eGFP and RFP657 allowed us to detect putatively clonal hematopoietic expansion in vivo (Figures 2C and 2D, Supplementary Figure 5), though some clonal, mutated populations were only GFP dim (Supplementary Figure 6 and 7).
For two mice, we purified populations from peripheral blood based on their eGFP/RFP657 density profile (red gated) by fluorescence-activated cell sorting (FACS) (Figure 2C and 2D). We examined the targeted genes by PCR amplification, sub-cloning and sequencing of targeted gene regions. In one mouse, a population expanded gradually over 12 weeks that had mutations in Tet2, Dnmt3a, Runx1 and Nf1 (Figure 2C, Supplementary Figure 8A), whereas no mutations in Ezh2 and Smc3 were detected. In a second mouse, a population expanded rapidly after transplantation with mutations in Tet2, Runx1, Nf1 and Ezh2 (Figure 2D, Supplementary Figure 8B), and no mutations in Dnmt3a and Smc3. In both cases, the detected mutational pattern (detection of two allele variants) suggests that a single cell was transduced with four separate sgRNAs, resulting in modification of one or both alleles of four tumor suppressor genes. Despite expansion of these clonal populations, no transformation to acute leukemia was observed in these mice during the observation period of up to 30 weeks.
Transformation to acute leukemia was observed in a different mouse transduced with the same pool of sgRNAs. A clonal population was detected at 4 weeks post-transplant, and the mouse was euthanized at 6.5 weeks post-transplant with mild leukocytosis (white blood cell (WBC) count: 15.3×106/ml; normal: 5–10×106/ml) and splenomegaly (436 mg; normal: 80–120mg). The cells were skewed to the myeloid lineage (Gr1+/CD11b+) (Figure 3A left panel) with 85% of cells expressing the sgRNA:Cas9 vectors (Figure 3A right panel). Analysis of the targeted genes revealed deletions in Dnmt3a, Ezh2, Smc3 and Nf1 (Supplementary Figure 9). Leukemic cells showed enhanced re-plating capacity and analysis of methylcellulose colonies from the leukemia confirmed that all mutations occurred within a single clone (Supplementary Figure 10, Supplementary Table 2). The peripheral blood from these mice revealed increased granulocytes and circulating blasts, indicative of leukemic transformation (Figure 3B, left panel). Bone marrow histology revealed greater than 20% blast cells and suppression of normal differentiation (Figure 3B, right panel, Supplementary Figure 11A, Supplementary Figure 15), consistent with acute myeloid leukemia (AML).
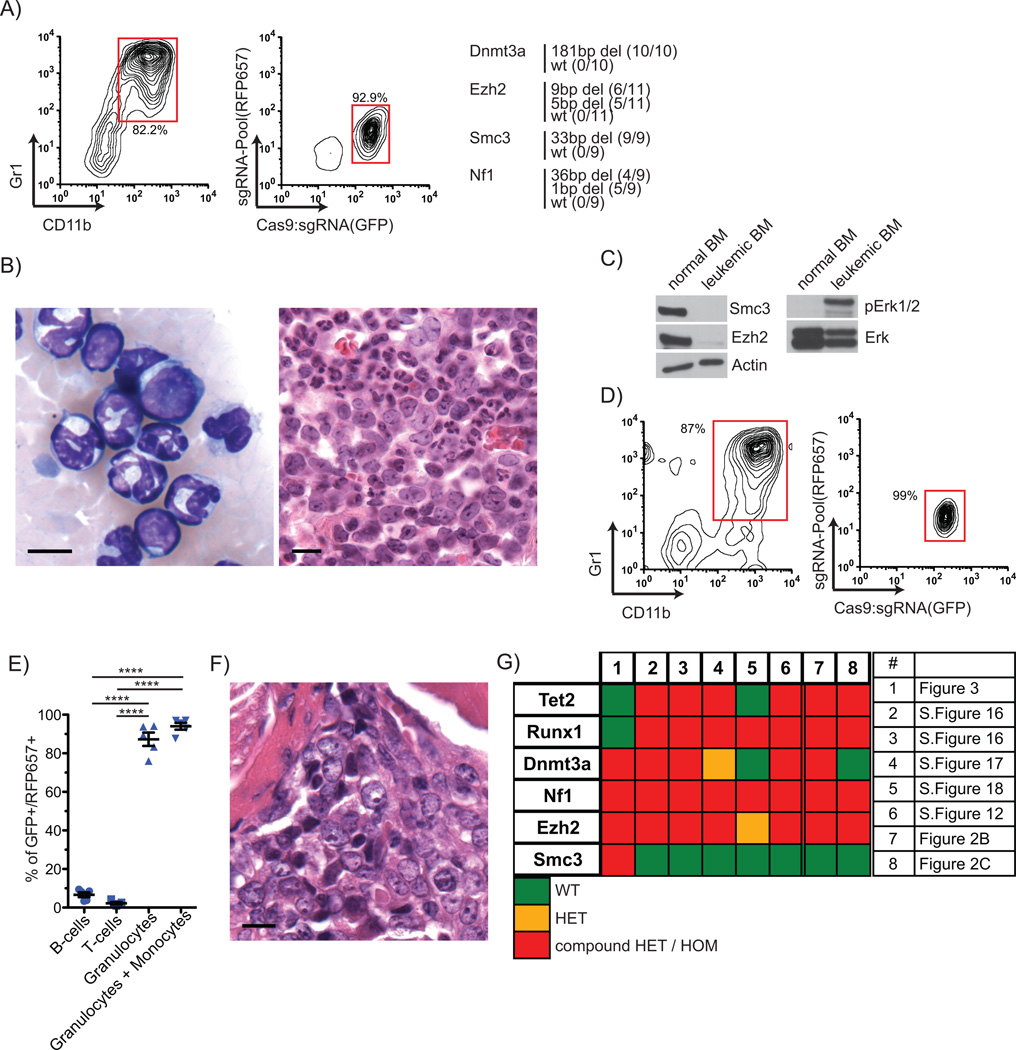
FIGURE 3. Myeloid malignancy modeling with multiplex genome editing.
A) Flow cytometric analysis of a diseased C57Bl/6 mouse for expression of the introduced Cas9:sgRNA vectors and for myeloid (Gr1+/CD11b+). Detected mutations are shown in the right panel (displayed in red box). Sequence alignments are shown in Supplementary Figure 9. B) Peripheral blood smear (left; May-Gruenwald/Giemsa (MGG); 1000x) and BM (right; Hematoxylin-Eosin(HE), 1000x) showing granulocytic cells and blasts. Scale bar (5µm) given in the lower left. C) Western blots of Ezh2, Smc3 and activated Erk1/2 (pErk) protein levels in the leukemic BM compared to normal C57Bl/6 wild-type BM. D) Representative flow cytometry of secondary transplant recipient mouse peripheral blood. E) Lineage distribution in peripheral blood from secondary mice 8 weeks post-transplant (n=5). F) Histopathology analysis of BM (MGG; 1000×) from a secondary mouse that succumbed to leukemia. Scale bar (5µm) given in the lower left. G) Heatmap summarizing mutations detected in all mice. Each column presents clone. Targeted genes are presented in rows. Color legend is given below. A table indicating where individual clones have been presented is given on the right (Supplementary Figure abbreviated as S. Figure).
We next confirmed that the detected mutations had an impact on protein expression and cellular phenotype. Both Ezh2 and Smc3 protein expression were markedly reduced, consistent with the bi-allelic mutations detected in both genes. Nf1 is a negative regulator of the Ras pathway, and we observed de-repression of the downstream targets, Erk1 and Erk2, as expected for cells with inactivation of Nf1 (Figure 3C). Serial re-plating of FACS-purified, double positive eGFP/RFP657-expressing cells indicates increased self-renewal capacity compared to wild-type C57Bl/6 BM cells (Supplementary Figure 10). Following transplantation into sub-lethally irradiated secondary recipient mice we observed robust engraftment of the GFP/RFP657 double positive mutated cells (Supplementary Figure 11B). At 8 weeks post-transplant two out of five recipient mice developed leukocytosis and succumbed to leukemia at 11–12 weeks post transplant (Supplementary Figure 11C/D). Mutated cells in all mice showed a strong differentiation bias, with >90% of the cells belonging to the myeloid lineage (Figure 3D and E). Bone marrow pathology confirmed AML in the secondary recipients (Figure 3F, Supplementary Figure 11E).
At 14 weeks following transplantation with the pool of tumor suppressor-targeting sgRNA pooled lentivirus, another primary mouse succumbed to a distinct myeloid leukemia characterized by splenomegaly (412mg), leukocytosis (WBC: 33.6×106/ml), excess myeloid cells in the BM (Supplementary Figure 12A), circulating blasts in peripheral blood and diagnostic features of AML in the bone marrow (>20% blasts) (Supplementary Figure 12B). Clonal mutations were identified in Tet2, Dnmt3a, Runx1, Nf1 and Ezh2. None of the mice transplanted with non-targeting sgRNA expressing cells developed disease during the course of the experiment (Supplementary Figure 13). These studies demonstrate that by using lentiviral delivered sgRNA:Cas9 components, we have engineered novel models of myeloid malignancy bearing mutations in 4–5 different genes that can be propagated and studied through serial transplantation.
To examine an alternative set of co-operative mutations, the same pool of sgRNAs targeting leukemia tumor suppressor genes was used to transduce LSK cells from mice with heterozygous knock-in of the Flt3-ITD oncogene allele. Targeting of Runx1 alone did not induce transformation of Flt3-ITD heterozygous expressing cells (Figure 1D). Consistent with the results of pooled tumor suppressor gene targeting in wild-type C57Bl/6 mice, expansion of sgRNA:Cas9-transduced, Flt3-ITD-expressing cells was observed over time. In some mice, expression of the sgRNA:Cas9 vector was GFP-dim at 8–10 weeks (Supplementary Figure 14). A total of five out of nine (55%) mice developed disease at 10–16 weeks with leukocytosis and splenomegaly (Supplementary Figure 15). Three of the mice were analyzed further. Bone marrow flow cytometry demonstrated that cells were myeloid biased in each of these mice (Supplementary Figures 16A, 17A, 18A). Diagnostic features of AML were evident with an excess of myeloblasts (>20%) in the bone marrow and circulating blast cells in the peripheral blood (Supplementary Figures 16B, 17B, 18B). Clonal mutations were detected in 3–5 genes in the three mice (Supplementary Figures 16C, 17C, 18C). Because the mutations were engineered in the Flt3-ITD heterozygous background, AML was generated from primary cells with mutations in 4 to 6 leukemia driver genes.
As off-target cleavage by Cas9:sgRNA could potentially contribute to leukemogenesis, we examined leukemias we generated for off-target genome editing.13, 17 We performed targeted sequencing of the top five off-target sites for each of the sgRNAs in each of 4 leukemias. None of the off-target sequences had any mutations in any of the leukemias examined, indicating that off-target cleavages are unlikely to be a significant contributor to leukemogenesis in our studies (Supplementary Table 3/4).
Taken together, these studies of lentivirus-delivered Cas9:sgRNA with multiplex sgRNAs demonstrate efficient genome editing in mouse HSPCs. We simultaneously inactivated multiple tumor suppressor genes causing outgrowth of a myeloid clone and transformation to acute leukemia. We describe models with up to 5 genes modified, with editing of up to 10 individual alleles, when a pool of sgRNAs targeting 6 genes was introduced in primary HSPCs (Figure 3G). Although genetic models have recently been generated using Cas9:sgRNA in whole organisms7–9, our system enables tissue specific interrogation of gene function in adult organisms whereas avoiding potential developmental defects. The genetic complexity thus generated is beyond the capacities of current approaches for engineering murine malignancies and will allow rapid generation of novel disease models beyond hematopoietic malignancies. Such models are critical for the functional interrogation of novel disease alleles and for the development and pre-clinical testing of targeted therapies.
The leukemia models generated reflect both the number and functional classes of co-occurring mutations that occur during the pathogenesis of human myeloid malignancies.1,18 All of the clones characterized had inactivating mutations in Nf1, a lesion that activates Ras signaling, and the only mutation in the screen that causes activated kinase signaling, a common feature of leukemias and other cancers.18 Although NF1 mutations are not frequent in AML patients, it was chosen as an amendable target for NHEJ mediated gene editing and representative for cytokine activation in AML. Nf1 inactivation in mice, however, is insufficient to cause AML and induces a long-latency myeloproliferativen disease.19 We also identified mutations in Ezh2, a gene recently shown to be involved in the differentiation block in AML20, in all mice that presented with myeloid transformation. Inhibition of myeloid differentiation has also been reported after deletion of Dnmt3a21, Tet222, 23 and Runx1,24 each of which were successfully mutated in our models.
Overall, our studies demonstrate a robust methodology to generate multiple genetic lesions via lentivirus-delivered Cas9:sgRNA in a broad range of cells, including the primary adult stem cells that are the disease-initiating population for leukemia. The use of viral-based Cas9:sgRNA will enable the rapid and efficient generation of cancer models that reflect the complex genetics of human disease.
Supplementary Material
Acknowledgments
The authors thank Cynthia Morton and Anita Hawkins form the Brigham and Women’s Cytogenetics Core and Roderick Bronson from the DF/HCC Rodent Pathology Core for technical assistance and discussions. The authors thank Feng Zhang for providing the CRISPR/Cas components. The authors also thank Axel Schambach, Emmanuelle Charpentier, Jan Kroenke and Ann Mullally for useful discussions, and thank Schraga Schwartz and Brian Haas for assistance with analysis of sequencing data. This work was supported by funding from the NIH (P01 CA108631), a Leukemia and Lymphoma Society Scholar Award, the SPARC consortium, a Center for Excellence in Genome Science grant (5P50HG006193-02 from the National Human Genome Research Institute) (A.R.) and Klarman Family Foundation at The Broad Institute (A.R.). DH was funded by the German Cancer Foundation (Mildred-Scheel Fellowship). MSK is an EMBO and European Hematology Association Fellow.
Footnotes
Author contribution
DH, MSK and BLE designed experiments. DH, MSK, DY, RB, RVP, MEC, and AT performed the experiments. DH, MSK, DY, RB, JCA, AR, and BLE analyzed and interpreted the data. DH and BLE wrote the manuscript.
Conflict of interest
A patent application relating to the work in this manuscript has been filed.
References
- 1.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scuoppo C, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nature biotechnology. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 11.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature biotechnology. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nature biotechnology. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee BH, et al. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer cell. 2007;12:367–380. doi: 10.1016/j.ccr.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annual review of genomics and human genetics. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Taylor BR, Shannon K, Clapp DW. Quantitative effects of Nf1 inactivation on in vivo hematopoiesis. The Journal of clinical investigation. 2001;108:709–715. doi: 10.1172/JCI12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S, et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120:1107–1117. doi: 10.1182/blood-2011-11-394932. [DOI] [PubMed] [Google Scholar]
- 21.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Mead AJ, et al. FLT3-ITDs instruct a myeloid differentiation and transformation bias in lymphomyeloid multipotent progenitors. Cell reports. 2013;3:1766–1776. doi: 10.1016/j.celrep.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 27.Zychlinski D, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 28.Schambach A, et al. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;13:391–400. doi: 10.1016/j.ymthe.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morozova KS, et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophysical journal. 2010;99:L13–L15. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.