Abstract
Ligands such as peptides, antibodies or other epitopes bind and activate specific cell receptors, and are employed for targeted cellular delivery of pharmaceuticals such as drugs, genes and imaging agents. Herein, we show that oxidized graphene nanoribbons, non-covalently functionalized with PEG-DSPE (1, 2-distearoyl-sn-glycero-3-phosphoethanolamine-N[amino(polyethyleneglycol)]) (O-GNR-PEG-DSPE) activate epidermal growth factor receptors (EGFRs). This activation generates predominantly dynamin-dependent macropinocytosis-like response, and results in significant O-GNR-PEG-DSPE uptake into cells with high EGFR expression. Cells with an integrated human papillomavirus (HPV) genome also show increased uptake due to the modulation of the activated EFGR by the viral protein E5. We demonstrate that this cell specific uptake of O-GNR-PEG-DSPE can be exploited to achieve significantly enhanced drug efficacies even in drug resistant cells. These results have implications towards the development of active targeting and delivery agents without ligand functionalization for use in the diagnosis and treatment of pathologies that overexpress EGFR or mediated by HPV.
1. Introduction
Many pharmaceutical formulations of drugs, genes and imaging agents show significant limitations that result in low therapeutic indices (ratio of therapeutic to toxic dose). These limitations stimulate the development of pharmaceutical delivery agents. There is now a large body of work that documents the tremendous progress in research and development of drug delivery agents [1–3]. A variety of micro- and nano-particles have been explored as delivery systems, including particles synthesized from carbon (fullerenes, metallofullerenes, carbon nanotubes and recently graphene[4–7]), ceramics, polymers, lipids, or metals and formed in a variety of configurations (i.e., spheres, tubes, branched structures, and shells)[8]. These systems serve as the scaffold onto which the active pharmaceutical ingredient (API) is covalently or non-covalently loaded. Multicomponent targeted delivery systems have also been developed, in which targeting moieties (e.g. antibodies or peptides) are covalently or non-covalently appended onto APIs, or API- loaded micro- or nano-particles. These systems typically target specific antigens on the cell surface to enhance the uptake of delivery systems into specific tumor cells to improve treatment efficacy. ‘Antibody mimics’ synthesized via imprinting of specific antigens onto polymeric scaffolds are used mainly in competitive binding assays[9]. Identification of materials that serve not only as scaffolds that efficiently load and deliver drugs, but also activate specific cell receptors without additional presence or functionalization of amino acid sequences or other epitopes would constitute a significant advance in targeted delivery system design.
Recently, we evaluated the cytotoxicity of oxidized graphene nanoribbon (O-GNR)-based formulations[10]. O-GNRs were synthesized in macroscopic amounts using an oxidative method pioneered by Kosynkin, Tour, and co-workers that longitudinally “unzips” carbon nanotubes[11]. O-GNR morphology is distinctly different from graphene oxide nanoplatelets, which are synthesized using graphite and are widely used in graphene-based cellular uptake and delivery studies [6, 12, 13]. O-GNRs (Figure 1A and B) water-solubilized with the amphiphilic polymer PEG-DSPE to form a supramolecular complex (O-GNR-PEG-DSPE) were incubated at various concentrations (0–400µg/ml) and time points (24–72 hours) in four different cell lines (HeLa, MCF7, SKBR 3 and NIH3T3). O-GNR-PEG-DSPEs were more cytotoxic to HeLa cells compared to other cell lines[10]. These studies provided preliminary indication that enhanced uptake of O-GNR-PEG-DSPEs into HeLa cells was an important reason for the observed differences in cytotoxicity. In this article, we further investigated O-GNR-PEG-DSPEs uptake mechanism, and report two surprising yet related phenomena. (1) O-GNR-PEG-DSPEs activate epidermal growth factor receptors (EGFRs) and are taken up in significant amounts in cells with high EGFR expression. (2) Cells with integrated human papilloma virus (HPV) genomes, which express EGFR (at normal or elevated levels), elicit enhanced O-GNR-PEG-DSPE uptake via the modulation of EFGR by the viral protein E5. We further demonstrate that these phenomena lead to differential and increased intracellular drug delivery efficacy even in drug resistant cells.
Figure 1.
(A) Representative atomic force microscopy (AFM) image of an O-GNR (Oxidized Graphene Nanoribbon); (B–H) Representative transmission electron microscopy (TEM) images. (B) O-GNR morphology (black arrows). (C) HeLa cell with membrane protrusions (white arrows) around O-GNR-PEG-DSPE DSPE (Oxidized Graphene Nanoribbons-1, 2-distearoyl-sn-glycero-3-phosphoethanolamine-N[amino(polyethylene glycol)]) aggregates (red arrows) after 15 minutes of incubation with O-GNR-PEG-DSPEs. (D) HeLa cell showing O-GNR-PEG-DSPE aggregates (red arrows) on the cell surface (blue arrows) and formation of membrane protrusions (white arrows) after 15 minutes of incubation with O-GNR-PEG-DSPEs. The image also shows smaller endosomal structures (yellow arrows). (E) HeLa cell showing significant uptake of O-GNR-PEG-DSPE aggregates (red arrows) into interconnected vesicular structures (blue arrows) localized around the nucleus (black arrow) after 30 minutes of incubation with O-GNR-PEG-DSPEs. (F) HeLa cell showing uptake of O-GNR-PEG-DSPE aggregates (red arrows) into large and small perinuclear vesicular structures (blue arrows; black arrow point to the nucleus) after 30 minutes of incubation with O-GNR-PEG-DSPEs. (G) HeLa cells, exposed to 1 µM gefitinib followed by O-GNR-PEG-DSPE treatment for 3 hours. Cells show no O-GNR-PEG-DSPE uptake (Brown arrows indicate individual cells). (H) Higher magnification of area within the black box in Panel (G) Large O-GNR-PEG-DSPE aggregates (red arrow) on surface of cells.
2. Methods
2.1 Synthesis and Characterization of O-GNR-DSPE-PEG formulations
Previously reported procedures were employed for the O-GNR synthesis from multi walled carbon nanotubes (Sigma-Aldrich,St Louis, MO) and preparation of O-GNR-DSPE-PEG dispersions.9,10 For atomic force microscopy (AFM) and TEM characterization, O-GNR-PEG-DSPE samples diluted to 5 µg/ml using a 1:1 ethanol water mixture were probe sonicated, (Cole Parmer Ultrasonicator LPX 750) and centrifuged at 2000 rpm for 30 minutes. The supernatant was collected and drop cast onto silicon wafers (AFM samples) or copper grids (Ted Pella) (TEM samples) and dried overnight. AFM was performed using a Nano Surf Easy Scan 2 AF microscope (NanoScience Instruments Inc, Phoenix, AZ), operating in tapping mode with a V-shaped cantilever. TEM was performed on a Tecnai Bio Twin G transmission electron microscope (FEI, Hillsboro, OR), at 80 kV. Digital images were acquired using an XR-60 CCD digital camera system (AMT, Woburn, MA).
2.2 Cell Culture
Eleven cell lines were used for the various experiments. All cell lines were obtained from ATCC (Manassas, VA, USA). HeLa, A549,MRC5,U251, A431 cells were grown in DMEM medium, SKBR3 cells were grown in McCoy’s medium, MCF7 and CaSki cells were grown in RPMI 1600 medium. SiHa, C33A and U87MG cells were grown in MEM media. All the media were supplemented with 10% fetal bovine serum and 1 % penicillin-streptomycin. Cells were incubated at 37°C in a humidified atmosphere of 5% CO2, and 95% air.
2.3 TEM of Cell Specimens
Six well plates with surfaces covered with ACLAR® film (Electron Microscopy Sciences, Hatford, PA) were plated with HeLa, C33A and A431 cells at a density of 5× 105 cells per plate, and exposed to O-GNR-PEG-DSPEs for 15 minutes, 30 minutes or 3 hours according to the specific assay. For inhibitor studies, cells were first incubated with 80 µM dynasore, 3 µg/ml filipin, 50 mM EIPA or 1µM Gefitinib for 30 minutes. Next, cells were treated with O-GNR-PEG-DSPE at 50 µg/ml concentrations for 15 minutes, 30 minutes or 3 hours. After 15 minutes, 30 minutes or 3 hours as per specific experiments, cells were fixed with 2.5% electron microscopy grade glutaraldehyde (Electron Microscopy Sciences, Hatford, PA) in 0.1 M PBS. After fixation, films containing fixed cells were placed in 2% osmium tetroxide in 0.1 M PBS, dehydrated through graded ethanol washes, and embedded in durcupan resin (Sigma-aldrich, St. Louis, USA). Areas with high cell densities were blocked, cut into 80 nm ultrathin sections using an Ultracut E microtome ( Reichert-Jung, Cambridge, UK), and placed on formvar-coated copper grids. The sections were then viewed with a Tecnai Bio Twin G transmission electron microscope (FEI, Hillsboro, OR) at 80 kV. Digital images were acquired using an XR-60 CCD digital camera system. (AMT, Woburn,MA). 15 cells of each cell line treated with O-GNR-PEG-DSPE was observed to come to a conclusion about uptake of O-GNR-PEG-DSPE in these cell lines
2.4 Confocal Microscopy
5× 105 cells were plated in glass bottom 35 mm plates and incubated for 18 hours. Following incubation, media was removed and replaced with serum-free media and cells were further incubated for 24 hours. Post-incubation, cells were treated with 50µg/ml O-GNR-PEG-DSPE for 15 minutes or 30 minutes. This step was followed by three washes with phosphate buffered saline and fixation for 30 minutes with 2.5% glutaraldehyde. Then, the fixed cells were treated with 0.5% Triton X-100, and washed 3 times with with PBS; this was followed by incubation with either phallodin rhodamine (for actin) (Invitrogen) or anti-phospho EGFR antibody with attached Alexa fluor 488(Millipore, Billerica, MA). Cells treated with gefitinib before O-GNR-PEG-DSPE treatment, EGF treated cells, and untreated cells were used as controls.
2.5 Doxorubicin loading on O-GNR-PEG-DSPE
The protocol used to load doxorubicin (Dox) onto O-GNR-PEG-DSPEs was adapted from the literature available for drug-loading for graphene. Briefly, 10 mg of Dox was mixed with 20 ml of 200µg/ml O-GNR-PEG-DSPE, bath sonicated for 60 minutes, and stirred for 24 hours. After 24 hours, the mixture was centrifuged at 13000 rpm for 60 minutes. The supernatant containing unloaded Dox was separated from the pelleted O-GNR-PEG-DSPEs containing loaded Dox by decantation. The amount of drug in the supernatant (i.e. drug not loaded) was calculated using the Dox absorbance spectra from the supernatant at 490 nm and the absorbance vs. Dox concentration standard curve. The amount of drug loaded on O-GNR-PEG-DSPEs was calculated by subtracting the drug in the supernatant from the starting amount of Dox (i.e., 1 mg). Drug loading efficiency (DLE) was calculated using the following formula: DLE = [(Drug Loaded in mg) / (Weight of O-GNR-PEG-DSPE in mg)]*100. The drug-loaded O-GNR-PEG-DSPE was resuspended in PEG-DSPE at 200µg/ml and 120 µg/ml (of O-GNR-PEG-DSPE) before being used as stock solutions for drug delivery and drug release experiments respectively.
2.6 Doxorubicin release from O-GNR-PEG-DSPE
Bipthhalate buffer (pH 4), phosphate buffered saline (pH 7), and borate buffered saline (pH 10) were used to assess drug release from Dox-loaded O-GNR-PEG-DSPEs. One ml of a 120 µg/ml Dox-loaded O-GNR-PEG-DSPE solution was re-suspended in 10 ml of the three different buffers to produce ~60 µg of Dox-loaded onto O-GNR-PEG-DSPE in each buffer solution. This solution was incubated at 37°C in a water bath placed on a horizontal shaker for 60 hours. One ml of buffer was collected every 12 hours, and the amount of Dox released was calculated using the absorbance of the collected buffer at 490 nm and the standard Dox absorbance vs. concentration standard curve.
2.7 Lactate Dehydogenase (LDH) Assay
This assay was conducted using a LDH release TOX-7 assay kit (Sigma-Aldrich, St Louis, MO). Cells were plated, at a density of 5 × 103 cells per well, in 96 well cell culture plates, and incubated for 18 hours. Following media changes, and cell treatment with Dox-loaded O-GNR-PEG-DSPE (50µg/ml), cells were incubated at 37°C for 24 hours. After 24 hours, media was collected from individual wells, and centrifuged at 1200 rpm for 5 minutes. Fifty µl of the media supernatant was added to a fresh 96 well plate along with LDH assay reagent, and incubated for 45 minutes. Absorbance values were recorded at 490 nm. The positive control was prepared by adding 10 µl of lysis solution to control cells, 45 min before centrifugation. LDH secretion (% of positive control) was calculated using the formula (ODtest – ODblank)/(ODpositive – ODblank), where ODtest is the optical density of control cells or cells exposed to O-GNR-PEG-DSPE, and ODpositive is the optical density of the positive control cells, and ODblank is the optical density of the wells without cells. Absorbance of culture media containing PEG-DSPE was used for baseline correction in all groups. LDH secretion (% of Dox-loaded O-GNR-PEG-DSPE treated cells) was calculated using the formula (ODtest – ODblank)/(ODtreatment – ODblank), where ODtest is the optical density of the inhibited cells exposed to Dox-loaded O-GNR-PEG-DSPE, ODtreatment is the optical density of the cells treated with only Dox-loaded O-GNR-PEG-DSPE, and ODblank is the optical density of the wells without cells. Absorbance of culture media containing PEG-DSPE was used for baseline correction in all groups.
2.8 Western Blots
HeLa, A431 cells and C33A cells were plated at cell density of 1 ×105 or 5 ×105 in six well plates and grown for 18 hours. Five ×105 A431 and C33A cells transfected with MSCV- FLAG -HPV16 E5 plasmid (Addgene, Cambridge,MA) in six well plates were also used. Cell lines were either left untreated or treated with O-GNR-PEG-DSPEs (50 µg/ml) for 15 minutes. Next, cells were lysed, and whole cell protein lysates were collected. Western blot analysis for EGFR expression before and after treatment with O-GNR-PEG-DSPEs was conducted using rabbit anti-EGFR primary antibody (Santa Cruz Biotechnology), and anti rabbit-HRP secondary antibody (Santa Cruz Biotechnology). Western blot analysis for activated EGFR expression before and after treatment with O-GNR-PEG-DSPE (and before and after gefitinib and O-GNR-PEG-DSPE treatment) was performed using mouse anti-phospho EGFR antibody (Millipore, Billerica, MA), and anti-mouse -HRP (Santa Cruz Biotechnology Santa Cruz, CA). Four different blots for each experiment were used for densitometry analysis using Image J and subsequent statistical analysis. Data obtained from densitometry analysis is represented as ratio of EGFR or p-EGFR band density and corresponding beta actin band density.
2.9 siRNA transfection of HeLa cells
One ×105 or 5 ×105 HeLa cells were plated in 96-well and 6-well plates and incubated for 18 hours. Cells were transfected with siRNA against EGFR (Santa Cruz Biotechnology, Santa Cruz, CA) and siRNA against HPV18E5 (Qiagen) using the Fugene 6 transfection reagent. Briefly, 1 µl of each siRNA against EGFR and siRNA against HPV18E5 were added to 489 µl of OPTI-MEM media and 10 µl of transfection reagent to prepare 500 µl of transfection reagent-DNA complex. One hundred µl of this complex was added to each well of 96 well plates and 500 µl of this complex was added to each well in 6 well plates. Transfection was allowed to proceed for 12 hours after which EGFR activation in the transfected cells was checked with confocal miscroscopy and flow cytometry using mouse anti-phospho EGFR primary and anti-mouse rhodamine as secondary antibody after exposing cells to 10 ng EGF per well. Drug delivery studies in siRNA transfected cells were conducted according to the method described in the LDH assay section. Ambion® Silencer® Negative Control #1 siRNA was used as the negative control and untreated cells (with and without siRNA treatment) were used as controls.
2.10 Transfection of HPV 16-E5 plasmid into A431 and C33A cells
MSCV-FLAG-HPV16-E5 plasmid was obtained from Addgene (Cambridge,MA), and transiently transfected into A431 and C33A cells using the Fugene 6 transfection reagent. Briefly, 2µg of the plasmid was mixed with 6 µl of Fugene 6 reagent and 94 µl of OPTI-MEM media to form 100 µl of transfection agent-plasmid mixture. 10 µl and 50 µl of this mixture was added to each well in 96 well plates containing 90 µl OPTI-MEM and cells and 6 well plates containing 450 µl OPTI-MEM media and cells respectively. The transfection was allowed to proceed for 12 hours following which successful transfection was confirmed by fluorescence microscopy and flow cytometry. Mouse anti-flag primary (Sigma-Aldrich,St Louis, MO) and anti-mouse-rhodamine (Sigma-Aldrich, St Louis, MO) secondary were used for fluorescence microcopy and flow cytometry. Drug delivery studies in the MSCV-FLAG-HPV16E5 transfected cells were conducted according to the method described under LDH assay. Untreated cells (transfected and untransfected) were used as controls.
2.11 Flow Cytometry
Flow cytometry was used toquantify the activated EGFR expression before and after siRNA transfection (against EGFR and HPV18 E5), and the transfection efficiency and expression of the MSCV-FLAG-HPV16 E5 plasmid in A431 and C33A cells. To prepare for flow cytometry, the cells were fixed using 2.5% glutaraldehyde, treated with 0.1% Triton X 100 for 5 minutes. Next, the cells were treated with appropriate antibodies, trypsinized, and resuspended in FACS buffer (Phosphate buffered saline containing 20% fetal bovine serum). Flow cytometry was performed immediately after all samples were prepared using a FACS Calibur Cell Sorter (BD Biosciences, San Jose, CA). BD FACS Diva 8.0 software was used for data analysis.
2.12 Statistics
All data are presented as mean ± standard deviation. Student‘t’ test was used to analyze the differences among groups. Each independent experiment (n=1) was an average of three wells done in parallel. One-way anova followed by Tukey Kramer post hoc analysis was used for multiple comparisons between groups. All statistical analyses were performed using a 95% confidence interval (p < 0.05).
3. Results
3.1. Water dispersed graphene nanoribbons activate epidermal growth factor receptors
We first conducted qualitative analysis of O-GNR-PEG-DSPE uptake into HeLa and three other cell lines (MCF7, A549 and MRC5) using transmission electron microscopy (TEM). These results suggest that HeLa cells take up O-GNR-PEG-DSPE aggregates into large vesicular cytoplasmic structures (resembling macropinosomes) (Figure 1C and D). Uptake seemed to be mediated through extensions from the plasma membrane, which engulfed O-GNR-PEG-DSPE on the cell surface (Figure 1 C and D, white arrows). We also observed large and small perinuclear vesicular structures within O-GNR-PEG-DSPE aggregates after 30 min of incubation (Figure 1 E and F) as well as a few endocytic vesicles, which formed before the macropinocytosis-like response, (Figure 1D, yellow arrows). In comparison, other cell lines (MCF7, MRC5 and A549) showed only small aggregates or O-GNR-PEG-DSPE uptake (Figure S1 A , B and C).
Next, we conducted inhibitor studies in HeLa cells to investigate the uptake mechanism at potentially safe concentrations of O-GNR-PEG-DSPE and inhibitors. Cellular analyses using TEM indicated that although very few endocytic vesicles were observed in non-inhibited HeLa cells treated with O-GNR-PEG-DSPE, dynasore (a dynamin inhibitor that prevents clathrin-mediated endocytosis) could completely prevent O-GNR-PEG-DSPE uptake (Figure S2 A and B) whereas filipin (a caveolae-mediated endocytosis inhibitor) does not show the same effect (Figure S2 C and D). Ethyl-isopropyl amiloride (EIPA), a macropinocytosis inhibitor largely prevented the uptake of larger aggregates, but in a few cases, smaller aggregates were found in endosomal vesicles even with EIPA inhibition (Figure S2 E and F). Based on these results, we hypothesized that the uptake mechanism for O-GNR-PEG-DSPE into HeLa cells is predominantly a dynamin-dependent macropinocytosis-like response although dynamin-dependent clathrin-mediated endocytosis may play a smaller role.
Investigation of actin polymerization of HeLa cells exposed to O-GNR-PEG-DSPE revealed the presence of circular dorsal ruffles (CDRs) 15 min post exposure (Figure S3B and C, white arrows). O-GNR-PEG-DSPE uptake was observed along CDR margins (Figure S3C, red arrows). Several reports demonstrated dynamin-dependent CDR formation, and a macropinocytosis-like uptake mechanism during activation and internalization of epidermal growth factor receptors (EGFRs),[14] involving plasma membrane protrusions that sequester a large number of ligand-bound (i.e., activated) EGFRs in large vesicular cytoplasmic structures. We observed similar protrusions in HeLa specimens treated with O-GNR-PEG-DSPE (Figure 1C and D). Activated EGFR uptake occurs via a complex network of connected vesicles unlike the spherical vesicles observed in classical macropinocytosis; localization of these vesicles is mainly perinuclear[14]. We noted O-GNR-PEG-DSPE in structures with similar features, such as connected vesicles with perinuclear localization (Figure 1E and F, blue arrows, black arrows point to nucleus). Thus, we performed additional inhibitory studies in HeLa cells with gefitinib (an EGFR kinase inhibitor) to ascertain whether O-GNR-PEG-DSPE uptake is dependent on EGFR activation and sequestration[15]. TEM results showed no observable nanoparticles inside the cells in cytoplasmic vesicles even after 3-hours exposure to the cells (Figure 1 G). O-GNR-PEG-DSPE aggregates were present on the membrane (Figure 1 H), but not CDRs (Figure S3D). Taken together, these results taken together indicated that gefitinib prevents cellular uptake of these nanoparticles (Figure 1 E).
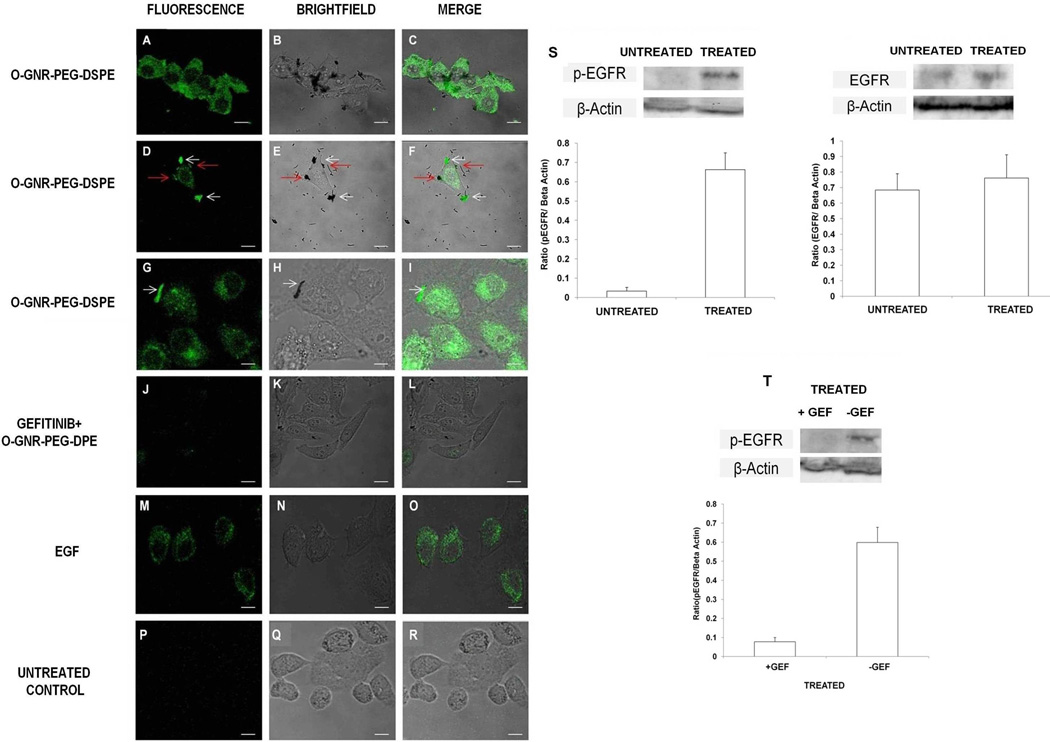
We next employed fluorescently tagged anti-phospho EGFR antibodies, and investigated whether O-GNR-PEG-DSPE activates EGFR in HeLa cells, and subsequently leads to O-GNR-PEG-DSPE uptake. HeLa cells grown in serum free media and treated with O-GNR-PEG-DSPE showed increased green fluorescence, which is indicative of increased EGFR activation (i.e. increased EGFR phosphorylation; Figure 2 A, B and C). O-GNR-PEG-DSPE activated cell surface EGFR (Figure 2 D, E and F, red arrows). Our results also indicated that O-GNR-PEG-DSPE aggregates co-localize with activated EGFR receptors in vesicles (Figure 2 D–I). HeLa cells exposed to gefitinib prior to O-GNR-PEG-DSPE treatment failed to show significant EGFR activation (Figure 2 J, K and L). A431 cells, which also overexpress EGFR showed activation, albeit at lower levels (Figure S4). MCF7 cells, which have low EGFR expression showed insignificant EGFR activation (Figure S4). Western blot analysis of unexposed and exposed HeLa cells showed that the number of activated EGFR receptors increased post exposure to O-GNR-PEG-DSPE. However, total EGFR content remained the same (Figure 2S). Gefitinib pre-treatment could decrease this phosphorylation (Figure 2T). These results provided additional corroboration that O-GNR-PEG-DSPE uptake is dependent on EGFR activation and sequestration.
Figure 2.
Representative fluorescence, bright field microscopy and merge images. All cells were exposed to 50µg/ml O-GNR-PEG-DSPE for 30 minutes. (A–C) Serum deprived HeLa cells exposed to O-GNR-PEG-DSPEs and anti-phospho epidermal growth factor receptor (EGFR) antibody that exhibit activated EGFR receptors (green fluorescence). (D–F) Activated surface EGFR receptors co-localized with O-GNR-PEG-DSPEs (red arrows), and activated receptors co-localized with O-GNR-PEG-DSPEs (white arrows) in vesicles. (G–I) activated receptors in vesicles colocalized with O-GNR-PEG-DSPEs (white arrows). (J–L) Gefitinib-pretreated HeLa cells show no significant EGFR activation. (M–O) HeLa cells exposed to EGF and anti-phospho EGFR antibody show activated EGFR (positive control). (P–R) Unexposed serum-deprived HeLa cells exposed to anti-phospho EGFR antibody show low activation of EGFR (baseline control). (S) Western blot and densitometric quantification for EGFR activation and total EGFR before and after treatment with O-GNR-PEG-DSPEs (T) Western blot and and densitometric quantification for EGFR activation with and without gefitinib treatment and treatment with O-GNR-PEG-DSPEs (Scale bar=20 µm for microscopy images).
3.2 Differential intracellular drug delivery and enhanced drug efficacies
We next performed drug delivery studies using O-GNR-PEG-DSPEs. The primary goals of these studies were twofold: 1) to further corroborate that O-GNR-PEG-DSPE uptake of into cells occurs via EGFR activation; 2) to determine possible reasons for higher O-GNR-PEG-DSPE uptake by HeLa cells. An ancillary goal was to investigate the capabilities of O-GNR-PEG-DSPEs as cell specific delivery agents. The FDA-approved anti-cancer drug doxorubicin (Dox) was chosen for these studies because it only enters cells through passive diffusion, and thus at low concentrations shows poor uptake into many cancer cells;[16] Dox typically requires a delivery agent to increase efficacy. Once inside the cell, membrane-bound p-glycoprotein (P-gp), expressed in certain cells (see Table 1), can ‘pump’ Dox out of the cells, [16] which may in turn lead to drug resistance. Cell clones resistant to low concentrations (treatment of cells with 20µg/ml Dox for 24 hours of Dox) were chosen. Thus, any increase in cytotoxic effects of similar concentration (20µg/ml) of Dox loaded onto O-GNR-PEG-DSPEs would imply increased delivery of Dox by O-GNR-PEG-DSPEs.
Table 1.
Summary of cell lines used, their EGFR and P-gp expression and drug delivery efficiency observed compared to free drug.
| Cell Line | Type | EGFR Expression |
Cytotoxic Activity |
P-glycoprotein expression |
| HeLa | HPV 18 infected cervical carcinoma |
Normal17 | 100% more than free drug |
Negative26 |
| CaSki | HPV 16 infected cervical carcinoma |
Over-expressed15 | 100% more than free drug |
Negative27 |
| SiHa | HPV 16 infected cervical carcinoma |
Normal15 | 75% more than free drug |
Positive27 |
| MDA-MB 231 |
Breast Adenocarcinoma | Over-expressed25 | 100% more than free drug |
Negative34 |
| A431 | Squamous Carcinoma of vulva |
Over-expressed18 | 20% more than free drug |
Positive28 |
| U251 | Glioblastoma | Over-expressed19 | 15% more than free drug |
Positive31 |
| U87MG | Glioblastoma | Normal20 | Same as free drug | Negative29 |
| MRC5 | Fibroblast cell line from fetal lung |
Normal16 | Same as free drug | Negative32 |
| A549 | Adenocarcinoma of alveolar epithelia |
Low-normal21 | Same as free drug | Positive30 |
| MCF7 | Breast Adenocarcinoma | Low22 | Same as free drug | Positive30 |
| C33A | Cervical carcinoma not infected by HPV |
Low23 | Same as free drug | Positive33 |
EGFR expression per cell: Low=less than 40000, Normal=40000-100000 24, Overexpressed=greater than 1000000
Dox was non-covalently loaded onto O-GNR-PEG-DSPE through simple pi-pi stacking interactions (Figure S5A)[5]. The optimum drug loading efficiency of the O-GNR-PEG-DSPE formulation was 40%, i.e., 0.4 mg of Dox could be loaded on 1 mg of O-GNR-PEG-DSPE (Figure S5B). Dox release from O-GNR-PEG-DSPE took place mainly in an acidic environment (pH, 4 to 6, 100% release in 3 days) with minimal drug release in a neutral or basic environment (pH ≥ 7, 10% release in 3 days). Drug release at acidic pH followed first order kinetics. Such a drug release profile is ideal for tumor drug delivery agents as the pH of tumors is predominantly acidic (Figure S5D)[17].
In vitro drug delivery studies were conducted using 50 µl of O-GNR-PEG-DSPE solution at a potential therapeutic dosage of 50 µg/ml (determined from previous cytotoxicity studies for O-GNR-PEG-DSPE[10]) loaded with 40% Dox by weight (20 µg/ml); treatment duration was 24 hours and the solution was studied in 11 cell lines (Table 1, cell line selection criteria for these studies were based on previous results). Since HeLa is a cervical cancer cell line with an integrated HPV genome, we chose cell lines comprised of cervical cancers cells with or without an HPV genome, and non-cervical cancer cells with low, normal or high EGFR expression[18–28]. Expression capability of P-gp, also known as multidrug resistant protein 1 (MDR1) expression status of the cell lines is also provided since it can also influence accumulation and efficacy of Dox inside the cells [29–37]. We used a lactate dehydrogenase (LDH) assay to assess cytotoxicity due to cellular delivery of Dox. LDH, a cytoplasmic marker for membrane integrity, provides an indirect means of assessing cytotoxicity[10]. Leaky membranes of damaged or dead cells show increased LDH release into surrounding media compared to normal intact cells. Our previous results validated this assay as a robust assessment of cytotoxicity without any interference from O-GNR-PEG-DSPE[10]. Under the same conditions, HeLa and CaSki cells showed ~100% greater LDH release, and SiHa cells showed ~75% greater LDH release when both were compared to cells treated with free Dox dispersed in PEG-DSPE (Figures 3B–F). HeLa and SiHa express normal levels of EGFR while CaSki cells over-express EGFR. At the same loading concentration, three cell types with high EGFR expression MDA-MB-231, A431 cells (a non cervical cancer squamous cell carcinoma cell line of the vulva) and U251 cells (a glioblastoma cell line) showed ~20% and ~15% higher LDH release respectively compared to cells treated with free Dox dispersed in PEG-DSPE. C33a, a cervical cancer cell line without the HPV genome, and other cell lines (U87MG, MCF7, A549 and MRC5 cells) did not show a statistically significant decrease in viability compared to free Dox (Table 1 and Supplementary Figure S6A–F). Six cell lines that showed increased LDH release (HeLa, CaSki, SiHa, MDA-MB-231, A431 and U251 cells) either over-express EGFR on their surface and/or have integrated HPV genome/genomes[18, 20–22, 28]. The other cell lines show low or normal EGFR expression [19, 23–26]. HeLa,CaSki and MDA-MB-231 cells, that either possess integarted HPV genome/s or overexpress EGFR or both, and do not express P-gp, exhibited the highest LDH release. Cell lines with an integrated HPV genome (SiHa) or high EGFR expression (A431and U251 cells) that also express P-gp, also exhibited higher LDH release compared to free drug. However, other cell lines with low or normal EGFR expression, but do not express P-gp (MRC5, U87MG, A549, MCF7, C33A), did not show a similar increase in LDH levels. These results indicate that cells with an HPV genome or EGFR overexpression exhibit increased drug efficacy, even in those with p-glycoprotein expression.
Figure 3.
(A–F) Lactic dehydrogenase (LDH) release after treatment with doxorubin (Dox)-loaded O-GNR-PEG-DSPEs in the following cells: (A) HeLa. (B) CaSki. (C)SiHa (D) MDA-MB-231 (E) A431. (F) U251 cells. Untreated cells, cells treated with free dox in PEG-DSPE, and lysed cells were additional controls. All data are normalized to LDH released from lysed control cells. Data are presented as mean ± SD (n = 6 per group). * indicates significant increase (p < 0.05) in LDH release compared to cells exposed to Dox in PEG-DSPE at the same concentration as loaded onto O-GNR-PEG-DSPEs. 1= Untreated Control 2= O-GNR-PEG-DSPE (50 µg/mL) 3= O-GNR-PEG-DSPE-Dox 4= DOX in PEG-DSPE (Same concentration as loaded) 5= DOX in PEG-DSPE (Double concentration as loaded)
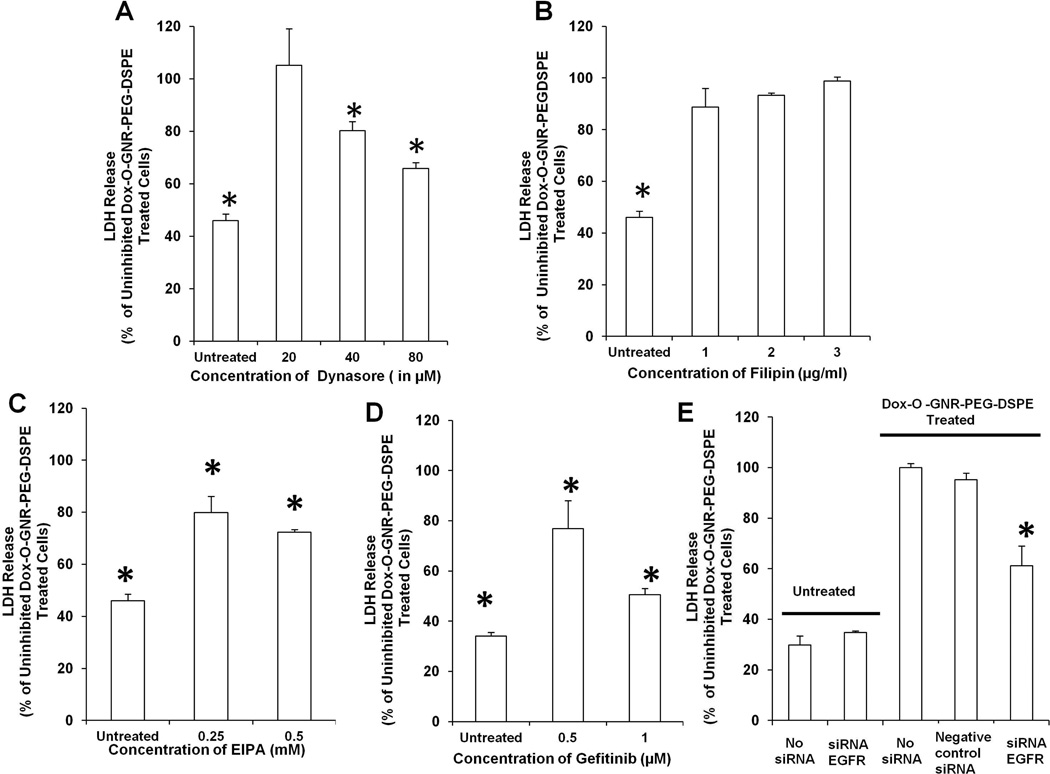
Next, drug delivery studies were performed on HeLa and CaSki cell lines after treatment with endocytosis and macropinocytosis inhibitors. We used very low non-toxic concentrations of cell inhibitors to ensure that any observed cell death was mainly due to Dox-loaded O-GNR-PEG-DSPE uptake. Figure 4A–D and Figure S7 A – D shows the salient results of these studies for HeLa cells and CaSki cells, respectively. In HeLa cells, unexposed control cells showed approximately 40% LDH release compared to cells exposed to the drug loaded nanoparticle (basal LDH release). HeLa cells treated with dynasore at 20µM showed no difference in LDH release. However, when HeLa cells were treated with dynasore at 80µM, LDH release decreased to ~65% of cells exposed to the drug loaded nanoparticle (Figure 4A). HeLa cells treated with filipin at various concentrations (1–3 µg/ml) did not show a significant decrease in LDH levels (Figure 4B). HeLa cells treated with EIPA, at 0.25mM and 0.5mM concentrations, showed a decrease to ~80% and ~72% LDH release, respectively (Figure 4C). HeLa cells treated with gefitinib, at 0.5µM concentration, decreased LDH release to ~ 76%, and at 1µM to ~50% (Figure 4D)
Figure 4.
LDH Release from Dox-loaded O-GNR-PEG-DSPE treated HeLa cells exposed to the following: (A) Dynasore. (B) Fillipin. (C) EIPA. (D) Gefitinib. (E) si RNA against EGFR. Data are presented as mean ± SD (n = 3 per group). * indicates significant decrease (p < 0.05) in LDH release compared to uninhibited cells exposed to Dox-O-GNR-PEG-DSPEs. All data are normalized to LDH release from non-inhibited Dox-O-GNR-PEG-DSPE-treated cells.
Dynasore inhibition of dynamin prevented O-GNR-PEG-DSPE uptake by both clathrin-mediated endocytosis and CDR-mediated vesicle formation. EIPA inhibits only macropinocytosis-like processes (CDR-mediated vesicle formation in this case). EIPA inhibition decreased LDH Release to ~72% of non-inhibited cells, similar to the ~65% LDH release observed after dynasore inhibition. This comparison provided additional indication that dynamin-dependent endocytosis might play a minor role in drug-loaded nanoparticle uptake. Filipin, which prevents caveolae-mediated endocytosis, did not show such an effect, thus ruling it out as an uptake mechanism. Gefitinib, a tyrosine kinase inhibitor also decreases LDH release in response to drug-loaded O-GNR-PEG-DSPEs to ~50% that of uninhibited cells. CaSki cells produced similar results indicating that the mechanism of O-GNR-PEG-DSPE uptake in both cell lines was similar although CaSki cells expressed more EGFR compared to HeLa cells (Figure S6 A – D). Further, transfection of small interfering ribonucleic acid (siRNA) against EGFR (Figure S9) into HeLa cells also significantly decreased cell death due to drug delivery, and corroborated the EGFR-mediated uptake mechanism of O-GNR-PEG-DSPEs (Figure 4E). Efficiency of EGFR inhibition in HeLa cells was confirmed by confocal microscopy and flow cytometry (Figure S9). Flow cytometry results indicated that si RNA transfected cells produced ~5 times less activated EGFR compared to uninhibited cells treated with EGF (activation evidenced by increase in fluorescence intensity from untreated control cells).
Similar drug delivery experiments on HeLa cells performed with Dox-loaded PEG-DSPE-coated multi-walled carbon nanotubes and graphene nanoplatelets as well as Dox-loaded Dextran or Pluronic F127 (routinely used to improve water dispersibility of carbon nanotube and graphene nanoparticles)-coated O-GNRs failed to show the same drug delivery response (Figure S8 A – D). O-GNRs themselves could not be used an experimental group due to poor dispersibility and stability in buffer and media solutions.
3.3 Role of human papillomavirus E5 protein
HeLa, CaSki and SiHa cells, which showed the highest cytotoxicity during drug delivery experiments are all squamous cervical carcinoma (SCC) cells with integrated human papillomavirus (HPV16 or HPV18) genomes[18]. Thus, we investigated the role of the HPV genome in mediating enhanced uptake of Dox-loaded O-GNR-PEG-DSPEs. HPV viral proteins E5, E6 and E7 are associated with major oncogenic factors in high-risk HPV [38]. E5, a transmembrane protein, increases ligand-dependent EGFR activation and signaling [39]. E5 functions by preventing acidification of endosomes containing internalized EGFR, which in turn prevents EGFR degradation, and results in recycling of activated EGFR back to the cell surface. The number of times that activated EGFRs are recycled by E5 before being degraded is still not known. Among the above three cell lines (HeLa, CaSki and SiHa), HPV genome incorporation is high in CaSki cells (500–600 copies), lower in SiHa cells (1–2 copies) and moderate in HeLa cells (40–50 copies) [40]. E5 expression in these cell lines is not proportional to the number of incorporated HPV genomes; HPV genome incorporation is random, and often results in loss of viral genome fragments encoding E5 [41]. Although, several studies showed the presence of E5 open reading frames or E5 encoding mRNA transcripts in all three cell lines [42–44] very few studies successfully quantified E5 protein expression in the three cell lines.
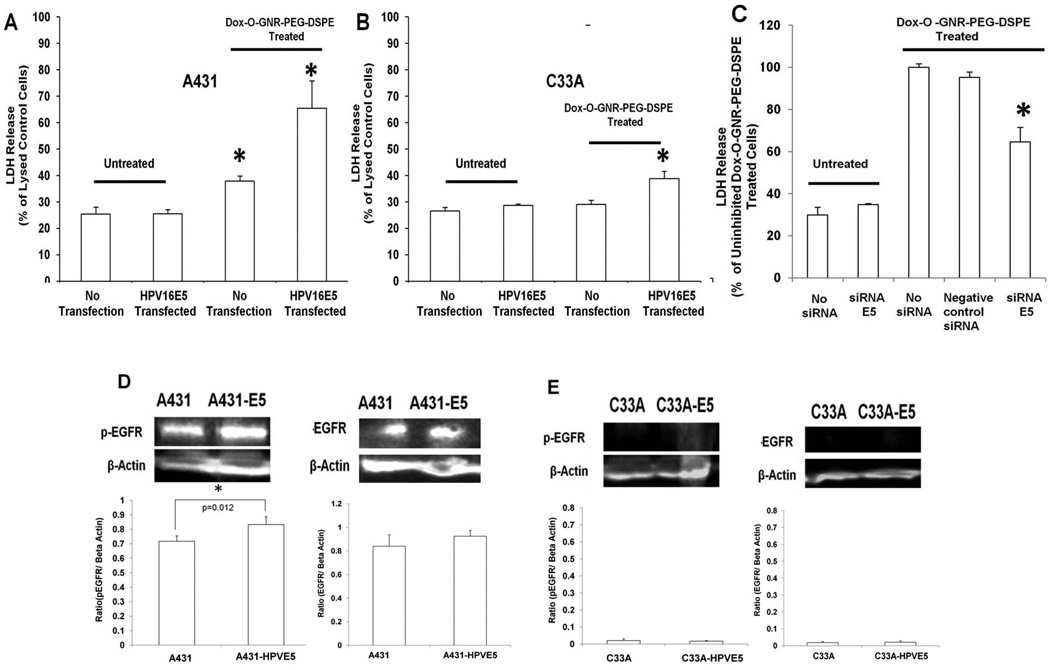
To investigate the role of E5 in O-GNR-PEG-DSPE uptake, we transiently transfected a plasmid containing a FLAG tagged HPV16E5 gene into A431 cells (which show high EGFR expression) and C33A cells (which show low EGFR expression); we conducted drug delivery experiments on these transfected cells and confirmed E5 expression and localization using confocal microscopy (with anti-FLAG antibodies) and quantification using flow cytometry (Figure S10). The transfection efficiency was ~99.8% in A431 cells and ~62% in C33A cells (calculated from flow cytometry results shown in Figure S10). Results indicated that E5 over-expression in A431 cells increased the drug delivery response (i.e., increased LDH release) by approximately 52 % (Figure 5A). We also observed a small increase in activated EGFR in EGF-treated and HPV16E5-transfected A431 cells (Figure 5D). C33A cells transfected with FLAG tagged HPV16E5 did not show the same response although we did see a small increase in LDH release (Figure 5B). This difference is probably due to low EGFR expression in C33A cells compared to A431 cells. We did not observe increased EGFR expression or activation in transfected C33A cells (Figure 5E). TEM images qualitatively confirmed that over-expression of HPV16E5 increased uptake of O-GNR-PEG-DSPEs in A431 cells but not in C33A cells (Figure S11A–D). siRNA transfection against E5 in HeLa cells (Figure S9) decreased the cytotoxic response upon treatment with Dox loaded O-GNR-PEG-DSPEs (Figure 5C). Flow cytometry results quantified the EGFR inhibition in response to transfection of siRNA against HPV 18 E5, and showed, upon treatment with EGF, ~ 4.5 times lower activation of EGFR in transfected cells compared to uninhibited cells (activation evidenced by increase in fluorescence intensity from untreated control cells) (Figure S9) Taken together, results shown in Figure 5 indicate that presence E5 protein by itself is insufficient for high uptake of O-GNR-PEG-DSPE. Normal or high EGFR expression is necessary to achieve significantly increased drug efficacy. These results corroborated our hypothesis that the E5 is involved in the increased uptake of Dox loaded O-GNR-PEG-DSPEs into HPV-mediated cells.
Figure 5.
(A–D) LDH Release from Dox-loaded O-GNR-PEG-DSPE treated with the following in A431, C33A or HeLacells: (A) A431 cells transfected with HPV16 E5. (B) C33A cells transfected with HPV16 E5. Untransfected and untreated cells were used as controls. Data are presented as mean ± SD (n = 3 per group). * indicates significant increase (p < 0.05) in LDH release compared to untreated control cells. All data are normalized to LDH release from lysed control cells. (C) HeLa cells transfected with siRNA against HPV18 E5. For A, B and C, data are presented as mean ± SD (n = 3 per group). * indicates significant decrease (p < 0.05) in LDH release compared to uninhibited cells treated with Dox-O-GNR-PEG-DSPE. All data are normalized to the LDH released from uninhibited Dox-O-GNR-PEG-DSPE treated cells. (D) Western blot and densitometric quantification of EGFR activation (phosphorylated EGFR) and expression in A431 cells before and after transfection with the HPV 16 E5 expressing plasmid. (E) Western blot and densitometric quantification depicting EGFR activation (phosporylated EGFR) and expression in C33A cells before and after transfection with the HPV 16 E5 expressing plasmid after treatment with Dox-loaded O-GNR-PEG-DSPE.
4. Discussion
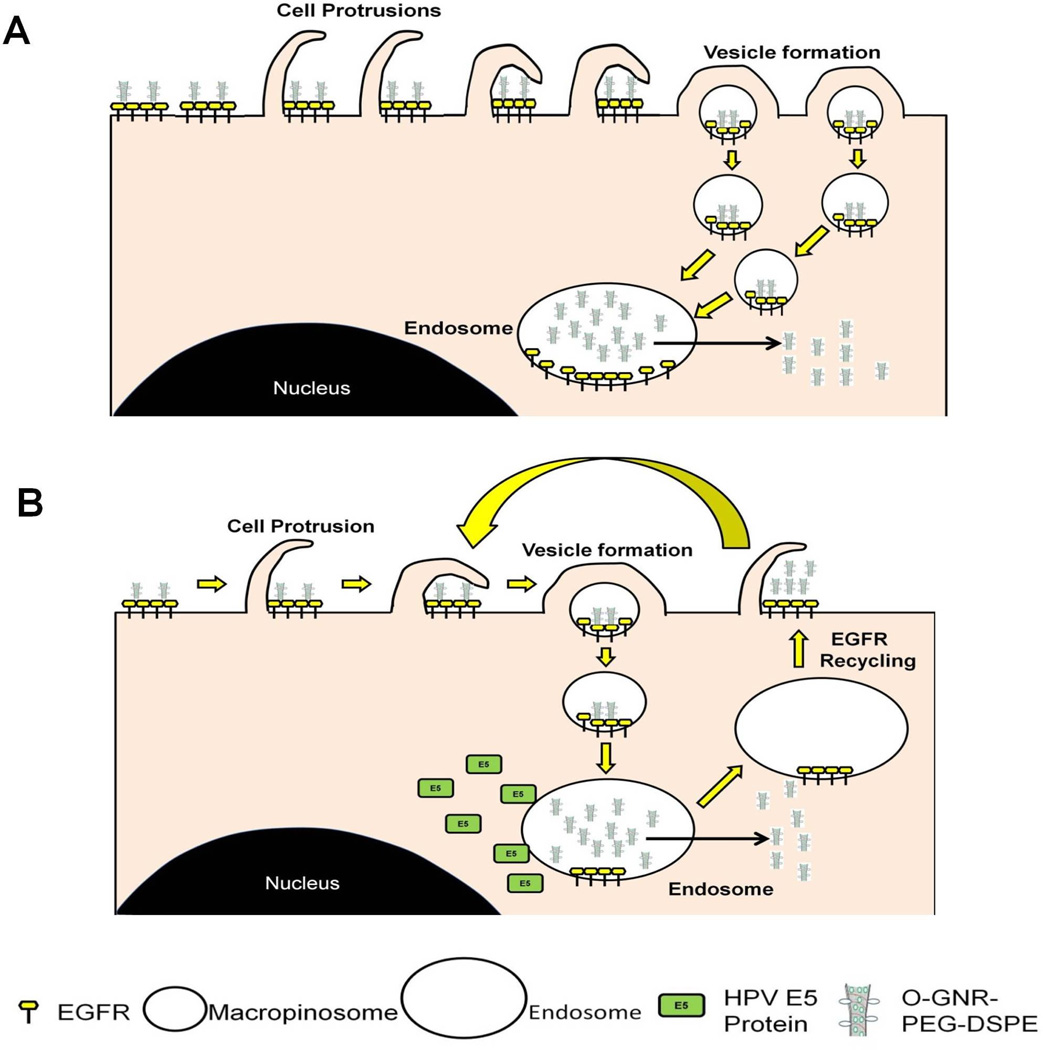
Based on above results and existing literature regarding EGFRs,[45] and HPV E5 protein[39, 41], we propose the following two models to explain O-GNR-PEG-DSPE uptake and drug delivery processes (Figure 6). For non-HPV cells (Figure 6A), the O-GNR-PEG-DSPEs activate dense EGFR clusters present at different locations on cell membrane surface. Simultaneous activation of these EGFRs results in a predominantly macropinocytotic response leading to O-GNR-PEG-DSPE uptake along with these receptors. Cell with high EGFR expression should contain a greater number of these EGFR clusters. In these cells, O-GNR-PEG-DSPEs are more likely to interact with and activate these EGFRs clusters on the membrane surface; thus, these cells show higher O-GNR-PEG-DSPE uptake and consequently increased drug delivery and efficacy compared to cells with normal or low EGFR expression. For cells with the HPV genome (Figure 6B), E5 prevents degradation of activated EGFR receptors, and recycles them onto the cell surface, which results in repetition of the uptake mechanism for nanoparticles on the cell surface or surrounding the cells without further EGFR activation. Consequently, these cells with normal or high EGFR expression have higher uptake capacity for Dox-loaded O-GNR-PEG-DSPEs, and show increased cell death compared to cells with only high EGFR expression.
Figure 6.
Schematic representation of mechanism of uptake postulated for O-GNR-PEG-DSPEs into (A) EGFR overexpressing cells. O-GNR-PEG-DSPEs activate dense EGFR clusters at different locations on cell membrane surface. Simultaneous activation of these EGFRs results in a predominantly macropinocytotic response for uptake of O-GNR-PEG-DSPEs along with these receptors. (B) Cells with an integrated HPV genome. E5 prevents degradation of activated EGFR receptors, instead recycling them onto the cell surface; this results in a repeated uptake process without further EGFR activation and additional uptake of on nanoparticles on the cell surface or the surrounding regions.
The exact mechanism by which O-GNR-PEG-DSPE activates EGFR requires further study. Interaction of nanoparticles with cell surface proteins could be influenced by a variety of physicochemical attributes including size, morphology including surface, and charge [46]. Certain nanoparticles interact with extracellular matrix components and these interactions, in turn, activate cell receptors [46]. Recently synthetic heteropolymers, comprising of PEG blocks non-covalently functionalized onto single walled carbon nanotubes, have also been reported to facilitate biomolecular recognition [47]. The ‘normal’ activators for EGFRs are EGF and transforming growth factor alpha (TGF-α),[48] EGFRs can also be activated by other mechanisms such as reactive oxygen species (ROS) generation, cellular stress and membrane depolarization [49]. Thus, additional experiments are underway to elucidate potential EFGR activation mechanism(s) by O-GNR-PEG-DSPEs. Further our results suggest that EGFR activation depends on yet to be determined physicochemical characteristic(s) of the O-GNR-PEG-DSPE complex and that O-GNRs are a critical component. Further investigation is needed to determine whether our results are unique to PEG-DSPE-coated O-GNRs[11] used in this study, or can be generalized to nanoribbons prepared by other methods [50].
5. Conclusions
O-GNR-PEG-DSPEs activate epidermal growth factor receptors (EGFRs) and are taken up in significant amounts in cells with high EGFR expression. This phenomena leads to differential and increased intracellular drug delivery efficacy. For cells with high EGFR expression, or with HPV genome, the intracellular delivery of the drug doxorubicin by O-GNR-PEG-DSPE increases its efficacy by 100% greater compared to drug alone. Even in cells with high EGFR expression, or with HPV genome, that express the multidrug resistant protein 1 (MDR1), the drug efficacies increase upto 75% compared to drug alone. Drug alone dispersed in PEG-DSPE at the same or twice the concentration loaded onto O-GNR-PEG-DSPEs did not show any statistically significant increase in its efficacy compared to untreated cells. Cells with integrated human papilloma virus (HPV) genomes, which express EGFR (at normal or elevated levels), elicit enhanced O-GNR-PEG-DSPE uptake via the modulation of EFGR by the viral protein E5. Nevertheless, the intriguing cell specific uptake of O-GNR-PEG-DSPEs together with its other attributes (i.e., high API loading efficiencies, API release at acidic pH, and other nanoparticle-related properties such as enhanced permeability-and-retention (EPR) effect[8]) suggest strong potential for O-GNR-PEG-DSPE use as a pharmaceutical delivery agent to detect or treat pathologies that overexpress EGFR or are mediated by HPV [51]. Such a delivery agent could mitigate the problem encountered with presently used agents such as drug resistance and toxicity to normal cells [52, 53].
Supplementary Material
Acknowledgements
This work was supported by NIH Director’s New Innovator Award 1DP2OD007394-01.The authors thank Susan Van Horn and Guo Wei Tian from Stony Brook Central Microscopy for help with TEM and confocal microscopy respectively. The authors also thank Renee Pessin of RDP Editorial Consulting Inc. for editorial assistance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. Competing Financial Interests
Stony Brook University and the investigators have filed patents related to the technology reported in this article. If licensing or commercialization occurs, the researchers are entitled to standard royalties
References
- 1.Sant S, Tao SL, Fisher OZ, Xu Q, Peppas NA, Khademhosseini A. Microfabrication technologies for oral drug delivery. Adv Drug Deliv Rev. 2012;64:496–507. doi: 10.1016/j.addr.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubbell JA, Langer R. Translating materials design to the clinic. Nat Mater. 2013;12:963–966. doi: 10.1038/nmat3788. [DOI] [PubMed] [Google Scholar]
- 3.Bae YH, Park K. Targeted drug delivery to tumors: Myths, reality and possibility. J Controlled Release. 2011;153:198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalwani G, Sitharaman B. Multifunctional Fullerene-and metallofullerene-based nanobiomaterials. Nano LIFE. 2013;03:1342003. [Google Scholar]
- 5.Liu Z, Tabakman S, Welsher K, Dai H. Carbon nanotubes in biology and medicine: in vitro and in vivo detection, imaging and drug delivery. Nano Res. 2009;2:85–120. doi: 10.1007/s12274-009-9009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitounis D, Ali-Boucetta H, Hong BH, Min D-H, Kostarelos K. Prospects and Challenges of Graphene in Biomedical Applications. Adv Mater. 2013;25:2258–2268. doi: 10.1002/adma.201203700. [DOI] [PubMed] [Google Scholar]
- 7.Frame MD, Dewar AM, Mullick Chowdhury S, Sitharaman B. Vasoactive effects of stable aqueous suspensions of single walled carbon nanotubes in hamsters and mice. Nanotoxicology. 2014;8:867–875. doi: 10.3109/17435390.2013.837209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haley B, Frenkel E. Nanoparticles for drug delivery in cancer treatment. Urol Oncol-Semin Ori. 2008;26:57–64. doi: 10.1016/j.urolonc.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Vlatakis G, Andersson LI, Müller R, Mosbach K. Drug assay using antibody mimics made by molecular imprinting. Nature. 1993;361:645–647. doi: 10.1038/361645a0. [DOI] [PubMed] [Google Scholar]
- 10.Mullick Chowdhury S, Lalwani G, Zhang K, Yang YJ, Neville K, Sitharaman B. Cell specific cytotoxicity and uptake of graphene nanoribbons. Biomaterials. 2013;34:283–293. doi: 10.1016/j.biomaterials.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosynkin DV, Higginbotham AL, Sinitskii A, Lomeda JR, Dimiev A, Price BK, Tour AM. Longitudinal unzipping of carbon nanotubes to form graphene nanoribbons. Nature. 2009;458:872–876. doi: 10.1038/nature07872. [DOI] [PubMed] [Google Scholar]
- 12.Kanakia S, Toussaint JD, Chowdhury SM, Lalwani G, Tembulkar T, Button T, et al. Physicochemical characterization of a novel graphene-based magnetic resonance imaging contrast agent. International journal of nanomedicine. 2013;8:2821. doi: 10.2147/IJN.S47062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury SM, Kanakia S, Toussaint JD, Frame MD, Dewar AM, Shroyer KR, et al. In Vitro Hematological and In Vivo Vasoactivity Assessment of Dextran Functionalized Graphene. Scientific reports. 2013:3. doi: 10.1038/srep02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orth JD, Krueger EW, Weller SG, McNiven MA. A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res. 2006;66:3603–3610. doi: 10.1158/0008-5472.CAN-05-2916. [DOI] [PubMed] [Google Scholar]
- 15.Kitazaki T, Oka M, Nakamura Y, Tsurutani J, Doi S, Yasunaga M, Takemura M, Yabuuchi H, Soda H, Kono S. Gefitinib, an EGFR tyrosine kinase inhibitor, directly inhibits the function of P-glycoprotein in multidrug resistant cancer cells. Lung Cancer. 2005;49:337–343. doi: 10.1016/j.lungcan.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 16.Arora HC, Jensen MP, Yuan Y, Wu A, Vogt S, Paunesku T, Woloschak GE. Nanocarriers enhance doxorubicin uptake in drug-resistant ovarian cancer cells. Cancer Res. 2012;72:769–778. doi: 10.1158/0008-5472.CAN-11-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerweck LE, Vijayappa S, Kozin S. Tumor pH controls the in vivo efficacy of weak acid and base chemotherapeutics. Mol Cancer Ther. 2006;5:1275–1279. doi: 10.1158/1535-7163.MCT-06-0024. [DOI] [PubMed] [Google Scholar]
- 18.Bachran D, Schneider S, Bachran C, Urban R, Weng A, Melzig MF, Hoffmann C, Kaufmann M, Fuchs H. Epidermal growth factor receptor expression affects the efficacy of the combined application of saponin and a targeted toxin on human cervical carcinoma cells. Int J Cancer. 2010;127:1453–1461. doi: 10.1002/ijc.25123. [DOI] [PubMed] [Google Scholar]
- 19.Home N. Wild type EGFR is stabilized by direct interaction with HSP90 in cancer cells and tumors. Neoplasia. 2012;14(8):670–7. doi: 10.1593/neo.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkers J, Henegouwen PvB en, Boonstra J. Three classes of epidermal growth factor receptors on HeLa cells. J Biol Chem. 1991;266:922–927. [PubMed] [Google Scholar]
- 21.Defize L, Arndt-Jovin D, Jovin T, Boonstra J, Meisenhelder J, Hunter T, Hay HTde, de Laat SW. A431 cell variants lacking the blood group A antigen display increased high affinity epidermal growth factor-receptor number, protein-tyrosine kinase activity, and receptor turnover. J cell biol. 1988;107:939–949. doi: 10.1083/jcb.107.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stea B, Falsey R, Kislin K, Patel J, Glanzberg H, Carey S, Ambrad AA, Meuillet EJ, Martinez JD. Time and dose-dependent radiosensitization of the glioblastoma multiforme U251 cells by the EGF receptor tyrosine kinase inhibitor ZD1839 (‘Iressa’) Cancer lett. 2003;202:43–51. doi: 10.1016/j.canlet.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, Tan P, Depinho RA, Cavenee W, Furnari F. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirotnak FM, Zakowski MF, Miller VA, Scher HI, Kris MG. Efficacy of cytotoxic agents against human tumor xenografts is markedly enhanced by coadministration of ZD1839 (Iressa), an inhibitor of EGFR tyrosine kinase. Clin Cancer Res. 2000;6:4885–4892. [PubMed] [Google Scholar]
- 25.Abourbeh G, Shir A, Mishani E, Ogris M, Rödl W, Wagner E, Levitzki A. PolyIC GE11 polyplex inhibits EGFR-overexpressing tumors. IUBMB life. 2012;64:324–330. doi: 10.1002/iub.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meira DD, Almeida VH, Mororó JS, Caetano MS, Nóbrega IP, Batista D, Sternberg C, Ferreira CG. Efficient Blockade of Akt signaling is a determinant factor to overcome resistance to Matuzumab. Mol cancer. 2011;10:151. doi: 10.1186/1476-4598-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong S-F. Cetuximab: an epidermal growth factor receptor monoclonal antibody for the treatment of colorectal cancer. Clin ther. 2005;27:684–694. doi: 10.1016/j.clinthera.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Cai Z, Chen Z, Bailey KE, Scollard DA, Reilly RM, Vallis KA. Relationship between induction of phosphorylated H2AX and survival in breast cancer cells exposed to 111In-DTPA-hEGF. J Nucl Med. 2008;49:1353–1361. doi: 10.2967/jnumed.108.051805. [DOI] [PubMed] [Google Scholar]
- 29.De Rosa MF, Sillence D, Ackerley C, Lingwood C. Role of multiple drug resistance protein 1 in neutral but not acidic glycosphingolipid biosynthesis. J Biol Chem. 2004;279:7867–7876. doi: 10.1074/jbc.M305645200. [DOI] [PubMed] [Google Scholar]
- 30.Biing J, Yang Y, Liu H, Ye C, Chao C. The induction of multidrug resistance in human cervical carcinoma cell lines by estrogenic hormones. Proc Natl Sci Counc Repub China B. 1994;18:64. [PubMed] [Google Scholar]
- 31.Li L, Xu J, Min T, Huang W. Up-regulation of P-glycoprotein expression by catalase via JNK activation in HepG2 cells. Redox Rep. 2006;11:173–178. doi: 10.1179/135100006X116682. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Kim Y-S, Yan X, Jacobson O, Chen X, Liu S. 64Cu-labeled Lissamine Rhodamine B: a promising PET radiotracer targeting tumor mitochondria. Mol pharm. 2011;8:1198–1208. doi: 10.1021/mp200025m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mechetner E, Kyshtoobayeva A, Zonis S, Kim H, Stroup R, Garcia R, Parker RJ, Fruehau JP. Levels of multidrug resistance (MDR1) P-glycoprotein expression by human breast cancer correlate with in vitro resistance to taxol and doxorubicin. Clin Cancer Res. 1998;4:389–398. [PubMed] [Google Scholar]
- 34.Jacobs VL, Valdes PA, Hickey WF, De Leo JA. Current review of in vivo GBM rodent models: emphasis on the CNS-1 tumour model. ASN neuro. 2011;3:171–181. doi: 10.1042/AN20110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villar VH, Vögler O, Martínez-Serra J, Ramos R, Calabuig-Fariñas S, Gutiérrez A, Barceló F, Martín-Broto J, Regina A. Nilotinib counteracts P-glycoprotein-mediated multidrug resistance and synergizes the antitumoral effect of doxorubicin in soft tissue sarcomas. PLoS ONE. 2012;7:e37735. doi: 10.1371/journal.pone.0037735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutterbach B, Sun D, Schuetz J, Hiebert SW. The MYND motif is required for repression of basal transcription from the multidrug resistance 1 promoter by the t (8; 21) fusion protein. Mol cell biol. 1998;18:3604–3611. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mutoh K, Tsukahara S, Mitsuhashi J, Katayama K, Sugimoto Y. Estrogen-mediated post transcriptional down-regulation of P-glycoprotein in MDR1-transduced human breast cancer cells. Cancer sci. 2006;97:1198–1204. doi: 10.1111/j.1349-7006.2006.00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulenouar S, Weyn C, Van Noppen M, Ali MM, Favre M, Delvenne PO, Bex F, Nöel A, Englert Y, Fontaine V. Effects of HPV-16 E5, E6 and E7 proteins on survival, adhesion, migration and invasion of trophoblastic cells. Carcinogenesis. 2010;31:473–480. doi: 10.1093/carcin/bgp281. [DOI] [PubMed] [Google Scholar]
- 39.DiMaio D, Mattoon D. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene. 2001;20:7866–7873. doi: 10.1038/sj.onc.1204915. [DOI] [PubMed] [Google Scholar]
- 40.Samama B, Plas-Roser S, Schaeffer C, Chateau D, Fabre M, Boehm N. DNA detection by in situ hybridization with catalyzed signal amplification on thin-layer cervical smears. J Histochem Cytochem. 2002;50:1417–1420. doi: 10.1177/002215540205001014. [DOI] [PubMed] [Google Scholar]
- 41.Tsai TC, Chen SL. The biochemical and biological functions of human papillomavirus type 16 E5 protein. Arch Virol. 2003;148:1445–1453. doi: 10.1007/s00705-003-0111-z. [DOI] [PubMed] [Google Scholar]
- 42.Baker CC, Phelps WC, Lindgren V, Braun MJ, Gonda MA, Howley PM. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer-Hofmann R, Borghouts C, Auvinen E, Bourda E, Rösl F, Alonso A. Genomic cloning and characterization of the nonoccupied allele corresponding to the integration site of human papillomavirus type 16 DNA in the cervical cancer cell line SiHa. Virology. 1996;217:33–41. doi: 10.1006/viro.1996.0090. [DOI] [PubMed] [Google Scholar]
- 44.Sherman L, Alloul N, Golan I, Durst M, Baram A. Expression and splicing patterns of human papillomavirus type-16 mrnas in pre-cancerous lesions and carcinomas of the cervix, in human keratinocytes immortalized by HPV 16, and in cell lines established from cervical cancers. Int J Cancer. 1992;50:356–364. doi: 10.1002/ijc.2910500305. [DOI] [PubMed] [Google Scholar]
- 45.Abulrob A, Lu Z, Baumann E, Vobornik D, Taylor R, Stanimirovic D, Johnston LI. Nanoscale imaging of epidermal growth factor receptor clustering effects of inhibitors. J Biol Chem. 2010;285:3145–3156. doi: 10.1074/jbc.M109.073338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF. Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat Nanotechnol. 2010;6:39–44. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Landry MP, Barone PW, Kim J-H, Lin S, Ulissi ZW, Lin D, Bin M, Boghossian AA, Hilmer AJ, Rwei A, Hinckley AC, Kruss S, Shandell MA, Nair N, S Blake S, F Sen F, Sen S, Croy RG, Li D, Yum K, Ahn J-H, Jin H, Heller DA, Essigmann JM, Blankschtein D, Strano MS. Molecular recognition using corona phase complexes made of synthetic polymers adsorbed on carbon nanotubes. Nat Nanotechnol. 2013;8(12):959–968. doi: 10.1038/nnano.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henriksen L, Grandal MV, Knudsen SLJ, van Deurs B, Grøvdal LM. Internalization Mechanisms of the Epidermal Growth Factor Receptor after Activation with Different Ligands. PLoS ONE. 2013;8:e58148. doi: 10.1371/journal.pone.0058148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CH, Cheng TH, Lin H, Shih NL, Chen YL, Chen YS, Cheng CF, Lian WS, Meng TC, Chiu WT, Chen JJ. Reactive oxygen species generation is involved in epidermal growth factor receptor transactivation through the transient oxidization of Src homology 2-containing tyrosine phosphatase in endothelin-1 signaling pathway in rat cardiac fibroblasts. Mol pharmacol. 2006;69:1347–1355. doi: 10.1124/mol.105.017558. [DOI] [PubMed] [Google Scholar]
- 50.Jiao L, Wang X, Diankov G, Wang H, Dai H. Facile synthesis of high-quality graphene nanoribbons. Nat Nanotechnol. 2010;5:321–325. doi: 10.1038/nnano.2010.54. [DOI] [PubMed] [Google Scholar]
- 51.Siddiqui S, Fang M, Ni B, Lu D, Martin B, Maudsley S. Central role of the EGF receptor in neurometabolic aging. Int J Endocrinol. 2012;2012:739428. doi: 10.1155/2012/739428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, Takigawa N, Takata M, Kiura K, Tanimoto M. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non-small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 53.TJ Lynch TJ, ES Kim ES, Eaby B, Garey J, West DP, Lacouture ME. Epidermal growth factor receptor inhibitor-associated cutaneous toxicities: an evolving paradigm in clinical management. Oncologist. 2007;12:610–621. doi: 10.1634/theoncologist.12-5-610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.