Abstract
Background
Lung cancer is the leading cancer killer. Early detection of lung cancer will reduce the related deaths. The objective of this study was to identify potential biomarkers in sputum supernatant for early stage lung cancer.
Materials and Methods
Using shotgun proteomics, we detected changes of protein profiles that were associated with lung cancer by analyzing sputum supernatants of six early stage lung cancer patients and five cancer-free controls. Using western blotting, we validated the proteomic results in 22 lung cancer cases and 22 controls. Using enzyme-linked immunosorbent assay (ELISA), we evaluated the diagnostic performance of the biomarker candidates in an independent set of 35 cases and 36 controls.
Results
Proteomics identified eight biomarker candidates for lung cancer. Western blotting validation of the candidates showed that enolase 1 (ENO1) displayed a higher expression level in cancer patients compared with cancer-free subjects (P=0.015). ELISA revealed that the assessment of ENO1 expression in sputum supernatant had 58.33% sensitivity and 80.00% specificity in distinguishing stage I lung cancer patients from cancer-free subjects.
Conclusion
the analysis of protein biomarkers in sputum may provide a potential approach for the early detection of lung cancer. Future validation of all the candidates defined by shotgun proteomics in a large cohort study may help develop additional biomarkers that can be added to ENO1 to have more diagnostic efficacy for lung cancer.
Introduction
Lung cancer is responsible for 29% of all cancer deaths in men and women, causing more deaths than breast, colon, and prostate cancers combined 1. Approximately 85% of lung tumors are non-small cell lung cancers (NSCLCs) 2. NSCLC comprises two major histological subtypes: squamous cell carcinoma (SCC) and adenocarcinoma (AC) 2. The development of easily performed and noninvasively approaches for early detection of NSCLC followed by suitable treatments can reduce the mortality1, 3, 4.
Sputum is one of the most noninvasively accessible body fluids. Numerous studies have shown that molecular genetic changes in the exfoliated respiratory epithelial cells of sputum could provide potential biomarkers for early stage lung cancer 5,6, 7, 8, 9, 10, 11. The exfoliated epithelial cells in sputum mainly comprise 1), bronchial epitheliums that are derived from centrally located tumors that mainly are SCC, 2), respiratory epitheliums that may share clonally molecular genetic lesions with SCC tumors, however not directly shed from the primary SCC tumors. Therefore, the bronchial epithelial cell-based analysis often provides higher accuracy for SCCs that are centrally located in the lungs compared with ACs that arise peripherally 5,6, 7,8, 9. 10, 11.
The previous studies of the exfoliated epithelial cells in sputum have mainly focused on genetic and epigenetic analysis of nucleic acids to measure the sequence, copy number, mutation, methylation, and expression changes of genes 5,6, 7,8, 9. 10, 11. However, few of the genetic and epigenetic analytic approaches in sputum have been integrated into clinical practice and shown an impact on reducing the mortality of NSCLC. Proteins are the ultimate products of gene expression and more diverse than DNA or RNA. Furthermore, alternative splicing and more than 100 unique post-translational modifications from each gene can create tens to hundreds of species of protein 12. In addition, many physiologic changes are mediated post-transcriptionally, and will not be revealed at the nucleic acid level 13. Moreover, proteins are more dynamic and reflective of cellular physiology, and carry more information than nucleic acids. Therefore, the analysis of protein changes in sputum may provide an alternative means for diagnosis of early stage lung cancer.
Differing from the epithelial cells exfoliated from local respiratory tract sites, sputum supernatant is a circulating cell-free body fluid, which may contains molecules originating from primary tumors either as a result of metastasizing cells or the leakage from the tumors into the circulation. The assessment of the circulating molecules, e.g., proteins, in sputum supernatants may present a potential approach to help diagnosis of lung cancer, particularly ACs that are difficult to be detected by studying the exfoliated epithelial cells.
Mass spectrometry (MS)-based proteomics represents an important technologic choice for arraying and characterizing constituent proteins. MS was used to characterize protein expressions in bronchoalveolar lavage fluid from individuals with cystic fibrosis and discover potential biomarkers for the disease 14-16. Of the proteomic techniques, shotgun proteomics combing liquid chromatography (LC) and MS can globally delineate proteome profiles in complex mixtures and rapidly identify biomarker candidates in clinical samples 17.
This study, which represents the first proteomic study using in-gel digestion coupled with LC/MS to address differential proteins of sputum supernatants in subjects with lung cancer versus control individuals, aims to identify protein biomarkers in sputum that my potentially be useful in the early detection of the disease.
Materials and Methods
Subjects and sputum collection and preparation
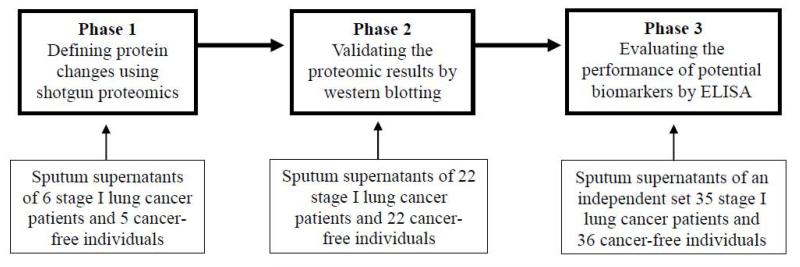
The diagram in Figure 1 describes the design for biomarker discovery and validation in this study, which was performed under a research protocol approved by the Institutional Review Board of University of Maryland Baltimore. As shown in Figure 1 and Table 1, a total of sixty-four lung cancer patients and 64 cancer-free smokers were enrolled. In phase 1, we discovered significant changes of protein profiles that were associated with lung cancer by analyzing sputum supernatants of six early stage lung cancer patients and five cancer-free controls. In phase 2, using western blotting, we validated the results in 22 lung cancer cases and 22 controls. In phase 3, using enzyme-linked immunosorbent assay (ELISA), we evaluated the diagnostic performance of the biomarker candidates in an independent set of 35 cases and 36 controls. Geographic and clinical characteristics of the cases and controls are shown in Table 1. The cancer patients were stage I NSCLC patients before receiving surgical treatment, preoperative adjuvant chemotherapy and radiotherapy. Inclusion criteria for cancer-free controls were individuals who had no a history of cancer in the last three years at the time of enrollment. Clinical diagnosis of lung cancer was made with histopathologic examinations of specimens obtained by CT-guided transthoracic needle biopsy, transbronchial biopsy, videotape-assisted thoracoscopic surgery, or surgical resection. The surgical pathologic staging was determined according to the TNM classification of the International Union Against Cancer with the American Joint Committee on Cancer and the International Staging System for Lung Cancer. Histopathological classification was determined according to the World Health Organization classification.
Figure 1.
Overall study design and patient flow.
Sixty-four lung cancer patients and 64 cancer-free smokers were enrolled. In phase 1, sputum supernatants of six early stage lung cancer patients and five cancer-free controls were analyzed by using shotgun proteomics to detect protein profiles of lung cancer in the specimens. In phase 2, sputum supernatants of 22 lung cancer cases and 22 controls were analyzed by using western blotting to validate the proteomic results. In phase 3, sputum supernatants of 35 cases and 36 controls were analyzed by using enzyme-linked immunosorbent assay (ELISA) to evaluate the diagnostic performance of the identified biomarker candidates.
Table 1.
Demographic and clinical data of 64 lung cancer patients and 64 cancer-free controls
| Parameter |
Lung Cancer Patients Mean |
% |
Cancer-free Controls Mean |
% |
|---|---|---|---|---|
| Age, years | 67.1 | 66.9 | ||
| Gender | ||||
| Men | 43 | 67.18 | 44 | 68.75 |
| Women | 21 | 32.82 | 20 | 31.25 |
| Race | ||||
| White American | 40 | 62.5 | 43 | 67.19 |
| African American | 24 | 37.5 | 21 | 32.81 |
| Smoking, pack-years | 48.6 | 39.6 | ||
| 26.9 | 18.6 | |||
| Stage | All are stage I NSCLC | |||
| Histology | ||||
| Adenocarcinoma | 36 | 56.25 | ||
| Squamous cell carcinoma | 28 | 43.75 | ||
| Location of lung cancer | ||||
| Peripheral | 35 | 54.69 | ||
| Central | 29 | 45.31 |
Abbreviations: NSCLC, non-small cell lung cancer.
Sputum was collected from the participants as described in our previous reports 10, 11, 18-23. Immediately after sputum sample was received, sputum supernatant was prepared as previously described 24. Briefly, four volumes of phosphate buffered saline (PBS) (Sigma-Aldrich Corporation, St Louis, MO) were added in each sample. The samples were incubated on a roller at room temperature for 15 minutes, and then filtered through 48-m nylon gauze. The sputum supernatant was collected after centrifugation at 400 × g for 10 minutes and stored at −80°C until use.
One-dimensional gel electrophoresis (1D-GE)
1D-GE analysis was performed as previously described 25. Briefly, a total of 20μg of protein from each sample was incubated at 70 °C for 10 min in lithium dodecyl sulfate (LDS)-sample buffer (Sigma-Aldrich Corporation) and electrophoresed on 4% to 12% Bis-Tris sodium dodecyl sulfate (SDS)-Polyacrylamide gel electrophoresis (PAGE) (Invitrogen, Carlsbad CA) gels according to the manufacture’s protocol. The gel was then stained with Coomassie blue R-250 (Merck Millipore, Billerica, MA) and destained as previously described 26.
In-gel preparation of proteins for MS
1X1-mm pieces were washed with 100 mM NH4HCO3, shrunken with acetonitrile (Burdick and Jackson, Muskegon, MI), and dried in a vacuum centrifuge (Labconco, Kansas City, MO). Reduction was carried out with 10 mM dithiothreitol in 100 mM NH4HCO3 for 1 h at 60 °C followed by alkylation with 55 mM iodoacetamide in 100 mMNH4HCO3 (Sigma-Aldrich Corporation) for 45 min at room temperature. Rehydration with digestion solution (50 μl of H2O, 50 μl of 100 mM NH4HCO3, 5 μl of CaCl2, and 1.5 μ g of trypsin (Promega, Madison, WI) was performed on ice for 45 min. Any remaining supernatant was removed, and 25 μl of digestion buffer (digestion solution without trypsin) was added for overnight enzymatic cleavage at 37 °C. Peptides were extracted at 37 °C for 15 min with shaking once with 50 mM NH4HCO3, pH 7.8, and twice with 5% formic acid/acetonitrile (Sigma-Aldrich Corporation).
LC-MS analysis and protein identification
Reversed phase separation of peptides was performed using a Surveyor liquid chromatography system (Thermo Scientific, Waltham, MA) as previously described 27. Briefly, peptides were loaded onto desalting peptide trap (Michrom Bioresources, Auburn, CA) using an autosampler (Thermo Scientific). All MS analyses were performed using an LCQ Deca mass spectrometer (Thermo Scientific) equipped with a nanospray ionization source. Peptides were introduced into the mass spectrometer via a 75 μm ID/15 μm tip ID C18-packed PicoFrit®column (New Objective, Woburn, MA). The spray voltage was 2.0 kV and the heated capillary temperature was 200 °C. MS data was acquired using a top 3 data-dependent acquisition method with dynamic exclusion enabled. MS spectra was searched against a human database (downloaded on Nov. 29, 2007 from NCBI; 88,334 sequences) by using Sorcerer-SEQUEST (SageN Research, Milpitas, CA). The quality of peptide and protein assignments was assessed using PeptideProphet and ProteinProphet. Proteins with probabilities of ≥ 0.9 and ≥ 2 unique peptides were accepted as confidently identified peptides.
Western blotting
A total of 40μg sputum supernatant protein was incubated at 95 °C for 2 min in SDS-sample buffer, electrophoresed on a 4-20% PAGE gel, and transferred to chemiluminescence membranes (Amersham-Pharmacia Biotech Piscataway, NJ). The membranes were blocked with 5% fat free milk in PBS-tween for 30 min at room temperature and incubated with murine anti-ENO1 monoclonal IgM (Abnova, Walnut, CA) overnight at 4 °C. After three washes with TBST and tween, the membranes were incubated with IgG-horseradish peroxidase (HRP) secondary antibody (Sigma-Aldrich Corporation) and visualized with chemiluminescence (Pierce, Rockford, lL). The band density was analyzed by Kodak 1D 3.6 imaging system (Kodak, New Haven, CT) and the ratio ENO1 to beta-actin was calculated.
ELISA for detection of ENO1 in sputum supernatant specimens
ELISA analysis for determining ENO1 level was carried out using a protocol established by Shih et al 28 with modifications. Briefly, 100 ml of 5 mg/ml murine anti-ENO1 monoclonal IgM (Abnova) in adsorption buffer was added to each well of a 96-well plate, which was incubated overnight at 4°C. The wells were washed with 200 ml of binding buffer. After adding 200 ml of blocking buffer, the plates were incubated at 37°C for 90 min and washed with 200 ml of binding buffer. Sputum supernatant samples were diluted with binding buffer at a final dilution of 1:10, and added 100 ml to each well. The plate was then incubated at room temperature for 30 min. After additional washing with binding buffer, 100 ml of 20 mg/ml rabbit anti-ENO1 polyclonal IgG (Abcam, San Francisco, CA) was added to each well and incubated for 30 min. 100 ml of goat anti-rabbit IgG-HRP (Abcam), diluted 1:5,000, was added to each well and then incubated at room temperature for 30 min. After washing again, bound antibody-HRP was detected by incubation with 2-2′-azino-di-(3ethylbenzthiazoline sulfonic acid) peroxidase substrate (KPL, Gaithersburg, MD). The reaction was stopped with 5% sodium dodecyl sulfate, and the optical density (OD) was measured in an ELISA reader (BMG LABTECH Inc, Cary, NC) at 405 nm. For the standard curve, serial dilutions of the ENO1 antibody were plotted with corresponding OD405 values. Each sample was analyzed in triplicate.
Statistical analysis
We used t-test to determine significant differences of values between groups. We applied Pearson’s correlation analysis to assess relationship between ENO1 level and demographic characteristics of the patients and cancer-free controls. We used clinical and pathologic diagnoses as reference standards to estimate sensitivity and specificity of the analysis of ENO1. We applied the receiver-operator characteristic (ROC) curve and area under the curve (AUC) analyses to determine the accuracy of using ENO1 as a potential biomarker in a given specimen. All P values shown were two sided, and a P value of <0.05 was considered statistically significant.
Results
Characteristics of sputum supernatant proteins by quantitative MS
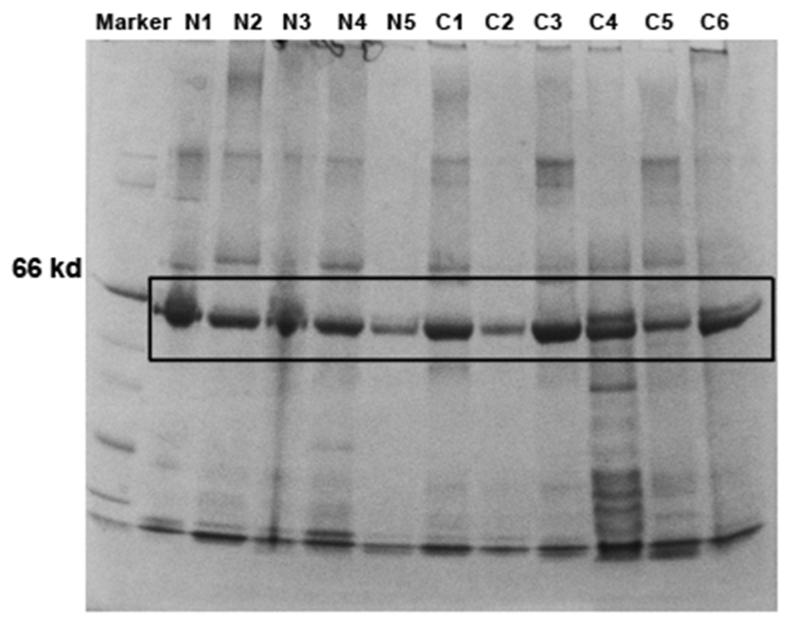
To identify potential biomarkers for lung cancer, proteins of sputum supernatants of six lung cancer patients and five healthy controls were separated by SDS-PAGE. As shown in Figure 2, all the samples had a major band around 50 kDa. The bands were then excised from all the samples and submitted for in-gel digestion and LC/MS analysis. After deconvolution of raw MS data, interrogation of human protein database produced a total of 29 unique proteins (Supplementary Table 1) in the specimens. Of the proteins, eight showed statistical difference between lung cancer and cancer-free groups (All P values less than 0.01) (Table 2). Of the eight proteins, five displayed a high level of expression, while three showed a low level in sputum supernatants of lung cancer patients compared with cancer-free individuals. The five proteins having a high level in sputum of lung cancer patients included ENO1, membrane protein DAP10, guanine nucleotide exchange factor, low density lipoprotein receptor related protein-deleted in tumor, and hemopexin. The three proteins showing a reduced level in sputum of lung cancer patients were transmembrane secretory component (poly-Ig receptor), lactoferrin precursor, and high molecular weight salivary mucin MG1. The expression level of ENO1 was approximately four times higher than that of the rest proteins in the samples of lung cancer patients. Because ENO1 presented the highest level in lung cancer patients compared with cancer-free individuals, it was used for follow-up validation in this present study.
Figure 2.
One-dimensional SDS-Polyacrylamide Gel Electrophoresis (1D SDS-PAGE) of protein samples of sputum supernatants.
1D SDS-PAGE analysis of protein samples of sputum supernatants of six early stage lung cancer patients (C1-6) and five cancer-free controls (N1-5).
Table 2.
Proteins showing difference in sputum supernatants between lung cancer patients and cancer-free controls
| Genbank Accession Definition | Log2 (cancer/control)* | |
|---|---|---|
| NP_001419 | enolase 1 | 19.18‡ |
| AAD47911 | membrane protein DAP10 | 4.13 ‡ |
| AAF36817 | guanine nucleotide exchange factor | 4.06 ‡ |
| AAF70379 | low density lipoprotein receptor related protein-deleted in tumor | 3.12 ‡ |
| AAA58678 | hemopexin | 2.53 1 |
| AAB20203 | transmembrane secretory component; poly-Ig receptor; | 0.68 α |
| AAG48753 | lactoferrin precursor | 0.56 α |
| AAB61398 | high molecular weight salivary mucin MG1 | 0.43 α |
All P values are less than 0.05.
the proteins having a high level in sputum of lung cancer patients.
,the proteins having a reduced level in sputum of lung cancer patients.
Validation of LC/MS data by western blotting
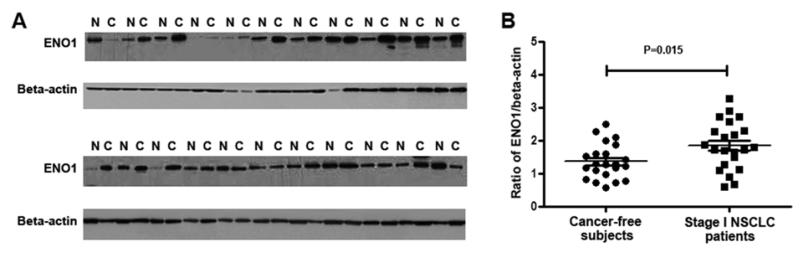
Western blots of sputum supernatants of 22 lung cancer patients and 22 cancer-free individuals were probed with specific antibody to ENO1 protein. As shown in Figure 3, after normalizing with beta-actin, sputum supernatants of lung cancer patients exhibited a higher level of ENO1 compared with cancer-free individuals (1.85 ± 0.156 vs. 1.37 ± 0.113, P=0.015). Therefore, the results of western blotting for the analysis of ENO1 in a different cohort of samples were consistent with the shotgun proteomics data.
Figure 3.
Validation of proteomic findings using western blotting.
A. Western blots of protein samples of sputum supernatants of early stage lung cancer patients (C) and cancer-free controls (N) were probed with antibodies specific for ENO1 and beta-actin. B, ENO1 expression was normalized with beta-actin. Sputum supernatants of lung cancer patients had a higher level of ENO1 compared with cancer-free individuals.
Evaluation of the diagnostic performance of ENO1 by ELISA in an independent set of 35 cases and 36 controls
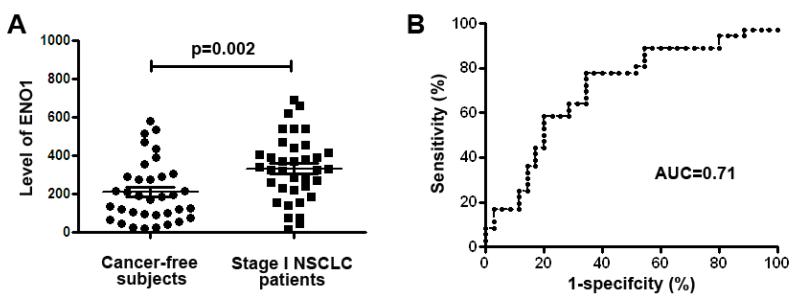
Level of ENO1 determined by ELISA in sputum supernatants of lung cancer patients was significantly higher than in cancer-free individuals (332.87 ± 28.10 vs. 210.53 ± 26.61, p=0.002) (Fig. 4A). Furthermore, the assessment of ENO1 generated AUC value of 0.71 (95% confidence interval: 0.59 to 0.83; P=0.002) in distinguishing NSCLC patients from the controls (Figure 4B). Given a specificity of 80.00%, analysis of ENO1 revealed a sensitivity of 58.33% in differentiating NSCLC patients from the cancer-free subjects. Furthermore, given a sensitivity of 77.78%, analysis of ENO1 produced a specificity of 65.71% in the set of cases and controls. The prevalence of ENO1 level in sputum was related with pack-years of smoking (P < 0.05), however, not associated with patient age and gender. In addition, there was no association of the changes of ENO1 expression with the histological tumor type and location of lung tumors (all P > 0.05). The observations imply that the assessment of ENO1 in sputum supernatants might be potentially useful in diagnosis of not only SCCs that is often centrally located, but also ACs that arise peripherally in the lungs.
Figure 4.
Expression level of ENO1 determined by ELISA in 35 stage I NSCLC patients and 36 controls.
ELISA to determine ENO1 expression in sputum supernatants of 35 stage I NSCLC patients and 36 controls was performed with antibody specific for ENO1. A, ENO1 had a significantly higher level in lung cancer patients than in cancer-free individuals (p=0.002). B, the ELISA analysis produced AUC value of 0.71 (95% confidence interval: 0.588 to 0.833; P=0.002) in distinguishing lung cancer patients from the controls.
Discussion
Using 1DE gel and LC/MS-based shotgun proteomics, we separated protein changes in sputum supernatants and found that ENO1 displayed a high expression level in samples of lung cancer patients. ENO1 is present on the surface of cells and plays an important role in tissue invasion and metastasis29, 30. The enzymatic role of ENO1 is to convert 2-phosphoglycerate to phosphoenolpyruvate 29, 30. Activated ENO1 has a direct coupling to glycolytic activity 29, 30. Furthermore, ENO1 could be upregulated by HIF1α in response to hypoxia 31. Dysregulation of ENO1 has been suggested to involve in tumorigenesis. For instance, using a functional genomic approach, we previously found that ENO1 was one of the genes with increases in both genomic copy number and transcript in lung cancer cells compared with normal cells 32. Furthermore, a meta-analysis of gene-expression and expressed sequence tags showed that ENO1 was overexpressed in up to 50% of 24 different types of human tumors, including lung cancer 33. In addition, we developed a panel of six genes (including ENO1) that could be detected in sputum via in situ technique. The assessment of the panel of genes produced 86.7% sensitivity and 93.9% specificity for diagnosis of lung SCCs 11. Moreover, plasma ENO1 level of NSCLC patients was significantly higher than that in plasma of healthy individuals 34. This present study extends the previous findings by demonstrating that testing protein level of ENO1 in sputum supernatant might have the potential to be used for lung cancer diagnosis.
Previous studies 5,6, 7,8, 9, 10, 11 suggested that cytological and molecular studies of exfoliated bronchial epitheliums in sputum were more accurate to identify SCC tumors that predominantly located in central areas of the lungs. However, the cell-based approaches have low accuracy for the diagnosis of ACs, which are the most common type of lung cancer and frequently occur in the peripheral region of the lungs. Interestingly, the analysis of ENO1 in sputum supernatant has similar sensitivity and specificity for diagnosis of SCCs and ACs, suggesting the potential biomarker could not only detect SCCs, but also discover ACs. These observations may be clinically important, as future use of biomarkers in sputum supernatant would improve the detection rate for ACs that are more difficult to be found by using the previous cell-based techniques.
There are some weaknesses in this study. First, the sample size is small. Second, although 1D-GE is lesser expensive and more easily to be performed compared with 2D-GE, it has limitation in separating proteins as opposed to 2D-GE. We are carrying out a study using 2D-GE to comprehensively profile protein changes in sputum that are specific for lung cancer patients. Third, the specificity and sensitivity of the ENO1 biomarker are far too poor for distinguishing lung cancer patients from cancer-free individual. Moreover, the performance of the protein biomarker is inferior to the prior gene analyses 8-12. Therefore, this potential biomarker is not now applicable in a clinical setting. We are extensively evaluating all the protein biomarker candidates defined by the shotgun proteomics in a large case-control study to identify additional biomarkers that can be added to ENO1 so that the diagnostic efficacy of the approach could be improved. Furthermore, future integrating the protein biomarkers with other types of biomarkers across sputum and plasma might provide an accurate and noninvasive approach for lung cancer diagnosis. The biomarkers might be useful to improve the accuracy of CT screening for lung cancer early detection. For example, using the biomarkers may help identify lung tumors from the indeterminate nodule identified by CT analysis. Moreover, we will compare the protein-biomarkers with cytological diagnosis in a future study.
Conclusion
Using shotgun proteomics together with western blotting and ELISA, we identify sputum ENO1 as a biomarker candidate for lung cancer. Although the one biomarker-based test is insufficiently discriminative to support undertaking a multicenter clinical trial, the findings from this study may lay the basis to perform a next step to develop multiple biomarkers that might have future clinical utility.
Clinical Practice Points
To develop biomarkers in sputum for lung cancer early detection, the previous studies have mainly focused on genetic and epigenetic analysis of nucleic acids to measure the sequence, copy number, mutation and methylation, and expression changes of genes in exfoliated respiratory epithelial cells of sputum. However, few of the biomarkers have been integrated into clinical practice for lung cancer early detection. Differing from the epithelial cells exfoliated from local respiratory tract sites, sputum supernatant is a circulating cell-free body fluid, which may contains molecules originating from primary tumors either as a result of metastasizing cells or the leakage from the tumors into the circulation. We hypothesized that the assessment of the circulating molecules in sputum supernatants may present a potential approach to help diagnosis of lung cancer. This study, which represents the first proteomic study by using in-gel digestion coupled with LC/MS to address differential proteins of sputum supernatants in lung cancer patients versus control individuals, aims to identify protein biomarkers for early stage lung cancer. We identify ENO1 as a potential biomarker in sputum supernatants that may help diagnose early stage lung cancer. Future studies for identifying additional biomarkers in sputum that can be added to ENO1 to improve lung cancer early detection are required.
Supplementary Material
Acknowledgments
Funding sources: This work was supported in part by National Cancer Institute grant R01CA161837 and VA merit Award I01 CX000512, LUNGevity/Upstage Foundation Early Detection Award (F. J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 3.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 4.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thunnissen FB. Sputum examination for early detection of lung cancer. J Clin Pathol. 2003;56:805–810. doi: 10.1136/jcp.56.11.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, Crowell RE. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 7.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 8.Varella-Garcia M, Kittelson J, Schulte AP, Vu KO, Wolf HJ, Zeng C, Hirsch FR, Byers T, Kennedy T, Miller YE, Keith RL, Franklin WA. Multi-target interphase fluorescence in situ hybridization assay increases sensitivity of sputum cytology as a predictor of lung cancer. Cancer Detect Prev. 2004;28:244–251. doi: 10.1016/j.cdp.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Varella-Garcia M, Schulte AP, Wolf HJ, Feser WJ, Zeng C, Braudrick S, Yin X, Hirsch FR, Kennedy TC, Keith RL, Baron AE, Belinsky SA, Miller YE, Byers T, Franklin WA. The detection of chromosomal aneusomy by fluorescence in situ hybridization in sputum predicts lung cancer incidence. Cancer Prev Res (Phila) 2010;3:447–453. doi: 10.1158/1940-6207.CAPR-09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Todd NW, Qiu Q, Fan T, Zhao RY, Rodgers WH, Fang HB, Katz RL, Stass SA, Jiang F. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin Cancer Res. 2007;13:482–487. doi: 10.1158/1078-0432.CCR-06-1593. [DOI] [PubMed] [Google Scholar]
- 11.Jiang F, Todd NW, Li R, Zhang H, Fang H, Stass SA. A panel of sputum-based genomic marker for early detection of lung cancer. Cancer Prev Res (Phila) 2010;3:1571–1578. doi: 10.1158/1940-6207.CAPR-10-0128. [DOI] [PubMed] [Google Scholar]
- 12.Aebersold R, Anderson L, Caprioli R, Druker B, Hartwell L, Smith R. Perspective: a program to improve protein biomarker discovery for cancer. J Proteome Res. 2005;4:1104–1109. doi: 10.1021/pr050027n. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Gharib SA, Vaisar T, Aitken ML, Park DR, Heinecke JW, Fu X. Mapping the lung proteome in cystic fibrosis. J Proteome Res. 2009;8:3020–3028. doi: 10.1021/pr900093j. [DOI] [PubMed] [Google Scholar]
- 15.MacGregor G, Gray RD, Hilliard TN, Imrie M, Boyd AC, Alton EW, Bush A, Davies JC, Innes JA, Porteous DJ, Greening AP. Biomarkers for cystic fibrosis lung disease: application of SELDI-TOF mass spectrometry to BAL fluid. J Cyst Fibros. 2008;7:352–358. doi: 10.1016/j.jcf.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Bai Y, Galetskiy D, Damoc E, Paschen C, Liu Z, Griese M, Liu S, Przybylski M. High resolution mass spectrometric alveolar proteomics: identification of surfactant protein SP-A and SP-D modifications in proteinosis and cystic fibrosis patients. Proteomics. 2004;4:2300–2309. doi: 10.1002/pmic.200400855. [DOI] [PubMed] [Google Scholar]
- 17.Takadate T, Onogawa T, Fukuda T, Motoi F, Suzuki T, Fujii K, Kihara M, Mikami S, Bando Y, Maeda S, Ishida K, Minowa T, Hanagata N, Ohtsuka H, Katayose Y, Egawa S, Nishimura T, Unno M. Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int J Cancer. 2013;132:1368–1382. doi: 10.1002/ijc.27797. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 20.Xie Y, Todd NW, Liu Z, Zhan M, Fang H, Peng H, Alattar M, Deepak J, Stass SA, Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer-J Iaslc. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anjuman N, Li N, Guarnera M, Stass SA, Jiang F. Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. Clin Transl Med. 2013:2–15. doi: 10.1186/2001-1326-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang F, Todd NW, Qiu Q, Liu Z, Katz RL, Stass SA. Combined genetic analysis of sputum and computed tomography for noninvasive diagnosis of non-small-cell lung cancer. Lung Cancer. 2009;66:58–63. doi: 10.1016/j.lungcan.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu Q, Todd NW, Li R, Peng H, Liu Z, Yfantis HG, Katz RL, Stass SA, Jiang F. Magnetic enrichment of bronchial epithelial cells from sputum for lung cancer diagnosis. Cancer. 2008;114:275–283. doi: 10.1002/cncr.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erin EM, Jenkins GR, Kon OM, Zacharasiewicz AS, Nicholson GC, Neighbour H, Tennant RC, Tan AJ, Leaker BR, Bush A, Jose PJ, Barnes PJ, Hansel TT. Optimized dialysis and protease inhibition of sputum dithiothreitol supernatants. Am J Respir Crit Care Med. 2008;177:132–141. doi: 10.1164/rccm.200603-311OC. [DOI] [PubMed] [Google Scholar]
- 25.Brown JK, Lauer KB, Ironmonger EL, Inglis NF, Bourne TH, Critchley HO, Horne AW. Shotgun proteomics identifies serum fibronectin as a candidate diagnostic biomarker for inclusion in future multiplex tests for ectopic pregnancy. PLoS One. 2013;8:e66974. doi: 10.1371/journal.pone.0066974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steely HT, Dillow GW, Bian L, Grundstad J, Braun TA, Casavant TL, McCartney MD, Clark AF. Protein expression in a transformed trabecular meshwork cell line: proteome analysis. Mol Vis. 2006;12:372–383. [PubMed] [Google Scholar]
- 27.Dai J, Shieh CH, Sheng QH, Zhou H, Zeng R. Proteomic analysis with integrated multiple dimensional liquid chromatography/mass spectrometry based on elution of ion exchange column using pH steps. Anal Chem. 2005;77:5793–5799. doi: 10.1021/ac050251w. [DOI] [PubMed] [Google Scholar]
- 28.Shih NY, Lai HL, Chang GC, Lin HC, Wu YC, Liu JM, Liu KJ, Tseng SW. Anti-alpha-enolase autoantibodies are down-regulated in advanced cancer patients. Jpn J Clin Oncol. 2010;40:663–669. doi: 10.1093/jjco/hyq028. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Zeng H, Yao Q, Yuan J, Zhang Y, Qiu D, Yang X, Yang H, Liu Z. Steinernema glaseri surface enolase: molecular cloning, biological characterization, and role in host immune suppression. Mol Biochem Parasitol. 2012;185:89–98. doi: 10.1016/j.molbiopara.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Miller DM. Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J Biol Chem. 2000;275:5958–5965. doi: 10.1074/jbc.275.8.5958. [DOI] [PubMed] [Google Scholar]
- 31.Ito T, Funamoto K, Sato N, Nakamura A, Tanabe K, Hoshiai T, Suenaga K, Sugawara J, Nagase S, Okamura K, Yaegashi N, Kimura Y. Maternal undernutrition induces the expression of hypoxia-related genes in the fetal brain. Tohoku J Exp Med. 2012;226:37–44. doi: 10.1620/tjem.226.37. [DOI] [PubMed] [Google Scholar]
- 32.Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL, Stass SA, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;25:2628–2635. doi: 10.1038/sj.onc.1209289. [DOI] [PubMed] [Google Scholar]
- 33.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Li M, Liu Y, Han N, Zhang K, Xiao T, Cheng S, Gao Y. ENO1 protein levels in the tumor tissues and circulating plasma samples of non-small cell lung cancer patients. Zhongguo Fei Ai Za Zhi. 2010;13:1089–1093. doi: 10.3779/j.issn.1009-3419.2010.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.