Abstract
Whereas research on CD1d has emphasized a few glycosyl ceramides, the broader family of four human CD1 antigen-presenting molecules binds hundreds of distinct self-lipids. Individual lipid types bind within CD1 grooves in different ways, such that they partially fill the groove, match the groove volume, or protrude substantially from the groove. These differing modes of binding can now be connected to differing immunological functions, as individual lipids can act as stimulatory antigens, inhibitory ligands, or space-filling scaffolds. Because each type of CD1 protein folds to produce antigen-binding grooves with differing sizes and shapes, CD1a, CD1b, CD1c, CD1d, and CD1e have distinct mechanisms of capturing self-lipids and exchanging them for foreign lipids. The size discrepancy between endogeneous lipids and groove volume is most pronounced for CD1b. Recent studies show that the large CD1b cavity can simultaneously bind two self-lipids, the antigen, and its scaffold lipid, which can be exchanged for one large bacterial lipid. In this review, we will highlight recent studies showing how cells regulate lipid antigen loading and the roles CD1 groove structures have in control of the presentation of chemically diverse lipids to T cells.
Keywords: CD1, Lipid antigens, Spacers, Scaffolds, Structures
Introduction
Nearly two decades ago, CD1 proteins were shown to mediate T cell autoreactivity and present lipid antigens to T cells [1–4]. Since that time, crystal structures of T cell receptors (TCRs) bound to CD1-lipid complexes have established the basic model of lipid antigen recognition by T cells [5–14] (Fig. 1). The aliphatic hydrocarbon chains of lipids insert into the hydrophobic pockets of CD1 grooves, allowing the carbohydrate, peptide, phosphate, or sulfate moieties to protrude through a portal positioned between two α-helices to the surface of CD1, where they directly contact TCRs. However, within the constraints of this general mechanism, there is considerable diversity in the efficiency of antigen capture, stringency of antigen release from the groove, or even the number of lipids bound in the groove at any time. Here we review recent studies that show how CD1 proteins act in cells to capture surprisingly hundreds of self-lipid ligands or antigens. This review focuses on the problem of how non-polymorphic CD1 proteins, which are hollow cavities with a finite and defined volume, can capture lipids of diverse size and structure. For major histocompatibility class I proteins (MHC I), this basic function is accomplished through generation of nonamer peptides by proteases from precursor proteins of highly divergent size. In contrast, cells do not typically trim lipids so that they are of one size that match the volume of CD1 grooves. The aliphatic hydrocarbon chains of CD1-presented antigens are highly resistant to covalent cleavage [15], and the CD1 system relies mainly on expressing many types of CD1 proteins (CD1a, CD1b, CD1c, CD1d, CD1e) with differing groove volumes, combined with a combination of scaffolds, spacers, portals, and escape hatches to match ligand size to groove volume.
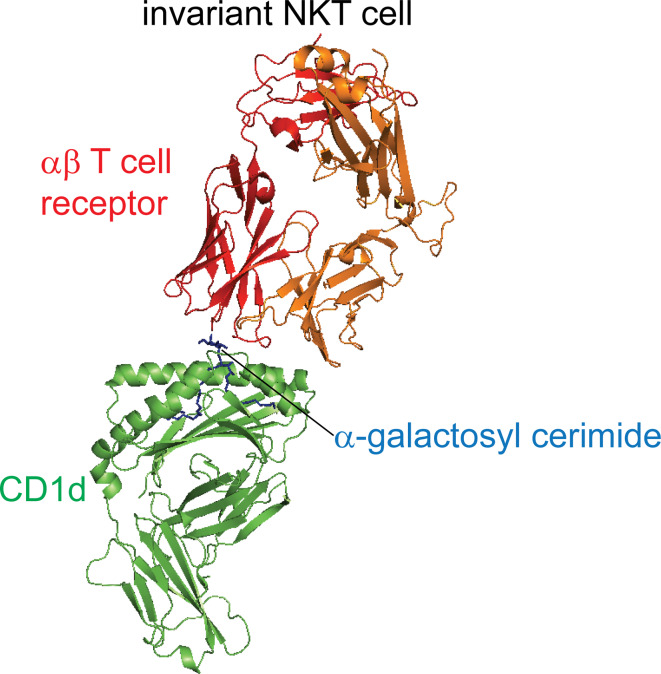
Fig. 1.
CD1d and TCR complexes. The invariant natural killer T cell TCR recognizes the ligand-CD1d complex (green) using predominantly the TCRα chain (red) [figure generated from RCSB protein data bank files 2PO6 (iNKT-CD1d)]
Structure of CD1
The first crystal structure of CD1 revealed a molecule resembling MHC class I [16]. All known CD1 antigen-presenting molecules consist of a heavy chain comprised of extracellular antigen-binding domain, a transmembrane domain, and an intracellular tail, which directs intracellular trafficking. The extracellular domain consists of an MHC fold with two anti-parallel α-helices (α1 and α2) flanking a floor composed of a six-strand α sheet. The α1 and α2 helices are supported by the α3 domain, which non-covalently interacts with β2-microglobulin (β2m) [16, 17]. CD1 proteins differ from the MHC peptide-binding grooves in at least two key aspects. The inner surface of the CD1 groove is largely lined by hydrophobic residues, and the α1 helices of CD1 proteins extend “upwards”, so that CD1 proteins have a larger vertical depth between the floor and the antigen entry portal.
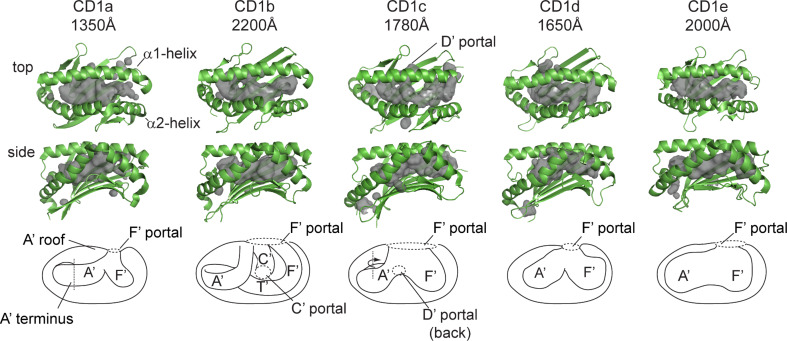
For example, the first CD1 crystal structure showed that murine CD1d has two deep pockets named the A′ and F′, based on the analogous locations of the A and F pockets of MHC I [16]. All four types of human CD1 antigen-presenting molecules, CD1a, CD1b, CD1c, and CD1d, have now been crystallized [16, 18–21]. CD1b differs most substantially from the rest in that it is larger (~2,200 Å3) and has two additional pockets, C′ and T′. The centrally located C′ pocket is named after the C pocket in MHC I, and the T′ pocket name is derived from the “tunnel” found only in the deepest point of the CD1b cavity. A crystal structure of human CD1b, which was the first solved structure bound to lipid ligands [18], showed that lipids are orientated such that hydrophobic alkyl chains are buried deep within the binding pockets of CD1, with exposure of hydrophilic head groups at the protein surface. Each CD1 structure reveals unique aspects of the antigen-binding grooves present in CD1a, CD1b, CD1c, and CD1d, which differ in volume and number of binding pockets (Fig. 2). Whereas all CD1 proteins have a main portal, located above the F′ pocket (F′ portal), CD1b and CD1c have additional portals (accessory portals) that are thought to allow large lipids to escape from the interior of the groove to the outer surface of the CD1 protein in ways that do not directly control TCR binding. The key points of structural divergence among individual CD1 isoforms are considered in turn.
Fig. 2.
CD1 isoforms each have unique-antigen binding groves and capacities. CD1 isoforms have differing groove architecture, as revealed by crystal structures of CD1 with ligands: CD1a with dideoxymycobactin, CD1b with C55 glucose monomycolate, CD1c with mannosyl-phosphomycoketide, CD1d with α-galactosyl-ceramide, and CD1e alone. CD1 structures are rendered in green with cavity surface highlighted in gray and schematic of cavity [figures were generated from RCSB protein data bank files for 1XZO (CD1a), 1UQS (CD1b), 3OV6 (CD1c), 2PO6 (CD1d), and 3S6C (CD1e)]
CD1a
CD1a crystal structures have been solved as liganded complexes with the self-antigen, sulfatide, and the mycobacterial lipopeptide, dideoxymycobactin. These structures show that CD1a has the smallest binding groove among human CD1 isoforms, with a volume of ~1,350 Å3 [19, 22]. CD1a has two binding pockets, the A′ and F′, and no known accessory portals, although a small gap in the lateral wall of the F′ pocket might allow lipids to protrude laterally so that they do not remain beneath and do not traverse the plane of TCR contact. As is the case for most CD1 proteins, the A′ pocket of CD1a is separated from the outer surface of the protein by a roof-like structure above the A′ pocket, known as the A′ roof (Fig. 2). The roof is formed by interdomain contacts between the α1 and α2 helices and likely limits direct access of antigens to the A′ pocket. Instead, the region just above the F′ pocket is open to solvent, so antigens likely access the groove above the F′ pocket, through the F′ portal (Fig. 2) [19]. Whereas the A′ pocket of other CD1 isoforms connect directly with other pockets, the A′ pocket of CD1a curves around a central pole in the A′ pocket and then abruptly terminates deep inside the groove at a molecular barrier formed by Val28. As such, the A′ pocket of CD1a likely acts as a “molecular ruler” for selective binding of alkyl chains of discrete length [22], a prediction confirmed by preferential presentation of dideoxymycobactins with the optimal chain length (~C20), which matches the volume of this pocket [23].
CD1b
Several CD1b-lipid complexes have been reported [18, 24–26]. Amongst all CD1 isoforms, CD1b has the largest binding groove (~2,200 Å3) [18]. The striking structural feature of the CD1b groove is its large volume comprised of four interconnected pockets (Fig. 2). Only CD1b is known to have a deeply buried T′ tunnel, a laterally oriented pocket, which connects the vertically oriented A′ and F′ pockets across the bottom of the groove to form a A′T′F′ super channel [18]. The A′T′F′ superchannel allows CD1b to bind extremely long meromycolate branches found in three types of CD1b-presented mycolyl lipids antigens: free mycolic acid, glucose monomycolate, and glycerol monomycolate [3, 27, 28]. Other CD1 isoforms lack this long channel and are not known to present long-chain mycolyl lipids. Additionally, it has been observed that an accessory portal, known as the C’ portal, connects the inner pocket of CD1b to the outer surface [18].
CD1c
CD1c has two hydrophobic antigen-binding pockets, A’ and F’, which create an antigen-binding volume capacity of ~1,780 Å3 [20]. This structure has been solved in complex with mannosyl phosphomycoketide antigens, which have a series of five methyl branches in the alkyl chains [20, 29]. This branched lipid is positioned in the toroidal A′ pocket of CD1c with the all (S) branches pointed outward toward the larger concave surface, as the lipid descends downward into the groove. This mechanism provides a precise fit for CD1c with two known mycoketide lipids [30]. The independent identification of mannosyl-phosphomycoketide and phosphomycoketide as two branched lipids that function as CD1c presented antigens suggests that the mycoketide backbone might be specialized to bind to CD1c.
Additionally, CD1c presents a lipopeptide with one alkyl chain [31]. Thus, the three known foreign or exogenous antigens for the CD1c system all have one alkyl chain. The A′ pocket is largely continuous with the F′ pocket, but opens to the exterior through an accessory portal known as the D′ portal, located under the α1 helix [20]. Unique amongst the CD1 isoforms, the F′ pocket of CD1c is the most exposed to solvent, potentially allowing for more promiscuous lipid occupancy and raising the possibility that antigens with lipid tails that are larger than those present in currently known antigens might also be presented by CD1c [32]. The crystallized CD1-lipid complex shows that the groove simultaneously binds two lipids. The A′ pocket captures the mycoketide backbone, and the F′ pocket contains a C12 hydrocarbon chain, which might be a detergent that was incidentally captured during crystallization and fills the space not occupied by the larger mycoketide lipid [20]. Because all three of the known CD1c antigens have only one alkyl chain that is smaller than the groove volume, spacer lipids might influence CD1c’s antigen-presenting function in ways that are analogous to their proven role in CD1b, as discussed below.
CD1d
Human and mouse CD1d represent the most extensively studied CD1 proteins, which have been crystallized with numerous self- and exogenous antigens, including the high affinity superagonists, α-galactosylceramides [5, 6, 8–11, 16, 33, 34]. CD1d contains A′ and F′ pockets, which provide total cavity volume of ~1,650 Å3, and no accessory portals are known (Fig. 2). The sphingosine chain of α-galactosylceramides binds within the F′ pocket with the fatty acyl tail occupying the A′ pocket, thereby positioning the galactosyl head group on the surface. For a detailed description of CD1d and its ligands, see recent reviews for additional reading [35–37].
CD1e
CD1e is the only isoform that is not expressed on the cell surface, but instead traffics exclusively within the endolysosomal network, where it is cleaved to become a soluble protein. In contrast to the antigen-presenting function of other CD1 proteins, CD1e functions as a lipid-binding protein [38]. The crystal structure of CD1e shows a wide, solvent-exposed antigen-binding domain (Fig. 2) with A’ and F’ pockets not clearly separated from one another [21]. This groove has a volume of ~2,000 Å3, and has a proposed in role in lipid transfer to CD1 antigen-presenting molecules [21, 38].
Accessory portals
In some [18] but not all [26] crystal structures, CD1b has been observed to have an accessory portal known as the C′ portal because it directly connects the interior of the C′ pocket to the outer surface of CD1b. Unlike the F′ portal, which is adjacent to the plane of TCR contact, the C′ portal is located beneath the α1 helix, distant from the site of TCR contact (Fig. 2). Therefore, even if present only transiently, this portal might represent a structural modification to allow the termini of particularly long mycolates to protrude slightly from the groove in ways that promote binding of lipids without affecting TCR contact [15]. Similarly, CD1c has at least one accessory portal known as the D′ portal located under the α1 helix, which exposes the A′ pocket to the outer surface of CD1c [20]. In crystal structures, it has not been possible to directly observe lipid protrusion to the outer surface of CD1 through these portals. The lack of electron density observed outside the portal does not suggest that lipids do not protrude, but instead more likely reflects the fact that such lipids would not take on an ordered structure. Supporting this general idea, sulfatides added to CD1a to form liganded crystal structures had longer alkyl chains than those observed as ordered density within the groove, a finding that was interpreted as likely lipid protrusion as unordered structures through the lateral wall of the F′ pocket [19]. Also, the CD1b groove is predicted to have the capacity for C72–76 lipids, yet it actually binds C80–86 glucose mycolates, a finding interpreted as lipids likely protruding through the C′ portal [15]. In summary, the interior grooves of CD1a, CD1c, and CD1d are similar in size, but differ in the nature of the accessory portals, which likely represent isoform-specific adaptations to carry lipids of differing chain length.
The size problem for CD1b
In the MHC system, cellular and viral proteins greatly exceed the volume of MHC I and II. Therefore, cellular processing of proteins into peptides of 8 to ~30 amino acids is nearly universally required for MHC binding and recognition. In contrast, the lipids eluted from CD1a [39], CD1c, and CD1d proteins [40, 41] show a good size match of most known ligands to the ~1,700 Å3 volume. Therefore, the simplest model for CD1 antigen presentation is that cells produce ligands that naturally approximate the size of the grooves that capture them, and so the aliphatic alkyl chains of lipids do not need to universally undergo antigen-processing reactions. Also, this simple model predicts that CD1 binds lipids with one-to-one stoichiometry. Although deglycosylation reactions that modify headgroups of sphingolipids and mycoketides are known [30, 42], the natural size match of the naturally occurring alkyl chains to CD1 groove volume, along with data that such antigens can be loaded onto CD1 proteins in APC-free systems, strongly support a working model that trimming of alkyl chains is not a usual antecedent to lipid-antigen loading.
However, the natural antigens for CD1b range in length from C30 to C80, and so include antigens that would not fully occupy the groove, which is estimated to hold C72–76 lipids, as well as those, like C80 mycolates, which apparently exceed the groove volume. Studies of long-chain mycolates bound to CD1b directly ruled out lipid trimming, suggesting that they bind in an intact form [43]. At the other end of the length range, most cellular lipids, which form the pool of self-lipid antigens, have a combined lipid length of about C32–C42, which is substantially smaller than the CD1b groove [44]. In fact, no common self-lipids approximate the expected C72 lipid length that fits within the CD1b groove, so there is a basic size mismatch between endogenous ligands and the groove volume of CD1b. Therefore, the question arises: what are the natural endogenous self-lipids for CD1b, and how could they fill this large groove volume? The problem of size was experimentally confirmed and highlighted by a study by Huang and colleagues that assess the average mass of ligands eluted from CD1a, CD1b, CD1c, and CD1d lipids [44]. Although CD1b has a larger volume, it bound a range of endogenous ligands that was similar in mass compared to ligands bound by CD1 proteins with smaller grooves. This study confirmed the size mismatch and showed that it is true for most or all ligands bound to CD1b.
Spacers and scaffolds
A theoretical solution to this problem would be that CD1a, CD1c, and CD1d typically bind self-lipids with 1–1 stoichiometry, but CD1b might bind two lipids at once. If portals allow concomitant capture of lipid antigen and a spacer lipid, then their combined length might match the groove volume. Direct evidence for such spacer lipids derived from early studies of CD1b, which bound phosphatidylinositol or GM2 ganglioside, whose lipid tails occupied only the A′ and C′ pockets. The T′ and F′ pockets were occupied by two electron densities of about ~C16 in length, which were assumed to be detergent molecules used during the protein refolding [18]. This observation led to the hypothesis that such artificial spacer lipids might have a naturally occurring equivalent that might normally fill partially empty grooves, starting the search for natural spacer lipids.
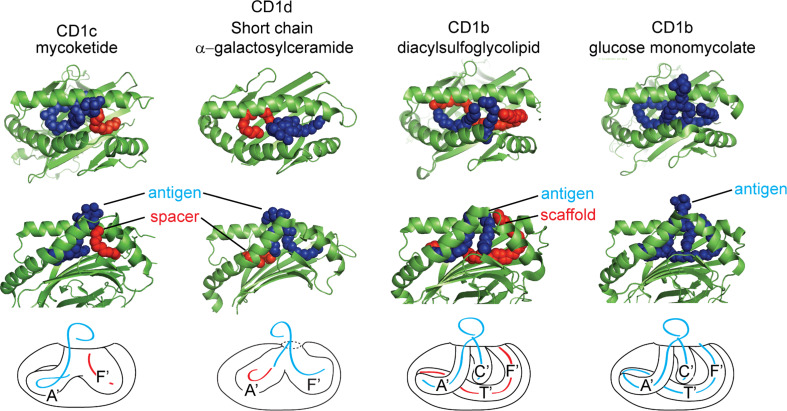
Several crystal structures of CD1 in complex with lipid antigens have revealed the presence of densities interpreted as aliphatic hydrocarbon chains within the binding pockets, which do not correspond to the alkyl chains of the antigens that protrude “upwards” through the F′ portal. In agreement with the earlier observations of Gadola et al. [18], studies have shown that naturally occurring spacer lipids were present when native refolding conditions were used to crystallize CD1d with short-chain α-galactosylceramide [45, 46] and CD1b with phosphatidylcholine [26]. Electron density measurements, as well as detection by native mass spectrometry revealed natural 16–40 carbon length “spacers” that serve to fill the remainder of the unoccupied CD1 pocket [26, 45, 46]. Additionally, lipid spacers have often been found in structures of CD1 in complex with monoacyl antigens. Structures of dideoxymycobactins in complex with CD1a and mannosyl-phosphomycoketide with CD1c (Fig. 3) identify small hydrocarbon chains that likely correspond to spacer lipids within the F′ pocket [20, 22]. Similarly lyso-phosphatidylcholine, in complex with CD1d, identifies a hydrocarbon chain in the unoccupied A′ pocket [8]. Dynamic modeling of CD1 molecules in lipid-bound and lipid-free states indicates that spacers may act to stabilize CD1 complexes from collapse. Unliganded CD1 proteins show collapse of hydrophobic pockets in the absence of lipids, resulting in closure of the helices. Surprisingly, the CD1a-binding cavity was interpreted as remaining preserved in the unliganded state [47].
Fig. 3.
Hydrophobic spacer lipids can fill the empty cavity of CD1 isoforms as shown in the structure CD1c with mannosyl-phosphomycoketide (MPM) and CD1d with short-chain α-galactosylceramide (PBS-25). When higher affinity lipids are present in permissive environments, ligands and spacers must both be displaced during antigen exchange reactions. To illustrate this, CD1 is rendered with antigenic lipids in blue with hydrophobic spacer lipids in red, with schematic [figures were generated from RCSB protein data bank files 3OV6 (CD1c-MPM), 1Z5L (CD1d-PBS-25), 3T8X (CD1b-sulfoglycolipid), and 1UQS (CD1b-GMM)]
Two studies independently identified the natural spacer lipids in CD1b as diacylglycerols and deoxyceramides [25, 44]. Natural sulfoglycolipids are a family of mycobacterial polyketide lipids that contain a heterogeneous mixture of fatty acids and polyketides linked to sulfotrehalose [48]. Sulfoglycolipid in complex with CD1b was found to release diacylglycerol in mass spectrometry. Further, the eluted spacer lipid corresponded in size to an electron density within the T′ and F′ pockets, which was positioned below the sulfoglycolipid (Fig. 3) [25]. The binding of sulfoglycolipids to CD1b results in a conformational change near the F′ pocket, allowing amino acids glutamate 80 within the α1 helix and tyrosine 151 within the α2 helix to interact. This interaction is thought to prevent the egress of diacylglycerol from the F’ pocket, thus stabilizing the CD1b-lipid complex [25].
A separate lipidomics study of all CD1b ligands found particularly hydrophobic lipids eluting selectively from CD1b, and identified diacylglycerol and deoxyceramides [44]. Further, adding diacylglycerols during the loading of CD1b-presented antigens resulted in increased T cell recognition of a short-chain form of glucose monomycolate, but abrogated recognition of long-chain glucose monomycolate. Thus, adding one large lipid, or instead adding two lipids whose total alkane chain length approximates the volume of the CD1b groove, leads to improved antigen recognition. Thus, the naturally occurring spacer lipids have an actual function in augmenting antigen recognition. Because crystal structures suggest that the natural spacer lipids are located in the T′ tunnel “beneath the antigen” the term “scaffold” emphasizes their upward lifting function [44]. Such scaffold lipids enhance recognition and so contrast with the side-by-side orientation of spacer lipids observed in CD1c, whose function, if any, on antigen recognition is unknown [20]. It is currently unknown what percentage of CD1b proteins are initially loaded with one or two lipids [44]. However, the relatively small average mass of all lipids eluted from CD1b relative to its large groove volume suggests that a large percentage of the initially formed CD1b-lipid complexes contain two or more lipid ligands [44]. With the broad range of structurally diverse ligands encountered in the endoplasmic reticulum and endosome, we speculate that scaffolding lipids might act to satisfy the energetic requirements for expelling water from hydrophobic surfaces that line the binding grooves of CD1. Stabilization of CD1 may influence antigen presentation. Studies using short-tail CD1 antigens have demonstrated that lipid length of the antigen will influence CD1-restricted T cell recognition glycolipid analogs [49, 50]. Though spacer lipids fill the remainder of unoccupied binding grooves, short-chain antigens may be less stable in vivo due to their inability to stably load within detergent-insoluble lipid rafts [51, 52].
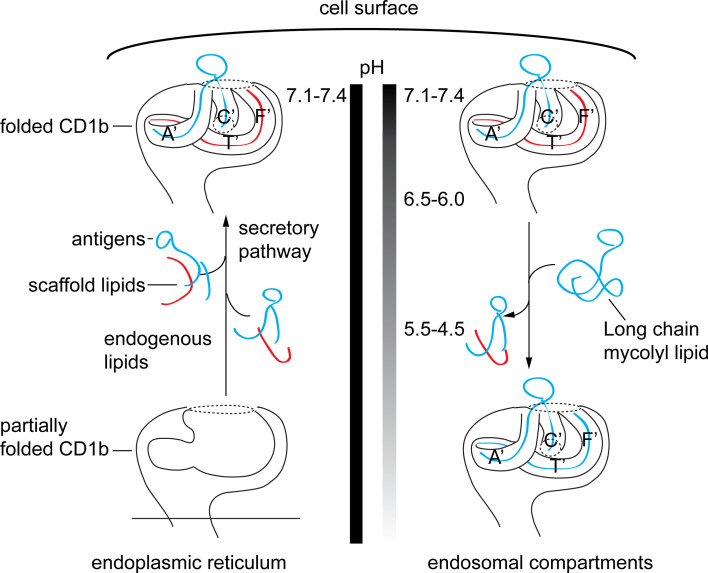
Spacers are likely displaced when large exogenous lipids are encountered (Fig. 4). A highly consistent observation in the CD1b system is that C32 glucose monomycolate can be loaded on CD1b proteins at the cell surface, whereas C80 glucose monomycolate antigens have stringent requirements for exchange in the low-pH environment of lysosomes [15, 43]. We speculate that the smaller C32 antigen might exchange for the antigen located in the “upper” region of the groove (Fig. 4, blue), whereas loading the C80 antigen likely requires unloading of the more deeply positioned scaffold lipids as well (Fig. 4, red and blue). Supporting this hypothesis, spacers are not detected in the structure of CD1b in complex with long-chain glucose monomycolate (Fig. 3), which fully accommodate the CD1b-binding channel [24]. Detailed mechanisms of lipid exchange and spacer displacement are not completely understood, but the reversible denaturation of CD1b at low pH promotes antigen loss from the groove and renders CD1b proteins able to bind large lipids [53]. Thus, several predictions of the scaffold loss hypothesis have been experimentally verified.
Fig. 4.
Endosomal recycling of CD1b and pH. CD1 is synthesized in the ER in the presence of a variety of lipid ligands, which help to stabilize the protein-lipid complex. CD1b then is shuttled to the cell surface within the secretory pathway, where further lipid exchange can take place with endogenous or exogenous lipids. CD1b is re-internalized and sorted based on isoform-specific sorting motifs into early endosomes or enter late endosome/lysosomes. Lipids entering with CD1b or from other endosomal compartments can be generated by endosomal co-factors. Acidic pH alters the physical properties of CD1b, which promotes lipid exchange in an editing process that allows lipids with higher affinity for CD1b to be loaded in late endosomes before recycling back to the cell surface. Whereas the ability of cell surface CD1b to bind only short-chain lipids has been long known, spacers now provide a candidate mechanism that explains why short-chain lipids have lower stringency loading requirements: exogenous short-chain lipids would only need to exchange with the superficially seated antigen on top, whereas long-chain lipids would require expulsion of antigens and deeply seated spacers
CD1 proteins load lipids in distinct compartments
CD1 proteins are assembled in the endoplasmic reticulum in association with chaperones calnexin, calreticulin, and with the thiol oxidoreductase ERp57, which facilitate the folding and formation of disulfide bonds within the glycoprotein [54]. Once assembled and matured, CD1 associates with β2m and traffics through the trans-Golgi network towards the plasma membrane [55]. CD1 proteins are subsequently internalized and enter the endosomal pathway. It is here that CD1 isoforms diverge and traffic into different endosomal compartments based on the amino acid sorting motifs of their cytoplasmic tails. CD1b, CD1c, and CD1d are targeted to the endosomal network by their expression of a tyrosine-based sorting motifs, YXXZ, where Y is tyrosine, X is any amino acid, and Z, is a bulky hydrophobic amino acid. This tail motif binds the adaptor protein complex 2 (AP2) to direct internalization to a variety of endosomal compartments via clathrin-coated pits [56]. CD1a does not contain any sorting motifs and is internalized through an AP-independent pathway, similar to MHC class I [57]. Human CD1b and mouse CD1d have tail motifs that mediate additional interactions with adaptor protein complex 3 (AP3), driving these antigen-presenting molecules into lysosomes. These interactions predict the observed localization of each CD1 isoform within the endolysosomal pathway. CD1a associates with the GTPases ADP-ribosylation factor 6, Ras-related protein Rab-22A, and ADP-ribosylation factor-like protein 13B, which regulate endocytic recycling traffic [58]. Lacking an endosomal localization motif, CD1a is mainly seen at the surface at steady state, whereas CD1b, CD1c, and CD1d substantially enter lysosomal-associated membrane protein 1 expressing compartments [59–61]. CD1e never reaches the cell surface, but traffics within the trans-Golgi network of immature dendritic cells and upon maturation reaches the late endosome lysosomal compartments where it is cleaved into its active form [38]. The isoform-specific trafficking patterns allow CD1 to sample and survey the endogenous and exogenous lipid contents that arrive from different cellular compartments.
ER assembly and lipid association
Throughout CD1 assembly and recycling, CD1 proteins sample and survey lipids from the ER and endosomal compartments [56]. Whereas many studies of trafficking focus on late events in endosomal recycling, the initial capture of self-ligands in the endoplasmic reticulum and egress to the surface are biologically important events that lead to the capture of self-lipids, so are emphasized here. The first endogenous lipids eluted from murine CD1d included phosphatidylinositol (PI) and phosphatidylinositol-glycans, which were assembled with CD1d in the ER [62, 63]. Whereas early studies suggested that phosphatidylinositol containing lipids might dominate the spectrum of endogenous CD1 ligands, later studies clearly show that the cellular ligands of CD1 proteins are diverse, and include neutral lipids, sphingolipids, and phospholipids [44].
Elution of secreted CD1d has shown that all major family members of glycerophospholipids and sphingolipids were found associated with CD1d [41, 64, 65]. The variety of glycerophospholipid head groups included unmodified phosphatidic acid (PA), phosphatidylglycerol (PG), phosphatidylserine (PS), phosphatidylethanolamine (PE), and phosphatidylcholine (PC) and previously identified PI. Sphingolipid species included sphingomyelin (SM), glycosphingolipids (GSL), and higher-order GM gangliosides [41, 64]. Diacyl species were predominant, but lyso-phospholipids, and tetra-acyl cardiolipins were also found. Further, analysis also revealed the presence of peroxisome-derived plasmalogen lipids [41, 65, 66]. In fact, under homeostatic conditions, CD1 molecules accumulate the most abundant lipids from both secretory and endosomal compartments [65]. For example, CD1d engineered with an ER retention signal is predominantly found associated with phosphatidylcholine, the most abundant lipid in the cell. In addition to phosphatidylcholine, CD1d that have access to secretory and endosomal compartments was found associated with sphingomyelin and lysophospholipids, dominant lipids within the trans-Golgi network and lysosomes, respectively [65]. Thus, it seems that CD1 is capable of associating promiscuously with a variety of endogenous lipids. In contrast to invariant chain model in which one type of peptide predominates in the initially formed MHC class II complex, dozens [41] or hundreds [44] of hydrophobic lipids act to stabilize the initial CD1 formed in cells.
The ER resident protein, microsomal transfer protein (MTP), which promotes the assembly and secretion of large apolipoprotein B lipoproteins, was also found to function in CD1 lipid association [67]. MTP facilitates the transfer of triacylglycerol and glycerophospholipids from sites of synthesis to the nascent apolipoprotein B polypeptide [68] and can also associate with CD1d to influence phospholipid association with CD1d in cell-free systems [69]. The role of microsomal transfer protein in CD1 lipid presentation is substantiated in patients with abetalipoproteinemia (ABP), an autosomal recessive disorder caused by mutations in microsomal transfer protein. Patients with abetalipoproteinemia have low levels of apolipoprotein B in the plasma, as well as APCs that have reduced ability to activate CD1-restricted T cells [70]. Microsomal transfer protein deficiency affects the presentation of secretory as well as endosomal antigens that require lysosomal loading. For example, self-reactive CD1-restricted T cells, as well as NKT cells recognizing the glycosphingolipid isoglobotrihexosylceramide 3 (iGb3), or CD1b-restricted T cells recognizing mycoyl lipids, have reduced activity when presented by microsomal transfer protein-deficient antigen-presenting cells [69–72]. The surprising finding that an ER protein may also affect endosomal loading of CD1 lipids could be explained by the observation that CD1 molecules from MTP-deficient antigen-presenting cells are more susceptible to lysosomal degradation [70] and show reduced endosomal recycling [71]. The distal effect of microsomal transfer protein deficiency suggests that microsomal transfer proteins play a role in maintaining stability of CD1 molecules, possibly by regulating lipid availability or by acting as a CD1 lipid “editor”, selecting for lipids only capable of stabilizing CD1 molecule during assembly and biosynthesis.
Regulation by CD1 structure and trafficking
With the capacity of CD1 molecules to bind a variety of structurally unrelated self-lipid ligands [41, 44], the biophysical properties of CD1 isoforms and their distinct trafficking into various endosomal compartments regulate availability of antigens for presentation to T cells. For example, the shallow antigen-binding groove, early endosomal trafficking, and the stability of the CD1a-binding cavity in the absence of ligand [47], all contribute to the general conclusion that CD1a may have the least stringent loading requirements amongst CD1 isoforms. Consistent with this hypothesis, CD1a binds shorter alkyl chains that load at neutral pH or on the cell surface [17, 73].
CD1b and CD1d traffic into late endosome/lysosomes where acidic pH and endosomal cofactors help regulate lipid availability and CD1 complex formation. The capacity of CD1b to bind very long chain mycoyl lipids of mycobacteria requires acid in vitro [15] and occurs more readily in cells with intact acidification mechanisms [43, 74]. Mechanistically, acid functions to neutralize acidic residues that would normally use their positively charged state to bind anionic residues and tether the α1 and α2 helices of CD1b together. Low pH interrupts these charge–charge interactions to promote partial unfolding to allow lipid exchange [53]. Besides pH, acidic compartments also contain cofactors such as saposins, which can catalyze the formation of CD1 lipid complexes by directly binding and solubilizing lipids or by destabilizing cellular membranes, thereby making lipid ligands available for CD1 capture [75–78].
Similar to CD1b, several examples exists that indicate the requirement of CD1d to traffic into late endosome/lysosomes where pH-dependent co-factors are required to process CD1d ligands to antigens [42, 79, 80]. The unique properties of CD1c also allow for a level of lipid regulation. The A′ pocket of CD1c binds branched alkyl chain lipids such as mycoketides [29, 30], and its promiscuous trafficking patterns allow it to bypass lysosomal compartments to present lipopeptides that would otherwise be degraded in lysosomes [31]. Thus, lipid complex formation is both regulated by the physical properties of CD1 isoforms and its ability to traffic into various endosomal compartments that promote more permissive loading environments (Fig. 4).
Concluding thoughts
The molecular immunology and cell biology have advanced since the discovery of CD1 and lipid reactive T cells over the last 20 years, such that an integrated model of cellular lipid capture and display is now emerging (Fig. 4). Developments in crystallography have revealed isoform-specific CD1-binding grooves, and show how lipids bind within CD1 isoforms. New methods in mass spectrometry and high-resolution analysis have shown that not only amphipathic lipids bind within CD1, but that highly hydrophobic spacer lipids also play important roles in maintaining CD1 stability and regulation. The ability of CD1 to ubiquitously associate with the majority of cellular self-lipids suggests a potential role of self-lipids in maintaining lipid-reactive T cell homeostasis or T cell selection. Indeed, data has shown that self-antigen reactive CD1a-restricted T cells are very common amongst individuals and may have roles in skin homeostasis [81, 82]. Emerging tools such as humanized mice [83] CD1 transgenic mice [84], and human CD1 tetramers [30, 85–87] will shed light on the role of T cell recognition of lipids in vivo.
Acknowledgments
This work was supported by the Burroughs Wellcome Fund for Translational Research, NIAID R01 AI04393 and R01 AR 048632.
References
- 1.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4–CD8-cytolytic T lymphocytes. Nature. 1989;341(6241):447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 2.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4–8-T lymphocytes to a microbial antigen. Nature. 1992;360(6404):593–597. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 3.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta + T cells. Nature. 1994;372(6507):691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 4.Beckman EM, Melian A, Behar SM, Sieling PA, Chatterjee D, Furlong ST, Matsumoto R, Rosat JP, Modlin RL, Porcelli SA. CD1c restricts responses of mycobacteria-specific T cells. Evidence for antigen presentation by a second member of the human CD1 family. J Immunol. 1996;157(7):2795–2803. [PubMed] [Google Scholar]
- 5.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448(7149):44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 6.Girardi E, Maricic I, Wang J, Mac TT, Iyer P, Kumar V, Zajonc DM. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat Immunol. 2012;13(9):851–856. doi: 10.1038/ni.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Sagaseta J, Kung JE, Savage PB, Gumperz J, Adams EJ. The molecular basis for recognition of CD1d/alpha-galactosylceramide by a human non-Valpha24 T cell receptor. PLoS Biol. 2012;10(10):e1001412. doi: 10.1371/journal.pbio.1001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Sagaseta J, Sibener LV, Kung JE, Gumperz J, Adams EJ. Lysophospholipid presentation by CD1d and recognition by a human natural killer T-cell receptor. EMBO J. 2012;31(8):2047–2059. doi: 10.1038/emboj.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, Patel O, Vivian JP, Matsuda JL, McCluskey J, Godfrey DI, Marrack P, Rossjohn J, Gapin L. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34(3):315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel O, Pellicci DG, Gras S, Sandoval-Romero ML, Uldrich AP, Mallevaey T, Clarke AJ, Le Nours J, Theodossis A, Cardell SL, Gapin L, Godfrey DI, Rossjohn J. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13(9):857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- 11.Pellicci DG, Clarke AJ, Patel O, Mallevaey T, Beddoe T, Le Nours J, Uldrich AP, McCluskey J, Besra GS, Porcelli SA, Gapin L, Godfrey DI, Rossjohn J. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12(9):827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wun KS, Cameron G, Patel O, Pang SS, Pellicci DG, Sullivan LC, Keshipeddy S, Young MH, Uldrich AP, Thakur MS, Richardson SK, Howell AR, Illarionov PA, Brooks AG, Besra GS, McCluskey J, Gapin L, Porcelli SA, Godfrey DI, Rossjohn J. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34(3):327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Valpha14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010;207(11):2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardi E, Yu ED, Li Y, Tarumoto N, Pei B, Wang J, Illarionov P, Kinjo Y, Kronenberg M, Zajonc DM. Unique interplay between sugar and lipid in determining the antigenic potency of bacterial antigens for NKT cells. PLoS Biol. 2011;9(11):e1001189. doi: 10.1371/journal.pbio.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng TY, Relloso M, Van Rhijn I, Young DC, Besra GS, Briken V, Zajonc DM, Wilson IA, Porcelli S, Moody DB. Role of lipid trimming and CD1 groove size in cellular antigen presentation. EMBO J. 2006;25(13):2989–2999. doi: 10.1038/sj.emboj.7601185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277(5324):339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 17.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5(5):387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 18.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol. 2002;3(8):721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 19.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4(8):808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 20.Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, Kazen AR, Shah S, Haddadian EJ, Gumperz JE, Saghatelian A, Faraldo-Gomez JD, Meredith SC, Piccirilli JA, Adams EJ. The 2.5-Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33(6):853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Alles LF, Giacometti G, Versluis C, Maveyraud L, de Paepe D, Guiard J, Tranier S, Gilleron M, Prandi J, Hanau D, Heck AJ, Mori L, De Libero G, Puzo G, Mourey L, de la Salle H. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci USA. 2011;108(32):13230–13235. doi: 10.1073/pnas.1105627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, Moody DB, Wilson IA. Molecular mechanism of lipopeptide presentation by CD1a. Immunity. 2005;22(2):209–219. doi: 10.1016/j.immuni.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T cell activation by lipopeptide antigens. Science. 2004;303(5657):527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 24.Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR, Besra GS, Cerundolo V, Jones EY. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2004;172(4):2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Alles LF, Collmann A, Versluis C, Lindner B, Guiard J, Maveyraud L, Huc E, Im JS, Sansano S, Brando T, Julien S, Prandi J, Gilleron M, Porcelli SA, de la Salle H, Heck AJ, Mori L, Puzo G, Mourey L, De Libero G. Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci USA. 2011;108(43):17755–17760. doi: 10.1073/pnas.1110118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Alles LF, Versluis K, Maveyraud L, Vallina AT, Sansano S, Bello NF, Gober HJ, Guillet V, de la Salle H, Puzo G, Mori L, Heck AJ, De Libero G, Mourey L. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25(15):3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De Libero G, Puzo G, Gilleron M. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol. 2009;16(1):82–92. doi: 10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278(5336):283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 29.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404(6780):884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 30.Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S, Bhatt A, van Summeren RP, Altman JD, Jacobs WR, Jr, Adams EJ, Minnaard AJ, Porcelli SA, Moody DB. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med. 2013;210(4):729–741. doi: 10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Rhijn I, Young DC, De Jong A, Vazquez J, Cheng TY, Talekar R, Barral DC, Leon L, Brenner MB, Katz JT, Riese R, Ruprecht RM, O’Connor PB, Costello CE, Porcelli SA, Briken V, Moody DB. CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. J Exp Med. 2009;206(6):1409–1422. doi: 10.1084/jem.20082480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams EJ. Diverse antigen presentation by the Group 1 CD1 molecule, CD1c. Mol Immunol. 2013;55(2):182–185. doi: 10.1016/j.molimm.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gomez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH, Kawakami K, Nizet V, Besra GS, Tsuji M, Zajonc DM, Kronenberg M. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12(10):966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, Kronenberg M, Zajonc DM. Lipid binding orientation within CD1d affects recognition of Borrelia burgdorferi antigens by NKT cells. Proc Natl Acad Sci USA. 2010;107(4):1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joyce S, Girardi E, Zajonc DM. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J Immunol. 2011;187(3):1081–1089. doi: 10.4049/jimmunol.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12(12):845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13(2):101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 38.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310(5752):1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 39.Jong AC, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar A, Rhijn I, Peña-Cruz V, Ruan DT, Altman JD, Rossjohn J, Moody DB. CD1a autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15(2):177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haig NA, Guan Z, Li D, McMichael A, Raetz CR, Xu XN. Identification of self-lipids presented by CD1c and CD1d proteins. J Biol Chem. 2011;286(43):37692–37701. doi: 10.1074/jbc.M111.267948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox D, Fox L, Tian R, Bardet W, Skaley M, Mojsilovic D, Gumperz J, Hildebrand W. Determination of cellular lipids bound to human CD1d molecules. PLoS ONE. 2009;4(5):e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291(5504):664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 43.Moody DB, Briken V, Cheng TY, Roura-Mir C, Guy MR, Geho DH, Tykocinski ML, Besra GS, Porcelli SA. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3(5):435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 44.Huang S, Cheng TY, Young DC, Layre E, Madigan CA, Shires J, Cerundolo V, Altman JD, Moody DB. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci USA. 2011;108(48):19335–19340. doi: 10.1073/pnas.1112969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zajonc DM, Cantu C, 3rd, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6(8):810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu D, Zajonc DM, Fujio M, Sullivan BA, Kinjo Y, Kronenberg M, Wilson IA, Wong CH. Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc Natl Acad Sci USA. 2006;103(11):3972–3977. doi: 10.1073/pnas.0600285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garzon D, Anselmi C, Bond PJ, Faraldo-Gomez JD. Dynamics of the antigen-binding grooves in CD1 proteins: reversible hydrophobic collapse in the lipid-free state. J Biol Chem. 2013;288(27):19528–19536. doi: 10.1074/jbc.M113.470179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis . J Exp Med. 2004;199(5):649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, Tarlton A, Reddy BG, Schmidt RR, Reiter Y, Griffiths GM, van der Merwe PA, Besra GS, Jones EY, Batista FD, Cerundolo V. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204(5):1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Jong A, Arce EC, Cheng TY, van Summeren RP, Feringa BL, Dudkin V, Crich D, Matsunaga I, Minnaard AJ, Moody DB. CD1c presentation of synthetic glycolipid antigens with foreign alkyl branching motifs. Chem Biol. 2007;14(11):1232–1242. doi: 10.1016/j.chembiol.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Im JS, Arora P, Bricard G, Molano A, Venkataswamy MM, Baine I, Jerud ES, Goldberg MF, Baena A, Yu KO, Ndonye RM, Howell AR, Yuan W, Cresswell P, Chang YT, Illarionov PA, Besra GS, Porcelli SA. Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity. 2009;30(6):888–898. doi: 10.1016/j.immuni.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan BA, Nagarajan NA, Wingender G, Wang J, Scott I, Tsuji M, Franck RW, Porcelli SA, Zajonc DM, Kronenberg M. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J Immunol. 2010;184(1):141–153. doi: 10.4049/jimmunol.0902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Relloso M, Cheng TY, Im JS, Parisini E, Roura-Mir C, DeBono C, Zajonc DM, Murga LF, Ondrechen MJ, Wilson IA, Porcelli SA, Moody DB. pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity. 2008;28(6):774–786. doi: 10.1016/j.immuni.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang SJ, Cresswell P. Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J Biol Chem. 2002;277(47):44838–44844. doi: 10.1074/jbc.M207831200. [DOI] [PubMed] [Google Scholar]
- 55.Odyniec AN, Barral DC, Garg S, Tatituri RV, Besra GS, Brenner MB. Regulation of CD1 antigen-presenting complex stability. J Biol Chem. 2010;285(16):11937–11947. doi: 10.1074/jbc.M109.077933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7(12):929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 57.Barral DC, Cavallari M, McCormick PJ, Garg S, Magee AI, Bonifacino JS, De Libero G, Brenner MB. CD1a and MHC class I follow a similar endocytic recycling pathway. Traffic. 2008;9(9):1446–1457. doi: 10.1111/j.1600-0854.2008.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barral DC, Garg S, Casalou C, Watts GF, Sandoval JL, Ramalho JS, Hsu VW, Brenner MB. Arl13b regulates endocytic recycling traffic. Proc Natl Acad Sci USA. 2012;109(52):21354–21359. doi: 10.1073/pnas.1218272110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Briken V, Jackman RM, Watts GF, Rogers RA, Porcelli SA. Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. J Exp Med. 2000;192(2):281–288. doi: 10.1084/jem.192.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity. 2002;16(5):697–706. doi: 10.1016/S1074-7613(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 61.Sugita M, van Der Wel N, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci USA. 2000;97(15):8445–8450. doi: 10.1073/pnas.150236797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joyce S, Woods AS, Yewdell JW, Bennink JR, De Silva AD, Boesteanu A, Balk SP, Cotter RJ, Brutkiewicz RR. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279(5356):1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 63.De Silva AD, Park JJ, Matsuki N, Stanic AK, Brutkiewicz RR, Medof ME, Joyce S. Lipid protein interactions: the assembly of CD1d1 with cellular phospholipids occurs in the endoplasmic reticulum. J Immunol. 2002;168(2):723–733. doi: 10.4049/jimmunol.168.2.723. [DOI] [PubMed] [Google Scholar]
- 64.Muindi K, Cernadas M, Watts GF, Royle L, Neville DC, Dwek RA, Besra GS, Rudd PM, Butters TD, Brenner MB. Activation state and intracellular trafficking contribute to the repertoire of endogenous glycosphingolipids presented by CD1d [corrected] Proc Natl Acad Sci USA. 2010;107(7):3052–3057. doi: 10.1073/pnas.0915056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182(8):4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Facciotti F, Ramanjaneyulu GS, Lepore M, Sansano S, Cavallari M, Kistowska M, Forss-Petter S, Ni G, Colone A, Singhal A, Berger J, Xia C, Mori L, De Libero G. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol. 2012;13(5):474–480. doi: 10.1038/ni.2245. [DOI] [PubMed] [Google Scholar]
- 67.Brozovic S, Nagaishi T, Yoshida M, Betz S, Salas A, Chen D, Kaser A, Glickman J, Kuo T, Little A, Morrison J, Corazza N, Kim JY, Colgan SP, Young SG, Exley M, Blumberg RS. CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med. 2004;10(5):535–539. doi: 10.1038/nm1043. [DOI] [PubMed] [Google Scholar]
- 68.Hussain MM, Rava P, Pan X, Dai K, Dougan SK, Iqbal J, Lazare F, Khatun I. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr Opin Lipidol. 2008;19(3):277–284. doi: 10.1097/MOL.0b013e3282feea85. [DOI] [PubMed] [Google Scholar]
- 69.Dougan SK, Salas A, Rava P, Agyemang A, Kaser A, Morrison J, Khurana A, Kronenberg M, Johnson C, Exley M, Hussain MM, Blumberg RS. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202(4):529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeissig S, Dougan SK, Barral DC, Junker Y, Chen Z, Kaser A, Ho M, Mandel H, McIntyre A, Kennedy SM, Painter GF, Veerapen N, Besra GS, Cerundolo V, Yue S, Beladi S, Behar SM, Chen X, Gumperz JE, Breckpot K, Raper A, Baer A, Exley MA, Hegele RA, Cuchel M, Rader DJ, Davidson NO, Blumberg RS. Primary deficiency of microsomal triglyceride transfer protein in human abetalipoproteinemia is associated with loss of CD1 function. J Clin Invest. 2010;120(8):2889–2899. doi: 10.1172/JCI42703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sagiv Y, Bai L, Wei DG, Agami R, Savage PB, Teyton L, Bendelac A. A distal effect of microsomal triglyceride transfer protein deficiency on the lysosomal recycling of CD1d. J Exp Med. 2007;204(4):921–928. doi: 10.1084/jem.20061568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaser A, Hava DL, Dougan SK, Chen Z, Zeissig S, Brenner MB, Blumberg RS. Microsomal triglyceride transfer protein regulates endogenous and exogenous antigen presentation by group 1 CD1 molecules. Eur J Immunol. 2008;38(8):2351–2359. doi: 10.1002/eji.200738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cernadas M, Cavallari M, Watts G, Mori L, De Libero G, Brenner MB. Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J Immunol. 2010;184(3):1235–1241. doi: 10.4049/jimmunol.0804140. [DOI] [PubMed] [Google Scholar]
- 74.Ernst WA, Maher J, Cho S, Niazi KR, Chatterjee D, Moody DB, Besra GS, Watanabe Y, Jensen PE, Porcelli SA, Kronenberg M, Modlin RL. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8(3):331–340. doi: 10.1016/S1074-7613(00)80538-5. [DOI] [PubMed] [Google Scholar]
- 75.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5(2):175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 76.Winau F, Schwierzeck V, Hurwitz R, Remmel N, Sieling PA, Modlin RL, Porcelli SA, Brinkmann V, Sugita M, Sandhoff K, Kaufmann SH, Schaible UE. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5(2):169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 77.Zhou D, Cantu C, 3rd, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303(5657):523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leon L, Tatituri RV, Grenha R, Sun Y, Barral DC, Minnaard AJ, Bhowruth V, Veerapen N, Besra GS, Kasmar A, Peng W, Moody DB, Grabowski GA, Brenner MB. Saposins utilize two strategies for lipid transfer and CD1 antigen presentation. Proc Natl Acad Sci USA. 2012;109(12):4357–4364. doi: 10.1073/pnas.1200764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darmoise A, Teneberg S, Bouzonville L, Brady RO, Beck M, Kaufmann SH, Winau F. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity. 2010;33(2):216–228. doi: 10.1016/j.immuni.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306(5702):1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 81.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nat Immunol. 2010;11(12):1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Lalla C, Lepore M, Piccolo FM, Rinaldi A, Scelfo A, Garavaglia C, Mori L, De Libero G, Dellabona P, Casorati G. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur J Immunol. 2011;41(3):602–610. doi: 10.1002/eji.201041211. [DOI] [PubMed] [Google Scholar]
- 83.Lockridge JL, Chen X, Zhou Y, Rajesh D, Roenneburg DA, et al. Analysis of the CD1 Antigen Presenting System in Humanized SCID Mice. PLoS ONE. 2011;6(6):e21701. doi: 10.1371/journal.pone.0021701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Felio K, Nguyen H, Dascher CC, Choi HJ, Li S, Zimmer MI, Colmone A, Moody DB, Brenner MB, Wang CR. CD1-restricted adaptive immune responses to mycobacteria in human group 1 CD1 transgenic mice. J Exp Med. 2009;206(11):2497–2509. doi: 10.1084/jem.20090898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kasmar AG, van Rhijn I, Cheng TY, Turner M, Seshadri C, Schiefner A, Kalathur RC, Annand JW, de Jong A, Shires J, Leon L, Brenner M, Wilson IA, Altman JD, Moody DB. CD1b tetramers bind alphabeta T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208(9):1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195(5):625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kasmar AG, Rhijn I, Magalhaes KG, Young DC, Cheng TY, Turner MT, Schiefner A, Kalathur RC, Wilson IA, Bhati M, Gras S, Birkinshaw RW, Tan LL, Rossjohn J, Shires J, Jakobsen S, Altman JD, Moody DB. Cutting Edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol. 2013;191(9):4499–4503. doi: 10.4049/jimmunol.1301660. [DOI] [PMC free article] [PubMed] [Google Scholar]