Summary
Background
Class II myosins generate contractile forces in cells by polymerizing into bipolar filaments and pulling on anchored actin filaments. Nonmuscle myosin II (NMII) plays central roles during cell adhesion, migration, cytokinesis, and tissue morphogenesis. NMII is present in virtually all mammalian cell types as tissue-specific combinations of NMIIA, NMIIB, and NMIIC isoforms. It remains poorly understood how the highly dynamic NMII-actin contractile system begins to assemble at new cellular locations during cell migration and how incorporation of different NMII isoforms into this system is coordinated.
Results
Using platinum replica electron microscopy in combination with immunogold labeling, we demonstrate that individual activated (phosphorylated on the regulatory light chain and unfolded) NMIIA and NMIIB molecules represent a functional form of NMII in motile cells and that NMIIA and NMIIB copolymerize into nascent bipolar filaments during contractile system assembly. Using subdiffraction STED microscopy together with a pharmacological block-and-release approach, we report that NMIIA and NMIIB simultaneously incorporate into the cytoskeleton during initiation of contractile system assembly, whereas the characteristic rearward shift of NMIIB relative to NMIIA is established later in the course of NMII turnover.
Conclusions
We show existence of activated NMII monomers in cells, copolymerization of endogenous NMIIA and NMIIB molecules, and contribution of both isoforms, rather than only NMIIA, to early stages of the contractile system assembly. These data change the current paradigms about dynamics and functions of NMII and provide new conceptual insights into organization and dynamics of the ubiquitous cellular machinery for contraction that acts in multiple cellular contexts.
Introduction
Class II myosins are responsible for generation of contractile forces in cells. The classic underlying mechanism of contraction involves polymerization of myosin II molecules into bipolar filaments, which then pull on anchored actin filaments. Nonmuscle myosin II (NMII) is present in virtually all mammalian cell types as tissue-specific combinations of NMIIA, NMIIB, and NMIIC isoforms, which play both unique and overlapping roles [1]. NMII is well-known to play key roles in cell adhesion, migration, cytokinesis, and tissue morphogenesis [1, 2]. Additionally, recent data increasingly point to unconventional roles of NMII in membrane organelle morphogenesis, such as exocytosis [3], endocytosis [4], post-Golgi and Golgi-to-ER trafficking [5, 6], and mitochondrion fission [7].
NMII molecules (also referred here as NMII monomers) consist of two heavy chains and two pairs of light chains. NMII activity is regulated by phosphorylation of the myosin regulatory light chain (MRLC). In the dephosphorylated state, NMII monomers are inactive and have a folded autoinhibitory conformation. When MRLC is monophosphorylated on Ser19 (pMRLC) or biphosphorylated on Thr18 and Ser19 (ppMRLC), NMII molecules unfold, acquire motor activity [8], and can polymerize into bipolar filaments [9]. Bipolar filaments can then pull on and reorganize actin filaments, thus inducing assembly of the contractile actin-NMII system, which consists of interconnected actin-NMII bundles and networks associated with cell-matrix and cell-cell adhesions [10–12]. In motile cells, the contractile system is highly dynamic and undergoes constant assembly near the cell leading edge, followed by disassembly in the cell body [11, 13] and recycling of inactive NMII monomers back to the cell periphery [14]. However, many questions remain about specifics of this cycle, especially at early stages of contractile system assembly, and about commonalities and differences between NMII isoforms in this process.
It is generally believed that bipolar NMII filaments are the only functional form of NMII in cells. Their assembly occurs in the distal lamellae of migrating cells behind lamellipodia [11–13], a phenomenon thought to be the first event of the NMII turnover cycle. However, we recently found that the earliest stage of focal adhesion formation depends on NMII activity under conditions where bipolar filaments are virtually absent and activated NMII monomers abundant [10]. Such conditions were created by treating REF52 fibroblasts with blebbistatin, a specific inhibitor of NMII motor activity, and then washing blebbistatin out. Blebbistatin treatment induced depolymerization of NMII bipolar filaments without inhibiting MRLC phosphorylation, thus generating a large pool of phosphorylated NMII monomers. Upon blebbistatin washout, when these monomers regain motor activity, small focal adhesions were formed before the formation of NMII bipolar filaments, thus suggesting a role of activated NMII monomers in adhesion initiation [10]. Since blebbistatin treatment-and-washout likely synchronizes the formation of the contractile system at nascent adhesions, a process that occurs stochastically in lamellae under normal conditions, activated NMII monomers might be present even in untreated cells, but direct evidence for this idea is currently missing.
In cells, NMIIB is often displaced toward the cell rear relative to NMIIA [15, 16], which is thought to reflect incorporation of NMIIB into pre-existing structures made by NMIIA [17]. Such model implies that NMIIA and NMIIB more likely form homotypic filaments that sequentially incorporate into the contractile system. However, existing data suggest that muscle and nonmuscle myosins II can copolymerize in vitro [18], truncated NMIIA and NMIIB proteins can heteropolymerize in vitro [19] and in cells [20], and endogenous NMII isoforms can be coimmunoprecipitated from lysates [21]. However, explicit evidence for copolymerization of endogenous NMII isoforms into heterotypic bipolar filaments is not available and significance of such copolymerization, if it exists, is not known.
In this study, we resolve key outstanding questions regarding the NMII turnover in motile cells. We provide direct evidence that (i) activated NMIIA and NMIIB monomers represent a functional form of NMII in cells; (ii) NMIIA and NMIIB copolymerize in bipolar filaments; and (iii) NMIIA and NMIIB simultaneously incorporate into the cytoskeleton during initiation of contractile system assembly, whereas the anterior-posterior gradient of their localization is established much later, likely, in the course of bipolar filament recycling.
Results
Direct evidence for existence of activated monomers in cells
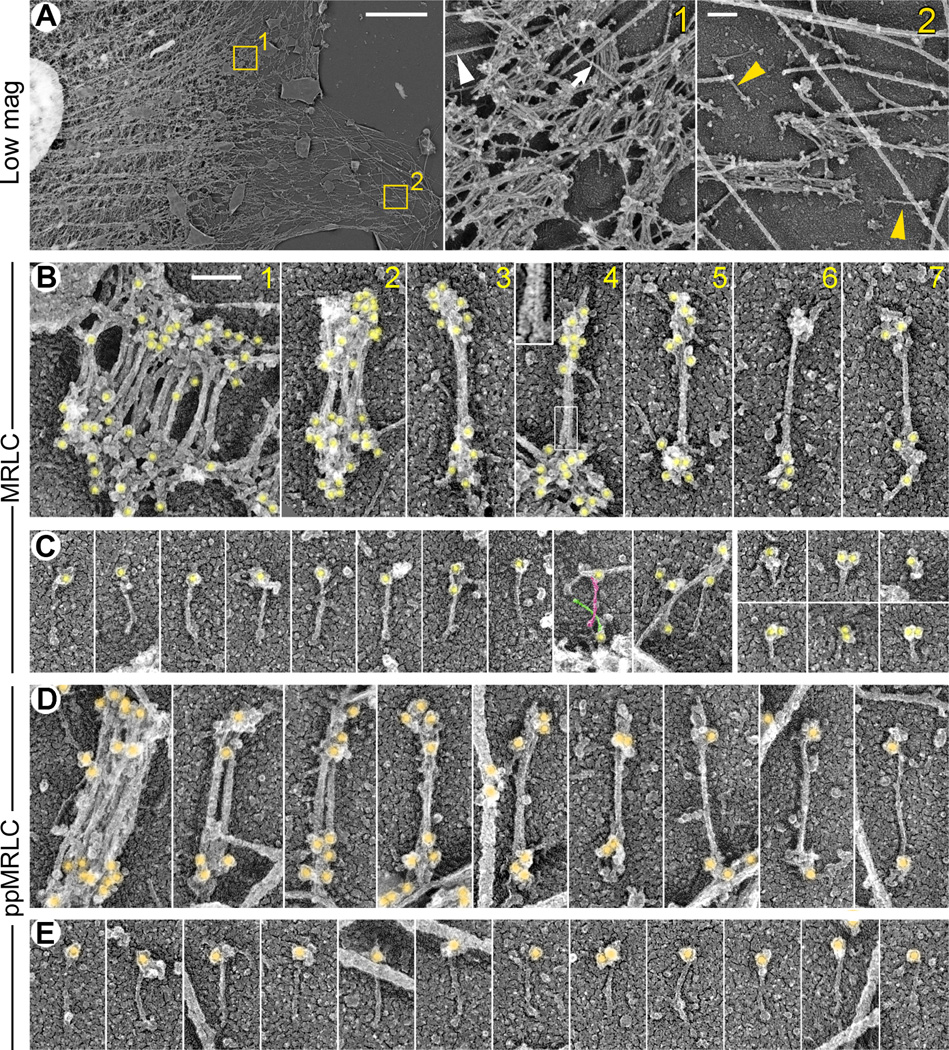
Activated NMII monomers have been proposed to represent functional species in cells and play a role in focal adhesion initiation [10], secretion [22], and Golgi dynamics [23]. To directly visualize activated NMII molecules in spreading REF52 fibroblasts, we applied platinum replica electron microscopy (PREM) in combination with immunogold staining of total MRLC, pMRLC, or ppMRLC (Fig. 1 and Fig. S1). To visualize morphology of immunogold-labeled NMII species, we depleted actin filaments from detergent-extracted cells by gelsolin treatment [24]. It has been previously shown by correlative PREM of cells injected with fluorescent myosin II that all discrete myosin structures visible by fluorescence microscopy remain intact after detergent extraction and gelsolin treatment [11, 12, 25]. The same protocol was used in this study. As expected, the majority of NMII species were in the form of bipolar filaments, which formed complex networks in the proximal lamella and cell body (Fig. 1A). Individual filaments could be observed primarily near the leading edge. They had a dumbbell shape of 330 ± 33 nm (mean ± SD, N=58) in total length and a bare zone width of 12 ± 2 nm (all measurements include a ∼2–4 nm thickness of platinum coating). The MRLC (Fig. 1B), pMRLC (Fig. S1), and ppMRLC (Fig. 1D) immunogold particles marked ends of bipolar filaments consistent with the MRLC position within the NMII motor domain. Unlabeled filament ends were also occasionally observed, most likely because of incomplete immunolabeling. In distal lamellae, bipolar filaments were often present individually or in small assemblies, such as stacks or networks. Some bipolar structures that superficially looked like a single filament, in fact, represented small stacks (Fig. 1B). Distinction could be made based on the number of bare zones. Whereas only one bare zone was present in single filaments, two or more were seen in the stacks (Fig. 1B,D).
Figure 1. NMII species revealed by immunogold PREM with MRLC antibodies in detergent-extracted and gelsolin-treated REF52 fibroblasts.
(A) Overall distribution of NMII in REF52 cells. Boxed regions in the left panel are zoomed at right. Stacks of several NMII filaments are seen in both proximal (#1) and distal (#2) lamellae; single NMII filaments (yellow arrowheads) are detected in distal lamella (#2). Microtubule (white arrowhead) and an intermediate filament (arrow) are marked in region 1.
(B, C) MRLC-labeled NMII species include bipolar filaments (B) and monomers (C). Gold particles (yellow) label heads of NMII filaments and monomers.
(B) NMII filaments form stacks of several (panels 1 and 2) or only two (panels 3 and 4) filaments, or exist as individual filaments (panels 5–7). Boxed region in panel 4 is zoomed in the inset to show bare zones of two filaments.
(C) MRLC-labeled NMII monomers have extended (left 10 panels) or folded (right 6 panels) configuration. Tails of two criss-crossing NMII monomers in 9th panel are highlighted in magenta and green.
(D, E) NMII species labeled with ppMRLC immunogold (orange) in untreated cells include bipolar filaments (D) and unfolded NMII monomers (E). Last 4 panels in D show single filaments. Bars, 5 µM (A, left panel), 200 nm (A, zoomed regions), and 100 nm (shown in B1 and applies to all panels in B–E).
See also Figure S1.
Importantly, we detected unfolded NMII monomers in distal cell lamellae (Fig. 1C,E and S1). Their dimensions (147 ± 13 nm in length and 6 ± 1 nm in width, mean ± SD, N=43), which were similar to dimensions of purified NMII monomers [26], and the labeling pattern exhibiting MRLC, pMRLC or ppMRLC immunogold at one end of the molecule strongly suggest that NMII molecules are unfolded and activated. In MRLC-labeled samples, in addition to extended molecules, we also observed compact gold-labeled structures (Fig. 1C, right), which could correspond to folded autoinhibited NMII monomers associated with unphosphorylated MRLC [26]. Although unfolded gold-labeled monomers were found with roughly similar frequencies in pMRLC- and ppMRLC-stained samples, it remains uncertain whether unfolded NMII monomers are equally efficiently produced by mono- and biphosphorylation, because staining efficiencies of the pMRLC and ppMRLC antibodies are likely different. The observed amount of NMII monomers, folded or unfolded, detected in PREM samples likely represented only a small fraction of their real abundance in the cytoplasm. Indeed, we previously showed biochemically that ppMRLC-positive NMII monomers could form up to 50% of the entire population of NMII associated with ppMRLC in these cells [10]. However, monomers are more likely to be solubilized by detergent extraction than NMII filaments or their assemblies, because NMII monomers have only two actin-binding sites compared with dozens in bipolar NMII filaments [9] and because NMII motors spend only a small fraction of time in an actin-bound state [2]. Together, these data provide direct evidence that unfolded activated NMII monomers represent a bona fide NMII population in cells and likely represent the first step of the NMII turnover cycle.
We have previously found that under blebbistatin treatment bipolar filaments undergo disassembly [10], probably, due to release of tension, because increased tension, conversely, promotes NMII polymerization [27]. Simultaneously, cells accumulated activated NMII monomers [10]. Consistent with these data, we found unfolded pMRLC- and ppMRLC-labeled NMII monomers in blebbistatin-treated cells, as well as few remaining bipolar filaments (Fig. S1). These filaments, however, often exhibited aberrant morphological features, probably, reflecting intermediate stages of bipolar filament disassembly (Fig. S1). First, we detected a statistically significant decrease (p=0.0025, unpaired Student’s t-test) in the average length of NMII filaments in blebbistatin-treated cells (310 ± 31 nm, mean ± SD, N=45) relative to that in untreated cells (330 ± 33 nm, N=58). Second, the immunogold-labeled head domains of bipolar filaments in blebbistatin-treated cells were often splayed apart or unusually broadly distributed along the filament length, whereas their bare zone was frequently bent, associated with additional material, and had signs of splaying. Finally, we found unusual structures that could be best described as ‘half-filaments’, because they were gold-labeled only at one pole and were about half as long as normal filaments, but their bare zone was too thick and the gold-labeled pole too complex to represent single NMII molecules (Fig. S1E). The unlabeled end of these structures was also often bulky, probably, due to interaction with additional proteins. We assume that these ‘half-filaments’ could reflect existence of a depolymerization mechanism that disentangles NMII monomers in the middle of a bipolar filament, for example, through binding of Mts1/S100A4 or phosphorylation of the heavy chain tail [28, 29]. Future studies should show whether this speculation is viable.
Activated monomers comprise both NMIIA and NMIIB isoforms
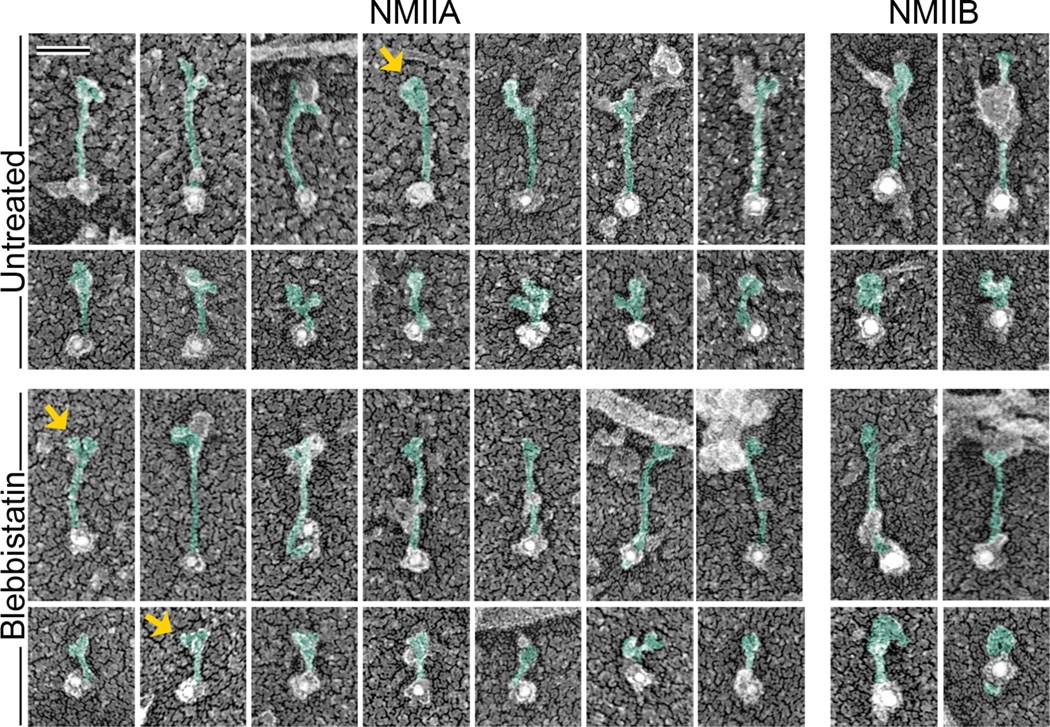
Since many cell types express a mixture of NMII heavy chain isoforms [1], we next asked which NMII isoform(s) constitute the population of unfolded monomers in REF52 cells. Among three mammalian NMII isoforms, NMIIA and NMIIB are broadly expressed, whereas NMIIC has restricted expression [1]. Both NMIIA and NMIIB are expressed in REF52 cells, but NMIIA was visually more abundant than NMIIB, which could reflect a higher expression level of NMIIA in these cells and/or greater efficiency of the NMIIA antibody. Immunogold staining of gelsolin-treated cytoskeletons revealed both NMIIA and NMIIB monomers, either compact or extended, in untreated or blebbistatin-treated cells (Fig. 2). The lengths of extended NMIIA-labeled (143 ± 15 nm, N=62) and NMIIB-labeled (150 ± 10 nm, N=15) structures were indicative of unfolded monomers. Since the isoform-specific antibodies used in these experiments recognize the extreme C-terminal region of NMII heavy chains, the position of gold particles at the tip of thin elongated structures was also consistent with their identification as unfolded NMII monomers. In favorable cases, two NMII motor domains (heads) could be discerned at the unlabeled end of the molecule. When heads were not clearly discernable, the unlabeled end was still enlarged or appeared to be associated with other structures (Fig. 2). Additional material was also occasionally observed along the NMII tail and probably corresponded to some of NMII interaction partners, whose list is constantly growing [6, 30–32]. Although the gold-labeled NMII monomers in cells less frequently exhibited clearly discernable heads, as compared with preparations of purified myosin II [26], the narrow range of length and thickness distributions of labeled structures, as well as reproducible labeling at the tips, support the idea that these structures are extended NMII monomers. Compact gold-labeled structures likely represented folded NMII monomers, as suggested by their dimensions similar to those observed in vitro [26]. These data show that unfolded monomers of both NMIIA and NMIIB are present in cells.
Figure 2. NMII monomers in REF52 fibroblasts comprise both NMIIA and NMIIB isoforms.
Immunogold PREM of NMIIA (12 nm gold) and NMIIB (18 nm gold) reveals unfolded (odd rows) and folded (even rows) NMII monomers in indicated experimental conditions. Gold particles associate with the tip of extended NMII tails consistent with the antibody specificity for C-termini of respective NMII isoforms. NMII heads can be occasionally recognized on the opposite end (arrows). Our best interpretation of the shape of NMII molecules is depicted by green pseudocolor, whereas associated material remains uncolored. Bar, 50 nm (applies to all panels).
NMIIA and NMIIB copolymerize into heterotypic bipolar filaments
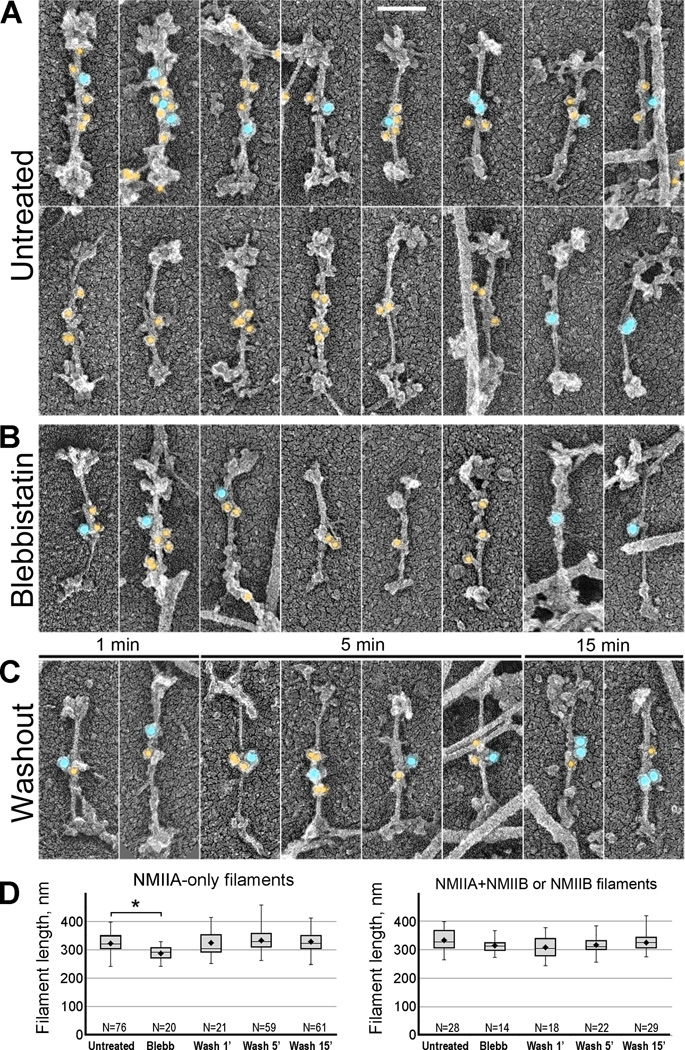
A long-standing question in the field is whether different NMII isoforms heteropolymerize in cells. To resolve this problem, we performed double immunogold labeling of NMIIA and NMIIB in spreading REF52 fibroblasts and human umbilical vein endothelial cells (HUVECs) using secondary antibodies conjugated with colloidal gold of different sizes (Fig. 3 and Fig. S2). We found a significant fraction (∼25–40%, N=348; see Table S1 for details) of individual bipolar filaments simultaneously labeled with NMIIA and NMIIB antibodies, indicating that NMIIA and NMIIB copolymerize in vivo. Individual filaments, which were carefully distinguished from small parallel stacks, displayed gold labeling in the bare zone. Thinner filaments that presumably contained fewer subunits were labeled by few gold particles residing in the middle of the bare zone, whereas thicker filaments that presumably contained more subunits were labeled throughout the bare zone, consistent with staggering of NMII molecules within filaments. Parallel stacks of bipolar filaments also could be labeled by one or both sizes of immunogold (not shown), indicating that they contain NMII heteropolymers and/or a mixture of NMIIA and NMIIB filaments. Bipolar filaments labeled with only one size immunogold, either NMIIA (∼60–75%) or NMIIB (∼2%), were also observed (Table S1). Because the labeling efficiency is likely less than complete, a fraction of singly labeled filaments might represent heteropolymers. These data demonstrate that NMIIA and NMIIB naturally copolymerize in vivo.
Figure 3. Immunogold PREM reveals hetero- and homopolymers of NMIIA and NMIIB isoforms in cell lamellae. NMIIA and NMIIB are labeled with 12 nm (yellow) and 18 nm (blue) immunogold, respectively.
(A–C) NMII filaments in untreated cells (A), cells treated with blebbistatin (B), and cells washed out of blebbistatin for indicated periods of time (C). Bar, 100 nm (applies to all panels in A–C).
(D) NMII filament lengths (including thickness of platinum coating) in indicated experimental conditions. Filaments labeled with NMIIA antibody only (left plot) are shorter in blebbistatin-treated cells, than in other conditions. Filaments containing NMIIB (right panel) have statistically indistinguishable lengths in all conditions. Top and bottom of a box indicate 75th and 25th quartiles, respectively; whiskers encompass all data; dot is the mean; and the middle line is the median. An asterisk indicates statistical significance (p<0.0001; unpaired Student’s t-test; N numbers are indicated on plots).
To further evaluate our finding that NMII filament length is decreased in the presence of blebbistatin, we tested whether such response depends on the isoform composition of bipolar filaments. For this analysis, we used cells treated with blebbistatin and cells recovering from blebbistatin treatment for different periods of time. Fewer NMII filaments were found in blebbistatin-treated cells and cells washed out of blebbistatin for just 1 min, than in untreated cells or cells washed out of blebbistatin for longer periods of time, consistent with our previous data [10]. We found that lengths of NMIIB-containing filaments (NMIIA+NMIIB or NMIIB only) were statistically indistinguishable across all conditions (Fig. 3D, right). They were also similar to the lengths of homotypic NMIIA filaments in all conditions except for blebbistatin-treated cells, where NMIIA filaments were slightly, but significantly shorter (Fig. 3D, left). Since NMIIA filaments represent the majority of all NMII filaments, their length changes could explain the decreased average length of the total population of NMII filaments in blebbistatin-treated samples labeled with MRLC antibodies. These results suggest that incorporation of NMIIB subunits could prevent shortening of NMIIA filaments in the presence of blebbistatin.
NMIIA and NMIIB jointly participate in initiation of contractile system assembly
The presence of a substantial fraction of NMIIB-containing bipolar filaments in close proximity to the cell leading edge was unexpected, because in polarized cells NMIIB is typically displaced toward the cell rear, whereas distal lamellae in various cell types are mostly populated by NMIIA [13, 16, 33]. Formation of this anterior-posterior NMIIA-NMIIB gradient was explained by sequential incorporation of NMIIA and NMIIB isoforms into contractile structures [17]. An alternative possibility suggests that both NMIIA and NMIIB can simultaneously begin polymerizing at the cell front and segregate into anterior and posterior locations, respectively, during subsequent turnover. To distinguish these possibilities, we took advantage of the blebbistatin washout strategy, which synchronizes the formation of the contractile system in cells [10]. In blebbistatin-treated cells, total NMII depolymerizes and becomes greatly enriched in membrane protrusions (mostly ruffles), but following blebbistatin washout it leaves protrusions and forms contractile structures in lamellae [10]. If NMIIA polymerizes first and NMIIB joins the NMIIA-formed structures later, NMIIA should be enriched in the lamella immediately after blebbistatin washout. However, if both NMII isoforms polymerize simultaneously, but become segregated at later stages, the gradient formation will be delayed.
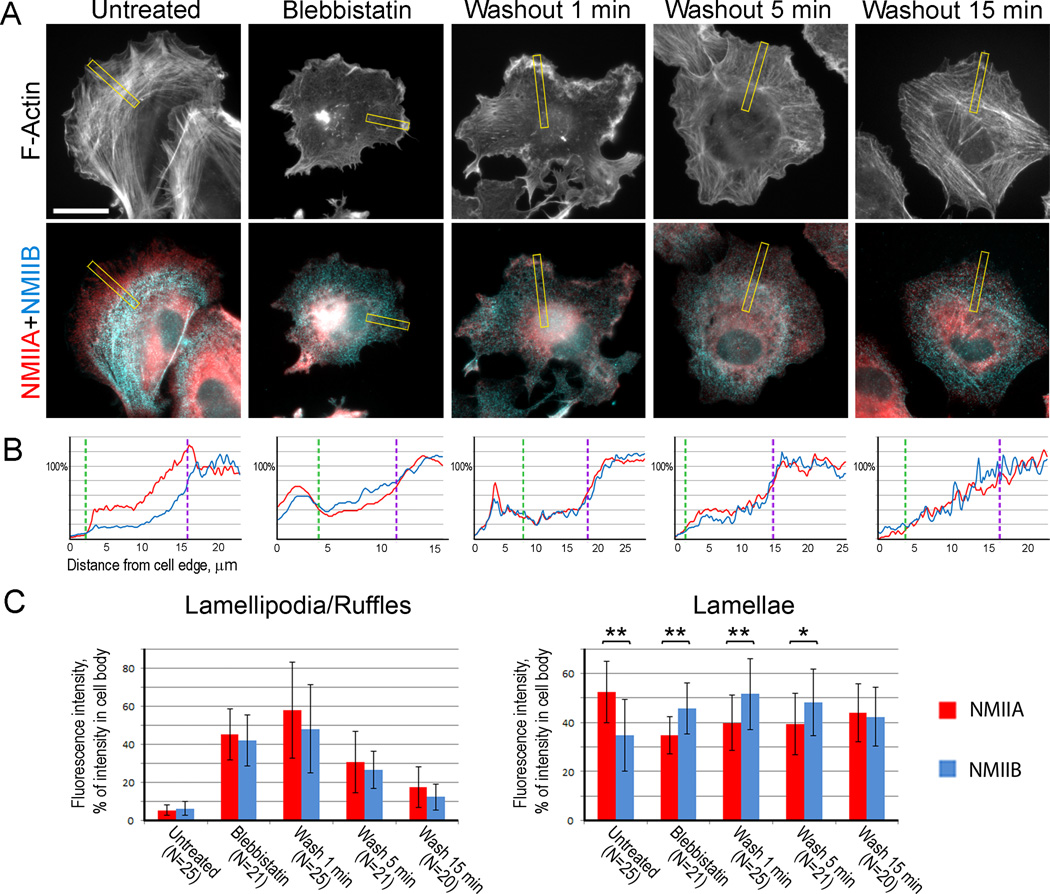
We fluorescently stained untreated, blebbistatin-treated, and blebbistatin-washout cells with NMIIA and NMIIB antibodies, as well as with phalloidin to detect actin filaments (Fig. 4). For this analysis, we used directly fixed cells (see Experimental Procedures) to visualize both soluble and cytoskeleton-associated NMII populations. To quantitatively evaluate the relative abundance of NMIIA and NMIIB in distal cell regions, we normalized the average fluorescence intensities of NMIIA and NMIIB in lamellipodia and lamellae to the average fluorescence intensity of the respective isoform in the cell body, where both NMIIA and NMIIB are abundant. In untreated cells, we found an expected distribution of NMIIA and NMIIB isoforms. Namely, both NMII had equally low levels in lamellipodia, but NMIIA was more abundant in lamellae, than NMIIB, consistent with the existence of anterior-posterior NMIIA-NMIIB gradient (53 ± 13% intensity for NMIIA vs 35 ± 15% for NMIIB, p<0.0001, paired Student’s t-test).
Figure 4. NMIIA and NMIIB have similar distribution in lamella during early stages of contractile system assembly.
(A) Wide-field Immunofluorescence microscopy of REF52 cells triple stained with phalloidin (top row) and NMIIA and NMIIB antibodies (merged in bottom row) in indicated experimental conditions. The yellow boxes are the regions used for the representative linescans shown in panel 4B. Bar, 20 µm.
(B) Representative linescans of NMIIA (red) and NMIIB (blue) fluorescence intensities (Y-axis) across lamellipodium, lamella, and cell body regions (X-axis) for cells shown in A. Dashed lines indicate lamellipodium-lamella (green) and lamella-cell body (purple) boundaries.
(C) Normalized average fluorescence intensities of NMIIA and NMIIB in lamellipodia (left) and lamellae (right) in different experimental conditions. Data are represented as mean ± SD. Asterisks indicate statistical significance (**, p<0.0001; *, p<0.01, paired Student’s t-test).
Consistent with the reported behavior of total NMII [10], both NMIIA and NMIIB became enriched in ruffles of blebbistatin-treated cells, but decreased in this region following blebbistatin washout (Fig. 4C, left). Importantly, we found that in lamellae of blebbistatin-treated cells or cells recovering from blebbistatin for 1 or 5 min, NMIIB was more abundant than NMIIA, and both NMII exhibited similar abundance after 15 min washout (Fig. 4C, right). The lack of NMIIA enrichment relative to NMIIB in the lamella for up to 15 min following blebbistatin washout indicated that the anterior-posterior NMIIA-NMIIB gradient was not yet established during this period of time, even though it existed in steady-state conditions. These data suggest that NMIIA and NMIIB exhibit similar behavior during early stages of the contractile system assembly, rather than assemble sequentially.
The model of simultaneous assembly and subsequent segregation of the NMIIA and NMIIB isoforms also predicts that NMIIA/B copolymers should be more abundant in lamellae of cells recovering from blebbistatin, than in lamellae of untreated cells at the steady state. To compare the degree of NMIIA/B colocalization in untreated, blebbistatin-treated, and blebbistatin-washout cells, we used STED microscopy (Fig. 5), which can resolve structures below the optical diffraction limit and is more suitable than PREM for quantification of large samples. In our STED imaging conditions, we achieved 70–80 nm resolution in the X-Y plane. To avoid interference from soluble NMII pools, we used cells that were detergent-extracted prior to fixation. To define subcellular domains, we also costained cells with phalloidin and imaged F-actin distribution in the same cells by confocal microscopy (Fig. S3).
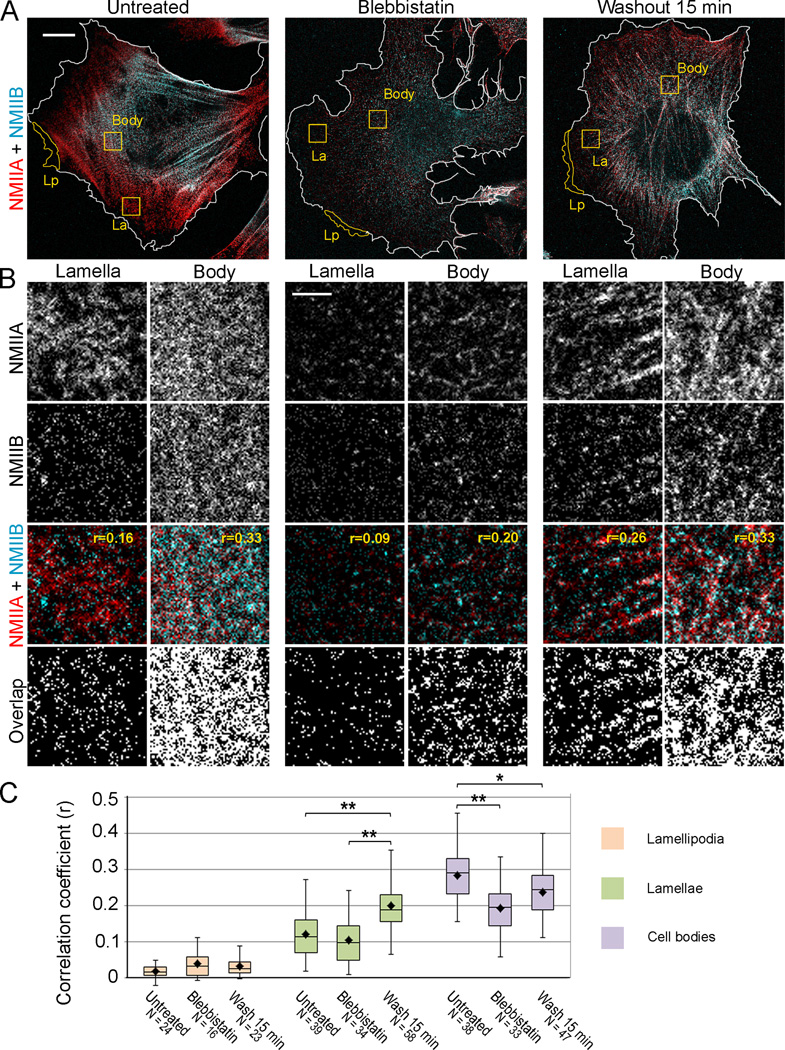
Figure 5. Colocalization of NMIIA and NMIIB in lamellae is greater at early stage of contractile system assembly than in steady-state conditions.
(A) STED microscopy of cells in indicated conditions stained with NMIIA and NMIIB antibodies. White outlines indicate cell boundaries based on phalloidin staining (see Fig. S3). Yellow outlines are examples of regions used for determination of correlation coefficients in lamellipodia (Lp), lamellae (La) and cell bodies (Body). Bar, 10 µm.
(B) Square regions from lamellae and cell bodies of each cell (yellow boxes in A) are zoomed as separate channels (NMIIA and NMIIB) and merged images (NMIIA+NMIIB). Correlation coefficient for NMIIA and NMIIB immunofluorescence (r) is indicated for each example on merged panels. “Overlap” panels show pixels exhibiting colocalization (white) of NMIIA and NMII signals that were thresholded to discard the lowest 5% intensity levels.
(C) Average correlation coefficients for NMIIA and NMIIB immunofluorescence in lamellipodia, lamellae, and cell bodies for the same experimental conditions, as in A. Top and bottom of a box indicate 75th and 25th quartiles, respectively; whiskers encompass all data; dot is the mean; and the middle line is the median. Asterisks indicate statistical significance (**, p<0.0001; *, p<0.01, unpaired Student’s t-test); the number of analyzed regions (N) from 11–14 cells for each condition is indicated on the plot. Bar, 2 µm.
See also Figure S3.
Consistent with wide-field fluorescence microscopy of non-extracted cells, STED microscopy of extracted cells in normal conditions revealed a prominent anterior-posterior gradient of NMIIA and NMIIB with the NMIIB distribution shifted toward the cell center relative to NMIIA. Accordingly, high magnification STED images showed a greater overlap of the NMIIA and NMIIB signals in the cell body, as compared with the distal lamella (Fig. 5B). In the presence of blebbistatin, the overall STED fluorescence signals for both NMII isoforms were very low (Fig. 5A), because of increased solubility of NMII in these conditions [10, 34], but an overlap of NMIIA and NMIIB fluorescence could be seen among remaining structures. After 15 min blebbistatin washout, overall immunofluorescence intensities of NMIIA and NMIIB increased compared with those of blebbistatin-treated cells, but their relative distribution did not exhibit a prominent gradient, as in untreated cells. At high magnification, frequent overlaps of NMIIA and NMIIB signals could be seen in lamellae and cell bodies.
To quantitatively evaluate proximity of NMIIA and NMIIB immunofluorescence signals within the STED resolution limit, we calculated correlation coefficients (r) for different cellular regions in untreated cells, cells treated with blebbistatin, and cells recovering from blebbistatin treatment for 15 minutes. Correlation coefficients vary between 1 (complete colocalization) and −1 (complementary distribution), where 0 indicates random overlap. We found virtually no colocalization between NMIIA and NMIIB in lamellipodia for all three experimental conditions (Fig. 5C), which is consistent with the idea that NMII species in lamellipodia are monomers. In contrast, partial colocalization between NMIIA and NMIIB was observed in the lamella and cell body (Fig. 5C). The overall magnitude of obtained correlation coefficients (in the order of 0.1–0.4) was not high, because the number of NMIIA-positive pixels greatly exceeded the number of NMIIB-positive pixels, which results in an overall decrease of the correlation coefficient. Yet, the obtained values were always significantly higher than control correlation coefficients for the same regions with one channel rotated 90 degrees (p<0.0001, paired Student’s t-test). Importantly, the average correlation coefficient between NMIIA and NMIIB localization in lamellae was significantly greater for blebbistatin-washout cells (r=0.20 ± 0.06), than for untreated cells (r=0.12 ± 0.06). These results suggest that when the contractile system assembles de novo from readily available pools of NMIIA and NMIIB monomers, both isoforms exhibit similar incorporation behavior into detergent-insoluble cytoskeleton.
Discussion
We provide high resolution characterization of endogenous NMII species in their natural context in motile nonmuscle cells and report several novel and unexpected features of NMII organization and dynamics in cells. We have found that activated monomers of both NMIIA and NMIIB isoforms represent an endogenous functional NMII species. This finding changes the current paradigm that NMII bipolar filaments are the only functional species of NMII. We also show that NMIIA and NMIIB naturally copolymerize in cells and propose that NMII copolymerization may affect the bipolar filament properties. Finally, we report that NMIIA and NMIIB simultaneously incorporate into nascent contractile structures, suggesting that they become segregated along the anterior-posterior axis at late stages of the contractile system assembly and turnover.
Roles of activated NMII monomers in cells
Existence of activated NMII monomers in cells suggests that unpolymerized NMII may play some roles in cell physiology, such as focal adhesion initiation, lamellipodial protrusion [10], and membrane organelle dynamics [22]. A compelling mechanism to generate activated NMII monomers for this purpose may rely on NMII heavy chain regulation, because phosphorylation of the NMIIA [28] or NMIIB [29, 35] heavy chain, or binding of Mts1/S100A4 to the NMIIA heavy chain [28], inhibit NMII polymerization without inhibiting NMII motor activity. Moreover, regulators of NMII polymerization at the heavy chain level are enriched in lamellipodia and affect protrusive activity [35–38], thus supporting an idea that NMII monomers are produced within lamellipodia and may provide traction for lamellipodial protrusion by inducing nascent adhesions. Activated NMII monomers can also account for unconventional functions of NMII in membrane organelle dynamics [3–7, 22, 30]. The role of NMII in these processes is often explained by contractile activity of bipolar filaments, which form structures analogous to cytokinetic contractile rings. However, the ∼300 nm long NMII filaments are too large to contract or constrict membrane organelles by such mechanism, because organelles are several-fold smaller than the bipolar filament length. In contrast, activated monomers appear to be appropriate functional species of NMII to perform these functions. Although NMII monomers are only two-fold shorter than bipolar filaments, they likely act by a totally different mechanism than bipolar filaments, for example, by binding actin filaments with the motor domain and using their tail to bind another cargo, similar to unconventional myosins. Consistent with this idea, monomeric NMIIA has been found to associate via its C-terminal tail with lytic granules in natural killer cells [22] and Golgi membranes isolated from the intestinal epithelium [23].
Heteropolymerization of NMIIA and NMIIB into bipolar filaments
Despite repeated efforts to answer the question of NMII heteropolymerization in vivo by biochemical or molecular biology approaches [19–21], the isoform composition of endogenous bipolar NMII filaments remained unknown. Our immunogold PREM data give an explicit answer to this question by showing individual double-labeled NMIIA/NMIIB bipolar filaments. While our manuscript was under review, coassembly of over-expressed tagged NMII isoforms visualized by subdiffraction fluorescence microscopy has been reported [39]. In this study, co-expression of one NMII isoform with a green tag on the head and another isoform with a red tag on the tail produced green-red-green triplets of fluorescent puncta, suggesting the formation of heterotypic NMII filaments. However, at the light microscopy resolution the triplets could also correspond to closely aligned filament stacks that are numerous in cells. In comparison, we visualized endogenous bipolar filaments naturally formed in cells, rather than those made of over-expressed truncated [19] or tagged [39] NMII constructs, and used the high resolution PREM approach that can reliably distinguish individual filaments and stacks of parallel filaments.
The significance of NMII heteropolymerization is not fully clear. It is interesting in this respect that the lengths of homotypic NMIIA or NMIIB filaments assembled in vitro from pure proteins [26] closely match the lengths of respective filaments in blebbistatin-treated cells. However, in conditions allowing NMII to exhibit actin-binding and motor activities, the length of homotypic NMIIA filaments, but not NMIIB-containing filaments, increases. This observation can potentially be explained by existence of tension applied to NMII filaments, because filaments in blebbistatin-treated cells, similar to filaments assembled in vitro, are not under tension, in contrast to filaments in untreated cells or cells recovering from blebbistatin treatment. Therefore, it seems that NMIIA filaments slightly extend under tension, whereas addition of NMIIB subunits could stabilize the length of resulting heterotypic filaments.
Formation of anterior-posterior NMIIA-NMIIB gradient
Using acute induction of NMII polymerization, we found that NMIIA and NMIIB simultaneously begin polymerizing at the cell front and segregate into anterior and posterior locations, respectively, at later stages. These findings are consistent with the recently reported decreased segregation of NMII isoforms in freshly plated cells, as compared with fully polarized cells [39], because spreading cells also exhibit de novo contractile system assembly. Both sets of data, however, contrast the previously proposed model of sequential assembly of NMII isoforms, where NMIIB joins relatively mature structures premade by NMIIA [17]. One possibility for this discrepancy is that early incorporation of NMIIB into nascent structures might be difficult to detect by fluorescence microscopy of live cells in the presence of soluble NMII pools [17]. It is also possible that the extraction procedure used in our experiments not only removes the soluble pool of NMII, but also a fraction of the cytoskeletal pool. In this case, NMIIA and NMIIB could be unevenly extracted from the samples [34], thus altering the proportion of the two isoforms compared with live cells. However, since we compare only relative amounts of NMIIA and NMIIB in different experimental conditions (control cells, blebbistatin-treated and blebbistatin-washout cells), the potentially lost fraction should be the same for NMIIA and NMIIB. Therefore, if a difference between samples exists after extraction, it should have existed in live cells.
One possibility of how the anterior-posterior NMIIA-NMIIB gradient can form at a slow rate following initial assembly of the contractile system is through different rates of bipolar filament recycling. Indeed, fluorescence recovery after photobleaching revealed a slower turnover rate and a smaller mobile fraction of NMIIB compared with NMIIA [34, 40]. It is not clear at present whether the fast NMIIA turnover reflects preferential disassembly of homotypic NMIIA filaments or preferential extraction of NMIIA subunits from heterotypic filaments. Regardless of the exact mechanism, slow recycling of NMIIB suggests that even if at starting conditions NMIIA and NMIIB are similarly distributed along the cell radius, polymeric NMIIB stays bound to the actin cytoskeleton longer than NMIIA and moves rearward with retrograde flow to eventually accumulate in the cell body. Simultaneously, this process is expected to cause depletion of the soluble pool of NMIIB monomers. In contrast, more dynamic NMIIA can quickly repopulate newly formed cellular regions and thus has more even distribution throughout the cell.
Since NMIIB, like NMIIA, begins its turnover cycle at the leading edge, NMIIB may have specific functions at the cell front, not only at the cell rear, as previously thought. For example, NMIIB may support lamellipodial protrusion. Indeed, NMIIB has been reported to localize to lamellipodia [16, 33, 41], whereas deficiency in NMIIB results in prominent impairment of lamellipodial protrusion [42–45]. Our data suggest that NMIIB may support lamellipodia in a monomeric form, thus providing a new framework to rethink and properly interpret existing data on this point.
In conclusion, we propose a model that both NMIIA and NMIIB begin their turnover cycle at the leading edge as activated monomers; then they equally efficiently assemble into homo- and heterotypic filaments, which jointly participate in the formation of nascent contractile networks and bundles, and only later NMIIA and NMIIB isoforms become sorted into anterior and posterior locations, probably, because of different turnover rates of NMIIA and NMIIB subunits. Together, our data provide new conceptual insights into organization and dynamics of the ubiquitous cellular machinery for force generation, which acts in multiple cellular contexts and critically relies on NMII activity.
Experimental Procedures
Reagents
Working concentration of blebbistatin (Toronto Research Chemical, Inc.) was prepared from 10 mM stock in DMSO. The following primary antibodies were used: mouse monoclonal antibodies to MRLC (Sigma, Cat. # M4401), NMIIA (Abcam, Cat.# ab55456), and NMIIB (Iowa Hybridoma Bank, Cat. # CMII 23, for Fig. S2), and rabbit polyclonal antibodies to pMRLC (Ser19 phospho-myosin light chain 2, Cell Signaling, Cat. # 3671), ppMRLC (Thr18/Ser19 phospho-myosin light chain 2, Cell Signaling, Cat. # 3674), NMIIA (BTI, Cat. # BT-567, for Fig. S2) and NMIIB (Cell Signaling, Cat. # 3404). The following secondary anti-mouse and anti-rabbit antibodies were used: 12 nm and 18 nm colloidal gold-conjugated antibodies (Jackson ImmunoResearch) for immunoelectron microscopy; AlexaFluor 488 and AlexaFluor 594 (Molecular Probes) for wide-field fluorescence microscopy, and Atto-425 (Rockland Immune Research) and Dylight-488 (Thermo Scientific) for STED microscopy. AlexaFluor 680 phalloidin was from Molecular Probes.
Cell culture and drug treatment
REF52 rat embryo fibroblasts [12] were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) and penicillin/streptomycin at 37°C and 5% CO2. For experiments, cells were plated on glass coverslips and allowed to spread for 3 h. HUVECs (Lonza) were maintained in Endothelial Cell Basal Medium (Lonza) and cultured for 6 passages maximum. For experiments, HUVECs were plated on collagen-coated coverslips applied at approximate concentration of 5 µg/cm2 (BD Biosciences). For blebbistatin treatment experiments, blebbistatin was added to cell cultures 3 h after plating at final concentration of 100 µM and cells were incubated for additional 1 h. For washout experiments, after 1h incubation with blebbistatin, cells were transferred to fresh blebbistatin-free medium, incubated for various periods of time, and fixed.
Platinum replica electron microscopy (PREM)
Detailed procedures for PREM, gelsolin treatment and immunogold staining were described previously [24, 46–48]. Briefly, cells were quickly rinsed with PBS, extracted for 2–3 min at room temperature with 1% Triton X-100 in PEM buffer (100 mM PIPES-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 2 µM paclitaxel (Sigma) and PhosStop (Roche) at concentration recommended by the manufacturer (1 tablet per 10 ml). Paclitaxel and PhosStop were also added at all subsequent steps before fixation. After 3 quick rinses with PEM buffer, detergent-extracted cells were incubated with 0.4 µg/ml full length gelsolin (gift of Dr. A. Weber, University of Pennsylvania) in G-buffer (pH 6.3, 50 mM MES-KOH, 0.1 mM CaCl2, 2 mM MgCl2, 0.5 mM DTT) for 15 min at room temperature. After an additional rinse with PEM buffer, samples were incubated with primary antibodies in PEM buffer for 30 min. For pMRLC or ppMRLC staining, primary antibodies were added to gelsolin solution. Samples were rinsed twice in PEM, incubated in the third change of PEM for 15 min and fixed with 0.2% glutaraldehyde in 0.1 M cacodylate buffer for 20 min. Samples were quenched with NaBH4 in PBS, incubated overnight with secondary gold-conjugated antibodies and fixed with 2% glutaraldehyde. For PREM, samples were then fixed with tannic acid and uranyl acetate; critical point dried; coated with platinum and carbon; transferred onto electron microscopic grids and analyzed using JEM 1011 transmission EM (JEOL USA, Peabody, MA) operated at 100 kV. Images were captured by ORIUS 832.10W CCD camera (Gatan, Warrendale, PA) and presented in inverted contrast. Gold particles in PREM samples were identified at 200K magnification after contrast enhancement to distinguish them from other bright objects. Gold particles were pseudocolored using a brush tool in Adobe Photoshop with 50% opacity. Color labeling of other structures of interest was performed using Hue/Saturation tool in Adobe Photoshop to avoid obscuring the structural details.
Fluorescence microscopy
For immunofluorescence staining, cells were quickly rinsed with PBS and either directly fixed with 4% formaldehyde in PBS for 15 min, or pre-extracted for 2–3 min with 1% Triton X-100 in PEM buffer containing 2% polyethelene glycol (35,000 KDa) and 2 µM unlabeled phalloidin. Directly fixed samples were permeabilized with 0.1% Triton X-100 in PBS and stained with primary and secondary antibodies. AlexaFluor 488 and AlexaFluor 594-conjugated secondary antibodies were used for wide-field microscopy. For STED microscopy, detergent-extracted cells were fixed and incubated with the mouse NMIIA and rabbit NMIIB primary antibodies followed by the addition of secondary Atto-425 goat anti-mouse and Dylight-488 goat anti-rabbit antibodies. For costaining of F-actin, fluorescently labeled phalloidin was added to the secondary antibody solution. Coverslips were extensively washed with PBS and mounted with Prolong Gold antifade reagent (Invitrogen).
Wide-field fluorescence microscopy was performed using Eclipse TE2000-U inverted microscope (Nikon) equipped with Plan Apo 100 × 1.3 NA objective and Cascade 512B CCD camera (Photometrics) driven by Metamorph imaging software (Molecular Devices). STED images were collected using a Leica SP5-STED-CW microscope (Leica Microsystem) using the sequential acquisition mode and the following excitation lasers: 458 nm and 514 nm for STED and 633 nm for confocal imaging. The depletion laser (592 nm) was used at 100% of the power to obtain maximum STED resolution, which was between 70–80 nm. Shown images were not deconvolved. Slides were analyzed in a blinded fashion.
Image analyses
Dimensions in PREM images were determined using Adobe Photoshop software. All presented measurements include the thickness of platinum coating (approximately 2 nm). Since the thickness of coating can vary depending on the Z-position of the structure, the actual structure dimensions are estimated to be 2–4 nm smaller. Immunogold-labeled structures were measured together with the gold particles, when they were localized at the tip of the structure.
Determination of relative enrichment of NMIIA and NMIIB in different cellular regions was performed using wide-field fluorescence images of cells directly fixed with PFA and triple stained with NMIIA and NMIIB antibodies and phalloidin. The linescan tool in Metamorph was used to obtain fluorescence intensity profiles from background-subtracted images along 3 µm wide lines drawn perpendicular the cell edge and crossing the lamellipodium, lamella, and cell body (perinuclear region). The lamellipodium-lamella and lamella-body boundaries were manually assigned based on the phalloidin channel (lamellipodium-lamella) or the levels of NMII fluorescence approaching a plateau (lamella-cell body). The average intensities of NMIIA and NMIIB in the lamellipodium, lamella, and cell body were calculated using corresponding segments of the intensity profile and expressed as a percentage of the average intensity of the respective NMII isoform signal in the cell body. Data were averaged from 20–25 profiles from 11–17 cells for each condition.
Determination of colocalization of NMIIA and NMIIB in different cellular regions was performed using single-plane STED images. Correlation coefficients for NMIIA and NMIIB immunofluorescence signals were calculated for background-subtracted regions of interest using Correlation Plot tool in Metamorph. Square 6×6 µm regions of interest were selected in lamellae and the cell body and irregularly shaped regions of approximately the same area were outlined in lamellipodia based on phalloidin staining. Regions of interest were selected from 11–14 cells for each condition. Statistical significance of correlation coefficients for square regions in lamellae and cell bodies was determined relative to correlation coefficients for the same regions with one channel rotated 90 degrees. The correlation coefficient for rotated images was invariably close to zero (random overlap) for all cell regions and treatment conditions. Statistical analyses were performed using Excel (Microsoft) or Instat (GraphPad Software) software packages.
Supplementary Material
Highlights.
Activated unpolymerized NMIIA and NMIIB molecules are directly visualized in cells
NMIIA and NMIIB copolymerize endogenously
NMIIA and NMIIB simultaneously join nascent contractile structures
Posterior shift of NMIIB relative to NMIIA develops during steady-state dynamics
Acknowledgments
We thank members of the Svitkina lab for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health grants GM095977 (T.S.) and the Pew Charitable Trusts (C.G.).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes three figures and one table.
Author Contributions
M.S. and T.S. conceived the project, designed research, and wrote the paper. M.S. performed majority of experiments and analyzed the data. W.S. and C.G. contributed to the design and execution of STED experiments.
References
- 1.Wang A, Ma X, Conti MA, Adelstein RS. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem Soc Trans. 2011;39:1131–1135. doi: 10.1042/BST0391131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heissler SM, Manstein DJ. Nonmuscle myosin-2: mix and match. Cell Mol Life Sci. 2013;70:1–21. doi: 10.1007/s00018-012-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nightingale TD, Cutler DF, Cramer LP. Actin coats and rings promote regulated exocytosis. Trends Cell Biol. 2012;22:329–337. doi: 10.1016/j.tcb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasekar I, Goeckeler ZM, Turney SG, Wang P, Wysolmerski RB, Adelstein RS, Bridgman PC. Nonmuscle myosin II is a critical regulator of clathrin-mediated endocytosis. Traffic. 2014;15:418–432. doi: 10.1111/tra.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duran JM, Valderrama F, Castel S, Magdalena J, Tomas M, Hosoya H, Renau-Piqueras J, Malhotra V, Egea G. Myosin motors and not actin comets are mediators of the actin-based Golgi-to-endoplasmic reticulum protein transport. Mol Biol Cell. 2003;14:445–459. doi: 10.1091/mbc.E02-04-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miserey-Lenkei S, Chalancon G, Bardin S, Formstecher E, Goud B, Echard A. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat Cell Biol. 2010;12:645–654. doi: 10.1038/ncb2067. [DOI] [PubMed] [Google Scholar]
- 7.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umemoto S, Bengur AR, Sellers JR. Effect of multiple phosphorylations of smooth muscle and cytoplasmic myosins on movement in an in vitro motility assay. J Biol Chem. 1989;264:1431–1436. [PubMed] [Google Scholar]
- 9.Niederman R, Pollard TD. Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J Cell Biol. 1975;67:72–92. doi: 10.1083/jcb.67.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shutova M, Yang C, Vasiliev JM, Svitkina T. Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS One. 2012;7:e40814. doi: 10.1371/journal.pone.0040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolega J, Taylor DL. Gradients in the concentration and assembly of myosin II in living fibroblasts during locomotion and fiber transport. Mol Biol Cell. 1993;4:819–836. doi: 10.1091/mbc.4.8.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breckenridge MT, Dulyaninova NG, Egelhoff TT. Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol Biol Cell. 2009;20:338–347. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolega J. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol Biol Cell. 2003;14:4745–4757. doi: 10.1091/mbc.E03-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci. 1994;107(Pt 11):3077–3090. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- 17.Vicente-Manzanares M, Newell-Litwa K, Bachir AI, Whitmore LA, Horwitz AR. Myosin IIA/IIB restrict adhesive and protrusive signaling to generate front-back polarity in migrating cells. J Cell Biol. 2011;193:381–396. doi: 10.1083/jcb.201012159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard TD. Electron microscopy of synthetic myosin filaments. Evidence for cross-bridge. Flexibility and copolymer formation. J Cell Biol. 1975;67:93–104. doi: 10.1083/jcb.67.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsuhashi M, Sakata H, Kinjo M, Yazawa M, Takahashi M. Dynamic assembly properties of nonmuscle myosin II isoforms revealed by combination of fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy. J Biochem. 2011;149:253–263. doi: 10.1093/jb/mvq134. [DOI] [PubMed] [Google Scholar]
- 20.Beach JR, Egelhoff TT. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J Biol Chem. 2009;284:27377–27383. doi: 10.1074/jbc.M109.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marini M, Bruschi M, Pecci A, Romagnoli R, Musante L, Candiano G, Ghiggeri GM, Balduini C, Seri M, Ravazzolo R. Non-muscle myosin heavy chain IIA and IIB interact and co-localize in living cells: relevance for MYH9-related disease. Int J Mol Med. 2006;17:729–736. [PubMed] [Google Scholar]
- 22.Sanborn KB, Mace EM, Rak GD, Difeo A, Martignetti JA, Pecci A, Bussel JB, Favier R, Orange JS. Phosphorylation of the myosin IIA tailpiece regulates single myosin IIA molecule association with lytic granules to promote NK-cell cytotoxicity. Blood. 2011;118:5862–5871. doi: 10.1182/blood-2011-03-344846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fath KR. Characterization of myosin-II binding to Golgi stacks in vitro. Cell Motil Cytoskeleton. 2005;60:222–235. doi: 10.1002/cm.20060. [DOI] [PubMed] [Google Scholar]
- 24.Svitkina TM, Borisy GG. Correlative light and electron microscopy of the cytoskeleton of cultured cells. Methods Enzymol. 1998;298:570–592. doi: 10.1016/s0076-6879(98)98045-4. [DOI] [PubMed] [Google Scholar]
- 25.Verkhovsky AB, Borisy GG. Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J Cell Biol. 1993;123:637–652. doi: 10.1083/jcb.123.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo T, Mohan K, Iglesias PA, Robinson DN. Molecular mechanisms of cellular mechanosensing. Nat Mater. 2013;12:1064–1071. doi: 10.1038/nmat3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–6876. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 29.Even-Faitelson L, Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol Biol Cell. 2006;17:2869–2881. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen CS, Watanabe S, Rasmussen HB, Schmitt N, Olesen SP, Frost NA, Blanpied TA, Misonou H. Specific sorting and post-Golgi trafficking of dendritic potassium channels in living neurons. J Biol Chem. 2014;289:10566–10581. doi: 10.1074/jbc.M113.534495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, Wang A, Conti MA, Adelstein RS. Nonmuscle myosin II is required for internalization of the epidermal growth factor receptor and modulation of downstream signaling. J Biol Chem. 2012;287:27345–27358. doi: 10.1074/jbc.M111.304824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosado LA, Horn TA, McGrath SC, Cotter RJ, Yang JT. Association between {alpha}4 integrin cytoplasmic tail and non-muscle myosin IIA regulates cell migration. J Cell Sci. 2011;124:483–492. doi: 10.1242/jcs.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley CA, Sellers JR, Gard DL, Bui D, Adelstein RS, Baines IC. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandquist JC, Means AR. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol Biol Cell. 2008;19:5156–5167. doi: 10.1091/mbc.E08-05-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg M, Ravid S. Protein kinase Cgamma regulates myosin IIB phosphorylation, cellular localization, and filament assembly. Mol Biol Cell. 2006;17:1364–1374. doi: 10.1091/mbc.E05-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahan I, Yearim A, Touboul Y, Ravid S. The tumor suppressor Lgl1 regulates NMII-A cellular distribution and focal adhesion morphology to optimize cell migration. Mol Biol Cell. 2012;23:591–601. doi: 10.1091/mbc.E11-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim EJ, Helfman DM. Characterization of the metastasis-associated protein, S100A4. Roles of calcium binding and dimerization in cellular localization and interaction with myosin. J Biol Chem. 2003;278:30063–30073. doi: 10.1074/jbc.M304909200. [DOI] [PubMed] [Google Scholar]
- 38.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 39.Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA., 3rd Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betapudi V. Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS One. 2010;5:e8560. doi: 10.1371/journal.pone.0008560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66:4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- 43.Diefenbach TJ, Latham VM, Yimlamai D, Liu CA, Herman IM, Jay DG. Myosin 1c and myosin IIB serve opposing roles in lamellipodial dynamics of the neuronal growth cone. J Cell Biol. 2002;158:1207–1217. doi: 10.1083/jcb.200202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281:35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 46.Svitkina TM, Borisy GG. Correlative light and electron microscopy studies of cytoskeletal dynamics. In: Celis J, editor. Cell Biology: A Laboratory Handbook. 3rd Edition. Vol. 3. Elsevier; 2006. pp. 277–285. [Google Scholar]
- 47.Svitkina T. Electron microscopic analysis of the leading edge in migrating cells. Methods Cell Biol. 2007;79:295–319. doi: 10.1016/S0091-679X(06)79012-4. [DOI] [PubMed] [Google Scholar]
- 48.Svitkina T. Gavin RH, editor. Imaging cytoskeleton components by electron microscopy. Methods Mol Biol - Cytoskeleton Methods and Protocols. 2009;586:187–206. doi: 10.1007/978-1-60761-376-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.