Abstract
The nuclear envelope in plant cells has long been known to be a microtubule organizing center (MTOC), but its influence on microtubule organization in the cell cortex has been unclear. Here we show that nuclear MTOC activity favors the formation of longitudinal cortical microtubule (CMT) arrays. We used green fluorescent protein (GFP)-tagged gamma tubulin-complex protein 2 (GCP2) to identify nuclear MTOC activity and GFP-tagged End-Binding Protein 1b (EB1b) to track microtubule growth directions. We found that microtubules initiate from nuclei and enter the cortex in two directions along the long axis of the cell, creating bipolar longitudinal CMT arrays. Such arrays were observed in all cell types showing nuclear MTOC activity, including root hairs, recently divided cells in root tips, and the leaf epidermis. In order to confirm the causal nature of nuclei in bipolar array formation, we displaced nuclei by centrifugation, which generated a corresponding shift in the bipolarity split point. We also found that bipolar CMT arrays were associated with bidirectional trafficking of vesicular components to cell ends. Together, these findings reveal a conserved function of plant nuclear MTOCs and centrosomes/spindle pole bodies in animals and fungi, wherein all structures serve to establish polarities in microtubule growth.
Keywords: Arabidopsis, Cytoskeleton, Microtubule, Polarity, Root, Root hair
Introduction
Microtubules (MTs) are tubulin polymers that undergo dynamic switches between states of growth (polymerization) and shortening (depolymerization). MTs are inherently polarized, possessing a highly dynamic plus end that undergoes rapid growth and shrinkage. In order to harness and translate MT polarity to a cell-wide scale, MT organizing centers (MTOCs) generate and anchor large groups of MTs, often forming highly polarized arrays with specialized functions. MTOCs are found in all eukaryotic cells, including centrosomes in animal cells, spindle pole bodies in fungi, and basal bodies in flagellated cells. Although MTOC structure varies across the kingdoms, the basic functions are conserved. These include mitotic spindle assembly/function, cell polarity and shape generation, and cell migration and motility.
In plants, the nuclear envelope can act as a MTOC. It contains MT nucleation components at its surface (Clayton et al. 1985, Wick 1985, Liu et al. 1993, Erhardt et al. 2002, Kumagai et al. 2003, Brown and Lemmon 2007, Seltzer et al. 2007) and has been shown to nucleate MTs at its surface in studies with isolated nuclei (Mizuno 1993, Stoppin et al. 1994), permeabilized cells (Wasteneys et al. 1989, Vantard et al. 1990) and intact cells (Falconer et al. 1988). Nuclear MT nucleating activity is most prominent in the actively cycling cells in the division zones of the shoot and root. Within these cells, nuclear MTOC activity is transient, being found in: (i) cells entering mitosis, during which time it contributes to formation of the pre-prophase band and the accompanying pre-spindle; and (ii) post-cytokinetic cells, where it persists until the onset of cell elongation (Nagata et al. 1994, Brown and Lemmon 2007, Ambrose and Wasteneys 2011).
In post-cytokinetic cells, nuclear MTOC activity generates endoplasmic MTs (EMTs) that extend through the cytoplasm in all directions, forming a radial MT array. These radial EMTs contact the cell cortex, to which they attach and incorporate into the cortical MT (CMT) array (Nagata et al. 1994, Kumagai et al. 2003). To achieve the highly directional (anisotropic) expansion seen in plant cells, CMTs form parallel arrays that interact with cellulose synthase complexes to generate tension-bearing cellulose microfibrils aligned in a similar direction to the CMTs. In post-cytokinetic cells, nuclear MTOC-derived mixed/longitudinally oriented CMT arrays presumably disfavor cell elongation by countering their inherent tendency toward transverse arrangement. This is supported by observations of the botero-1 mutant, which exhibits swollen root tips, and retains EMTs and disorganized CMTs for a longer time than in wild-type cells due to lack of MT-severing activity (Bichet et al. 2001). Beyond this, our understanding of how nuclear MTOC activity controls CMT organization is limited by the technical difficulty in studying radial EMT arrays, which are highly dynamic in all three spatial dimensions and appear for a short time only in small, cytoplasmically dense cells.
In the current study, we show how the activity and positioning of the nuclear MTOC influence the organization and polarity of CMTs in tip-growing root hairs and in recently divided cells. Specifically, nucleus-derived MTs enter the cell cortex and splay in two directions along the long axis of the cell, creating a bipolar CMT array with the nucleus as the split point. As the nucleus changes its position over time, the accompanying bipolarity split point follows along, thereby guiding the organization of the CMT array. This nuclear MTOC mechanism may provide an explanation for previously described bipolar arrays in plant cells (Sambade et al. 2012, Pietra et al. 2013, Vineyard et al. 2013). Additionally, we found that CMT bipolarity corresponds with a bi-directional transit and accumulation of vesicles to the ends of cells, suggesting a functional role for bipolarized CMT arrays in cell polarization. We observed other patterns of polarity in cells with more complex shapes, indicating that an interplay exists between MTOC patterns and cellular geometry.
Results
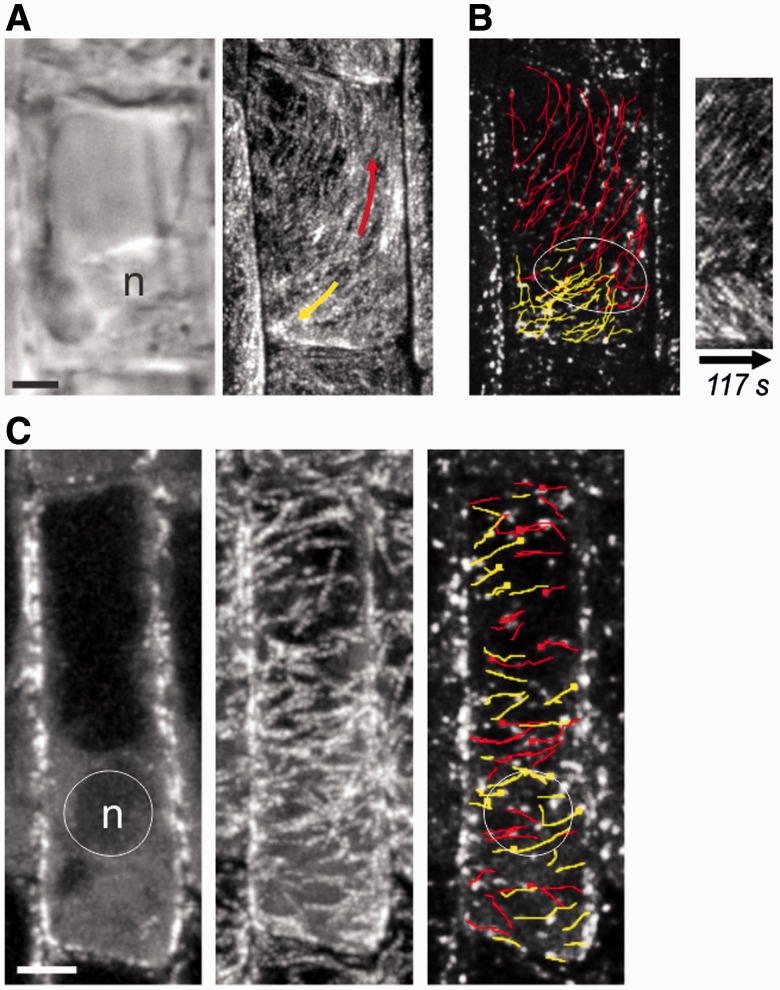
Cortical MTs grow bi-directionally away from the nucleus in root hairs
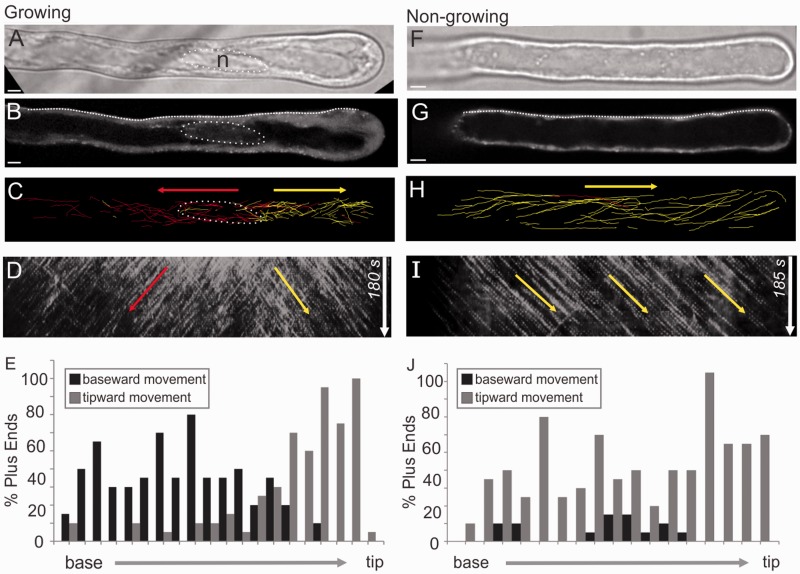
Using the MT plus-end marker EB1b–green fluorescent protein (GFP) as a reporter for MT growth direction, we were able to observe CMTs and EMTs in actively growing Arabidopsis thaliana root hairs. CMTs run lengthwise along the root hair. In assessing the directions of plus-end growth, we found a clear two-way growth polarity, wherein those within the distal portion grew toward the tip, while those in the proximal region grew toward the hair base (Fig. 1A–E; Supplementary Movie S1). This MT growth bipolarity encompassed the entire circumference of the cortex, thus being observable at any depth within a hair. The nucleus consistently resided at the split point of these two opposing growth polarities. This nucleocentric MT growth bipolarity is demonstrated clearly from kymograph analysis as two sets of diagonal lines mirrored about a central region, which corresponds to the position of the nucleus (Fig. 1D). Quantification of the direction of EB1b–GFP dot movement as a function of the length of the root hair shows a split distribution corresponding to the two opposing polarities (Fig. 1E). As the nucleus changed position along the length of the hair, the two-way split point moved along with it. Thus, in older root hairs, in which the nucleus has retreated to the base, unidirectional tipward MT growth polarity was observed throughout the entire hair (Fig. 1F–H; Supplementary Movie S2; Supplementary Fig. S1). Kymographs along these cells show a single set of sloped lines (Fig. 1I), and quantification shows more uniform distribution of tipward growth polarity along the length of the hair (Fig. 1J). Similarly, in newly emerging root hairs, the nuclei still demarcated a polarity split point, but this resided at the junction of the base of the root hair with the rest of the cell (Supplementary Fig. S1) Thus, these cells exhibited tripolar MT growth with respect to the nucleus, such that CMTs within the main cell region were longitudinal, splitting in two directions at the nucleus, and with a third group going out into the root hair. Using the MT marker GFP–MBD (microtubule-binding domain), we observed the same nucleocentric MT polarities as with EB1b–GFP (Supplementary Fig. S2).
Fig. 1.
Microtubule growth polarity corresponds to nuclear position in root hairs. Arabidopsis root hairs expressing EB1b–GFP in growing (A–E) and non-growing (F–J) hairs. (A and F) Differential interference contrast (DIC) images showing the nuclear position (circle). The nucleus is to the left of the imaged area in F. (B and G) Midplane confocal slices showing the nuclear position (circle). The nucleus is to the left of the imaged area in G. (C and H) Paths of manually tracked EB1b–GFP dots colored red for baseward or yellow for tipward movement. The nuclear position is shown with a dotted outline. (D and I) Kymographs corresponding to the midplane line drawn on a root hair. Arrows indicate dominant growth directions. (E and J) Histograms showing percentages of baseward vs. tipward MT growth polarities along the length of the root hairs. Scale bars, 5 µm.
Endoplasmic MTs spawn on the nuclear surface and enter the cortex bi-directionally
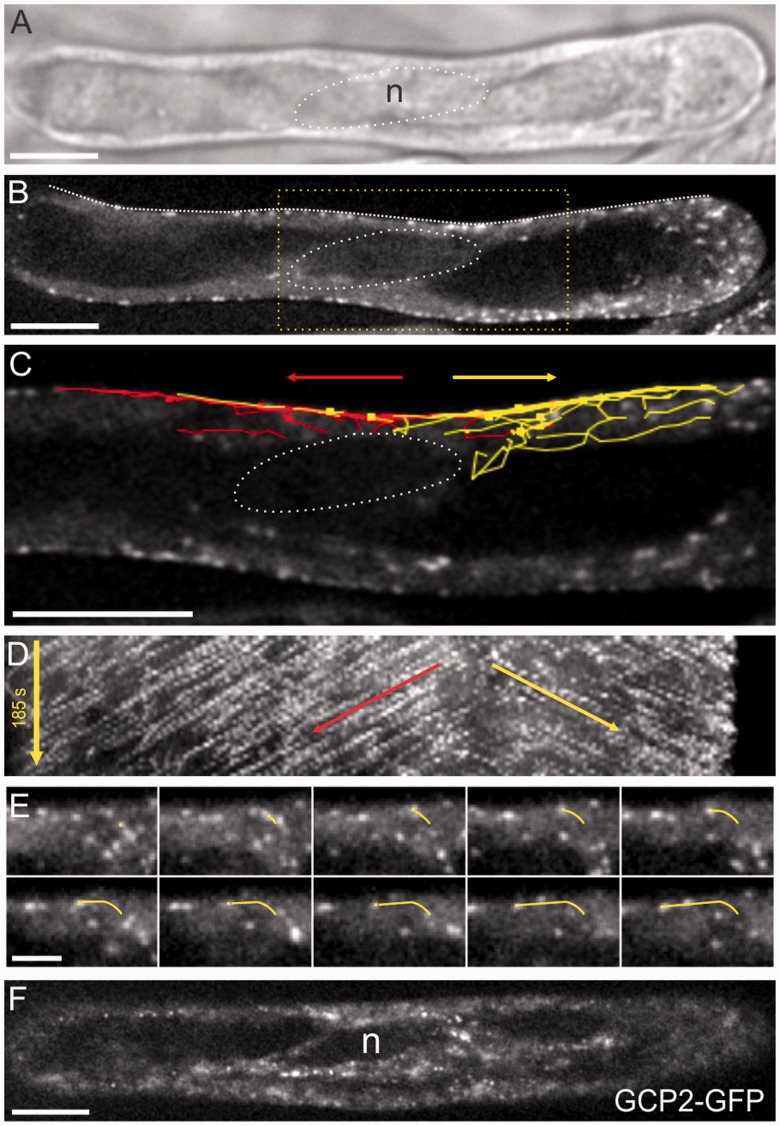
In order to confirm that the nuclear surface is the source of the bipolar cortical arrays in root hairs, we assessed the growth directionality of EMT bundles by means of EB1–GFP. We found that the bulk of the EB1–GFP within cytoplasmic strands moved away from the nucleus toward the cortex. By imaging the median plane of the root hair, we found that EB1b–GFP dots moved from the nuclear surface toward the cortex within cytoplasmic strands and, upon encountering the cortex, changed course to continue growing parallel to the long axis (Fig. 2A–E). This is illustrated using paths drawn from manually tracked EB1b–GFP dots (Fig. 2C), by kymographic analysis (Fig. 2D) and individual frames following the path of a single dot (Fig. 2E).
Fig. 2.
Microtubules initiate from the nucleus in root hairs and enter the cortex in two directions (A) DIC image showing the nuclear position (circle). (B) Single EB1b–GFP midplane image showing the nuclear position (dotted circle). (C) Higher magnification of the boxed region in B, showing paths of manually tracked EB1b–GFP dots overlaid on a single time point entering the cortex in two directions above the nucleus. Red is baseward movement and yellow is tipward movement. Arrows indicate dominant growth directions. (D) Kymograph corresponding to the line drawn on the root hair in B. Arrows indicate dominant growth directions. (E) Montage from time series tracking a single EB1b–GFP as it enters the cortex and then grows along the cortex. (The yellow line is shown for reference of the track.) Intervals are 5 s between frames; total time is 50 s. (F) GCP2–3×GFP is localized to the nuclear surface and surrounding the cytoplasmic strand. Shown is an image of the confocal midplane of a root hair. Scale bars, 10 µm for all panels except E, which is 5 µm.
To test whether MT nucleation factors were situated at the nuclear surface, we observed the distribution of a translational reporter of gamma tubulin-complex protein 2 (GCP2–3×GFP) (Nakamura et al. 2010). As shown in Fig. 2F, GCP2–3×GFP distributed to small punctae at the nuclear surface and endoplasmic strands. GCP2–3×GFP was also detected in the cortex of root hairs, consistent with MT initiation also occurring at the cortex. Taken together, the location of GCP2 at the nuclear surface and the movement of EB1b–GFP dots away from it demonstrate nuclear MTOC activity.
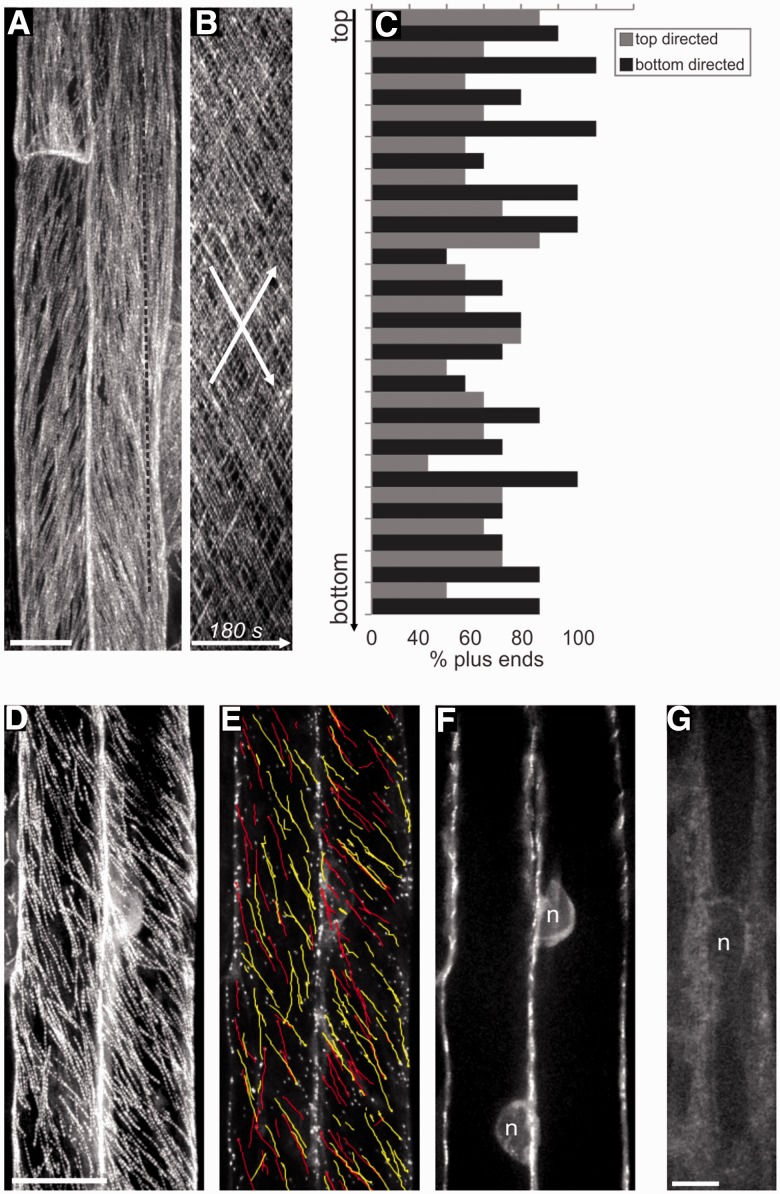
Atrichoblast cells lack bipolarity
In contrast to hair-forming cells, hairless cells at maturity did not exhibit CMT growth bipolarity (Fig. 3A–C). Instead these cells contained dense longitudinal or oblique CMTs that grew in both directions throughout the cell, regardless of nuclear position. Kymographs show this as uniform cross-hatching of two oppositely sloped lines (Fig. 3B). We also did not see nuclear MTOC activity in cells lacking MT array bipolarity (Fig. 3D–G). This was evident by the lack of EMTs running from nucleus to cortex in these cells (Fig. 3F) and the lack of perinuclear GFP–GCP2 accumulation (Fig. 3G).
Fig. 3.
Atrichoblast cells lack bipolarity. (A) ZT projections (180 s) of EB1b–GFP in atrichoblast cells. The dotted line refers to the kymograph in B. (B) Kymograph of the cell in A corresponding to the dotted line drawn along the long axis of the cell. Arrows indicate the opposite but overlapping MT polarities. (C) Histogram showing percentages of baseward vs. tipward MT growth polarities along the length of the cell in A (the one used for the kymograph). (D) ZT projections (180 s) of EB1b–GFP in atrichoblast cells. (E) Growth paths of manually tracked EB1b–GFP dots overlaid on a single time point from cells in D. Yellow is top-directed movement and red is bottom-directed movement. (F) ZT projection of the cell midplane from cells in D and E, showing the nuclear position and lack of perinuclear MTs. (G) GFP–GCP2 localization in an atrichoblast cell. Signal does not accumulate in the perinuclear region. Scale bars, 10 µm.
Taken together, the observations described so far show a correlation between nuclear position, nuclear MT initiation and CMT growth polarities.
CMT bipolarity occurs in other cells with nuclear MTOC activity
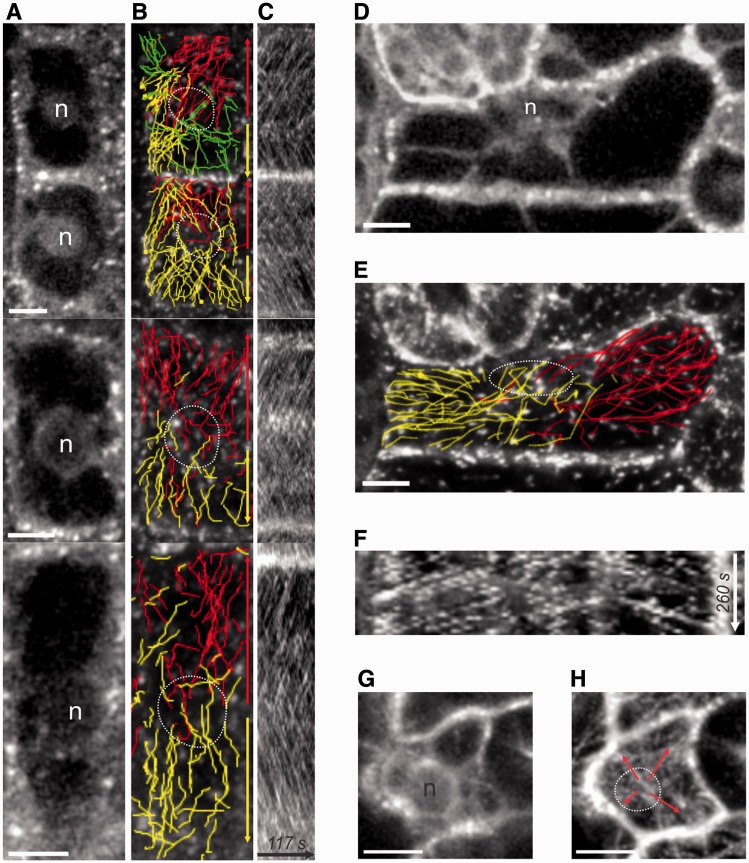
Based on the presence of nucleocentric bipolar CMT arrays in root hairs, we hypothesized that other cell types with nuclear MTOC activity would have similar CMT polarities. In order to test this, EB1b–GFP movement patterns were analyzed in various cell types. We identified nucleocentric bipolar CMT arrays in both post-cytokinetic root epidermal cells and young, recently formed leaf epidermal cells (Fig. 4). Several examples in root epidermal cells are shown in Fig. 4A–C, which shows nuclear position (Fig. 4A), overlaid tracks of EB1b–GFP dots (Fig. 4B) and accompanying kymographs drawn along the longitudinal axis of these cells (Fig. 4C). In contrast to root hairs, wherein the CMT bipolarity encircles the entire tubular wall, CMT polarities in these polyhedral cells typically arose from a central point specifically within the outer periclinal cell face, then spread out into the internal faces as MTs grew around cell edges. Bipolar arrays typically ran in a longitudinal or somewhat oblique direction with respect to the elongation axis of the cell, although, depending on nuclear position and cell geometry, other nucleocentric-based patterns could be observed, including three- and four-way polarities, and occasional transverse bipolarities in cells in which perinuclear MTOCs were still present.
Fig. 4.
Nucleocentric CMT polarities in the root division zone and leaf epidermal cells. (A) Midplane confocal slices showing the nuclear position for four representative cells with CMT bipolarities. (B) Paths of manually tracked EB1b–GFP dots colored red (topward movement) or yellow (bottomward movement) and overlaid on a single time point from the series. Dotted circles indicate the nuclear position. (C) Kymographs corresponding to lines drawn along the longitudinal axis in the middle of each cell. (D–F) Young leaf epidermal cell that still contains an active nucleus and exhibits nucleocentric CMT growth polarities. (D) Midplane confocal slice to show the nuclear position. (E) Paths of manually tracked EB1b–GFP dots colored red (rightward movement) or yellow (leftward movement) and overlaid on a single time point from the series. (F) Kymograph corresponding to the line drawn along the long axis in the middle of the cell. (G, H) Young polyhedral leaf epidermal cell showing EB1b–GFP tracks emanating from a central point in a radial manner. (G) Midplane showing the nuclear position. (F) Time projection of cell surface showing EB1b–GFP tracks. Arrows indicate growth directions. n, nucleus. Scale bars, 5 µm.
In young leaf epidermal cells we also observed nucleocentric CMT growth polarities. The large variation in cellular geometry of these cells corresponded to a variety of nucleocentric CMT polarities. Narrow, rectangular cells (geometries similar to root cells) showed bipolarity (Fig. 4D–F), while the typical polyhedral cells of the young leaf epidermis more frequently showed uniform radiation from a centralized point (Fig. 4G, H). These data show that complex CMT polarity patterns develop in cells with nuclear MTOC activity, and that variations in cell shape are associated with variations in polarity.
Nuclear position affects CMT growth polarity
Based on the strong correspondence between nuclear position and the split point of bipolar cortical arrays, we hypothesized that experimental manipulation of the nuclear position would lead to a corresponding change in the position of the CMT bipolar split point. To test this, we centrifuged roots gently in order to relocate the nuclei to the ends of cells. Briefly, roots were taken from plates, wrapped in a moist paper towel and placed in microcentrifuge tubes with the root tips oriented downward. We found that 20–30 min at 1,200×g was sufficient to move most nuclei to the ends of the cells with no obvious damage, in agreement with previous studies (Pickett-Heaps 1969, Mineyuki and Gunning 1990, Murata and Wada 1991). Nuclei typically returned to their central position within 30–60 min (Supplementary Fig. S3).
To ensure the sedimented nuclei had nucleation activity, we used cells from the early elongation zone, which show perinuclear GFP–GCP and initiate EMTs that radiate toward the cortex (Ambrose and Wasteneys 2011) (Supplementary Fig. S4). By assessing CMT plus-end growth polarity using time-lapse imaging of EB1b–GFP, we found that in cells with nuclear MTOC activity, CMTs were often aligned longitudinally, growing away from the sedimented nucleus (Fig. 5A, B). As with nucleocentric bipolarities, growth away from sedimented nuclei was most obvious in the elongate/rectangular cells of the early elongation zone. In contrast, in smaller/isodiametric cells with sedimented nuclei, a range of more complex patterns was observed, presumably due to influences of cell edges, which contain GCP proteins and nucleate MTs (Ambrose and Wasteneys 2011), and act as barriers to oncoming MT plus ends (Ambrose et al. 2011). In the case of cells from centrifuged roots where the nucleus retained their centralized nucleus, normal nucleocentric CMT growth bipolarity was observed.
Fig. 5.
CMT bipolarity split point is the over nucleus in centrifuged root cells. (A) Epidermal division zone cell with sedimented nucleus (N). Left panel is a DIC image corresponding to the right panel, which shows a ZT projection of an EB1b-GFP time series. (B) Dual colored overlay of several EB1b-GFP dots in the cell from A. Red lines indicate upward transit, and yellow lines indicate downward transit. To the right is a kymograph drawn from this cell showing split polarity. (C) Lack of CMT growth polarities in cells with sedimented nuclei that lack MTOC activity. An elongated lateral root cap cell is shown. Left panel: the cellular midplane showing the nuclear position at the bottom of the cell. Middle panel: ZT projection of the same cell. Right panel: overlay of several EB1b–GFP growth paths. Red lines indicate right transit, and yellow lines indicate left transit. n, nucleus. Arrows indicate EB1b–GFP growth polarities. Time intervals are 5 s between time points. Scale bars, 5 µm.
Elongating lateral root cap cells provide a convenient internal control for the centrifugation experiments since they contain transverse CMTs, lack nuclear MT initiation and provide cellular geometries that are comparable with similarly shaped cells that have nuclear MTOC activity. In these cells, GCP2–GFP was undetectable on nuclei (Supplementary Fig. S4), consistent with the lack of nuclear MT initiation. Elongating lateral root cap cells with sedimented nuclei retained transverse CMT alignment, thus excluding the possibility that sedimentation of the cytoplasm was the cause of the polarity shift (Fig. 5C). These results show that when perinuclear MT nucleation is active, CMT growth polarity and array organization is governed by nuclear position.
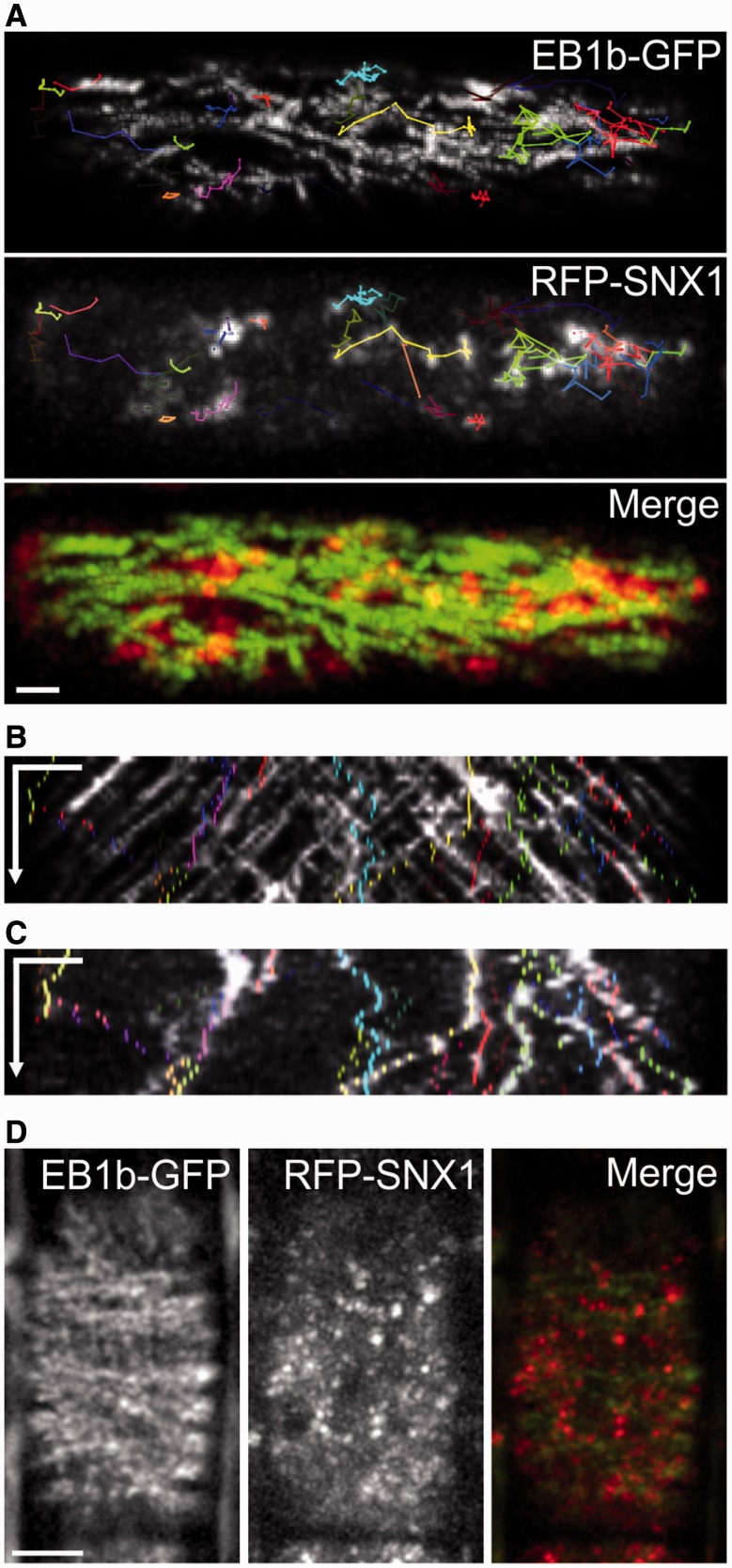
Bidirectional vesicle motility in young cells with bipolar CMTs
Given our observation of bipolar CMT arrays in young epidermal cells, we asked if bipolar distribution of microtubule-associated organelles could be seen in these cells. To this end we tracked the endosomal component Sorting Nexin 1 (SNX1), which has recently been shown to associate with CMTs via the MT-associated protein CLASP (Clip-associated protein; Ambrose et al. 2013). SNX1 was tagged with red fluorescent protein (RFP), and cells with bipolarized CMT arrays were identified by tracking EB1b–GFP. In agreement with previous data, SNX1 vesicles that associated with CMTs showed movement and deformations along the direction of the CMT (Ambrose et al. 2013). In young cells with bipolarized CMT arrays, we observed that the jiggling behavior of SNX1 vesicles has a directional bias along the axis of the bipolar CMT array (Fig. 6). This bidirectional pole-ward movement of SNX1–RFP vesicles is shown in Fig. 6A, wherein a time projection of EB1b–GFP tracks is overlaid with track paths of several RFP–SNX1 vesicles. The kymographs in Fig. 6B and C were drawn from the cell in Fig. 6A, and show correlation between vesicle paths and the EB1b–GFP channel (Fig. 6B). Fig. 6C shows the RFP channel alone to visualize vesicle movement, and is overlaid with the same SNX1–RFP tracks. Cells with transverse MT arrays do not show this bipolar transit (Fig. 6D).
Fig. 6.
Directional motility of sorting endosomes to cell poles in cells with bipolar CMTs. (A) Track paths of several RFP–SNX1 vesicles overlaid onto time projections of EB1b–GFP (top panel) and RFP–SNX1 (bottom panel). The bottom panel shows a merged image of GFP and RFP channels. (B) Kymograph showing correlation between vesicle paths (colored dots) and the EB1b–GFP channel. (C) Kymograph showing vesicle paths (colored dots) overlaid on the RFP–SNX1 channel. Time intervals are 2.5 s between time points, 60៎ s total. (D) Time projection of EB1b–GFP and RFP–SNX1 from the cell with the transverse CMT array. The right panel is the merged image of the first two panels. Total time is 120 s with 10 s intervals between acquisitions. Scale bars, 5 µm.
Discussion
Cortex and nuclear envelope are sources of CMTs
The orientation of parallel CMT arrays plays a major role in determining the axis of cell elongation and thus has been the subject of intense interest since their identification in 1963 (Ledbetter and Porter 1963). Live-cell imaging studies of CMTs over the past two decades (Wasteneys et al. 1993, Yuan 1994, Chan et al. 2003, Shaw et al. 2003, Dixit and Cyr 2004, Chan et al. 2007, Wightman and Turner 2007, Ambrose and Wasteneys 2008, Allard et al. 2010, Tindemans et al. 2010) along with recent mathematical modeling studies (Baulin et al. 2007, Allard et al. 2010, Eren et al. 2010, Tindemans et al. 2010, Eren et al. 2012) have identified and validated a wide variety of MT dynamic behaviors that contribute to the parallel orientation of CMTs. Most of the studies from which these observations have been made used large vacuolated cells, such as epidermal cells from plant hypocotyls and leaves, or algal internodal cells, in which nucleation at the cell cortex is the predominant source of CMTs (Wasteneys and Williamson 1987, Wasteneys and Williamson 1989,Wasteneys et al. 1989, Cleary and Hardham 1990, Shaw et al. 2003, Murata et al. 2005). Our current study shows that CMT organization and polarity can also be affected by nuclear envelope-derived MTs. Specifically, in both tip-growing root hairs and recently formed diffusely expanding cells, the activity and positioning of the nuclear MTOC can influence CMT orientation and polarity along the long axis of the cell.
Self-organizational establishment of polarized CMT arrays
The absence of well-defined MTOCs in the cortical array has led to the general consensus that CMTs establish parallel arrays predominantly through self-organization, which describes any system wherein global order emerges from local interactions between individual elements. MT–MT interactions influencing CMT self-organization include angle-dependent bundle formation, as well as nucleation of new MTs from pre-existing ones (Wasteneys and Ambrose 2009, Fishel and Dixit 2013). An apparent outcome of MT self-organization is the emergence of large groups of parallel MTs within the CMT array that grow with a unidirectional bias, often changing position and orientation over time (Dixit et al. 2006, Chan et al. 2007, Sambade et al. 2012, Vineyard et al. 2013). Of particular pertinence to the current study are recent studies that found bipolar longitudinal arrays in the outer periclinal face of hypocotyl epidermal cells (Sambade et al. 2012, Vineyard et al. 2013), and in the outer periclinal face of trichoblasts just prior to hair formation (Pietra et al. 2013). Our study directly confirms those of Pietra et al., showing that nuclear MTOC activity may drive these bipolarities in trichoblasts. Both hypocotyl studies followed the transition from longitudinal/oblique/mixed CMTs into transverse configuration upon initiation of light- or hormone-induced cell elongation, and suggest that self-organizational mechanisms alone may be sufficient to generate CMT bipolarity. Specifically, both studies showed that transversely oriented CMTs in the anticlinal faces enter the outer periclinal face during reorganization and contribute to the signal-induced switch to transverse order. These data are consistent with the observation that while the CMT orientations can be highly variable at the outer periclinal face of hypocotyl epidermal cells, those on the inner periclinal and anticlinal faces tend to remain transversely aligned (Chan et al. 2010, Crowell et al. 2011). Sambade et al. (2012) further showed that self-organizational mechanisms can be sufficient to generate bipolar arrays by restricting entry of CMTs from lateral cell faces in computational simulations.
How is the initial bipolarity established in these large, vacuolated cells? From a self-organizational standpoint, these polarities have been suggested to result from biased stability of MTs growing in the dominant orientation within a domain (Dixit et al. 2006, Eren et al. 2010), and directional nucleation of MTs from pre-existing MTs (Chan et al. 2009, Wasteneys and Ambrose 2009). Could transient nuclear MTOC activity in these cells also contribute to the initial establishment of bipolarity by over-riding CMT self-organization? This is the case in animal cells, where self-organizational mechanisms are also present but masked by MTOC-based inputs (Reilein et al. 2005).
Interplay between cellular geometry and nuclear MTOC
The CMT polarities reported here vary based on cellular geometry. Root hair cells are tubular, and show CMT bipolarity around the entire cortex. In contrast, bipolar arrays appear to be restricted to the outer periclinal face in the post-cytokinetic cells we observed, as well as in hypocotyl cells (Sambade et al. 2012, Vineyard et al. 2013). Thus, in all cases, CMT bipolarities are present in regions of the cell that are not in contact with other cells. Since the patterns of mechanical stress in an organ correlate with global CMT orientations on the outer periclinal faces (Hamant et al. 2008), it will be interesting to determine how cell–cell contact plays a role in transducing mechanical information to different faces of the cell. It is also possible that the observed enrichment of CLASP and GCP2/3 specifically to outer cell edges in post-cytokinetic cells plays a role in generating these outer face bipolarities.
Post-cytokinetic CMT polarity switching
A 1994 study by Nagata and colleagues of tobacco BY-2 suspension culture cells showed that immediately after cytokinesis, nuclear envelope-associated EMTs enter the outer cortex next to the new wall, and orient longitudinally, pointing toward the opposite cell pole (Nagata et al. 1994). This is consistent with our previous finding that newly formed cell edges accumulate GCPs and act as nucleation centers for CMTs (Ambrose and Wasteneys 2011), which confirmed Gunning’s hypothesis that cell edges can act as MTOCs (Gunning et al. 1978), and raises the new question of how the cell edge MTOC and the nuclear MTOC function together.
The presence of nuclear MTOC activity prior to cell plate fusion suggests a possible role in the delivery of GCPs and other nucleation factors early on to cell edges. Edge nucleation generates longitudinal CMTs that run perpendicular to the new edge, and which show strong cell-wide unipolarity pointing away from this nucleating edge. Shortly after this stage, there is a polarity reversal. The MT-associated protein CLASP accumulates along the newly formed sharp edges, where it acts to stabilize incoming MT plus ends (Ambrose et al. 2011). In the current study, we show how nucleocentric CMT bipolarity favors growth away from the cell center. The CMT array remains longitudinal, but is now bipolar instead of the edge-based GCP nucleation that creates unipolar arrays. Bipolar longitudinal arrays inherently direct CMTs toward the top and bottom cross-walls. If CLASP is present at these sharp edges, they will be stabilized and CMT longitudinality will be maintained. If CLASP is absent, MT catastrophe will occur, favoring the establishment of a transverse CMT array.
Nuclear MTOC effect on directional vesicle transit
In highly polarized animal cells such as neurons, vesicle movement is MT based and highly directional. In these cells, MTs extend along the length of the axon with their plus ends toward the synapse, to which vesicle transport is carried out by motor proteins. In plants, vesicle movement is primarily actomyosin based and typically not polarized. However, a growing body of evidence suggests an additional role for MTs in vesicle movement (Brandizzi and Wasteneys 2013). In cell wall formation, Golgi bodies may transiently associate with CMTs, during which time they assist in the delivery of smaller compartments carrying cellulose synthase components (Crowell et al. 2009, Gutierrez et al. 2009). Interestingly, these bodies known as smaCCs or MASCs can associate with shortening MT plus ends (Gutierrez et al. 2009). Our group recently showed that vesicles containing the retromer component SNX1 associate with MTs in a CLASP-dependent manner to facilitate recycling of the auxin transporter Pin-formed 2 (PIN2) (Ambrose et al. 2013). The results presented in the current study show that SNX vesicle movement has a directional bias in the direction of growing MT plus ends. It will be interesting to determine if the mechanism behind this is motor based or MT dynamics based, and if this bipolar vesicle flux contributes to cellular polarity in any way.
MTOC-based bipolar MT arrays are common in eukaryotic cells
Our finding here of nuclear MTOC-based MT bipolarity in plant cells reveals a remarkable similarity to previous findings in other systems. Bipolar MT arrays are common in eukaryotic cells, with a tendency to form in elongated cells that have centrally located MTOCs. The rod-shaped cells of fission yeast Schizosaccharomyces pombe contain interphase MTs that are oriented along the long axis of the cell with the plus ends pointing toward the poles and the minus ends overlapping at MTOCs on the centralized nucleus (Sawin and Nurse 1998). These bipolar MT arrays are involved in nuclear positioning (Tran et al. 2001) and trafficking of polarity factors to cell ends (Chang and Martin 2009). When animal cells are restricted to grow in length but not in width, the centrally positioned nucleus and associated centrosome generate MTs that form bipolarized arrays along the long axis of the cell to control cell length (Picone et al. 2010). The data of our current study indicate that conserved mechanisms are at work in bipolarized plant MT arrays, and that they share common functions with other cell types, including cell length control and polarization.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana Columbia ecotype plants were grown in continuous light conditions on vertical agar plates containing Hoagland’s medium as previously reported (Ambrose et al. 2007). Young expanding cotyledon cells were imaged at 3–4 d. Root tips were used for dividing and expanding root cells. Root hairs were visualized by growing roots along the coverglass within Nunc™ Lab-Tek™ coverglass chambers containing Hoagland’s medium. We used GCP2:GCP2-3xGFP lines to visualize MT nucleation sites (Nakamura et al. 2010), and 35S-driven EB1b–GFP lines were a gift from Takashi Hashimoto.
Centrifugation experiments
Seedlings were taken from plates and wrapped in a moist paper towel, which was then inserted into a microcentrifuge tube, with the root tip facing downward. Samples were then gently spun in a tabletop microcentrifuge (Sorvall Microcentrifuge 17) at 1,200×g for 20 min, and viewed within 10 min.
Microscopy and image analysis
Images were acquired on a Zeiss Pascal scanning confocal microscope, or with a Perkin-Elmer spinning disk microscope. Images were processed using imageJ software (http://rsb.info.nih.gov/ij/), and figures were assembled using Corel Draw software. For trafficking of SNX1–RFP vesicles, the imageJ plugin MtrackJ was used.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the National Science and Engineering Research Council [Discovery Grant 298264-2009]; the Canadian Institutes of Health Research [MOP-86675 to G.O.W.]; the UBC Bioimaging Facility.
Supplementary Material
Acknowledgments
We thank Takashi Hashimoto for the generous gifts of 35S:EB1b–GFP and GCP2–3×GFP lines.
Glossary
Abbreviations
- ABC
ATP-binding cassette
- CLASP
Clip-associated protein
- CMT
cortical microtubule
- EB1b
End Binding Protein 1b
- EMT
endoplasmic microtubule
- GCP2
gamma tubulin-complex protein 2
- GFP
green fluorescent protein, MBD, microtubule-binding domain
- MT
microtubule
- MTOC
microtubule organizing center
- RFP
red fluorescent protein
- SNX1
Sorting Nexin 1
Disclosures
The authors have no conflicts of interest to declare.
References
- Allard JF, Wasteneys GO, Cytrynbaum EN. Mechanisms of self-organization of cortical microtubules in plants revealed by computational simulations. Mol. Biol. Cel. 2010;21:278–286. doi: 10.1091/mbc.E09-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2011;2:430. doi: 10.1038/ncomms1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Ruan Y, Gardiner J, Tamblyn LM, Catching A, Kirik V, et al. CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell. 2013;24:649–659. doi: 10.1016/j.devcel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Ambrose C, Wasteneys GO. Cell edges accumulate gamma tubulin complex components and nucleate microtubules following cytokinesis in Arabidopsis thaliana cells. PLoS One. 2011;6:e27423. doi: 10.1371/journal.pone.0027423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Shoji T, Kotzer AM, Pighin JA, Wasteneys GO. The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell. 2007;19:2763–2775. doi: 10.1105/tpc.107.053777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose JC, Wasteneys GO. CLASP modulates microtubule–cortex interaction during self-organization of acentrosomal microtubules. Mol. Biol. Cell. 2008;19:4730–4737. doi: 10.1091/mbc.E08-06-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulin VA, Marques CM, Thalmann F. Collision induced spatial organization of microtubules. Biophys. Chem. 2007;128:231–244. doi: 10.1016/j.bpc.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Bichet A, Desnos T, Turner S, Grandjean O, Hofte H. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 2001;25:137–148. doi: 10.1046/j.1365-313x.2001.00946.x. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Wasteneys GO. Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J. 2013;75:339–349. doi: 10.1111/tpj.12227. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. The pleiomorphic plant MTOC: an evolutionary perspective. J. Integr. Plant Biol. 2007;49:1142–1153. [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C. Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat Cell Biol. 2007;9:171–175. doi: 10.1038/ncb1533. [DOI] [PubMed] [Google Scholar]
- Chan J, Calder GM, Doonan JH, Lloyd CW. EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 2003;5:967–971. doi: 10.1038/ncb1057. [DOI] [PubMed] [Google Scholar]
- Chan J, Crowell E, Eder M, Calder G, Bunnewell S, Findlay K, et al. The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J. Cell Sci. 2010;123:3490–3495. doi: 10.1242/jcs.074641. [DOI] [PubMed] [Google Scholar]
- Chan J, Sambade A, Calder G, Lloyd C. Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell. 2009;21:2298–2306. doi: 10.1105/tpc.109.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Martin SG. Shaping fission yeast with microtubules. Cold Spring Harb. Perspect. Biol. 2009;1:a001347–a001347. doi: 10.1101/cshperspect.a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton L, Black CM, Lloyd CW. Microtubule nucleating sites in higher plant cells identified by an auto-antibody against pericentriolar material. J. Cell Biol. 1985;101:319–324. doi: 10.1083/jcb.101.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AL, Hardham AR. Reinstatement of microtubule arrays from cortical nucleating sites in stomatal complexes of Lolium rigidum following depolymerization of microtubules by oryzalin and high-pressure. Plant Cell Physiol. 1990;31:903–915. [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof Y-D, Schumacher K, et al. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Timpano H, Desprez T, Franssen-Verheijen T, Emons AM, Hofte H, et al. Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell. 2011;23:2592–2605. doi: 10.1105/tpc.111.087338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Chang E, Cyr R. Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol. Biol. Cell. 2006;17:1298–1305. doi: 10.1091/mbc.E05-09-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Cyr R. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell. 2004;16:3274–3284. doi: 10.1105/tpc.104.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren EC, Dixit R, Gautam N. A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol. Biol. Cell. 2010;21:2674–2684. doi: 10.1091/mbc.E10-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren EC, Gautam N, Dixit R. Computer simulation and mathematical models of the noncentrosomal plant cortical microtubule cytoskeleton. Cytoskeleton (Hoboken) 2012;69:144–154. doi: 10.1002/cm.21009. [DOI] [PubMed] [Google Scholar]
- Erhardt M, Stoppin-Mellet V, Campagne S, Canaday J, Mutterer J, Fabian T, et al. The plant Spc98p homologue colocalizes with gamma-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 2002;115:2423–2431. doi: 10.1242/jcs.115.11.2423. [DOI] [PubMed] [Google Scholar]
- Falconer M, Donaldson G, Seagull R. MTOCs in higher plant cells: an immunofluorescent study of microtubule assembly sites following depolymerization by APM. Protoplasma. 1988;144:46–55. [Google Scholar]
- Fishel EA, Dixit R. Role of nucleation in cortical microtubule array organization: variations on a theme. Plant J. 2013;75:270–277. doi: 10.1111/tpj.12166. [DOI] [PubMed] [Google Scholar]
- Gunning BES, Hardham AR, Hughes JE. Evidence for initiation of microtubules in discrete regions of cell cortex in azolla root-tip cells, and an hypothesis on development of cortical arrays of microtubules. Planta. 1978;143:161–179. doi: 10.1007/BF00387788. [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jonsson H, Krupinski P, Uyttewaal M, Bokov P, et al. Developmental patterning by mechanical signals in Arabidopsis. Science. 2008;322:1650–1655. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- Kumagai F, Nagata T, Yahara N, Moriyama Y, Horio T, Naoi K, et al. Gamma-tubulin distribution during cortical microtubule reorganization at the M/G1 interface in tobacco BY-2 cells. Eur. J. Cell Biol. 2003;82:43–51. doi: 10.1078/0171-9335-00292. [DOI] [PubMed] [Google Scholar]
- Ledbetter MC, Porter KR. A ‘microtubule’ in plant cell fine structure. J. Cell Biol. 1963;19:239–250. doi: 10.1083/jcb.19.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Marc J, Joshi HC, Palevitz BA. A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 1993;104:1217–1228. doi: 10.1242/jcs.104.4.1217. [DOI] [PubMed] [Google Scholar]
- Mineyuki Y, Gunning BES. A role for preprophase bands of microtubules in maturation of new cell-walls, and a general proposal on the function of preprophase band sites in cell-division in higher-plants. J. Cell Sci. 1990;97:527–537. [Google Scholar]
- Mizuno K. Microtubule-nucleation sites on nuclei of higher plant cells. Protoplasma. 1993;173:77–85. [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, et al. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- Murata T, Wada M. Effects of centrifugation on preprophase-band formation in Adiantum protonemata. Planta. 1991;183:391–398. doi: 10.1007/BF00197738. [DOI] [PubMed] [Google Scholar]
- Nagata T, Kimagai F, Hasezawa S. The origin and organization of cortical microtubules during the transition between M and G1 phases of the cell cycle as observed in highly synchronized cells of tobacco BY-2. Planta. 1994;193:567–572. [Google Scholar]
- Nakamura M, Ehrhardt DW, Hashimoto T. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 2010;12:1064–1070. doi: 10.1038/ncb2110. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD. Preprophase microtubules and stomatal differentiation: some effects of centrifugation on symmetrical and asymmetrical cell division. J. Ultrastruct. Res. 1969;27:24–44. [PubMed] [Google Scholar]
- Picone R, Ren X, Ivanovitch KD, Clarke JDW, McKendry RA, Baum B. A polarised population of dynamic microtubules mediates homeostatic length control in animal cells. PLoS Biol. 2010;8:e1000542. doi: 10.1371/journal.pbio.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra S, Gustavsson A, Kiefer C, Kalmbach L, Hörstedt P, Ikeda Y, et al. Arabidopsis SABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nat. Commun. 2013;4:2779. doi: 10.1038/ncomms3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilein A, Yamada S, Nelson WJ. Self-organization of an acentrosomal microtubule network at the basal cortex of polarized epithelial cells. J. Cell Biol. 2005;171:845–855. doi: 10.1083/jcb.200505071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade A, Pratap A, Buschmann H, Morris RJ, Lloyd C. The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell. 2012;24:192–201. doi: 10.1105/tpc.111.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Nurse P. Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer V, Janski N, Canaday J, Herzog E, Erhardt M, Evrard J-L, et al. Arabidopsis GCP2 and GCP3 are part of a soluble γ-tubulin complex and have nuclear envelope targeting domains: targeting of γ-tubulin complex proteins. Plant J. 2007;52:322–331. doi: 10.1111/j.1365-313X.2007.03240.x. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- Stoppin V, Vantard M, Schmit AC, Lambert AM. Isolated plant nuclei nucleate microtubule assembly—the nuclear-surface in higher-plants has centrosome-like activity. Plant Cell. 1994;6:1099–1106. doi: 10.1105/tpc.6.8.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindemans SH, Hawkins RJ, Mulder BM. Survival of the aligned: ordering of the plant cortical microtubule array. Phys. Rev. Lett. 2010;104:58103. doi: 10.1103/PhysRevLett.104.058103. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoué S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantard M, Levilliers N, Hill AM, Adoutte A, Lambert AM. Incorporation of Paramecium axonemal tubulin into higher plant cells reveals functional sites of microtubule assembly. Proc. Natl Acad. Sci. USA. 1990;87:8825–8829. doi: 10.1073/pnas.87.22.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineyard L, Elliott A, Dhingra S, Lucas JR, Shaw SL. Progressive transverse microtubule array organization in hormone-induced Arabidopsis hypocotyl cells. Plant Cell. 2013;25:662–676. doi: 10.1105/tpc.112.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO, Ambrose JC. Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 2009;19:62–71. doi: 10.1016/j.tcb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Gunning BES, Hepler PK. Microinjection of fluorescent brain tubulin reveals dynamic properties of cortical microtubules in living plant-cells. Cell Motil. Cytoskel. 1993;24:205–213. [Google Scholar]
- Wasteneys GO, Jablonsky PP, Williamson RE. Assembly of purified brain tubulin at cortical and endoplasmic sites in perfused internodal cells of the alga Nitella tasmanica. Cell Biol. Int. Rep. 1989;13:513–528. [Google Scholar]
- Wasteneys GO, Williamson RE. Microtubule orientation in developing internodal cells of Nitella—a quantitative analysis. Eur. J. Cell Biol. 1987;43:14–22. [Google Scholar]
- Wasteneys GO, Williamson RE. Reassembly of microtubules in Nitella-tasmanica— assembly of cortical microtubules in branching clusters and its relevance to steady-state microtubule assembly. J. Cell Sci. 1989;93:705–714. [Google Scholar]
- Wick S. Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. III. Transition between mitotic/cytokinetic and interphase microtubule arrays. Cell Biol. Int. Rep. 1985;9:357–371. doi: 10.1016/0309-1651(85)90031-1. [DOI] [PubMed] [Google Scholar]
- Wightman R, Turner SR. Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 2007;52:742–751. doi: 10.1111/j.1365-313X.2007.03271.x. [DOI] [PubMed] [Google Scholar]
- Yuan M, Shaw PJ, Warn RM, Lloyd CW. Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc. Natl Acad. Sci. USA. 1994;91:6050–6053. doi: 10.1073/pnas.91.13.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.