Abstract
In plants, miRNAs and siRNAs, such as transacting siRNAs (ta-siRNAs), affect their targets through distinct regulatory mechanisms. In this study, the expression profiles of small RNAs (smRNAs) in Arabidopsis plants subjected to drought, cold, and high-salinity stress were analyzed using 454 DNA sequencing technology. Expression of three groups of ta-siRNAs (TAS1, TAS2, and TAS3) and their precursors was downregulated in Arabidopsis plants subjected to drought and high-salinity stress. Analysis of ta-siRNA synthesis mutants and mutated ARF3-overexpressing plants that escape the tasiRNA-ARF target indicated that self-pollination was hampered by short stamens in plants under drought and high-salinity stress. Microarray analysis of flower buds of rdr6 and wild-type plants under drought stress and nonstressed conditions revealed that expression of floral development- and auxin response-related genes was affected by drought stress and by the RDR6 mutation. The overall results of the present study indicated that tasiRNA-ARF is involved in maintaining the normal morphogenesis of flowers in plants under stress conditions through fine-tuning expression changes of floral development-related and auxin response-related genes.
1. Introduction
In order to adapt and survive the exposure to biotic and abiotic stress, plants have evolved various molecular responses for fine-tuning the control of adaptive responses that involve posttranscriptional regulatory mechanisms, as well as epigenetic and posttranslational modifications [1, 2]. Recent genome-wide transcriptome analyses using tiling arrays and next generation sequencing (NGS) have revealed a large number of stress-responsive noncoding RNAs (ncRNAs) [3–5].
Several small RNAs (smRNAs), such as miRNAs and siRNAs, were shown to function in development and stress responses in plants [6–9]. In plants, smRNAs exhibit a high level of complexity in their biogenesis and function. At the moment, smRNAs are classified into microRNAs (miRNAs) and three classes of small interfering RNAs (siRNAs) [6–9]. Transacting siRNAs (ta-siRNAs) are derived from TAS ncRNAs that are targeted by miR173 or miR390 [10–14]. Double-stranded RNAs (dsRNAs) are generated from cleaved ncRNAs by RNA-dependent RNA polymerase 6 (RDR6) and dsRNAs are processed into 21nt ta-siRNAs. ARF2, ARF3 (ETT), and ARF4 were demonstrated to be targets of TAS3 ta-siRNA (tasiRNA-ARF) [10–14].
In the present study, Arabidopsis deep smRNA sequencing was used to identify novel roles for smRNAs in abiotic stress response and it was discovered that ta-siRNAs and their precursors (TAS1, TAS2, and TAS3) were downregulated by drought and high-salinity stress treatments. Analysis of ta-siRNA synthesis mutants subjected to drought and high-salinity stresses revealed a short stamen phenotype and changes in the expression of floral development-related and auxin response-related genes, which was enhanced by the RDR6 mutation. These results demonstrate that the tasiRNA-ARF pathway functions in maintaining normal flower morphogenesis under environmental stress.
2. Materials and Methods
2.1. 454 Sequencing of Small RNAs
Two-week-old wild-type plants (Arabidopsis thaliana ecotype Columbia), grown on MS medium [3], were transferred to drought, cold (4°C), and high-salinity (250 mM NaCl) stressas previously reported [3]. The treated plants were harvested hourly from 1 to 10 hrs after the treatment was initiated. The 1–5 hr stress-treated samples and the 6–10 hr stress-treated samples were pooled into two groups. Total RNAs were prepared using an ISOGEN kit (Nippon Gene) and precipitated with 2 M LiCl and equal volume of ethanol. RNAs were resuspended in RNase-free water at 65°C and extracted twice with an equal volume of phenol for 30 min on ice. The RNAs were then precipitated with 2 M LiCl and equal volume of ethanol. They were resuspended in RNase-free water at 65°C and extracted twice with an equal volume of phenol for 30 min on ice and precipitated again by adding 1/10 volume of 3 M sodium acetate and 3 volume of ethanol. Subsequently, 17–30 nt smRNAs were extracted using flashPAGE (Life Technologies). A cDNA library was then constructed using a small RNA Cloning Kit (Takara). First, smRNAs were ligated with a 5′ adapter F: and a 3′ adapter to generate cDNAs. The cDNAs were electrophoresed in 8 M urea and 7.5% acrylamide gel, and 60–80 nt cDNAs were recovered. The cDNAs were amplified by 12–15 cycles of PCR and subjected to 454 DNA sequencing according to the manufacturer's instructions. To eliminate RNA degradation fragments, the number of RNA sequences was normalized against the total number of miRNA sequences obtained. The normalized number of smRNAs was then subjected to data analysis. Data sets of 454 sequencing are available in DDBJ (http://www.ddbj.nig.ac.jp/index-e.html) under the accession number AB948670-AB967973.
2.2. Stress Treatments Applied to tasiRNA-ARF Pathway-Related Mutants
rdr6-15 (SAIL_617), ago7 (SALK_095997), sgs3 (SALK_039005), dcl4-2 (GK-160G05), ARF3pro;ARF3 [15], ARF3pro:ARF3mut [15], arf3 (ett-15) [10], arf4-2 (SALK_070506), and wild-type Arabidopsis plants were grown for two weeks in pots containing 30 g of vermiculite soil. The drought stress treatment consisted of subjecting the two-week-old plants to water depletion for one week. After the one-week period, the watering of plants was then reinitiated. The high-salinity stress treatment consisted of watering three-week-old plants that started to bolt, with 100 mM NaCl-containing water for five days.
2.3. RNA Extraction
Total RNA was extracted with a Plant RNA Isolation Reagent (Life Technologies) and treated with DNase I (Life Technologies). The RNAs were then subjected to RT-quantitative PCR (RT-qPCR), microarray, and Northern analyses.
2.4. RT-qPCR Analysis
cDNAs were prepared from 1 μg of total RNA using Superscript III (Life Technologies). The target RNA concentration was obtained by measuring 1/10 of the cDNA using an ABI Prism 3100 (Life Technologies) and SYBR Premix Ex Taq II kit (Takara). For detecting small RNAs, a SYBR Advantage qPCR Kit (Takara) was used. The relative expression was calculated using the delta-delta CT method. U2 and ACT2 for smRNA and mRNA, respectively, were used as a reference gene for normalization. Three independent biological replicates were used in all of the RT-qPCR analyses.
2.5. Microarray Analysis
Two-week-old rdr6 mutants and wild-type plants were subjected to a drought stress treatment consisting of withholding water for one week. Microarray experiments using flower buds subjected to a drought stress or nonstressed treatment were carried out according to the manufacturer's (Agilent) preferred protocol using three biological replicates [16]. Fluorescent-labeled cRNAs were prepared from each total RNA sample using a Low Input Quick Amp Labeling Kit and were then hybridized to an Agilent Arabidopsis V4 microarray. The microarrays were scanned using an Agilent DNA Microarray Scanner G2539A ver. C. 75 percentile normalization was performed for the signals generated by the microarray probes according to the Agilent data analysis protocol. For microarray analysis, R program ver. 2.12.1 was used. Significant differentially expressed genes were identified by 2-way ANOVA analysis (FDR < 0.075) [17, 18]. The data set derived from the microarray analysis is available in GEO (http://www.ncbi.nlm.nih.gov/geo/info/linking.html) under the accession number GSE57174.
3. Results
3.1. RNA Sequencing of Arabidopsis Small RNAs in Plants under Abiotic Stress
Two-week-old wild type Arabidopsis plants were subjected to drought, cold, and high-salinity stress as described in Section 2. Six smRNA libraries were prepared from pooled samples of stress-treated plants and one pooled sample from nonstressed control plants using 454 DNA sequencing technology as described in Section 2. A total of 480,343 reads were obtained from these seven libraries. The smRNA sequences (17–30 nt) were used for further analysis. After the smRNA sequence data were assembled to unique sequences in each library and they were mapped to the Arabidopsis genome, resulting in 59,284 sequences that were perfectly matched to at least one locus (Table 1). The sequences represented 12,028 unique signatures in whole 7 libraries. Approximately 39% (4,681) of the unique signatures were represented by a single sequence. The smRNAs were classified based on their mapped genomic position. The composite profiles of the types of identified smRNAs were different in each stress treatment (Supplemental Figure 1; Supplementary Material available online at http://dx.doi.org/10.1155/2014/303451). The percentile of smRNAs mapped to the sense strand of AGI code genes was higher in drought-stress and high-salinity-stress treated samples than in the nonstressed control sample, suggesting that mRNA degradation occurs preferentially under these stress conditions.
Table 1.
Number of smRNA-seqs and smRNA loci in each treatment.
| Treatment | Number of sequencesa | Number of locib |
|---|---|---|
| Drought | ||
| 1–5 h | 17,758 | 4,574 |
| 6–10 h | 10,367 | 3,138 |
| Cold | ||
| 1–5 h | 2,104 | 1,595 |
| 6–10 h | 2,869 | 2,018 |
| High salinity | ||
| 1–5 h | 10,247 | 3,645 |
| 6–10 h | 6,148 | 2,417 |
| No treatment | 9,791 | 4,162 |
asmRNA sequences were mapped on Arabidopsis genome. Sequences of tRNAs, rRNAs snoRNAs, and snRNAs were eliminated. bsmRNAs mapped within less than 150 nt distance were grouped as the same loci.
3.2. Identification of Stress-Responsive miRNAs
Deep RNA sequencing analysis of smRNAs identified signatures of various miRNAs, including those previously reported to be stressresponsive miRNAs in Arabidopsis (Supplemental Figure 2). Expression of miR169 was downregulated in response to drought stress (Supplemental Figure 2). Downregulation of miR169 by drought has also been previously reported and demonstrated to be required for the acquisition of drought stress tolerance [19]. Expression of miR156 and miR319 was upregulated by salinity stress and miR408 expression was upregulated by cold stress (Supplemental Figure 2). These results are also consistent with a previous report [20].
3.3. Accumulation of ta-siRNA Expression
A further analysis of smRNA sequences identified in the present study revealed that the number of ta-siRNAs was reduced in response to the stress treatments when compared to the number of ta-siRNAs identified in the nonstressed control (Figure 1(a) and Supplemental Figure 2). The precursors of tasiRNA are transcribed as ncRNAs and processed into siRNAs after a miRNA cleavage event [10–14]. An analysis of Arabidopsis expression profiling data obtained from a previous study utilizing a tiling array [3] indicated that the expression of the TAS1/2/3 family was downregulated under drought and high-salinity stress (Figure 1(b)). Semiquantitative RT-PCR analysis, using primer sets that span the miRNA cleavage sites, showed that the expression of TAS1/2/3 precursors decreased under drought and high-salinity stress (Figure 1(c)). These results are consistent with the ta-siRNA expression profiling data obtained from smRNA sequencing. Collectively, the data indicate that abiotic stress signaling regulates ta-siRNA production through transcriptional regulation of their precursors.
Figure 1.
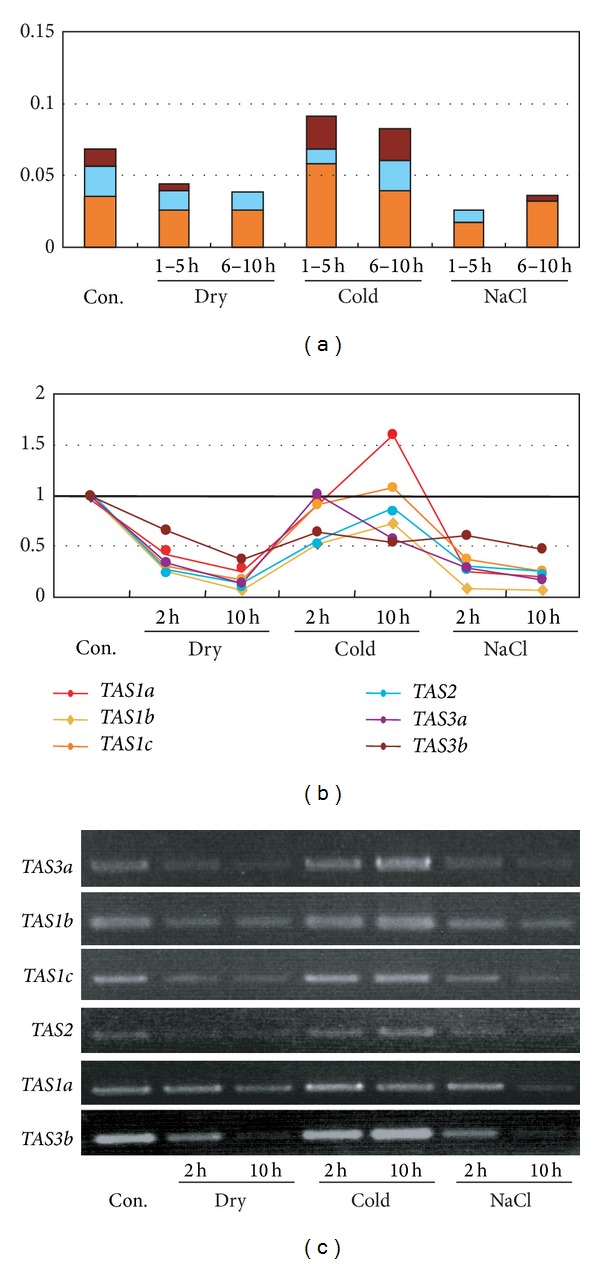
Downregulation of ta-siRNAs and TAS precursors in plants under drought and high-salinity stress. (a) Relative number of ta-siRNA sequences obtained by 454 DNA sequencing of cDNA libraries prepared from small RNAs. Orange, blue, and red boxes represent the relative number of TAS1 ta-siRNA, TAS2 ta-siRNA,and TAS3 ta-siRNA, respectively. (b) Relative expression of TAS precursors was analyzed using previous data from a tiling array [3]. Two-week-old Arabidopsis plants were subjected to drought (2 hr, 10 hr), cold (2 hr, 10 hr), and high-salinity (2 hr, 10 hr) (see Section 2). (c)PCR primer sets (Supplemental Table 1) spanning miRNA cleavage site were designed to determine the expression profiles of TAS precursors. RNA samples were isolated from drought (2 hr, 10 hr), cold (2 hr, 10 hr), and high-salinity (2 hr, 10 hr) treated wild-type plants and nonstress controls. Expression of TAS precursors was analyzed by semiquantitative RT-PCR.
It is known that ta-siRNA-ARF guides the cleavage of ARF3 and ARF4 mRNAs [10, 11, 14, 15]. Therefore, the accumulation of TAS3 precursor, ta-siRNA-ARF, MIR390, ARF3, and ARF4 in wild-type plants and a ta-siRNA synthesis mutant, rdr6, in response to a five-hour-drought stress treatment was measured using RT-qPCR in order to better understand the function of ta-siRNA in abiotic stress response (Figure 2). Expression of the TAS3a precursor and RDR6 was downregulated under drought stress (Figure 2). The TAS3a expression data is consistent with the tiling array data (Figure 1(b)) [3] and results obtained by semiquantitative RT-PCR (Figure 1(c)). Expression of ARF3 and ARF4 was downregulated under drought stress in wild-type plants but the level of downregulation in rdr6 mutants was much less (Figure 2). These results suggest that effective downregulation of ARF3/ARF4 mRNAs under drought stress occurs by degradation activity of tasiRNA-ARF (Figure 2) and transcriptional repression of ARF3/ARF4 mRNAs (Figure 2) under drought stress. Downregulation of tasiRNA-ARF might function in fine-tuning quantitative expression of ARF3/ARF4 under drought stress.
Figure 2.
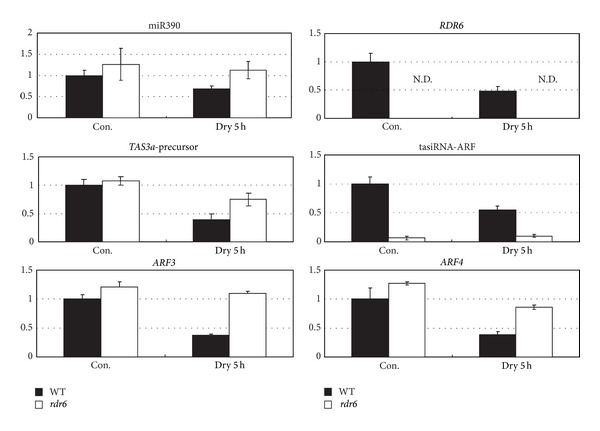
Expression profiles of miR390, TAS3 precursor, ARF3, and ARF4 in plants under drought stress. Expression profiles of tasiRNA-ARF pathway-related genes were analyzed in rdr6 mutants and wild-type plants subjected to a 5 hr drought stress and nonstressed conditions by RT-qPCR. The bar graphs indicate relative expression compared to ACT2. Values represent the mean and standard deviation of three experiments.
3.4. ta-siRNA Generation-Related Mutants Fail to Self-Pollinate due to Modifications in Flower Architecture
In order to identify the biological function of ta-siRNA under environmental stress, a moderate drought stress was applied to a variety of Arabidopsis mutants that are deficient in ta-siRNA biosynthesis (rdr6, sgs3, dcl4, and ago7). The moderate drought stress was applied to two-week-old plants grown in soil by withholding water for one week, resulting in an approximate 20% decrease in soil water content (Supplemental Figure 3(a)). Following the drought treatment, the plants were rewatered. After recovery, wild-type plants produced a number of seeds that was similar to the numbers produced in nonstressed control plants (Figures 3(a) and 3(b)). On the other hand, ta-siRNA mutants, such as rdr6, sgs3, dcl4, and ago7, produced a lower number of seeds after recovery from the drought stress compared to the numbers produced in nonstressed ta-siRNA mutants (Figures 3(a) and 3(b) and Supplemental Figure 3(b)).
Figure 3.

Modification of flower architecture and reduction in seed number in the ta-siRNA biosynthesis mutant, rdr6, under drought stress. (a) Two-week-old rdr6 and wild-type plants were subjected to drought stress treatment for one week and then rewatered. Seed number in the 1st through the 5th siliques was counted (n = 12). Siliques were numbered starting at the basal end of the main shoot. (b) Siliques in plants subjected to drought stress followed by rewatering. (c) Floral architecture in drought-stress and nonstressed wild-type, rdr6, ARF3pro:ARF3, and ARF3pro:ARF3mut plants. Flowers in rdr6 and ARF3pro:ARF3 plants have a slightly exerting stigma phenotype under nonstressed conditions. In contrast, flowers in rdr6 and ARF3pro:ARF3mut plants under drought stress exhibit short stamens, as shown as white arrows. (d) Average length of the stigmas and stamens in drought-stressed and nonstressed WT, rdr6, ARF3pro:ARF3, and ARF3pro:ARF3mut plants.
The reproduction failure phenotype was further investigated and it was found that the reproduction failure of rdr6 plants under drought stress could be rescued by artificial pollination (Supplemental Figure 3(c)). These data suggested that the reproduction failure under drought stress was due to insufficient contact between the anther and the stigma. Abnormal floral architecture was observed in stage 13 flowers of rdr6 plants, which had shorter stamens relative to the length of the stigma (Figure 3(c)). The length of stigmas and stamens in stage 13 flowers was examined in rdr6 and wild-type plants under drought stress and nonstressed conditions. Stigma lengthin rdr6 and wild-type plants was similar in plants under drought stress and nonstressed conditions. In contrast, the length of rdr6 stamens was shorter in plants under drought stress compared to the length of stamens in wild-type plants under drought stress (Figures 3(c) and 3(d)). Although rdr6 plants exhibited a tendency to produce slightly shorter stamens, relative to wild-type plants, even in nonstressed conditions, the difference was not statistically significant (Figures 3(c) and 3(d)). These data were consistent with a previous report [21].
It has been demonstrated that tasiRNA-ARF negatively regulates ARF3 and ARF4, both of which are genes that control flower organ identity [22–24]. The ability of tasiRNA-ARF to regulate correct stamen development under drought stress was examined using ARF3pro:ARF3mut. This transgenic genotype has a mutated ARF3 sequence that confers the ability of ARF3 to avoid tasiRNA-ARF targeted degradation and still translate ARF3 protein [15]. Similar to rdr6 plants, the ARF3pro:ARF3mut exhibited a short stamen phenotype in plants subjected to drought stress (Figures 3(c) and 3(d)), suggesting that the tasiRNA-ARF pathway has an important role in regulating stamen length under drought stress. Additionally, reproductive failure and the short stamen phenotype were also observed in rdr6 mutants and ARF3pro:ARF3mut plants that were subjected to high-salinity stress (Supplemental Figures 3(d), 3(e), and 3(f)). These results suggest that the tasiRNA-ARF pathway plays a critical role in the regulation of floral organ development under both drought and high-salinity stress.
3.5. RDR6 Functions in the Stabilization of Stress-Dependent Changes in the Expression of Floral Development-Related and Auxin-Related Genes in Plants under Drought Stress
Gene expression profiles in flower buds of Arabidopsis plants under moderate drought stress and nonstressed conditions were analyzed using a microarray in order to better understand the regulation of the ta-siRNA-mediated network in response to drought stress. Results identified 513 (30.3%) genes whose level of expression was significantly different in rdr6 plants compared to wild-type plants. The list of genes overlapped with drought stress-responsive genes (Figure 4(a), Supplemental Table 2). No correlation (R 2 = 0.09) was observed between the expression ratios of drought/control and those of rdr6/wild-type for the GO category of water deprivation response-related genes (GO:0009414) (Figure 4(b)). rdr6 and wild-type plants showed similar drought stress-responsive expression in the water deprivation response-related genes (Supplemental Figure 4(a)). These results suggest that a similar level of drought stress was applied to rdr6 and wild-type plants.
Figure 4.
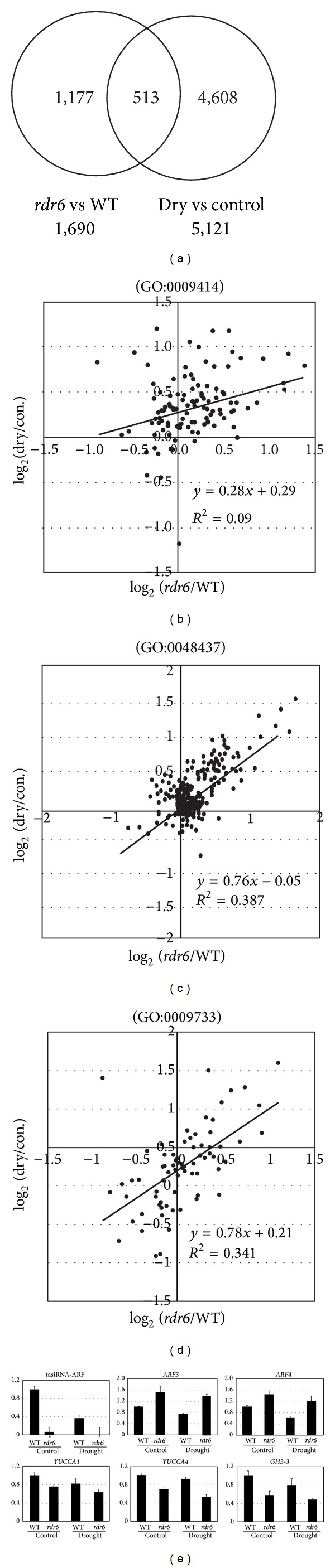
Microarray analysis of flower buds in plants under drought stress. (a) Venn diagram of differentially expressed genes in RDR6 plants compared to wild-type plants and drought stress-responsive genes. Statistically significant differentially expressed genes were identified based on the following criteria: FDR of 2-way ANOVA (rdr6 versus WT or drought-stressed versus nonstressed) < 0.075. ((b)–(d)) Scatter plot analysis of genes from different categories of GO terms. Horizontal axis represents log2 ratio of (rdr6 non-stressed + rdr6 drought)/(WT nonstressed + WT drought). Vertical axis represents log2 ratio of (WT drought+ rdr6 drought)/(WT nonstressed + rdr6 non-stressed). (b) Scatter plot analysis of water deprivation response-related genes. (c) Scatter plot analysis of floral organ development-related genes. (d) Scatter plot analysis of auxin response-related genes. (e) RT-qPCR expression profiles of tasiRNA-ARF, ARF3, ARF4, GH3-3, YUCCA1, and YUCCA4 in flower buds of rdr6 mutant and wild-type plants subjected to drought stress or nonstressed control.
Among the differentially expressed genes in rdr6 plants were a number of floral development-related genes (Supplemental Table 3). Expression levels of the C-class homeotic gene AGAMOUS [25] and AGAMOUS downstream genes that promote the development of stigma, style, and medial tissue of ovules, such as SHATTERPROOF 1 and SHATTERPROOF 2 [26], and the stigma and stamen identity gene, SUPERMAN [27], were upregulated under drought stress and their upregulation was affected in rdr6 mutants (Supplemental Table 3). In contrast, expression of the E-class organ identity gene, SEPALLATA 4 [28], and the petal identity gene PETALLOSS [29] was downregulated in response to drought stress in both wild-type and rdr6 mutants; however, their expression was significantly lower in rdr6 mutants relative to wild-type plants (Supplemental Table 3). These results indicate that the RDR6 mutation affects organ whorl identity genes and their downstream genes. It seems that tasiRNA-ARF regulation represses central floral organ development and enhances peripheral floral development under drought stress.
To better understand the relationship between drought stress response and the RDR6 mutation on flower development-related genes (GO:0048437), the expression ratios of drought/control and those of rdr6/wild-type were compared. A moderate positive correlation (R 2 = 0.387) was observed between the drought response and RDR6 mutation (Figure 4(c)). These data suggest that tasiRNA-ARF is involved in fine-tuning the expression of floral development-related genes in plants subjected to drought stress.
4. Discussion
The results of the present study demonstrate that tasiRNA-ARF functions in keeping correct flower architecture which is critical to self-pollination under drought and high-salinity stress. Various abiotic stresses, such as heat, high-salinity, drought, and cold, induce reproductive failure in plants [30–35]. This failure is the result of morphological abnormalities that arise during various stages of floral development. Molecular mechanisms responsible for the abortion of pollen development have been well-characterized [30–35]. Although defects in stamen development in plants under drought, high-salinity, and heat stress have been reported [32, 34, 35], the molecular mechanisms that protect floral development from the adverse effects of abiotic stress, however, remain unclear.
tasiRNA-ARF is required for the normal development of lateral organs, such as leaves, lateral roots, and flowers [14, 21, 36, 37]. For example, rdr6 mutant exhibits altered stamen and pistil elongation that results in variable seed production and supports the premise that reproduction in rdr6 plants is sensitive to growth conditions [22]. Interestingly, a mutation of RDR6 enhanced self-incompatibility in a transgenic, self-incompatible, Arabidopsis thaliana system [38]. These results suggest that tasiRNA-ARF functions as a key mediator for maintaining the correct pattern of flower architecture, as well as their development. ARF3 and ARF4 function in central organ identity in flowers and apical-basal patterning defects in the gynoecium [22–24].
ARFs regulate the expression of auxin-responsive genes by binding specifically to auxin response elements (AuxRE) [39]. Regarding auxin response-related genes (GO:0009733), a moderate positive correlation was observed between drought stress response and the effect of the RDR6 mutation (Figure 4(d)). The genes downregulated by both the drought stress treatment and the RDR6 mutation included the auxin-induced conjugating enzymes, GH3-2, GH3-3, GH3-6, GH3-10, and an auxin-responsive GH3-like protein (AT1G48660) (Supplemental Table 2). In addition, microarray analysis also identified that an expression of auxin biosynthesis-related gene, YUCCA 4, was significantly downregulated by drought stress and the RDR6 mutation (Supplemental Table 2). After conducting a closer study of 4 YUCCA genes that are mainly expressed in flowers [40], YUCCA 1 and YUCCA 4 were downregulated by drought stress and RDR6 mutation and YUCCA 2 and YUCCA 6 were downregulated by RDR6 mutation (Supplemental Figure 4(b)). The regulation of tasiRNA-ARF and mRNAs of ARF3, GH3-3, YUCCA 1, and YUCCA 4 in flower buds was confirmed by RT-qPCR (Figure 4(e)). The previous report showed that expression of an auxin reporter, DR5:GUS, was decreased in ARF3pro:ARF3mut plants [41]. These results suggest that the auxin biosynthesis and auxin response were attenuated in floral development by drought stress and loss of tasiRNA-ARF regulation. It was known that auxin signaling was important for floral development. The pin-shaped flower of yucca1/4 was similar to the flowers produced in ARF3- or ARF4-overexpressing plants [40]. Experiments utilizing an auxin transport inhibitor indicate that ARF3 functions as a modulator of auxin response during floral development [42]. There is a possible hypothesis that tasiRNA-ARF controls floral development by maintaining the proper level of auxin signaling under drought stress (Figure 5).
Figure 5.
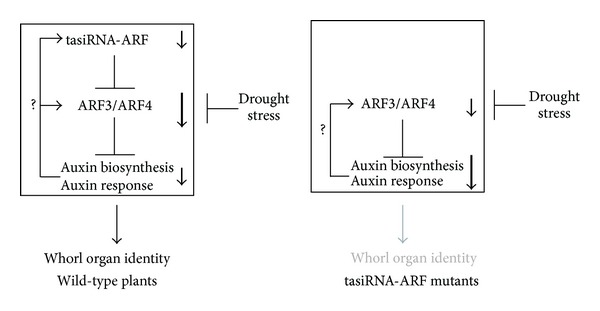
Proposed model for the fine-tuning of whorl architecture in flowers subjected to environmental stress via tasiRNA-ARF. tasiRNA-ARF is required for normal morphogenesis of floral whorl organs in plants subjected to drought stress. tasiRNA-ARF acts by modulating the expression of floral development-related genes in plants subjected to drought stress. Expression of YUCCA4 is downregulated by drought stress and in the RDR6 mutation, suggesting that auxin biosynthesis is modulated by tasiRNA-ARF.
Both tasiRNA-ARF and ARF3/4 were downregulated under drought stress, suggesting that tasiRNA-ARF are required for quantitative adjustment of ARF3/4 expression. Positive feedback regulation of auxin signaling might also function in the regulation of these genes (Figure 5). It is known that initiation of lateral roots modulated by positive and negative feedback regulation between tasiRNA-ARF and ARF2/3/4 through auxin signaling [38, 39]. ARF3 expression was also induced by increased auxin biogenesis through upregulation of YUCCA 4 in shoot initiation [43]. These previous reports invoke that drought stress affects tasiRNA-ARF regulatory network, but it remained unclear.
To search candidate genes connecting abiotic stress and tasiRNA-ARF regulatory network, microarray coexpression analysis of ARF3 (P < 0.01) and ARF4 (P < 0.01) was performed. 155 genes coexpressed with ARF3 and ARF4 involved twenty-five abiotic stress response-related genes, such as DREB2C [44], DWD (DDB1-binding WD40 protein) [45], ascorbate peroxidase 1 [46], glutathione S-transferase [47], and RD21B [48] (Supplemental Table 4). Thegenes also included ta-siRNA pathway-related genes, such as RDR6, TAS3, other TAS genes, and ta-siRNA target genes [49]. Sixty-four of the 155 genes possessed an ARF binding motif (tgtctc) [50] in the promoter region within 1kb upstream of the start codon (Supplemental Table 4). These genes may represent candidates that connect tasiRNA-ARF regulatory network and drought stress signaling pathway.
In conclusion, this study demonstrates that tasiRNA-ARF acts as a central modifier, negatively regulating changes in the expression of floral development-related genes in plants under drought and assists in maintaining normal floral morphogenesis.
Supplementary Material
Supplemental Figure 1. Distribution of smRNAs in abiotic stress-treated plants. smRNA sequences were classified based on where they mapped in the Arabidopsis genome (TAIR8). The circular graphs illustrate the distribution of mapped smRNA sequences obtained from plants subjected to each different treatment. The miRNAs and ta-siRNAs indicate that smRNA sequences are derived from MIRNA loci and TAS loci. Small RNAs mapped to regions coding for proteins were classified into two groups, smRNAs mapped to the sense strand and smRNAs mapped to the antisense strand of protein-coding genes. Transposable element (TE) genes and pseudogenes are AGI coded genes annotated as TEs and pseudo genes, respectively. Intergenic TEs represent smRNAs mapped to the TEs in intergenic regions.
Supplemental Figure 2. Deep sequencing of miRNAs and ta-siRNAs in non-stressed and stress-treated plants. The number of RNA sequences was normalized by dividing by the total number of obtained miRNA sequences. P-values were calculated using an enrichment test on each miRNA fraction, based on their hypergeometric distribution. Asterisk in the table indicates the p-value was lower than 10-3. Previously reported stress- upregulated or downregulated smRNAs are shown in red or blue, respectively.
Supplemental Figure 3. Seed production in ta-siRNA biosynthesis mutants, rdr6, sgs3, dcl4 and ago7, subjected to drought stress, and in rdr6 and ARF3pro:ARFmut mutants subjected to high-salinity stress. (a) Soil water content during the course of the imposed drought treatment. (b) Seed numbers in the 1st silique (n =12). (c) Seed number in the 1st silique of plants subjected to a drought stress after artificial pollination and self-pollination (n =12). (d) Seed numbers in the 1st silique of three-week-old plants grown in soil and treated with 100 mM NaCl for one week (n =12). (e) Short siliques were observed in rdr6 and ARF3pro:ARF3mut plants subjected to high-salinity stress. (f) Short stamens were observed in rdr6 and ARF3pro:ARF3mut plants subjected to high-salinity stress.
Supplemental Figure 4. Scatter plot analysis for drought stress-responsive expression in rdr6 and wild-type plants. (a) Scatter plot analysis for drought stress-responsive expression of water deprivation response-related genes (GO:0009414) in rdr6 and wild-type plants. Horizontal axis represents log2 ratio of (drought / non-stressed) in wild-type plants. Vertical axis represents log2ratio of (drought / non-stressed) in rdr6 mutants. (b) Line graph for expressions of YUCCA genes. Horizontal axis represents normalized intensities of microarray probes targeting YUCCA genes.
Supplemental Table 1. List of primer sequences.
Supplemental Table 2. Differentially expressed genes in flower buds of rdr6 and wild-type under drought stress treatment and untreatment.
Supplemental Table 3. Expression profiles of floral organ development-related genes (GO:0048437)
Supplebmental Table 4. Expression profiles of 155 genes co-expressed with ARF3 and ARF4.
Acknowledgments
The authors would like to thank Dr. James C. Carrington for providing seeds of ARF3pro:ARF3 and ARF3pro:ARF3mut and Dr. R. Scott Poethig for providing seeds of ett-15 and arf4-2. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Challenging Exploratory Research 24657041), Japan Science and Technology Agency (JST), and Core Research for Evolutionary Science and Technology (CREST) and grants from the RIKEN, Japan, to Motoaki Seki.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L. Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Science. 2008;174(4):420–431. [Google Scholar]
- 2.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant Journal. 2010;61(6):1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 3.Matsui A, Ishida J, Morosawa T, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant and Cell Physiology. 2008;49(8):1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 4.Rymarquis LA, Kastenmayer JP, Hüttenhofer AG, Green PJ. Diamonds in the rough: mRNA-like non-coding RNAs. Trends in Plant Science. 2008;13(7):329–334. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Laporte P, Merchan F, Amor BB, Wirth S, Crespi M. Riboregulators in plant development. Biochemical Society Transactions. 2007;35(6):1638–1642. doi: 10.1042/BST0351638. [DOI] [PubMed] [Google Scholar]
- 6.Matsui A, Nguyen AH, Nakaminami K, Seki M. Arabidopsis non-coding RNA regulation in abiotic stress responses. International Journal of Molecular Sciences. 2013;14(11):2642–2654. doi: 10.3390/ijms141122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khraiwesh B, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta—Gene Regulatory Mechanisms. 2012;1819(2):137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lima JC, Loss-Morais G, Margis R. MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genetics and Molecular Biology. 2012;35(4):1069–1077. doi: 10.1590/s1415-47572012000600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunkar R, Chinnusamy V, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig S. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis . Development. 2006;133(15):2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa M, Peragine A, Mee YP, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes and Development. 2005;19(18):2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America. 2005;102(36):12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adenot X, Elmayan T, Lauressergues D, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7 . Current Biology. 2006;16(9):927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 15.Fahlgren N, Montgomery TA, Howell MD, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta- siRNA affects developmental timing and patterning in Arabidopsis . Current Biology. 2006;16(9):939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 16.To TK, Nakaminami K, Kim J, et al. Arabidopsis HDA6 is required for freezing tolerance. Biochemical and Biophysical Research Communications. 2011;406(3):414–419. doi: 10.1016/j.bbrc.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B. Methodological. 1995;57(1):289–300. [Google Scholar]
- 18.Chambers JM, Freeny A, Heiberger RM. Statistical Models in S. chapter 5. 1992. Analysis of variance; designed experiments; pp. 145–193. [Google Scholar]
- 19.Li W, Oono Y, Zhu J, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20(8):2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana . RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes and Development. 2004;18(19):2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124(22):4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 23.Sessions RA, Zambryski PC. Arabidopsis gynoecium structure in the wild type and in ettin mutants. Development. 1995;121(5):1519–1532. doi: 10.1242/dev.121.5.1519. [DOI] [PubMed] [Google Scholar]
- 24.Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17(11):2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. The Plant cell. 1989;1(1):37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo M, Brambilla V, Marcheselli R, Caporali E, Kater MM, Colombo L. A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Developmental Biology. 2010;337(2):294–302. doi: 10.1016/j.ydbio.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 27.Sakai H, Medrano LJ, Meyerowitz EM. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature. 1995;378(6553):199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- 28.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Current Biology. 2004;14(21):1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Griffith ME, da Silva Conceiçao A, Smyth DR. PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development. 1999;126(24):5635–5644. doi: 10.1242/dev.126.24.5635. [DOI] [PubMed] [Google Scholar]
- 30.Koonjul PK, Minhas JS, Nunes C, Sheoran IS, Saini HS. Selective transcriptional down-regulation of anther invertases precedes the failure of pollen development in water-stressed wheat. Journal of Experimental Botany. 2005;56(409):179–190. doi: 10.1093/jxb/eri018. [DOI] [PubMed] [Google Scholar]
- 31.Cheptou PO, Berger A, Blanchard A, Collin C, Escarre J. The effect of drought stress on inbreeding depression in four populations of the mediterranean outcrossing plant Crepis sancta (Asteraceae) Heredity. 2000;85(part 3):294–302. doi: 10.1046/j.1365-2540.2000.00759.x. [DOI] [PubMed] [Google Scholar]
- 32.Sulpice R, Tsukaya H, Nonaka H, Mustardy L, Chen THH, Murata N. Enhanced formation of flowers in salt-stressed Arabidopsis after genetic engineering of the synthesis of glycine betaine. Plant Journal. 2003;36(2):165–176. doi: 10.1046/j.1365-313x.2003.01873.x. [DOI] [PubMed] [Google Scholar]
- 33.Satake T, Hayase H. Male sterility caused by cooling treatment at the young microspore stage in rice plants. V. Esti-mation of pollen developmental stage and the most sensitive stage to coolness. Proceedings of the Crop Science Society of Japan. 1970;39:468–473. [Google Scholar]
- 34.Sakata T, Oshino T, Miura S, et al. Auxins reverse plant male sterility caused by high temperatures. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8569–8574. doi: 10.1073/pnas.1000869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su Z, Ma X, Guo H, et al. Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell. 2013;25(10):3785–3807. doi: 10.1105/tpc.113.115428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin E, Jouannet V, Herz A, et al. mir390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22(4):1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon EK, Yang JH, Lim J, Kim SH, Lee WS. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Research. 2009;38(4):1382–1391. doi: 10.1093/nar/gkp1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tantikanjana T, Rizvi N, Nasrallah ME, Nasrallah JB. A dual role for the s-locus receptor kinase in self-incompatibility and pistil development revealed by an arabidopsis rdr6 mutation. Plant Cell. 2009;21(9):2642–2654. doi: 10.1105/tpc.109.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15(2):533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis . Genes & Development. 2006;20(13):1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tantikanjana T, Nasrallah JB. Non-cell-autonomous regulation of crucifer self-incompatibility by Auxin Response Factor ARF3. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(47):19468–19473. doi: 10.1073/pnas.1217343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemhauser JL, Feldman LJ, Zambryski PC. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development. 2000;127(18):3877–3888. doi: 10.1242/dev.127.18.3877. [DOI] [PubMed] [Google Scholar]
- 43.Cheng ZJ, Wang L, Sun W, et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiology. 2013;161(1):240–251. doi: 10.1104/pp.112.203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SJ, Kang JY, Park HJ, et al. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiology. 2010;153(2):716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JH, Yoon HJ, Terzaghi W, et al. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell. 2010;22(6):1716–1732. doi: 10.1105/tpc.109.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiology. 2007;144(4):1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sappl PG, Carroll AJ, Clifton R, et al. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. The Plant Journal. 2009;58(1):53–68. doi: 10.1111/j.1365-313X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana . Molecular & General Genetics. 1993;238(1-2):17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]
- 49.Meng C, Chen J, Ding S, Peng J, Wong S. Hibiscus chlorotic ringspot virus coat protein inhibits trans-acting small interfering RNA biogenesis in Arabidopsis. Journal of General Virology. 2008;89(9):2349–2358. doi: 10.1099/vir.0.2008/002170-0. [DOI] [PubMed] [Google Scholar]
- 50.Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant Journal. 1999;19(3):309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Distribution of smRNAs in abiotic stress-treated plants. smRNA sequences were classified based on where they mapped in the Arabidopsis genome (TAIR8). The circular graphs illustrate the distribution of mapped smRNA sequences obtained from plants subjected to each different treatment. The miRNAs and ta-siRNAs indicate that smRNA sequences are derived from MIRNA loci and TAS loci. Small RNAs mapped to regions coding for proteins were classified into two groups, smRNAs mapped to the sense strand and smRNAs mapped to the antisense strand of protein-coding genes. Transposable element (TE) genes and pseudogenes are AGI coded genes annotated as TEs and pseudo genes, respectively. Intergenic TEs represent smRNAs mapped to the TEs in intergenic regions.
Supplemental Figure 2. Deep sequencing of miRNAs and ta-siRNAs in non-stressed and stress-treated plants. The number of RNA sequences was normalized by dividing by the total number of obtained miRNA sequences. P-values were calculated using an enrichment test on each miRNA fraction, based on their hypergeometric distribution. Asterisk in the table indicates the p-value was lower than 10-3. Previously reported stress- upregulated or downregulated smRNAs are shown in red or blue, respectively.
Supplemental Figure 3. Seed production in ta-siRNA biosynthesis mutants, rdr6, sgs3, dcl4 and ago7, subjected to drought stress, and in rdr6 and ARF3pro:ARFmut mutants subjected to high-salinity stress. (a) Soil water content during the course of the imposed drought treatment. (b) Seed numbers in the 1st silique (n =12). (c) Seed number in the 1st silique of plants subjected to a drought stress after artificial pollination and self-pollination (n =12). (d) Seed numbers in the 1st silique of three-week-old plants grown in soil and treated with 100 mM NaCl for one week (n =12). (e) Short siliques were observed in rdr6 and ARF3pro:ARF3mut plants subjected to high-salinity stress. (f) Short stamens were observed in rdr6 and ARF3pro:ARF3mut plants subjected to high-salinity stress.
Supplemental Figure 4. Scatter plot analysis for drought stress-responsive expression in rdr6 and wild-type plants. (a) Scatter plot analysis for drought stress-responsive expression of water deprivation response-related genes (GO:0009414) in rdr6 and wild-type plants. Horizontal axis represents log2 ratio of (drought / non-stressed) in wild-type plants. Vertical axis represents log2ratio of (drought / non-stressed) in rdr6 mutants. (b) Line graph for expressions of YUCCA genes. Horizontal axis represents normalized intensities of microarray probes targeting YUCCA genes.
Supplemental Table 1. List of primer sequences.
Supplemental Table 2. Differentially expressed genes in flower buds of rdr6 and wild-type under drought stress treatment and untreatment.
Supplemental Table 3. Expression profiles of floral organ development-related genes (GO:0048437)
Supplebmental Table 4. Expression profiles of 155 genes co-expressed with ARF3 and ARF4.


