Abstract
ERCC2 is indispensable for nucleotide excision repair pathway, and its functional polymorphisms may be associated with cancer risk. In a large case-control study of 1126 esophageal squamous cell carcinomas (ESCC) patients and 1131 controls, we genotyped two SNPs in ERCC2 (rs238406 G > T and rs13181 T > G) and assessed their associations with ESCC risk. We found a significantly elevated ESCC risk associated with the rs238406 T variant genotypes (adjusted OR = 1.30 and 1.24, 95% CI = 1.02–1.66 and 1.03–1.49 for TG and TG/TT, respectively, compared with GG), particularly in the subgroup of those smoked more than 16 pack-years. Multivariate logistic regression analysis suggested a possible multiplicative gene-environment interaction between rs238406 genotypes and smoking (Pinteraction = 0.026) on ESCC risk. Although no significant risk associations were observed for rs13181, further mini meta-analysis with our and 18 other published studies of 5,012 cases and 8,238 controls found evidence of an association between the rs13181 variant G allele and esophageal cancer risk (TG/GG vs. TT, OR = 1.17; 95% CI = 1.02–1.33). Interestingly, we consistently found a significant correlation between variant genotypes of these two SNPs and ERCC2 mRNA expression. These findings suggest that potentially functional SNPs in ERCC2 may contribute to ESCC risk.
China is one of the countries with the highest incidence and mortality of esophageal cancer in the world. Esophageal squamous cell carcinomas (ESCC) account for 90% of all the cases in China1. Epidemiological studies have revealed that tobacco smoking, alcohol intake, nutritional deficiencies and dietary carcinogen exposure may contribute to the etiology of ESCC1, but only a small proportion of exposed individuals actually develop esophageal cancer, suggesting that genetic factors may also play a vital role in susceptibility to ESCC.
Internal and external environmental exposures, including physical, chemical, and biological carcinogens, can damage cellular DNA, resulting in changes of DNA structure and sequences, thus increasing genomic instability of proliferating cells2. At least four main, partly overlapping DNA repair pathways exist in humans for repairing DNA damage2. In mammals, nucleotide excision repair (NER) is the most versatile DNA repair mechanism responsible for removing a wide variety of helix-distorting lesions that intervene in base pairing and generally destruct transcription and normal replication2. Therefore, reduced DNA repair capacity (DRC) may confer susceptibility to cancer. It is widely recognized that the variation of individual DRC is determined by genetic factors, such as functional single nucleotide polymorphisms (SNPs) of the core genes in DNA repair pathways, which may be the molecular mechanisms underlying the inter-individual variation of DRC in the general population.
Excision repair cross complementing group 2 (ERCC2, also known as xeroderma pigmentosum complementation D, XPD), is one of the core genes involved in transcription-coupled NER2. The protein encoded by this gene has evolutionarily conserved ATP-dependent DNA helicase activity, responsible for unwinding DNA around the lesion site, a crucial step to initiate the NER process3. The ERCC2 protein also participates in DNA transcription as an integral member of the basal transcription factor BTF2/TFIIH complex4. The importance of ERCC2 is highlighted by the existence of three different disorders that are caused by hereditary defects in this protein, including cancer-prone syndrome xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy4.
Since the association of the ERCC2 Lys751Gln (rs13181) polymorphism with ESCC risk was first reported in 20025, there are additional investigations of the association between Lys751Gln and risk of ESCC among different ethnicities6,7,8, but the results have been mixed or conflicting, likely due to a relatively small sample size in each of the published studies. Interestingly, published genome-wide association studies (GWASs) of ESCC in Chinese populations did not identify rs13181 SNP as a susceptible locus9,10,11,12, perhaps owing to the stringent P values required to avoid false-positive findings, which dramatically decrease the possibility to reveal the modest effect of some common SNPs on risk of cancer, particularly for those SNPs that are potentially functional. Therefore, in the present study, we further investigated the association of two potentially functional ERCC2 SNPs, including rs13181, with ESCC risk in a large study of an Eastern Chinese population. In addition, we also explored the molecular mechanisms underlying the positive associations.
Results
Population characteristics
Population characteristics were described previously13. In brief, the cases and controls were adequately matched by age and sex. However, there were a higher proportion of smokers, drinkers and BMI < 25.0 in the cases than in the controls, which were further adjusted for in later multivariate logistic regression analyses (Supplemental Table 1).
Association between ERCC2 SNPs and ESCC risk
All the observed genotype frequencies for ERCC2 SNPs agreed with the Hardy-Weinberg equilibrium in the controls (P > 0.05). In the single-locus analyses, we found a significantly elevated ESCC risk associated with the rs238406 T variant genotypes (adjusted odds ratio (OR) = 1.30 and 1.24, 95% confidence interval (CI) = 1.02–1.66 and 1.03–1.49 for TG and TG/TT, respectively, compared with GG). However, these significant risk associations were not observed for the rs13181 SNP (Table 1).
Table 1. Genotype frequencies of the ERCC2 SNPs and their association with risk of ESCC.
| Cases a | Controls a | Crude OR | Adjusted OR | |||
|---|---|---|---|---|---|---|
| Variants | No. (%) | No. (%) | P b | (95% CI) | (95% CI) b | P c |
| ERCC2 rs238406, HWE = 0. 066g | ||||||
| GG | 325 (28.97) | 374 (33.66) | 0.057d | 1.00 | 1.00 | |
| TG | 558 (49.73) | 515 (46.35) | 1.25 (1.03–1.51) | 1.22 (1.00–1.48) | 0.053 | |
| TT | 239 (21.30) | 222 (19.98) | 1.24 (0.98–1.57) | 1.30 (1.02–1.66) | 0.038 | |
| TG/TT | 797 (71.03) | 737 (66.34) | 0.017e | 1.24 (1.04–1.49) | 1.24 (1.03–1.49) | 0.024 |
| ERCC2 rs13181, HWE = 0. 413g | ||||||
| TT | 937 (83.51) | 954 (85.87) | 0.300d | 1.00 | 1.00 | |
| TG | 175 (15.60) | 149 (13.41) | 1.20 (0.94–1.51) | 1.16 (0.42–1.48) | 0.229 | |
| GG | 10 (0.89) | 8 (0.72) | 1.27 (0.50–3.24) | 1.10 (0.42–2.89) | 0.844 | |
| TG/GG | 185 (16.49) | 157 (14.13) | 0.122e | 1.20 (0.95–1.51) | 1.16 (0.91–1.47) | 0.227 |
| No. of at-risk genotypes f | ||||||
| 0 | 248 (22.10) | 297 (26.73) | 0.010 | 1.00 | ||
| 1 | 766 (68.27) | 734 (66.07) | 1.25 (1.03–1.52) | 1.23 (1.00–1.50) | 0.050 | |
| 2 | 108 (9.63) | 80 (7.20) | 1.62 (1.16–2.26) | 1.56 (1.10–2.21) | 0.012 | |
| Ptrend = 0.007 | ||||||
| Dichotomized groups | ||||||
| 0 | 248 (22.10) | 297 (26.73) | 0.011 | 1.00 | ||
| ≥1 | 874 (77.90) | 814 (73.27) | 1.28 (1.06–1.56) | 1.26 (1.03–1.54) | 0.024 | |
aThe numbers of each single nucleotide polymorphism were less than the total number of subjects because some genotyping data were unavailable.
bTwo sides Chi-square test for genotype distributions between cases and controls.
cAdjusted for age, sex, BMI, smoking status, and drinking status in logistic regression models.
dFor additive genetic models.
eFor dominant genetic models.
fThe risk-genotypes used for the calculation were ERCC2 rs238406 TG/TT + rs13181 TG/GG
gA goodness-of-fit χ2 test for Hardy Weinberg equilibrium (HWE) for genotype distribution in controls.
In the combined analysis, we categorized all putative risk (OR > 1.0) genotypes from each SNP into a new variable according to the number of risk genotypes (i.e., ERCC2 rs238406 TG/TT + rs13181 TG/GG). As a result, we found that individuals carrying “≥1” risk unfavorable genotypes exhibited an increased ESCC risk (adjusted OR = 1.26, 95% CI = 1.03–1.54), compared with those carrying “0” unfavorable genotypes. Such a cumulative effect was dose-dependent, because the risk of ESCC significantly increased with an increasing number of the observed risk genotypes (adjusted OR = 1.23, 95% CI = 1.00–1.50 for one risk genotype; adjusted OR = 1.56, 95% CI = 1.10–2.21 for two risk genotypes; Ptrend = 0.007) (Table 1).
In the stratified analysis, we found that those who carried the rs238406 variant TG/TT genotypes had a significant increased risk, particularly in males (adjusted OR = 1.26, 95% CI = 1.02–1.55), subjects with the cumulative smoking dose >16 pack-years (adjusted OR = 1.58, 95% CI = 1.17–2.13) and subjects with BMI ≥ 25.0 (adjusted OR = 1.40, 95% CI = 1.06–1.84). We also found that the increased risk was more evident in older subjects who carried the rs13181 TG/GG variant genotypes (adjusted OR = 1.44, 95% CI = 1.03–2.01). Likewise, we found similar results for those carrying “≥1” risk genotypes among subgroups of older subjects, males, never-drinkers and subjects with the cumulative smoking dose >16 pack-years. However, further homogeneity tests suggested that there were no differences among all strata, except for subjects with the cumulative smoking dose >16 pack-years by the rs238406 TG/TT genotypes (P for homogeneity = 0.036) (Table 2), suggesting a possible interaction. Indeed, we did find a statistical evidence for a multiplicative gene-environment interaction between rs238406 genotypes and smoking (Pinteraction = 0.026) on ESCC risk.
Table 2. Stratification analysis for associations between ERCC2 SNPs and ESCC risk.
| rs238406 | rs13181 | Combined effect of risk genotypes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cases/controls) | (cases/controls) | (cases/controls) | ||||||||||
| Variables | GG | TG/TT | Adjusted OR (95% CI) a | P b | TT | TG/GG | Adjusted OR (95% CI) a | P b | 0 | ≥1 | Adjusted OR (95% CI) a | P b |
| Age, yr (median) | ||||||||||||
| ≤60 | 158/178 | 402/380 | 1.27 (0.97–1.66) | 0.573 | 477/476 | 83/82 | 0.89 (0.63–1.26) | 0.158 | 128/141 | 432/417 | 1.17 (0.87–1.56) | 0.218 |
| >60 | 167/196 | 395/357 | 1.26 (0.97–1.63) | 460/478 | 102/75 | 1.44 (1.03–2.01) | 120/156 | 442/397 | 1.39 (1.05–1.85) | |||
| Sex | ||||||||||||
| Males | 259/290 | 646/571 | 1.26 (1.02–1.55) | 0.708 | 757/740 | 148/121 | 1.16 (0.88–1.52) | 0.943 | 199/231 | 706/630 | 1.26 (1.01–1.58) | 0.820 |
| Females | 66/84 | 151/166 | 1.24 (0.81–1.89) | 180/214 | 37/36 | 1.18 (0.69–2.02) | 49/66 | 168/184 | 1.31 (0.83–2.08) | |||
| Pack-years | ||||||||||||
| 0 | 133/155 | 295/356 | 0.94 (0.70–1.25) | 0.036 | 363/444 | 65/67 | 1.20 (0.82–1.76) | 0.717 | 107/126 | 321/385 | 0.97 (0.71–1.31) | 0.050 |
| ≤16 (mean) | 37/85 | 115/156 | 1.53 (0.95–2.47) | 126/210 | 26/31 | 1.38 (0.77–2.50) | 27/69 | 125/172 | 1.60 (0.95–2.70) | |||
| >16 (mean) | 155/134 | 387/225 | 1.58 (1.17–2.13) | 448/300 | 94/59 | 1.06 (0.73–1.55) | 114/102 | 428/257 | 1.58 (1.14–2.20) | |||
| Drinking status | ||||||||||||
| Never | 173/245 | 451/504 | 1.26 (0.99–1.60) | 0.935 | 525/648 | 99/101 | 1.21 (0.89–1.65) | 0.797 | 131/197 | 493/552 | 1.33 (1.02–1.72) | 0.782 |
| Ever | 152/129 | 346/233 | 1.25 (0.92–1.69) | 412/306 | 86/56 | 1.03 (0.70–1.52) | 117/100 | 381/262 | 1.20 (0.87–1.66) | |||
| BMI | ||||||||||||
| <25.0 | 209/155 | 505/335 | 1.14 (0.89–1.47) | 0.253 | 591/427 | 123/63 | 1.38 (0.99–1.93) | 0.165 | 154/124 | 560/366 | 1.25 (0.95–1.64) | 0.748 |
| ≥25.0 | 116/219 | 292/402 | 1.40 (1.06–1.84) | 346/527 | 62/94 | 1.01 (0.71–1.43) | 94/173 | 314/448 | 1.31 (0.98–1.75) | |||
aObtained in logistic regression models with adjustment for age, sex, BMI, smoking status and drinking status.
bP for heterogeneity test using the Chi-square-based Q test.
We finally calculated the false-positive report probability (FPRP) values for all observed significant findings. With the assumption of a prior probability of 0.1, the FPRP values were 0.136, 0.192, 0.091, 0.096 and 0.094, respectively, for associations of rs238406 TG/TT genotypes, 1 combined risk genotype, 2 combined risk genotypes and “≥1” combined risk genotypes with an increased risk of ESCC in all subjects, as well as in the subgroups of the cumulative smoking dose >16 pack-years. The FPRP values were all <0.20, suggesting that these significant associations were noteworthy (Table 3).
Table 3. False-Positive Report Probability Values for associations between ESCC risk and genotypes of ERCC2 polymorphisms.
| Prior probability | ||||||||
|---|---|---|---|---|---|---|---|---|
| Genotypes | Positive OR (95% CI) a | P value b | Statistical Power c | 0.2500 | 0.1000 | 0.0100 | 0.0010 | 0.0001 |
| rs238406 | ||||||||
| TT vs. GG | 1.24 (0.98–1.57) | 0.075 | 0.988 | 0.185 | 0.406 | 0.883 | 0.987 | 0.999 |
| TG/TT vs. GG | 1.24 (1.04–1.49) | 0.017 | 0.975 | 0.050 | 0.136 | 0.633 | 0.946 | 0.994 |
| Pack-years > 16 | 1.49 (1.12–1.98) | 0.006 | 0.523 | 0.033 | 0.094 | 0.532 | 0.920 | 0.991 |
| Combined risk genotypes | ||||||||
| No. at-risk genotypes d | ||||||||
| 1 vs. 0 | 1.25 (1.03–1.52) | 0.026 | 0.984 | 0.073 | 0.192 | 0.723 | 0.964 | 0.996 |
| 2 vs. 0 | 1.62 (1.16–2.26) | 0.005 | 0.452 | 0.032 | 0.091 | 0.523 | 0.917 | 0.991 |
| ≥1 vs. 0 | 1.28 (1.06–1.56) | 0.011 | 0.930 | 0.034 | 0.096 | 0.539 | 0.922 | 0.992 |
aThe crude OR reported in Tables 2 and 3.
bThe omnibus chi-square test of the genotype frequency distributions reported in Tables 2 and 3.
cStatistical power was calculated using the number of observations in the study and the OR and P values in Tables 2 and 3.
dCombined risk genotypes were to referred to ERCC2 rs238406 TG/TT + rs13181 TG/GG.
Meta-analysis of ERCC2 SNPs with ESCC risk
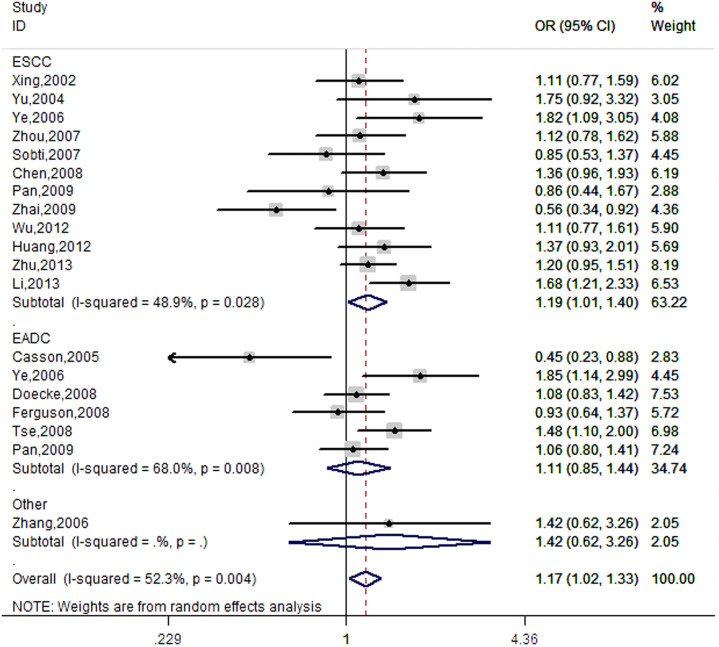
In addition to the present association study, we identified 18 other published case-control studies on ERCC2 rs13181 SNP6,7,8,14,15,16,17, with the sample sizes ranging between 151 and 1600, for which we performed a mini meta-analysis to assess the association of this SNP with ESCC risk (Supplemental Table 2). We found that the rs13181 variant G allele was significantly associated with an increased risk of esophageal cancer (OR = 1.17; 95% CI = 1.02–1.33 for TG/GG vs. TT) based on 5,012 cases and 8,238 controls in the pooled analysis as well as in subgroup analysis of these ESCC studies (OR = 1.19; 95% CI = 1.01–1.40 for TG/GG vs. TT) (Table 4, Figure 1). The leave-one-out sensitivity analysis validated the stability of the results (data not shown). The shapes of the funnel plots seemed symmetrical, and Egger's test further showed no publication bias (Table 4). However, we were not able to do a similar mini meta-analysis for rs238406, because there was only one published case-control study on esophageal adenocarcinoma cancer in a European population18.
Table 4. Meta-analysis of the association between ERCC2 rs13181 polymorphism and esophageal cancer risk.
| No. of studies | GG vs. TT | TG vs. TT | GG vs. TG/TT | TG/GG vs. TT | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | OR (95% CI)c | PORa | Phetb | OR (95% CI)d | PORa | Phetb | OR (95% CI)c | PORa | Phetb | OR (95% CI)d | PORa | Phetb | |
| All | 19 | 1.28 (1.08–1.52) | 0.005 | 0.075 | 1.15 (1.01–1.30) | 0.030 | 0.025 | 1.19 (1.01–1.40) | 0.034 | 0.231 | 1.17 (1.02–1.33) | 0.020 | 0.004 |
| Ethnicity | |||||||||||||
| Asian | 11 | 1.36 (0.97–1.91) | 0.078 | 0.165 | 1.16 (1.00–1.35) | 0.050 | 0.141 | 1.28 (0.92–1.80) | 0.140 | 0.176 | 1.18 (1.01–1.39) | 0.115 | 0.052 |
| Non-Asian | 8 | 1.26 (1.03–1.54) | 0.025 | 0.071 | 1.14 (0.90–1.43) | 0.276 | 0.019 | 1.16 (0.97–1.40) | 0.110 | 0.331 | 1.14 (0.90–1.45) | 0.267 | 0.007 |
| Histological type | |||||||||||||
| Squamous Cell | 12 | 1.33 (0.98–1.79) | 0.064 | 0.152 | 1.17 (1.01–1.37) | 0.039 | 0.083 | 1.20 (0.90–1.60) | 0.216 | 0.200 | 1.19 (1.01–1.40) | 0.034 | 0.028 |
| Adenocarcinoma | 6 | 1.26 (1.02–1.55) | 0.036 | 0.048 | 1.09 (0.85–1.41) | 0.500 | 0.023 | 1.18 (0.97–1.44) | 0.095 | 0.215 | 1.11 (0.85–1.44) | 0.448 | 0.008 |
| Other | 1 | 3.13 (0.13–77.85) | 0.486 | 0.076 | 1.33 (0.57–3.08) | 0.507 | -- | 3.03 (0.12–75.19) | 0.499 | -- | 1.42 (0.62–3.26) | 0.404 | -- |
| Publication Bias e | 0.955 | 0.414 | 0.970 | 0.497 | |||||||||
aP value of the Z-test for odds ration test.
bP value of the Q-test for heterogeneity test.
cFixed-effects model.
dRandom-effects model.
eP value of Egger's test for publication bias.
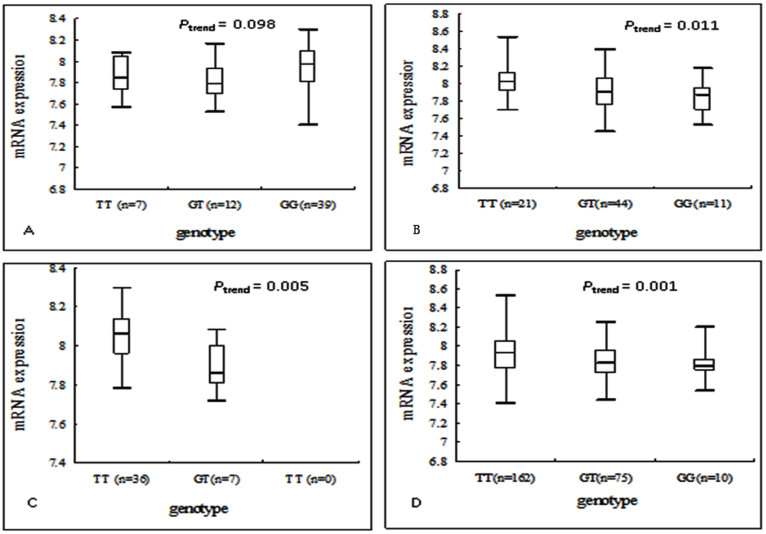
Figure 1. Effect of two SNPs on ERCC2 mRNA expression for different populations in EBV-transformed lymphoblastoid cell lines.
(A) the effect of rs238406 on mRNA expression for 58 Asians; (B) the effect of rs238406 on mRNA expression for 76 Europeans; (C) the effect of rs13181 on mRNA expression for 43 Chinese Han, Beijing (CHB); (D) the effect of rs13181 on mRNA expression for 247 subjects with different ethnicities.
Correlation analysis for ERCC2 mRNA expression levels and variant genotypes
To explore possible underlying molecular mechanisms, we performed the ERCC2 genotype-phenotype correlation analysis using the available data on the genotypes and mRNA expression levels of lymphoblastoid cell lines derived from peripheral lymphocytes from 270 people (ref. 43). As a result, we found a statistical significance for rs13181 allele T → G on gene transcription expression (Ptrend = 0.005 and 0.001 for CHB and all populations, respectively) (Figure 2), which supports the finding of an association between this SNP and ESCC risk. For rs238406, however, we found a statistically significant trend for the allele G → T effect on ERCC2 mRNA expression in Europeans (Ptrend = 0.011) but a borderline significance in CHB (Ptrend = 0.098).
Figure 2. Forest plot of overall esophageal cancer risk associated with ERCC2 rs13181 polymorphism (TG/GG vs. TT) by the random-effects for each of the 19 studies.
For each study, the estimates of OR and its 95% CI were plotted with a box and a horizontal line. The symbol filled diamond indicates pooled OR and its 95% CI.
Discussion
In the present study, we found that two potentially functional ERCC2 SNPs (i.e. rs238406 G > T and rs13181 T > G) were individually or collectively associated with ESCC risk. We also observed a multiplicative gene-environment interaction between rs238406 genotypes and smoking on ESCC risk. Because many studies have investigated the associations between ERCC2 rs13181 T > G SNP and risk of ESCC cancer, we also performed a mini meta-analysis that provided additional statistical evidence of such an association.
The ERCC2 gene, mapped to chromosome 19q13.3, comprises 23 exons and encodes 760 amino acids. Acting as a single-strand DNA-dependent ATPase, and also a 5′-3′ DNA helicase, the ERCC2 protein participates in both DNA unwinding during NER and transcription initiation by binding to the transcription factor IIH via p4419,20. Mutations in ERCC2 could result in transcription defects and abnormal apoptosis by reducing the BTF2/TFIIH activity, thus leading to a severe but variable depression of NER21. ERCC2 also has a second function that is involved in base excision repair (BER) of oxidative base damage of the transcribed strand of transcriptionally active genes21. It has been demonstrated that an increased risk for ESCC in Chinese populations was associated with reduced mRNA expression of ERCC2 as detected in peripheral blood mononuclear cells22. ERCC2 mRNA expression levels may also be of importance, as it has been shown that mRNA levels of ERCC2 are correlated with the DNA repair capacity phenotype in primary lymphoblasts23.
Given the role of the ERCC2 gene in the DNA repair pathway, it is believed that some subtle alterations in ERCC2 gene functions are more readily tolerated. Individuals with inherited ERCC2 defects display their disease phenotype in a recessive genetic model, because only the homozygotes are prone to accumulate genetic damage and may have a marked predisposition to cancer. However, the present study found some dominant effects of ERCC2 SNPs on ESCC risk, perhaps owing to complex gene-gene or gene-environment interactions that may have some impact on genetic models for disease risk24. This hypothesis needs to be tested in additional larger studies.
Previous studies found that ERCC2 rs238406 conferred susceptibility to cancers of the bladder25, lung26 and other organs. Only one reported study focused on esophageal cancer and found no association between rs238406 and esophageal adenocarcinoma risk with only 56 European patients and 95 healthy controls18. In contrast, the present study included over 1000 cases and 1000 controls with a much improved study power and thus found a statistical evidence for a weak association between the rs238406 variant genotypes and increased ESCC risk. SNP rs238406, a silent polymorphism in codon 156 of exon 6, does not change the amino acid residue (Arg156Arg). It is therefore unlikely that the enzymatic function of ERCC2 is affected by this SNP. However, it is likely that these SNPs may influence ERCC2 protein levels through an effect on mRNA splicing as predicted by the SNPinfo software. Such a possible molecular mechanism is biologically plausible. For example, a silent A → T substitution at codon 399 (Val399 Val) of the phenylalanine hydroxylase (PAH) gene is the major determinant for exon 11 skipping during the pre-mRNA processing step that thereby results in a phenylketonuria (PKU) phenotype27. Indeed, we found a trend for the rs238406 allele G → T effect on ERCC2 mRNA expression for different populations, indicating that it may be a potentially functional SNP. Alternatively, the SNP may be in LD with other untyped functional polymorphisms or with an adjacent susceptibility gene. For example, we found two SNPs (rs3810366 and rs2097215) located in 5′ near gene are in high LD (r2 > 0.8) with rs238406. Furthermore, the region of chromosome 19q13.3, comprising the ERCC2 gene, encodes several other DNA repair genes, such as ERCC1, XRCC1 and LIG28. Therefore, additional mechanistic studies are needed to unravel the molecular mechanisms underlying this observed association.
Some meta-analyses have reported that individuals carrying the SNP rs13181 (Lys751Gln) G variant allele had an increased risk of cancers of the lung29, stomach30, skin31 and esophagus14,32. Our meta-analysis not only confirmed the positive association with risk of esophageal cancer but also provided additional statistical evidence that the G variant allele was associated with an increased risk of esophageal cancer in Chinese populations. The rs13181 SNP, i.e., the T → G base substitution in exon 23, causes an amino acid alteration from lysine to glycine at codon 751, which leads to a complete change in electronic configuration of the amino acid and thus reduces DNA repair efficiency33. For example, the TT genotype was reported to be associated with a sub-optimal repair of chromatid aberrations induced by X-irradiation in breast cancer34. In the present study, we also found a significant trend for the rs13181 allele T → G effect on ERCC2 transcript expression levels in different ethnic populations, indicating that the ERCC2 rs13181 SNP may be a underlying genetic determinant of esophageal cancer risk.
The present study also identified that rs238406 SNP had a significant interaction with tobacco consumption in ESCC risk. NER is an indispensable pathway for the repair of bulky and helical distorting DNA adducts generated by tobacco smoke. Therefore, light smokers may have less DNA damage and less risk for developing advanced tumors stage and grade35, while in heavy smokers, the genetic effect may be accelerated by accumulated DNA damage induced by tobacco smoking. This hypothesis needs to be tested in larger studies that allow one to perform sufficient stratified analysis and to explore gene-gene and gene-environment interactions.
However, in recently published GWASs for ESCC9,10,11,12,36, these two ERCC2 SNPs were not among the reported top-hits, nor any SNPs in other DNA repair genes. There are some explanations for this. First, GWAS is bases on the theory that the common genetic variants are associated with common human diseases37. It is clear that most of the functional SNPs are believed not to be common, nor is cancer. Second, GWASs have an inherent “multiple testing problems” that often require the stringent P values to avoid false-positive findings. In that case, it would markedly decrease the power to detect the influence of common SNPs that may have modest impact on risk of cancer. Third, the lack of statistical power makes it difficult to interpret negative findings, such as for those DNA repair genes known to be involved in cancer etiology. These underscore the need to continue the search for such missing etiological factors in the GWAS dataset or in the combined analysis of several published GWASs in the future to identify additional causal but rare or functional variants. Forth, most published GWASs do not include comprehensive information about exposures for additional adjustment or stratification, so the actual associations may be either biased or masked. Indeed, Wu et al recently extended their GWAS findings by additional analyses for gene-environment interactions, in which they have discovered some new genetic contribution to ESCC through the analysis of interaction with alcohol consumption by performing a genome-wide gene-environment interaction analysis9. Moreover, GWAS is based on common tagging SNPs with the aim of not to capture all the “causative” but the “representative” SNPs. Therefore, the present study is another extension to the investigation of functional SNPs in cancer etiology and to the needs of identifying additional evidence of biological mechanisms underlying the positive association findings.
In conclusion, the present study demonstrated that potentially functional ERCC2 SNPs may confer susceptibility to ESCC, possibly by the effects on ERCC2 mRNA expression levels, suggesting an important role of functional ERCC2 SNPs in the etiology of ESCC in Chinese populations. Nevertheless, some limitations should be addressed. First, because of tissue access constraint, we did not have the opportunity to examine the levels of ERCC2 mRNA in the target tissue from the study population. It is likely that the exact mechanisms by which the ERCC2 variant regulates the gene transcription activity in vivo are not the same as those observed in cell lines in vitro, which warrants additional in vivo mechanistic studies. Second, insufficient study power may be another concern in regard to some negative findings, particularly for the analyses of subgroups with small sample sizes, or for SNPs with a rare variant allele, such as the rs13181 G variant allele. For example, we failed to find any statistical evidence of an association between rs13181 and ESCC risk, but we did find such a statistical evidence in a subsequent meta-analysis of a much larger sample size. Third, we only investigated two potentially functional SNPs in the present study due to financial constraints. As we know, cancer is a complex disease for which any single SNP may not be sufficient for the prediction of the overall risk. Future studies should include more genes and more SNPs with more rigorous study designs for mechanistic studies.
Methods
Study subjects
The study populations were described in details previously13. Briefly, all subjects were genetically unrelated ethnic Han Chinese from Eastern China. The present study included 1126 cases who had newly diagnosed and histopathologically confirmed primary ESCC from Fudan University Shanghai Cancer Center (FUSCC) between March 2009 and September 2011. We also included 1131 cancer-free controls who were frequency matched to the cases by age (±5 years) and sex, and who were randomly selected from those recruited at the same time period in the Taizhou Longitudinal Study (TZL)38. The overall response rate was 93% and 90% for cases and controls, respectively. Having signed a written informed consent, each participant provided information about demographics and environmental exposure history, including age, sex, ethnicity, body mass index (BMI), tobacco and alcohol consumption, and donated 10 mL venous blood sample, of which 1 mL was used for genomic DNA extraction. This research protocol was approved by the Institutional Review Board of FUSCC and the experiment on humans was performed in accordance with relevant guidelines and regulations.
SNP selection and genotyping
We selected two potentially functional SNPs located in the coding region (i.e., rs13181 T > G and rs238406 G > T) for genotyping, because rs13181 is one of the most reported non-synonymous SNPs that result in an amino acid change, while rs238406 is a synonymous SNP that has a putative function of splicing predicted by the SNP function prediction (FuncPred) software (SNPinfo, http://snpinfo.niehs.nih.gov/). In addition, these two SNPs also satisfy the following criteria: (1) minor allele frequency (MAF) ≥ 5% in Chinese Han, Beijing (CHB) descendants; (2) with low linkage disequilibrium (LD) of an r2 < 0.8 for each paired SNPs. The LD analysis suggested that these two potentially functional SNPs also captured other 11 untyped SNPs (r2 ≥ 0.8) within the ERCC2 gene (Supplemental Table 3). We extracted genomic DNA from blood samples and performed genotyping by using the Taqman real-time PCR method, as described previously13. The successful genotyping rate for both SNPs was greater than 95%.
Meta-analysis of ERCC2 SNPs with ESCC risk
To summarize the published data on these two SNPs, we searched two electronic databases (MEDLINE and EMBASE) for all relevant articles. We defined the search terms, inclusion and exclusion criteria as previously reported39. The last search update was on May 20, 2013. We chose the fixed-effects (Mantel–Haenszel method) or random-effects model (DerSimonian and Laird method) based on the heterogeneity test40,41. When P value of the heterogeneity test was <0.05, the random-effects model was used, which indicates the existence of heterogeneous effect sizes across all studies; otherwise, the fixed-effects model was more appropriate. We also conducted sensitivity analyses to evaluate the effect of an individual study on the overall risk of cancer and used the funnel plot and the Egger's linear regression test to examine potential influence of publication bias42. Because there were few reported studies on the association of rs238406 G > T SNP in the ERCC2 gene with ESCC risk, we could not perform its meta-analysis.
Correlation analysis for ERCC2 mRNA expression levels and variant genotypes
To provide possible underlying mechanisms, we analyzed the correlation between ERCC2 mRNA expression levels and variant genotypes. The genotyping data were from the HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) consisting of 3.96 million SNP genotypes from 270 individuals of four ethnic groups (CEU, 90 Utah residents from northern and western Europe; CHB, 45 unrelated Han Chinese in Beijing; JPT, 45 unrelated Japanese in Tokyo; YRI, 90 Yoruba in Ibadan, Nigeria), and the data on transcript expression levels were from EBV-transformed B lymphoblastoid cell lines from the same 270 individuals (http://app3.titan.uio.no/biotools/tool.php?app=snpexp)43.
Statistical methods
We examined the Hardy-Weinberg equilibrium for genotype distribution in controls by a goodness-of-fit χ2 test. We also used the χ2 test to assess the differences in demographic variables, risk factors and genotype frequency distributions between the cases and controls. We computed OR and their 95% CI from univariate and multivariate logistic regression models to estimate the associations of ERCC2 SNPs with ESCC risk, which were also evaluated in subgroup analyses stratified by demographic variables and risk factors. Finally, we performed linear regression model-trend test for the genotype–phenotype association analysis.
For significant findings observed in the present study, we used FPRP to assess false-positive associations. We calculated FPRP with a favorite prior probability of 0.1 to detect an OR of 0.67/1.50 (protective/risk effects) for an association with genotypes under investigation. A FPRP value < 0.20 was considered a noteworthy association44. We performed the meta-analysis with the STATA software, version 11.0 (Stata Corporation, College Station, TX), and other statistical analyses with the SAS software (version 9.1; SAS Institute, Cary, NC). All statistical tests were two-sided with a statistical significance level of P < 0.05.
Author Contributions
Conceived and designed the experiments: Q.Y.W. and J.Q.X. Performed the experiments: M.L.Z. and J.H. Analyzed the data: M.L.Z. and M.Y.W. Contributed reagents/materials/analysis tools: L.J., Y.J.Y., J.C.W., M.H.S., X.F.W. and L.Z.Z. Wrote the paper: M.L.Z. and Q.Y.W. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
This study was supported by funding from China Recruitment Program of Global Experts at Fudan University, Shanghai Committee of Science and Technology, China (Grant No. 12DZ2260100), Ministry of Science and Technology (2011BAI09B00) and Ministry of Health (201002007), China.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J. H. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001). [DOI] [PubMed] [Google Scholar]
- Egly J. M. The 14th Datta Lecture. TFIIH: from transcription to clinic. FEBS Lett 498, 124–128 (2001). [DOI] [PubMed] [Google Scholar]
- Lehmann A. R. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev 15, 15–23 (2001). [DOI] [PubMed] [Google Scholar]
- Xing D. et al. Polymorphisms of DNA repair genes XRCC1 and XPD and their associations with risk of esophageal squamous cell carcinoma in a Chinese population. Int J Cancer 100, 600–605, 10.1002/ijc.10528 (2002). [DOI] [PubMed] [Google Scholar]
- Chen M. R. et al. Polymorphism of DNA repair gene XPD and XRCCl and its relationship with esophageal squamous cell carcinoma. Fudan Univ J Med Sci 35, 273–277, 281 (2008). [Google Scholar]
- Huang C. G. et al. Analysis of XPD genetic polymorphisms of esophageal squamous cell carcinoma in a population of Yili Prefecture, in Xinjiang, China. Mol Biol Rep 39, 709–714, 10.1007/s11033-011-0789-z (2012). [DOI] [PubMed] [Google Scholar]
- Li R. Z. & Sun J. Association between XPD gene polymorphisms and esophageal squamous cell carcinoma. Mol Med Rep 7, 674–678, 10.3892/mmr.2012.1215 (2013). [DOI] [PubMed] [Google Scholar]
- Wu C. et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet 44, 1090–1097 (2012). [DOI] [PubMed] [Google Scholar]
- Wu C. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat Genet 43, 679–684 (2011). [DOI] [PubMed] [Google Scholar]
- Wang L. D. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet 42, 759–763 (2010). [DOI] [PubMed] [Google Scholar]
- Abnet C. C. et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet 42, 764–767 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. L. et al. Polymorphisms in the ERCC5 gene and risk of esophageal squamous cell carcinoma (ESCC) in Eastern Chinese populations. PLoS One 7, e41500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D. P., Ma W. L., He X. F. & Zhang Y. XPD Lys751Gln polymorphism and esophageal cancer susceptibility: a meta-analysis of case-control studies. Mol Biol Rep 39, 2533–2540 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang W. C. et al. Relationship between polymorphisms of DNA repair genes and esophageal cancer susceptibility. Chin J Public Health 22, 557–558 (2006). [Google Scholar]
- Wu X. B. et al. Association of XPD gene polymorphisms with susceptibility of esophageal squamous cell carcinoma in Henan province. Chin J Public Health 28, 446–449 (2012). [Google Scholar]
- Zhou R. M. et al. Correlation between single nucleotide polymophism of DNA repair gene XPD and the risks of esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. Tumor 27, 118–122,133 (2007). [Google Scholar]
- Casson A. G. et al. Polymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinoma. Carcinogenesis 26, 1536–1541 (2005). [DOI] [PubMed] [Google Scholar]
- Sung P. et al. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365, 852–855, 10.1038/365852a0 (1993). [DOI] [PubMed] [Google Scholar]
- Coin F. et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet 20, 184–188, 10.1038/2491 (1998). [DOI] [PubMed] [Google Scholar]
- Friedberg E. C. How nucleotide excision repair protects against cancer. Nat Rev Cancer 1, 22–33, 10.1038/35094000 (2001). [DOI] [PubMed] [Google Scholar]
- Liu R., Yin L. H. & Pu Y. P. Reduced expression of human DNA repair genes in esophageal squamous-cell carcinoma in china. J Toxicol Environ Health A 70, 956–963 (2007). [DOI] [PubMed] [Google Scholar]
- Vogel U., Dybdahl M., Frentz G. & Nexo B. A. DNA repair capacity: inconsistency between effect of over-expression of five NER genes and the correlation to mRNA levels in primary lymphocytes. Mutat Res 461, 197–210 (2000). [DOI] [PubMed] [Google Scholar]
- Minelli C., Thompson J. R., Abrams K. R., Thakkinstian A. & Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol 34, 1319–1328 (2005). [DOI] [PubMed] [Google Scholar]
- Shao J., Gu M., Xu Z., Hu Q. & Qian L. Polymorphisms of the DNA gene XPD and risk of bladder cancer in a Southeastern Chinese population. Cancer Genet Cytogenet 177, 30–36 (2007). [DOI] [PubMed] [Google Scholar]
- Yin J. et al. The DNA repair gene ERCC2/XPD polymorphism Arg 156Arg (A22541C) and risk of lung cancer in a Chinese population. Cancer Lett 223, 219–226 (2005). [DOI] [PubMed] [Google Scholar]
- Chao H. K., Hsiao K. J. & Su T. S. A silent mutation induces exon skipping in the phenylalanine hydroxylase gene in phenylketonuria. Hum Genet 108, 14–19 (2001). [DOI] [PubMed] [Google Scholar]
- Vogel U., Hedayati M., Dybdahl M., Grossman L. & Nexo B. A. Polymorphisms of the DNA repair gene XPD: correlations with risk of basal cell carcinoma revisited. Carcinogenesis 22, 899–904 (2001). [DOI] [PubMed] [Google Scholar]
- Feng Z., Ni Y., Dong W., Shen H. & Du J. Association of ERCC2/XPD polymorphisms and interaction with tobacco smoking in lung cancer susceptibility: a systemic review and meta-analysis. Mol Biol Rep 39, 57–69, 10.1007/s11033-011-0710-9 (2012). [DOI] [PubMed] [Google Scholar]
- Xue H. et al. The effect of XPD/ERCC2 polymorphisms on gastric cancer risk among different ethnicities: a systematic review and meta-analysis. PLoS One 7, e43431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Zhuang L. & Ma W. Comprehensive assessment of the association of ERCC2 Lys751Gln polymorphism with susceptibility to cutaneous melanoma. Tumour Biol 34, 1155–1160 (2013). [DOI] [PubMed] [Google Scholar]
- Yuan L. et al. XPD Lys751Gln polymorphism and esophageal cancer risk: a meta-analysis involving 2288 cases and 4096 controls. World J Gastroenterol 17, 2343–2348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou S. & Sarasin A. ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis 17, 463–469 (2002). [DOI] [PubMed] [Google Scholar]
- Lunn R. M. et al. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis 21, 551–555 (2000). [DOI] [PubMed] [Google Scholar]
- Rouissi K. et al. The effect of tobacco, XPC, ERCC2 and ERCC5 genetic variants in bladder cancer development. BMC Cancer 11, 101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R. et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 137, 1768–1775 (2009). [DOI] [PubMed] [Google Scholar]
- Bodmer W. & Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 40, 695–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Rationales, design and recruitment of the Taizhou Longitudinal Study. BMC Public Health 9, 223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. L. et al. Association between the ERCC5 Asp1104His polymorphism and cancer risk: a meta-analysis. PLoS One 7, e36293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959). [PubMed] [Google Scholar]
- Egger M., Smith G. D., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. Brit Med J 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K., Melum E., Franke A. & Karlsen T. H. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC bioinformatics 11, 600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L. & Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer I 96, 434–442 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.