Abstract
Transcription factors (TFs) are responsible for decoding and expressing the information stored in the genome, which dictates cellular function. Creating artificial transcription factors (ATFs) that mimic endogenous TFs is a major goal at the interface of biology, chemistry, and molecular medicine. Such molecular tools will be essential for deciphering and manipulating transcriptional networks that lead to particular cellular states. In this minireview, the framework for the design of functional ATFs is presented and current challenges in the successful implementation of ATFs are discussed.
Keywords: chemical biology, synthetic biology, transcription factor mimics, gene regulation, cooperative binding, protein-DNA interactions
1. Introduction
Most cells in multicellular organisms carry the same genome, yet are able to produce a wide range of phenotypes which gives rise to sets of specialized cells that differ in morphology and function. This diversity is in part attributed to differences in tightly regulated gene expression patterns, with some genes being actively transcribed and others repressed. Transcription factor (TF) proteins are active participants in the regulation of specific geneexpression programs in response to cellular needs. Therefore, it is not surprising that the malfunctioning of TFs has been directly linked to many disease states [1]. This link has turned TFs into attractive therapeutic targets for treating a wide range of diseases, including cancer [2– 4].
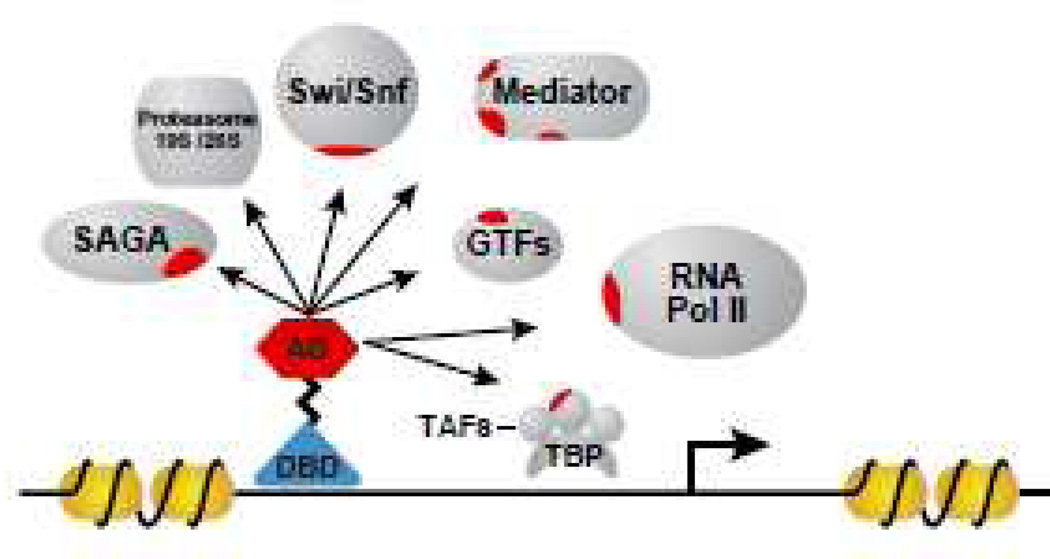
In response to specific signal, TFs target particular genes within the genome. Once localized to the targeted genes, TFs recruit macromolecular machines to modify chromatin and initiate transcription [5]. Over several decades, much effort has been invested in the identification of the components of the transcriptional machinery targeted by TFs [6, 7]. Transcription factors have been shown to interact with RNA polymerase II, the general transcription factors (GTFs) [5], coactivators, such as components of the Mediator protein complex [8, 9], and TBP-associated factors [10, 11]. TFs also recruit nucleosome remodeling complexes such as the Swi/Snf complex and histone acetyltransferases, such as the SAGA complex [12, 13]. Components of the proteasome have also been identified as targets of transcriptional activators (Figure 1) [7].
Figure 1. Transcription activation by transcription factors.
TFs are minimally composed of a DNA binding domain (DBD) and an activator domain (AD). The DBD recognize and binds to a DNA sequence to activate the targeted gene(s). The AD recruits the transcriptional machinery components through interactions with RNA polymerase II (RNApol), general transcription factors, (GTFs), (TBP)-associated factors (TAFs), the Mediator complex, chromatin remodeling complexes such as SAGA and Swi/Snf complexes and/or the 19S and 26S components of the proteasome.
Natural transcription factors can be minimally composed of two functional domains: a DNA-binding domain (DBD) and a regulatory domain (RD) [5]. The DBD determines which genes will be activated or repressed by selectively targeting specific DNA sequences within the cis-regulatory motifs associated with the target genes; the RD dictates whether to activate or repress transcription by recruiting components associated with the transcriptional machinery or the repression machinery, respectively. The magnitude of the response is encoded within the regulatory domain.
An important feature of natural TFs is that the DBD and the RD function independently from each other, as demonstrated by domain swapping experiments in yeast and other eukaryotes [14]. The modular nature of TFs highlights the possibility of exchanging the DBD and RD for synthetic counterparts to engineer artificial transcription factors (ATFs). Engineering replacements for the DBD and RD has been the most used strategy for creating TF mimics (Figure 1) [15].
The potential benefits of implementing ATF-based tools are extensive [16]. These molecular tools could be used to dissect genome-wide transcriptional cascades, yielding fundamental insights on developmental processes. Diseases based on malfunctioning transcription factors could be treated or prevented with ATFs. The metabolic pathways of an organism could be engineered to produce valuable compounds. ATFs would also be invaluable tools for the emerging field of synthetic biology, as they could be used to control synthetic cellular circuits [17].
2. DNA binding domains
The information contained within the DBD dictates which DNA sequence is targeted and therefore determines which genes are regulated by a given transcription factor. Similarly DBD confers specificity on a given ATF. Different types of binding domains have been employed in ATFs to target specific DNA sequences. Examples of DNA binding domains used for ATF construction include protein-based zinc fingers, oligonucleotides and oligonucleotide analogs, as well as synthetic small molecules (Figure 2).
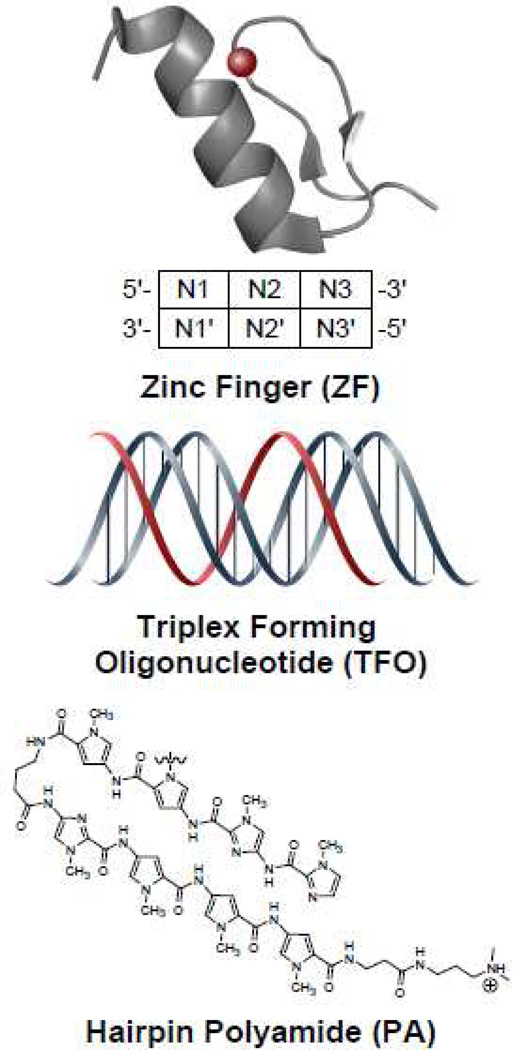
Figure 2. DNA binding domains commonly used in ATFs.
Zinc fingers (ZF) recognize and bind to 3bp (N1-3) in dsDNA; triplex forming oligonucleotides (TFOs); hairpin polyamides (PAs).
The zinc finger (ZF) domain is one of the most represented DBD in the human genome [18, 19]. A zinc finger module is composed of 30 amino acids assembled in a ββα fold stabilized by a zinc ion. Each ZF recognizes and binds to three base pairs in the target DNA (Figure 2). ZF modules can be strung together to recognize larger unique sequences in the genome. For example, three consecutive ZFs target a 9 bp sequence, and a polydactyl ZF consisting of six ZFs targets an 18 bp sequence [20]. The complexity of sequences that can be recognized by ZFs has been expanded through a variety of strategies, including structure-guided methods, phage display screens, and the bacterial one-hybrid system [21–23]. The most successful artificial ZF modules target sequences containing GNN triplets [20, 24]. A detailed protocol for the modular construction of ZF libraries was recently published by the Barbas group [25]. Zinc fingers have been widely employed as DNA-binding domains in the construction of ATFs [26]. However, it has been shown that in some cases the binding sequences of individual ZFs are not completely separable and that DNA binding is influenced by the neighboring ZFs as well [27, 28].
Two recent reports described the DNA recognition “code” of the transcription activator-like (TAL) effectors of bacteria from the genus Xanthomonas [29, 30]. TAL effectors are DNA binding proteins from plant pathogenic bacteria [31]. Members of the TAL effectors family posses a characteristic central domain of tandem repeats of 34 amino acids. In each repeat, the amino acids located in positions 12 and 13 are hypervariable and referred to as the repeat-variable diresidue (RVD). The DNA binding specificity of TAL effectors is determined by the tandem repeat region [32]. Specifically, a one-to-one correspondence was found between the identity of the RVD and target DNA [29, 30].
The deciphering of the DNA binding code of TAL effectors highlights the possibility of engineering TAL effectors with custom DNA sequence specificity. However, the molecular details on how the repeat domain of TAL effectors recognizes targeted DNA are currently lacking. Although more work is needed to support the generality of the proposed DNA binding code, TAL effectors could potentially be utilized as DBD in designing transcription factor mimics.
DBDs have also been constructed from oligonucleotides [33, 34] as well as oligonucleotide analogs, such as locked-nucleic acids (LNAs) [35] and peptide nucleic acids (PNAs) [36]. These molecules recognize and bind to DNA by forming a triple helix DNA strand (referred to as triplex-forming oligos (TFO)), or by strand invasion of double-stranded DNA [37]. An ATF consisting of a triplex-forming oligonucleotide DBD linked to a minimal VP16 peptide AD was first reported by Kuznetsova et al. [38]. This work was later extended by Young and colleagues to create TFO-based ATF that induced the expression of a reporter gene in tissue culture cells [39].
The most effective small-molecule DBDs to date are based on N-methylpyrrole and N-methylimidazole polyamides (PA). These molecules bind in the minor groove of dsDNA [40]. When engineered to form hairpins, PAs are capable of binding to targeted DNA sequences, based on a set of pairing rules, with nanomolar affinity [28, 41–43]. Due to this high affinity, a PA can modify gene expression by competitively inhibiting binding of endogenous TFs [44–47]. An artificial activation domain (AD) attached to a hairpin PA was shown to activate transcription in vitro [48, 49]. Applications using PA-based ATFs are often limited due to poor cell permeability of PAs; research efforts aimed at improving the cellular permeability of PA-based compounds are ongoing [50–53].
For an ATF to work properly, its DNA binding domain must find and bind to the targeted DNA sequence in the cellular context. In the cell, the accessibility of an ATF binding site is in part dictated by its chromatin state. However, genome-wide maps of nucelosome positions have highlighted that the regions near transcription start sites are often depleted of nucleosomes [54– 56]. These nucleosome-free regions are potential binding sites for ATFs. In addition, studies have shown that polyamide DBDs are able to bind to targeted DNA sequences in nucleosome particles [57] and nuclear chromatin [58]. In addition, strategies have been designed to alter the accessibility of DNA binding site by chromatin modification. Snowden et al fused a ZF DNA binding domain to histone modifying enzymes to (i.e., a histone deacetylase and a histone methyltransferae) [59, 60]. More recently a DNA methylase enzyme was fused to TFO DNA binding domain and shown to specifically methylate the targeted promoter in a reporter plasmid [61].
3. Regulatory domains
3.1. Activation domains
Most of the activation domains used in ATFs are derived from peptide sequences inspired by the architecture of natural activation domains [7]. Natural ADs are usually composed of unstructured peptides with potential to form amphipathic helices. Based on the peptide sequences of ADs and on structures of natural ADs bound to their protein partners, it appears that many ADs form an amphipathic α-helix upon interaction with the transcriptional machinery, with the hydrophobic face of the helix contacting the binding partner [62–64]. However, extended conformations with a buried hydrophobic surface are also observed [65, 66]. Potent short peptides that function as ADs have been indentified from screening libraries of random peptides [67, 68] and from peptide libraries that targeted components of the transcriptional machinery [69, 70]. Peptide-based ADs have the disadvantage of short lifetime in vivo, likely due to the unstructured nature of the peptide that alerts the cellular surveillance machinery (e.g. proteases) to degrade the peptide. Nevertheless, recent studies have highlighted that the potency of peptide-based ADs can be enhanced by engineering intramolecular interactions between the AD and the DBD (see section 6.2) [71, 72].
Proteolysis of the AD can be avoided by using peptoids or small molecules (Figure 3). A novel regulatory module was found by screening a combinatorial library of ~100,000 peptoids for binding to the KIX domain of the mammalian co-activator CBP. The most promising peptoid from the screen was delivered into cells as a dexamethasone conjugate that bound to a glucocorticoid receptor-Gal4 chimera DBD. This ATF activated transcription of a reporter luciferase gene in HeLa cells [73].
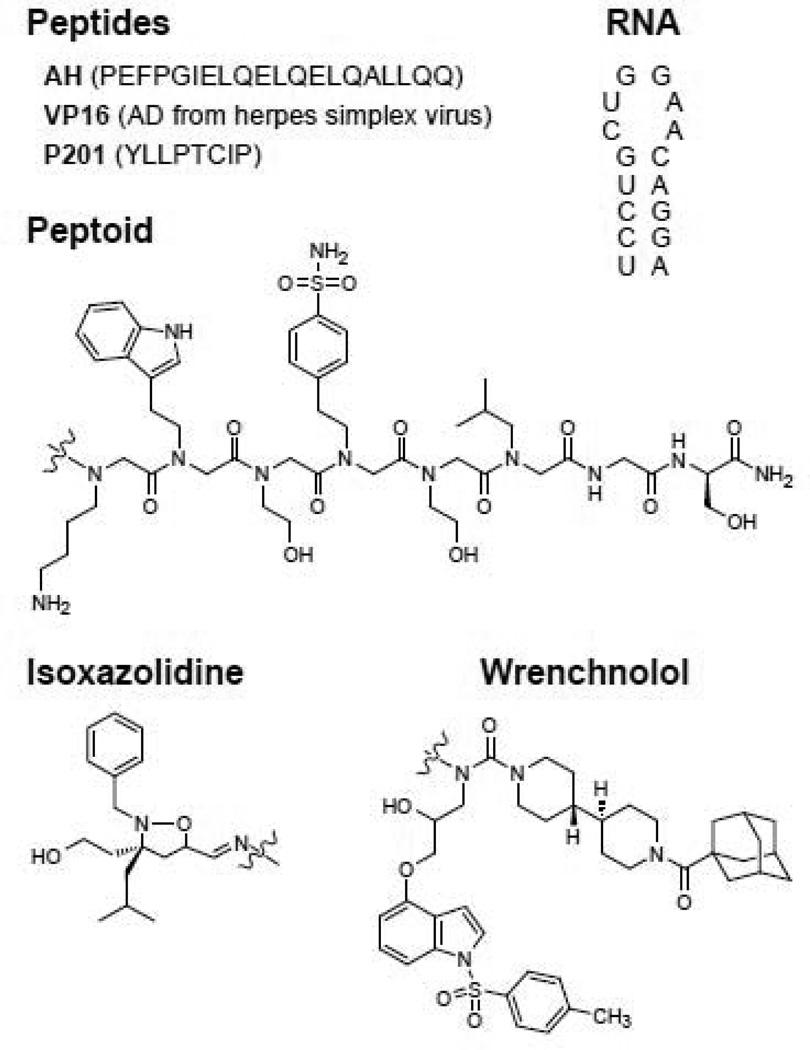
Figure 3. Activation domains commonly used in ATFs.
Peptides: amphipathic helix (AH) [62], VP16 from herpes simples virus [112], and P201 [68]. Peptoid [73]. RNA [75] Smallmolecule: isoxazolidine [78] and wrenchnolol [82].
In addition to peptides, RNA can activate transcription when conjugated to a DBD. In an adaptation of the three-hybrid system used to screen for RNA ligands that interacted with the yeast protein Snp1, it was observed that some RNA sequences activated transcription [74]. This finding was further supported by research from Saha et al., where a library of RNA hairpins with a randomized 10 nucleotides loop conjugated to a DBD was screened for transcriptional activation [75]. The authors found a consensus sequence of 6 nucleotides in the hairpin loop that activates transcription (Figure 3). RNA-based ADs have also been designed through in vivo evolution in yeast [76]. In vivo evolution can also be used to select for RNA-DBD conjugates that repress transcription [77].
An early small molecule AD was reported by Minter et al. [78]. Isoxazolidine derivatives displaying functional groups commonly found in natural ADs were synthesized (Figure 3). The functional groups were chosen to mimic the amphipathic character of natural ADs. These isoxazolidines, when tethered to a DBD, activated transcription both in vitro [78] and in vivo [79]. It was later shown that isoxazolidine ADs can bind to different components of the transcriptional machinery. Specifically, isoxazolidines have been shown to interact with the KIX domain of the co-activator CREB binding protein (CBP), TRRAP/Tra1 (a component of the SAGA complex), and the components of Mediator complex, Med15/Gal11 and MED23/Sur2 [80].
Another small molecule AD was discovered by targeting specific components of the transcriptional machinery. Wrenchnolol, a “wrench-shaped” molecule, previously shown to bind tightly to the transcriptional coactivator MED23/Sur2 [81], show modest transcription activation in vitro when conjugated to a hairpin polyamide DBD [82] (Figure 3). The function of the wrenchnolol-based ATF was further extended to modulate transcription activation in cells [83].
In principle, molecules that interact with the transcriptional machinery may function as ADs by increasing the local distribution of the machinery and its functional engagement at the targeted promoter.
3.2. Repressor domains
In addition to transcription activation, ATFs can be designed to repress the transcription of targeted genes. Repressor domains have been used less frequently in ATFs than their activator counterparts. Early attempts at artificial repressor modules revealed peptides enriched in positively charged residues [84]. Much of the recent work on repressor domains has been based on peptides derived from natural repressors (e.g., Kruppel-associated box (KRAB) domain) [15]. The Barbas group achieved transcriptional repression of the protooncogene erbB-2 by fusing a zinc finger DBD to the natural repressor domains KRAB, ERD repressor domain, or mSIN3 interaction domain (SID) [85]. The advantage of using repressor domains lies in the ability to actively repress gene expression, rather than doing so by competitive inhibition, as in the case of TF displacement [44], or by nucleic acid decoys [86, 87]. In principle, the repression domain bypasses the need for competitively displacing endogenous transcription factors and would use the cellular repression machinery to down regulate targeted genes.
4. Controlling the activity of ATFs
A desirable characteristic of a TF mimic would be the ability to externally regulate its function at desired times and locations. Also, the utility of an ATF would be greatly increased by coupling its function to endogenous signaling cascades. A first step toward this goal relied on the use of the ligand-binding domain (LBD) of nuclear receptors. Fusing the LBD to a zinc finger ATF allowed control of the ATF activity by external delivery of its hormone ligand [88]. In this example, the LBDs of the estrogen and progesterone receptors were used. In a more direct and elegant approach, the ZF DBD itself was engineered to dock a small molecule “prosthetic”. The resulting ATF required the small molecule ligand for DNA binding activity and transcriptional activity in cells [89].
A similar strategy was used for an RNA-based activation domain. The AD was rendered ligand-dependent by including an aptamer sequence in the AD that recognized tetramethylrosamine (TMR). In the absence of TMR, the ATF promoted transcription of a reporter gene in Saccharomyces cerevisiae. Conversely, in the presence of TMR, the conformational changes of the TMR-bound aptamer resulted in an inactive conformation of the RNA activation domain [90].
Using a novel approach to control ATF activity, Hauschild et al develop a temperature controlled ATF [91]. A hairpin PA was conjugated to a peptide that interacted with the endogenous transcription factor Exd. This PA-peptide conjugate efficiently recruited Exd to bind to DNA. The activity of the PA-peptide conjugate was made sensitive to temperature by optimizing the length of the linker connecting the PA and the peptide hook. One of the linkers tested efficiently recruited Exd to its DNA binding site between 4°C and 23°C, but above 30°C the ATF was no longer functional [91].
5. Current challenge: Improving cellular uptake
For ATFs to be more broadly effective, they must go through the cell membrane, enter the nucleus, find the targeted sequence in the genome and recruit the cellular machinery for either transcription activation or repression. Through all of these steps, the ATF must circumvent various surveillance mechanisms of the cell. Therefore, it is not surprising that efficient delivery of ATFs remains an obstacle.
One approach towards improving cellular uptake of molecules is the use of cell penetrating peptides (CPPs) [92, 93]. CPPs are often derived from proteins that naturally translocate across cell membranes, such as the Tat protein from HIV [94] and the homeodomain Antennapedia (AtnHD) from Drosophila [95]. Also, molecules enriched in arginine, inspired by the Tat protein, have been developed as synthetic CPPs [96, 97]. In an interesting application of this strategy, a zinc finger ATF that upregulates VEGF-A was rendered cell-permeable by conjugating it to a 10-residue fragment from the HIV Tat protein [98, 99]. In addition, Mascareñas and colleagues improved the nuclear localization of a tripyrrole DNA binding molecule by conjugating the compound to an arginine octapeptide [100].
In another approach, the Dervan lab has improved nuclear localization of PAs by conjugating isophthalic acid derivates to the C-terminus of the PA [51, 53]. Conversely, the Kodadek lab improved nuclear localization by using an ethylene diamine turn for the hairpin PA in an ATF [101]. It should be pointed out that in the latter example, the hairpin PA is conjugated to a lipophylic steroid, which may enhance the permeability of the PA through hydrophobic membranes. In fact, covalent attachment of a steroid to a PNA has been shown to increase cellular uptake [102]. Improving the cellular uptake of TF mimics remains an active area of research.
6. Future directions
6.1. Cooperativity
Attempts to target unique sites in the genome have relied on expanding the number of DNA binding modules. For example, polydactyl ZFs have been engineered to target DNA sites of 18 bp. In the case of PAs, tandem hairpins were synthesized to target larger sites [103]. While reasonable, both examples were accompanied by a significant drop in selectivity due to increased non-specific binding [24, 103]. Natural transcription factors overcome the difficulties associated with finding unique targets in large genomes by forming non-covalent complexes through cooperative binding at the DNA target site [5]. Binding sites for multiple activators are commonly found in the gene promoter regions of higher eukaryotes. In principle, incorporation of cooperative binding between ATFs and other transcription factors should enhance the functionality of ATFs [104].
A new class of ATFs that incorporates cooperative binding was developed by Arndt et al. The ATF functions by nucleating the assembly of natural transcription factors on promoters/enhancers [105]. In this particular case, a synthetic mimic of the Hox family of transcription factors was generated. Hox proteins are developmental regulators, and have poor DNA affinity and sequence specificity on their own. However, cooperative binding with partner proteins, such as Exd, can increase their DNA binding affinity and specificity [106]. A polyamide was conjugated to a dipeptide derived from the Hox protein known to interact with Exd. The Hox mimic was able to cooperatively interact with Exd and bind to specific DNA sequences with nanomolar affinity [105, 107].
The principle of cooperative binding to DNA was incorporated in the ATF designed by the Ptashne and Dervan groups [48]. Specifically, the dimerization domain of Gcn4, a yeast transcription factor, was incorporated into a polyamide-based ATF [48]. Similarly, cooperative binding to DNA was incorporated in a DBD designed by Blanco et al. [108]. In this work, a tripyrrole-cyclodextrin (CD) conjugate that binds to an A/T rich DNA region was used to recruit a 23 amino acid peptide derived from the basic region of a bzip TF conjugated to an adamantane group. In the absence of the tripyrrole-CD conjugate, the BR peptide does not bind to DNA. This work adds to previous studies by Schepartz and coworkers, that achieved the dimerization of bZip DNA binding domains by incorporating transition-metal binding groups into the DBDs [109].
6.2. “Molecular blinking” of activation domains
An unusually potent AD, P201, was discovered by screening random sequences of 8 amino acid residues attached to Gal4(1–100), a DNA binding domain [68]. Like natural TFs, P201 also targets Gal11/Med15, a component of the transcriptional machinery [110]. This small artificial AD is as potent as the natural AD, Gal4, and it is far stronger than artificial ADs of similar size [68]. Mechanistic studies of this potent ATF identified that hydrophobic intramolecular interactions between the dimerization domain of the DBD Gal4(1–100) and the AD were crucial for the potency of the ATF [71]. Mutations which disrupted or further stabilized this interaction significantly lowered the activity of the ATF.
It was hypothesized that this transient intramolecular interaction between domains “masks” the unstructured peptide AD from unproductive interactions that could lead to ATF degradation (Figure 4). In support of this “molecular blinking” hypothesis, the cellular potency of natural and unnatural peptide ADs was found to significantly increase when this “masking” interaction was engineered into the ATF [72]. The potency of the ATF decreased when the interaction between the AD and DBD was disrupted. This was completely unexpected, and it revealed that a fine balance between “exposure” and “masking” significantly affected the potency of the ATF. Engineering such new properties will aid in the development of potent ATFs.
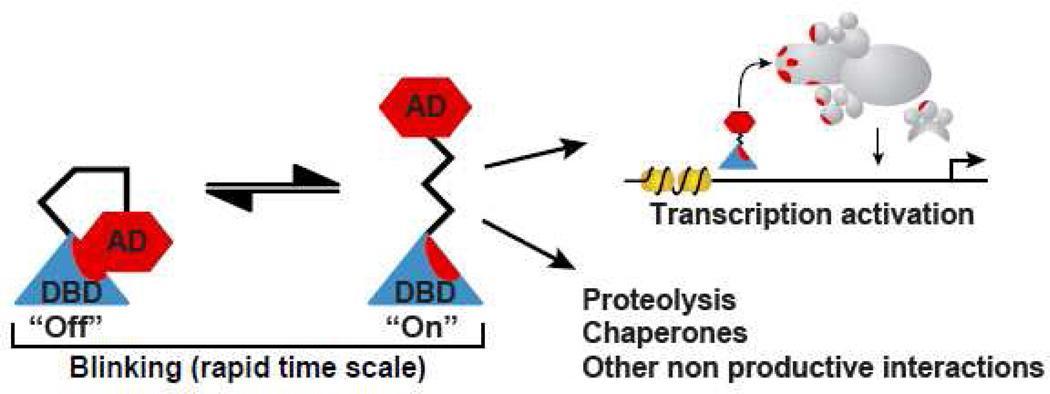
Figure 4. Model of ATF blinking.
In the blinking model, the ATF is in rapid equilibrium between an ‘off’ (masked) state and an ‘on’ (exposed). The interconversion between these two states is mediated by intermolecular interactions between the DBD and the AD. In the ‘off’ state, the ATF is masked from the cellular milieu that could lead to degradation; whereas in the ‘on’ state the AD is transiently exposed and is able to recruit the transcriptional machinery.
7. Conclusion
During the last decade, many advances have been made in the design of ATFs, with some ATFs currently undergoing clinical trials [111]. However, some obstacles must be overcome in order to realize the full potential of ATFs. Spatial and temporal control of the ATF activity and incorporating the ATF into cell signaling pathways are also highly desirable goals. Surmounting these challenges will require the collaboration of chemists, biologists, computational scientists, and bioengineers.
Acknowledgements
The authors would like to thank past and present members of the Ansari Lab. This work was supported by the NIH, NCI, NSF, March of Dimes and the Greater Milwaukee Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hauschild KE, Carlson CD, Donato LJ, Moretti R, Ansari AZ. Transcription Factors. In: Begley T, editor. Wiley Encyclopedia of Chemical Biology. Vol. 4. New York: John Wiley & Sons, Inc; 2008. pp. 566–584. [Google Scholar]
- 2.Darnell JE. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 3.Brennan P, Donev R, Hewamana S. Targeting transcription factors for therapeutic benefit. Mol. Biosys. 2008;4:909–919. doi: 10.1039/b801920g. [DOI] [PubMed] [Google Scholar]
- 4.Arora PS, Ansari AZ. A Notch above other inhibitors. Nature. 2009;462:171–173. doi: 10.1038/462171a. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2002. [Google Scholar]
- 6.Green MR. Eukaryotic transcription activation: Right on target. Mol. Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Mapp AK, Ansari AZ. A TAD further: Exogenous control of gene activation. ACS Chem. Biol. 2007;2:62–75. doi: 10.1021/cb600463w. [DOI] [PubMed] [Google Scholar]
- 8.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 11.Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 12.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 14.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 15.Ansari AZ, Mapp AK. Modular design of artificial transcription factors. Curr. Op. Chem. Biol. 2002;6:765–772. doi: 10.1016/s1367-5931(02)00377-0. [DOI] [PubMed] [Google Scholar]
- 16.Ansari AZ. Regulating gene expression: The design of synthetic transcriptional regulators. Curr. Org. Chem. 2001;5:903–921. [Google Scholar]
- 17.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavletich N, Pabo C. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 19.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 20.Beerli RR, Barbas CF. Engineering polydactyl zinc-finger transcription factors. Nat. Biotech. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 21.Segal DJ, Barbas CF. Custom DNA-binding proteins come of age: polydactyl zinc-finger proteins. Curr. Op. Biotech. 2001;12:632–637. doi: 10.1016/s0958-1669(01)00272-5. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys(2)His(2) zinc finger proteins. Ann. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 23.Meng XD, Brodsky MH, Wolfe SA. A bacterial one-hybrid system for determining the DNAbinding specificity of transcription factors. Nat. Biotech. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF. Modular system for the construction of zinc-finger libraries and proteins. Nat. Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv. Drug Del. Rev. 2009;61:513–526. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson CD, Warren CL, Hauschild KE, Ozers MS, Qadir N, Bhimsaria D, Lee Y, Cerrina F, Ansari AZ. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4544–4549. doi: 10.1073/pnas.0914023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 30.Moscou MJ, Bogdanove AJ. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science. 2009;326:1501–1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 31.Kay S, Hahn S, Marois E, Hause G, Bonas U. A Bacterial Effector Acts as a Plant Transcription Factor and Induces a Cell Size Regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 32.Gurlebeck D, Thieme F, Bonas U. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 2006;163:233–255. doi: 10.1016/j.jplph.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Doan TL, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Helene C. Sequence-specific recognition, photo-cross-linking and cleavage of the DNA double helix by oligo-alphathymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 35.Beane RL, Ram R, Gabillet S, Arar K, Monia BP, Corey DR. Inhibiting gene expression with locked nucleic acids (LNAs) that target chromosomal DNA. Biochemistry. 2007;46:7572–7580. doi: 10.1021/bi700227g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Han Y, Ferdous A, Corey DR, Kodadek T. Transcription activation by a PNA-peptide chimera in a mammalian cell extract. Chem. Biol. 2003;10:909–916. doi: 10.1016/j.chembiol.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Simon P, Cannata F, Concordet J-P, Giovannangeli C. Targeting DNA with triplex-forming oligonucleotides to modify gene sequence. Biochimie. 2008;90:1109–1116. doi: 10.1016/j.biochi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Kuznetsova S, Ait-Si-Ali S, Nagibneva I, Troalen F, Le Villain J, Harel-Bellan A, Svinarchuk F. Gene activation by triplex-forming oligonucleotide coupled to the activating domain of protein VP16. Nucleic Acids Res. 1999;27:3995–4000. doi: 10.1093/nar/27.20.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanojevic D, Young RA. A Highly Potent Artificial Transcription Factor. Biochemistry. 2002;41:7209–7216. doi: 10.1021/bi015906b. [DOI] [PubMed] [Google Scholar]
- 40.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Op. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 41.Wemmer DE, Dervan PB. Targeting the minor groove of DNA. Curr. Op. Struct. Biol. 1997;7:355–361. doi: 10.1016/s0959-440x(97)80051-6. [DOI] [PubMed] [Google Scholar]
- 42.Dervan PB, Poulin-Kerstien AT, Fechter EJ, Edelson BS. DNA Binders and Related Subjects. Vol. 253. Berlin: Springer-Verlag Berlin; 2005. Regulation of gene expression by synthetic DNA-binding ligands; pp. 1–31. [Google Scholar]
- 43.Puckett JW, Muzikar KA, Tietjen J, Warren CL, Ansari AZ, Dervan PB. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 2007;129:12310–12319. doi: 10.1021/ja0744899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 45.Gearhart MD, Dickinson L, Ehley J, Melander C, Dervan PB, Wright PE, Gottesfeld JM. Inhibition of DNA binding by human estrogen-related receptor 2 and estrogen receptor alpha with minor groove binding polyamides. Biochemistry. 2005;44:4196–4203. doi: 10.1021/bi047872o. [DOI] [PubMed] [Google Scholar]
- 46.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muzikar KA, Nickols NG, Dervan PB. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16598–16603. doi: 10.1073/pnas.0909192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Activation of gene expression by small molecule transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3930–3935. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansari AZ, Mapp AK, Nguyen DH, Dervan PB, Ptashne M. Towards a minimal motif for artificial transcriptional activators. Chem. Biol. 2001;8:583–592. doi: 10.1016/s1074-5521(01)00037-0. [DOI] [PubMed] [Google Scholar]
- 50.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao X, Yu P, Lim H-S, Sikder D, Kodadek T. A Cell-Permeable Synthetic Transcription Factor Mimic. Angew. Chem. Int. Ed. 2007;46:2865–2868. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]
- 53.Jacobs CS, Dervan PB. Modifications at the C-Terminus To Improve Pyrrole-Imidazole Polyamide Activity in Cell Culture. J. Med. Chem. 2009;52:7380–7388. doi: 10.1021/jm900256f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S-cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 55.Mavrich TN, Jiang CZ, Ioshikhes IP, Li XY, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF. Nucleosome organization in the Drosophila genome. Nature. 2008;453 doi: 10.1038/nature06929. 358-U327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, Wei G, Zhao K. Dynamic Regulation of Nucleosome Positioning in the Human Genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 58.Dudouet B, Burnett R, Dickinson LA, Wood MR, Melander C, Belitsky JM, Edelson B, Wurtz N, Briehn C, Dervan PB, Gottesfeld JM. Accessibility of nuclear chromatin by DNA binding polyamides. Chem. Biol. 2003;10:859–867. doi: 10.1016/j.chembiol.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-Specific Targeting of H3K9 Methylation Is Sufficient for Initiating Repression In Vivo. Curr. Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 60.Snowden AW, Zhang L, Urnov F, Dent C, Jouvenot Y, Zhong XH, Rebar EJ, Jamieson AC, Zhang HS, Tan SY, Case CC, Pabo CO, Wolffe AP, Gregory PD. Repression of vascular endothelial growth factor A in glioblastoma cells using engineered zinc finger transcription factors. Cancer Res. 2003;63:8968–8976. [PubMed] [Google Scholar]
- 61.van der Gun BTF, Maluszynska-Hoffman M, Kiss A, Arendzen AJ, Ruiters MHJ, McLaughlin PMJ, Weinhold E, Rots MG. Targeted DNA Methylation by a DNA Methyltransferase Coupled to a Triple Helix Forming Oligonucleotide To Down-Regulate the Epithelial Cell Adhesion Molecule. Bioconjugate Chem. 2010;21:1239–1245. doi: 10.1021/bc1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic alpha-helix linked to a DNA-binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 63.Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 64.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 65.Vanhoy M, Leuther KK, Kodadek T, Johnston SA. The acidic activation domains of the Gcn4 and Gal4 proteins are not alpha-helical but form beta-sheets. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- 66.Lee C, Chang JH, Lee HS, Cho YJ. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Gen. Dev. 2002;16:3199–3212. doi: 10.1101/gad.1046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 68.Lu X, Ansari AZ, Ptashne M. An artificial transcriptional activating region with unusual properties. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1988–1992. doi: 10.1073/pnas.040573197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frangioni JV, LaRiccia LM, Cantley LC, Montminy MR. Minimal activators that bind to the KIX domain of p300/CBP identified by phage display screening. Nat. Biotech. 2000;18:1080–1085. doi: 10.1038/80280. [DOI] [PubMed] [Google Scholar]
- 70.Wu ZQ, Belanger G, Brennan BB, Lum JK, Minter AR, Rowe SP, Plachetka A, Majmudar CY, Mapp AK. Targeting the transcriptional machinery with unique artificial transcriptional activators. J. Am. Chem. Soc. 2003;125:12390–12391. doi: 10.1021/ja036685v. [DOI] [PubMed] [Google Scholar]
- 71.Lu Z, Rowe SP, Brennan BB, Davis SE, Metzler RE, Nau JJ, Majmudar CY, Mapp AK, Ansari AZ. Unraveling the mechanism of a potent transcriptional activator. J. Biol. Chem. 2005;280:29689–29698. doi: 10.1074/jbc.M504895200. [DOI] [PubMed] [Google Scholar]
- 72.Lum JK, Majmudar CY, Ansari AZ, Mapp AK. Converting inactive peptides into potent transcriptional activators. ACS Chem. Biol. 2006;1:639–643. doi: 10.1021/cb600363n. [DOI] [PubMed] [Google Scholar]
- 73.Liu B, Alluri PG, Yu P, Kodadek T. A Potent Transactivation Domain Mimic with Activity in Living Cells. J. Am. Chem. Soc. 2005;127:8254–8255. doi: 10.1021/ja0515295. [DOI] [PubMed] [Google Scholar]
- 74.Sengupta DJ, Wickens M, Fields S. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA. 1999;5:596–601. doi: 10.1017/s1355838299002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saha S, Ansari AZ, Jarell KA, Ptashne M. RNA sequences that work as transcriptional activating regions. Nucleic Acids Res. 2003;31:1565–1570. doi: 10.1093/nar/gkg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buskirk AR, Kehayova PD, Landrigan A, Liu DR. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 77.Kehayova PD, Liu DR. In vivo evolution of an RNA-based transcriptional silencing domain in S. cerevisiae. Chem. Biol. 2007;14:65–74. doi: 10.1016/j.chembiol.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 78.Minter AR, Brennan BB, Mapp AK. A small molecule transcriptional activation domain. J. Am. Chem. Soc. 2004;126:10504–10505. doi: 10.1021/ja0473889. [DOI] [PubMed] [Google Scholar]
- 79.Rowe SP, Casey RJ, Brennan BB, Buhrlage SJ, Mapp AK. Transcriptional up-regulation in cells mediated by a small molecule. J. Am. Chem. Soc. 2007;129:10654–10655. doi: 10.1021/ja0736865. [DOI] [PubMed] [Google Scholar]
- 80.Buhrlage SJ, Bates CA, Rowe SP, Minter AR, Brennan BB, Majmudar CY, Wemmer DE, Al-Hashimi H, Mapp AK. Amphipathic small molecules mimic the binding mode and function of endogenous transcription Factors. ACS Chem. Biol. 2009;4:335–344. doi: 10.1021/cb900028j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shimogawa H, Kwon Y, Mao Q, Kawazoe Y, Choi Y, Asada S, Kigoshi H, Uesugi M. A wrenchshaped synthetic molecule that modulates a transcription factor - Coactivator interaction. J. Am. Chem. Soc. 2004;126:3461–3471. doi: 10.1021/ja038855+. [DOI] [PubMed] [Google Scholar]
- 82.Kwon Y, Arndt HD, Mao Q, Choi Y, Kawazoe Y, Dervan PB, Uesugi M. Small Molecule Transcription Factor Mimic. J. Am. Chem. Soc. 2004;126:15940–15941. doi: 10.1021/ja0445140. [DOI] [PubMed] [Google Scholar]
- 83.Jung DJ, Shimogawa H, Kwon Y, Mao Q, Sato S, Kamisuki S, Kigoshi H, Uesugi M. Wrenchnolol Derivative Optimized for Gene Activation in Cells. J. Am. Chem. Soc. 2009;131:4774–4782. doi: 10.1021/ja900669k. [DOI] [PubMed] [Google Scholar]
- 84.Saha S, Brickman JM, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:648–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- 85.Beerli RR, Segal DJ, Dreier B, Barbas CF. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cassiday LA, Maher LJ. In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- 87.Borgatti M, Finotti A, Romanelli A, Saviano M, Bianchi N, Lampronti I, Lambertini E, Penolazzi L, Nastruzzi C, Mischiati C, Piva R, Pedone C, Gambari R. Peptide nucleic acids (PNA)-DNA chimeras targeting transcription factors as a tool to modify gene expression. Curr. Drug Targets. 2004;5:735–744. doi: 10.2174/1389450043345155. [DOI] [PubMed] [Google Scholar]
- 88.Beerli RR, Schopfer U, Dreier B, Barbas CF. Chemically regulated zinc finger transcription factors. J. Biol. Chem. 2000;275:32617–32627. doi: 10.1074/jbc.M005108200. [DOI] [PubMed] [Google Scholar]
- 89.Lin Q, Barbas CF, Schultz PG. Small-molecule switches for zinc finger transcription factors. J. Am. Chem. Soc. 2003;125:612–613. doi: 10.1021/ja028408e. [DOI] [PubMed] [Google Scholar]
- 90.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 91.Hauschild KE, Metzler RE, Arndt HD, Moretti R, Raffaelle M, Dervan PB, Ansari AZ. Temperature-sensitive protein-DNA dimerizers. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5008–5013. doi: 10.1073/pnas.0501289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Snyder EL, Dowdy SF. Cell penetrating peptides in drug delivery. Pharm. Res. 2004;21:389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- 93.Joliot A, Prochiantz A. Transduction peptides: from technology to physiology. Nat. Cell Biol. 2004;6:189–196. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- 94.Frankel AD, Pabo CO. Cellular uptake of the Tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 95.Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. The design of guanidinium-rich transporters and their internalization mechanisms. Adv. Drug Del. Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tachikawa K, Schröder O, Frey G, Briggs SP, Sera T. Regulation of the endogenous VEGF-A gene by exogenous designed regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15225–15230. doi: 10.1073/pnas.0406473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yun CO, Shin HC, Kim TD, Yoon WH, Kang YA, Kwon HS, Kim SK, Kim JS. Transduction of artificial transcriptional regulatory proteins into human cells. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vazquez O, Blanco-Canosa JB, Vazquez ME, Martinez-Costas J, Castedo L, Mascarenas JL. Efficient DNA binding and nuclear uptake by distamycin derivatives conjugated to octa-arginine sequences. ChemBioChem. 2008;9:2822–2829. doi: 10.1002/cbic.200800345. [DOI] [PubMed] [Google Scholar]
- 101.Liu B, Kodadek T. Investigation of the Relative Cellular Permeability of DNA-Binding Pyrrole-Imidazole Polyamides. J. Med. Chem. 2009;52:4604–4612. doi: 10.1021/jm9002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boffa LC, Scarfi' S, Mariani MR, Damonte G, Allfrey VG, Benatti U, Morris PL. Dihydrotestosterone as a Selective Cellular/Nuclear Localization Vector for Anti-Gene Peptide Nucleic Acid in Prostatic Carcinoma Cells. Cancer Res. 2000;60:2258–2262. [PubMed] [Google Scholar]
- 103.Kers I, Dervan PB. Search for the optimal linker in tandem hairpin polyamides. Bioorg. Med. Chem. 2002;10:3339–3349. doi: 10.1016/s0968-0896(02)00221-3. [DOI] [PubMed] [Google Scholar]
- 104.Moretti R, Ansari AZ. Expanding the specificity of DNA targeting by harnessing cooperative assembly. Biochimie. 2008;90:1015–1025. doi: 10.1016/j.biochi.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 105.Arndt HD, Hauschild KE, Sullivan DP, Lake K, Dervan PB, Ansari AZ. Toward artificial developmental regulators. J. Am. Chem. Soc. 2003;125:13322–13323. doi: 10.1021/ja0371395. [DOI] [PubMed] [Google Scholar]
- 106.Mann RS, Chan S-K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 107.Stafford RL, Arndt HD, Brezinski ML, Ansari AZ, Dervan PB. Minimization of a protein-DNA dimerizer. J. Am. Chem. Soc. 2007;129:2660–2668. doi: 10.1021/ja067971k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blanco JB, Dodero VL, Vazquez ME, Mosquera M, Castedo L, Mascarenas JL. Sequencespecific DNA binding by noncovalent peptide-tripyrrole conjugates. Angew. Chem. Int. Ed. 2006;45:8210–8214. doi: 10.1002/anie.200603115. [DOI] [PubMed] [Google Scholar]
- 109.Cuenoud B, Schepartz A. Altered specificity of DNA-binding proteins with transition metal dimerization domains. Science. 1993;259:510–513. doi: 10.1126/science.8424173. [DOI] [PubMed] [Google Scholar]
- 110.Lu Z, Ansari AZ, Lu X, Ogirala A, Ptashne M. A target essential for the activity of a nonacidic yeast transcriptional activator. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8591–8596. doi: 10.1073/pnas.092263499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Ann. Rev. Biochem. 2010;79 doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 112.Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the transactivator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]