Abstract
Dysregulated usage of pre-mRNA splicing sites contributes to the progression of cancer, neurodegenerative diseases, and viral infections. Serine/arginine-rich (SR) proteins play major roles in the splice site recognition and are largely regulated by phosphorylation. This provides an option for the pharmacological correction of aberrant splicing by inhibiting the relevant kinases. Cdc2-like kinases (Clks) and dual specificity tyrosine phosphorylation-regulated kinases (Dyrks) were both reported to phosphorylate numerous SR proteins in vitro and in vivo. In this study, we describe the discovery of new selective dual Clk/Dyrk1A/1B inhibitors, which are able to modulate alternative pre-mRNA splicing of model gene transcripts in cells with submicromolar potencies. The optimization process yielded a dual Clk and Dyrk inhibitor with exceptionally high ligand efficiency. Our results suggested that dual inhibition of both Clk1 and Dyrk1A increased the efficacy of pre-mRNA splicing modulation.
Keywords: Dual kinase inhibitor, cdc2-like kinases, dual specificity tyrosine (Y) phosphorylation regulated kinases, alternative splicing modifiers
Alternative splicing of pre-mRNAs creates several mRNA isoforms that code for proteins having distinct functions. This versatile mechanism is involved in the control of embryonic development, cell growth, and apoptosis.1 However, dysregulated or nonfunctional splicing contributes to the development and progression of neurodegenerative disorders,2 cancer, and viral infections, e.g., by HIV-1.3,4 The splice site recognition is mainly determined by the interactions of serine-arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins with specific exonic and intronic pre-mRNA sequences.5 SR proteins mostly promote the inclusion of exons via binding to exonic splicing enhancers.5,6 Their activity is largely regulated by multisite phosphorylations.5,7 To date, several kinases have been reported to phosphorylate SR proteins, including Clk1, SRPK1, topoisomerase I, and Dyrk1A.5 For instance, Clk1 activity promotes the expression of HIV-1 Gag proteins by altering the splicing of the viral RNA8 and mediates rephosphorylation of SR proteins after initial heat shock response to viral infections, enabling the resumption of viral RNA processing.9 Given the multisite phosphorylation of SR proteins, co-operative effects of the relevant protein kinases on the splicing activity can be expected. Several SR proteins were reported to be substrates of both Clk1 and Dyrk1A, including AF2/ASF, SC35, and SRp55,10−12 suggesting that it might be possible to co-operatively enhance the modulation of alternative splicing by coinhibition of the structurally related kinases (both belonging to the CMGC family of the kinome); however, this was not investigated so far. Such a simultaneous inhibition may have a therapeutic utility, e.g., for the treatment of neurodegenerative tauopathies, which are partly caused by an imbalanced 3R/4R tau isoform ratio resulting from dysregulated alternative splicing;10−12 both Dyrk1A and Clk1 were reported to be causally involved due to hyperphosphorylation of SR proteins.10−14
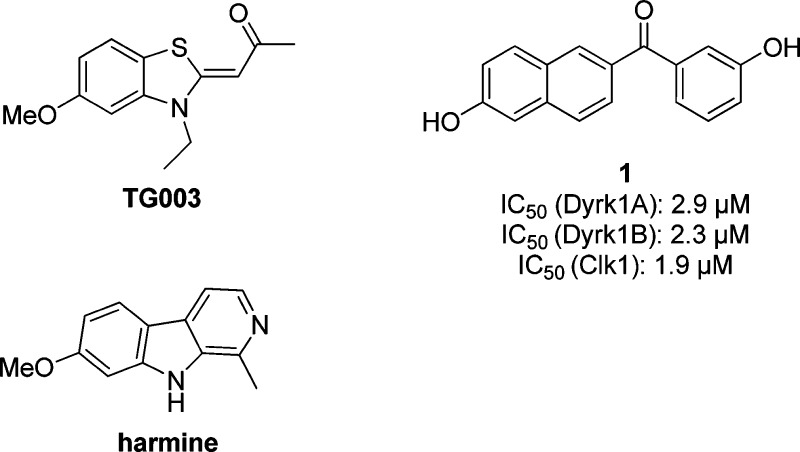
Many natural and synthetic compounds were described to interfere with splicing, some of which are general splicing inhibitors, such as spliceostatin (reviewed in15). Compounds that were reported to modulate alternative splicing of particular genes are mostly pleiotropic effectors and/or act via poorly defined mechanisms; they include cytostatic agents such as camptothecin, cardiac glycosides, flavonoids, ethanol, dietary supplements and some indole derivatives.15 In contrast, very few compounds originated from a target−oriented development of alternative splicing modulators.15 This is also true for kinase inhibitors, in spite of the known importance of SR protein phosphorylations in the regulation of alternative splicing; some of the few examples are TG003,16,17 KH-CB19,18 and NB-506.19 TG003, which is the best studied of these compounds, is a dual inhibitor of Clk’s and Dyrk1A (cf. Table S1),20 like harmine (Figure 1) and some other compounds.14 However, the majority of these compounds was not analyzed for the potential effect on alternative splicing, and the coinhibition of the Dyrk2 isoform, which functions as a negative regulator of cell proliferation in vitro and suppressor of tumor progression in vivo was a further drawback.21 This was also noted for TG003, limiting its usefulness.22
Figure 1.
Reported Clk1 and Dyrk1A inhibitors and the newly identified dual Clk/Dyrk inhibitor 1.
Herein we describe the discovery of a new class of potent and selective dual Clk and Dyrk1A/1B inhibitors. Additionally, we provide evidence that dual inhibition of Clk1 and Dyrk1A was more effective in the modulation of alternative splicing of two different model gene transcripts in cells than inhibition of Clk1 alone. Screening of our in-house collection of diverse enzyme inhibitors revealed a 6-hydroxynaphthalene ketone 1 (Figure 1) as a moderate Dyrk1A and Clk1 inhibitor. Importantly, 1 did not affect the “untouchable” Dyrk2 isoform. Therefore, we optimized 1 in terms of potency as well as selectivity. Our initial modification, the introduction of small substituents with distinct electronic effects at the phenyl ring did not have any significant impact on the biological activities (2 and 3, bearing para-methyl and para-fluorine, respectively; cf. Table S2, Supporting Information). Therefore, we replaced the central aromatic core by bioisosteric benzothiophene, benzofuran, and indole heterocycles, considering in each case the two possible hydroxyl positions (Tables 1 and S3, Supporting Information). The 6-hydroxyl-substituted compounds 4 to 9 (Table S3, Supporting Information) exhibited no improvement of potency compared to the naphthalene-based inhibitors. However, swapping the hydroxyl group from the 6- to the 5-position on the benzothiophene core led to a 10-fold increase of the activity toward the target kinases (compound 10, Table 1). Inhibition of Clk1 was slightly preferred, displaying an IC50 of about 100 nM. Because such a beneficial effect was not observed for any of the benzofuran- and indole-based analogues (11, 12, 13, and 15), further optimization efforts focused on the 5-hydroxybenzothiophene ketones. To explore the effect of the phenyl ring substitution pattern, the 3-hydroxyl group was moved to the para-position in 14, which significantly decreased the Dyrk1A inhibition by about 10-fold; however, the Clk1 inhibitory potency was less affected. Introduction of an additional para-methyl group (16) doubled the potency toward Dyrk1A and Clk1 compared with 10. While 16 was the most potent compound of this series, the complete loss of activity with 17 suggested a steric clash of the meta-methyl group with the ATP binding pocket. Interestingly, the introduction of fluorine in the phenyl moiety (18) dramatically increased the selectivity for Clk1 (IC50 = 50 nM) over Dyrk1A (IC50 = 2.0 μM). Most likely, the conformational constraints effected by the ortho-substitution were better tolerated by the Clk1 binding site. A comparison of 21 and 22 with 10 revealed that the hydroxyl group at the benzothiophene but not at the phenyl moiety was essential to the inhibitory activity. We wanted to investigate further if the phenyl ring was at all necessary for the biological activity by synthesizing compound 23, in which it was replaced by methyl. Strikingly, 23 exhibited the highest inhibitory potency besides 16, indicating that the phenyl ring might even impede an optimum protein–ligand interaction. Considering its low molecular mass (192.2 Da), 23 probably represented an optimal ligand for the ATP binding pockets of Dyrk1A and Clk1, as indicated by its exceptionally high ligand efficiency of 0.74. Since the phenyl ring was not needed for activity, it was straightforward to investigate whether an amide function was also tolerated in place of the methyl ketone. The Weinreb amide 25 turned out to be nearly as potent against Clk1 as 23, but it was considerably less active against Dyrk1A and Dyrk1B. Compound 25 was thus identified as a potential new lead for the development of selective Clk1 inhibitors.
Table 1. Inhibitory Potencies of 5-Hydroxybenzoheterocyclic Inhibitorsa.

| IC50 [μM]/% inhibition
@ 5 μM |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| compd | X | R1 | R2 | R3 | Dyrk1A | Dyrk1B | Dyrk2 | Clk1 | Ck2α |
| 10 | s | OH | H | H | 0.4 | 0.2 | 49% | 0.1 | 12% |
| 11 | 0 | OH | H | H | 3.1 | 6.1 | 34% | 1.9 | 21% |
| 12 | NH | OH | H | H | 15 | 41% | 24% | 2 | 22% |
| 13 | NMe | OH | H | H | 3.4 | 2.7 | 25% | 2.1 | 31% |
| 14 | S | H | OH | H | 3.4 | 0.8 | 25% | 0.3 | 33% |
| 15 | O | H | OH | H | 9.7 | 1.3 | 38% | 1–5 | 17% |
| 16 | S | OH | Me | H | 0.2 | 0.2 | 50% | 0.05 | 10% |
| 17 | S | Me | OH | H | 11 | 7 | 8% | 25% | |
| 18 | S | OH | H | F | 2 | 0.4 | 30% | 0.05 | 20% |
| 19 | S | OH | F | H | 1.2 | 0.4 | 27% | 0.3 | 15% |
| 20 | S | CN | H | H | 6 | 2.3 | 6% | 0.4 | 10% |
| 21 | S | H | H | H | 2 | 1 | 37% | 0.2 | 13% |
| 22 | S | 42 | 6% | 15% | 24% | ||||
| 23 | S | 0.2 | 0.1 | 46% | 0.06 | 9% | |||
| 24 | S | 10% | 40% | –7% | 47% | ||||
| 25 | S | 0.9 | 1.6 | 41% | 0.1 | ||||
| TG003 | 0.83 | 0.17 | |||||||
[ATP] = 100 μM.
A 6-hydroxybenzothiazole isomer of 10 had been previously described as a highly potent inhibitor of 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1).23 Therefore, we tested our structurally similar benzothiophene−based inhibitors for this potential off-target activity. Indeed, several compounds were identified as 17β-HSD1 inhibitors, including 16 (IC50 = 195 nM); however, 23 and 25 were completely inactive toward this enzyme (Table S4, Supporting Information), thus excluding side effects on the steroidal biosynthesis by our most active compound 23.
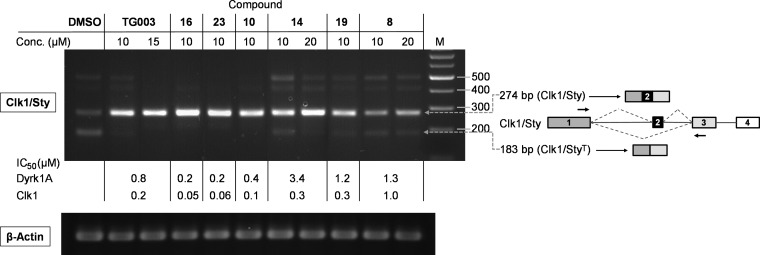
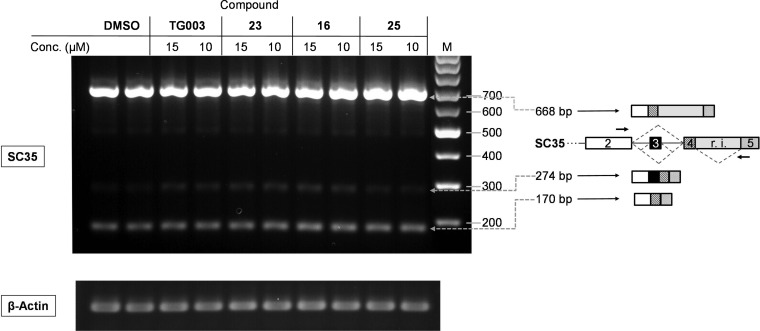
We expected good cellular activity of 16 and 23, considering that they were reasonably potent in our cell-free assays at 100 μM ATP, which was significantly above the reported KM value of Dyrk1A (cf. Table S5, Supporting Information). Thus, we decided to determine the general potency of the compounds to modulate pre-mRNA splicing in intact cells by analyzing the transcript pattern of two model genes, which are known to undergo SR protein phosphorylation−dependent alternative splicing, SC35 and Clk1 itself.24,25 Importantly, alteration of the splicing pattern of these transcripts after treatment with the reference compound TG003 had been reported.9,16 Catalytically active Clk1/Sty promotes skipping of exon 2 from its own pre-mRNA via hyperphosphorylation of SR proteins, thus producing a mRNA, which encodes the kinase-negative Clk1/StyT.16,24Vice versa, treatment of the model cell line, immortal embryonic fibroblasts (STO), with Clk1 inhibitors should lead to enhanced exon 2 inclusion. Indeed, as detected by reverse transcription (RT)-PCR, the compounds with the highest cell-free potency, 10, 16, and 23, strongly induced the production of the mature Clk1 mRNA retaining exon 2 (Figure 2). It became evident that the dual inhibitors 10, 16, 23, and TG003 were more potent in altering the splicing pattern than compounds 14 and 19, which were less active against Dyrk1A (Table 1). At the chosen concentration (10 μM), 23 led to a complete disappearance of the incompletely and alternatively spliced transcripts, while such were still visible in the case of TG003. The weakest effect among the hydroxybenzothiophenes was noted for 8, possessing the lowest potency for both Dyrk1A and Clk1. Hence, the efficacy of the Clk1 splicing modulation correlated with the potency to simultaneously inhibit Dyrk1A and Clk1. To corroborate our findings, we also analyzed the effects of the compounds on the splicing pattern of SC35, another substrate of Clk1 and Dyrk1A,11,26 which regulates splicing of its own pre-mRNA dependent on its phosphorylation status.9,16 Administration of compounds 16 and 23 into the culture medium of STO cells changed the splicing profile of SC35 in the same manner as previously shown for the reference compound TG003 (Figure 3).16 The specific enhancement of a splicing product lacking the terminal intron (represented by the 274 bp product) was slightly weaker in the case of the less potent Dyrk1A inhibitors TG003 and 25, suggesting again that dual inhibition of both Dyrk1A and Clk1 results in a stronger modulation of the alternative splicing process. The dynamic range of the visible changes in the splicing profiles was considerably higher in the case of the Clk1 mRNA (Figure 2), possibly supported by an enhanced splicing of larger, intron-retaining premature Clk1 mRNA transcripts, which were not amplified under the PCR conditions.9 Therefore, we chose the Clk1 mRNA as a PCR target for the development of a qPCR assay in order to quantify the strength of splicing modulation in cells. The results for the best compounds are shown in Table 2: 23 triggered a significant response of the Clk1 pre-mRNA splicing already at submicromolar concentrations (C5-fold = 1.1 μM), followed by 16. Again both compounds, being efficient dual Dyrk1A/Clk1 inhibitors, were more potent than the amide analogue 25, which was equally active against Clk1 but not against Dyrk1A in the cell free assay (Table 1). Similarly, significantly higher concentrations of TG003 than of 16 or 23 were needed to elicit a clear response, correlating with the less potent inhibition of Dyrk1A (IC50 = 830 nM, cf. Table 1). Assuming that there were no major differences in cell permeation between the compounds, our data suggested an at least additive effect of simultaneous Dyrk1A and Clk1 inhibition on the modulation of alternative pre-mRNA splicing. Interestingly, the maximum achievable effect on Clk1 pre-mRNA splicing was a compound−specific variable; hence, the calculated EC50 values appeared to be of limited significance. Next we examined the selectivity toward a carefully selected panel of kinases representing frequently reported off-targets and further kinases from each subfamily of the kinome. Compounds 16 and 23 showed good selectivity for the Clks and Dyrks in this panel; 23 was only slightly less selective than 16 (Table S6, Supporting Information) exhibiting a similar inhibitory potency among the closely related cdc2-like kinases Clk1–4 and, in addition, a moderate potency against the atypical kinase haspin (IC50 = 0.8 μM; Table S7, Supporting Information). Notably, our new inhibitors were clearly more selective against some frequent off-targets than TG003 (Table S8, Supporting Information). In conclusion, we developed a new dual inhibitor of Clks and Dyrks, 23, with exceptionally high ligand efficiency and notable cellular efficacy. Because of the low molecular weight, the good aqueous solubility and the selectivity among the panel of frequent off-target kinases, it might become a valuable tool for the in vitro and in vivo pharmacological validation of Dyrk1A and Clk1 coinhibition in missplicing-related diseases, including Alzheimer’s,10−14 viral infections,8,9 and congenital genetic disorders such as Duchenne muscular dystrophy.27
Figure 2.
Modulation of the Clk1/Sty splicing pattern by the Dyrk1A/Clk1 inhibitors in STO cells. The test compounds were added to the cell medium at the indicated concentrations for 4 h. DMSO was used as control (0.05%). Total RNA was purified, and the splicing pattern was analyzed by RT-PCR. The splicing variants and their expected PCR products using the primers indicated by arrows are depicted on the right. For comparison, IC50 values against the recombinant enzymes are also given; for a more detailed description, see Supporting Information, 1. Biology.
Figure 3.
Modulation of SC35 pre-mRNA splicing by the Dyrk1A/Clk1 inhibitors in STO cells. The test compounds were added to the cell medium at the indicated concentrations for 4 h. Total RNA was purified, and the splicing pattern was analyzed by RT-PCR. The splicing variants and their expected PCR products using the primers indicated by arrows are depicted on the right (for a more detailed description, see Supporting Information, 1. Biology).
Table 2. Modulation of Clk/Sty Pre-mRNA Splicing by 16, 23, and 25 in Comparison to TG003.
| TG003 | 23 | 16 | 25 | |
|---|---|---|---|---|
| C5-fold (μM)a | 3.1 | 1.1 | 1.9 | 2.4 |
| EC50 (μM)b | 6.6 | 8.9 | 1.8 | 2.1 |
| max. inductionc | 17 | 21.4 | 9.5 | 8.1 |
Concentration required to induce a 5-fold increase of the mature Clk1 transcript levels as determined by qPCR; SD ≤ 10%.
Concentration required for half-maximal generation of the mature Clk1 mRNA splicing product; SD ≤ 10%.
Maximum increase of mature Clk1 transcripts achievable with the inhibitors (fold increase over DMSO-treated samples); SD< 20%. Shown are means of three independent experiments performed in triplicates.
Acknowledgments
The authors thank Mrs. Nadja Weber and Tamara Paul for their assistance in performing splicing assays.
Glossary
ABBREVIATIONS
- CMGC
cyclin-dependent kinases, mitogen-activated protein kinases, glycogen-synthase kinases, and CDC-like kinases
- SR proteins
serine- and arginine-rich family of splicing proteins
- qPCR
quantitative real-time polymerase chain reaction
Supporting Information Available
Experimental details for all assays and syntheses; supplementary Tables S1–S8 and Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
C.S, P.M., M.M., and M.E. designed and performed research. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
We are grateful for the financial support by the DFG to M.E. (EN 381/3-1).
The authors declare no competing financial interest.
Supplementary Material
References
- Kelemen O.; Convertini P.; Zhang Z.; Wen Y.; Shen M.; Falaleeva M.; Stamm S. Function of alternative splicing. Gene 2013, 514, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey J. R.; Wang Z.; Hortobágyi T.; Witten J. T.; Zarnack K.; Kayikci M.; Clark T. A.; Schweitzer A. C.; Rot G.; Curk T.; Zupan B.; Rogelj B.; Shaw C. E.; Ule J. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011, 21, 1572–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D.; Nasr-Esfahani S.; Tan C.; O’Brien K.; Howard J.; Jans D.; Purcell D.; Stoltzfus C. M.; Sonza S. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology 2008, 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara T.; Hosoya T.; Shimizu S.; Sumi K.; Oshiro T.; Yoshinaka Y.; Suzuki M.; Yamamoto N.; Herzenberg L. A.; Herzenberg L. A.; Hagiwara M. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 11329–11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Fu X.-D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S.; Ben-Ari S.; Rafalska I.; Tang Y.; Zhang Z.; Toiber D.; Thanaraj T. A.; Soreq H. Function of alternative splicing. Gene 2005, 344, 1–20. [DOI] [PubMed] [Google Scholar]
- Misteli T.; Cáceres J. F.; Clement J. Q.; Krainer A. R.; Wilkinson M. F.; Spector D. L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998, 143, 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.; Balachandran A.; Mao A.; Dobson W.; Gray-Owen S.; Cochrane A. Differential effect of CLK SR Kinases on HIV-1 gene expression: potential novel targets for therapy. Retrovirology 2011, 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya K.; Kataoka N.; Hagiwara M. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 2011, 195, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Zhang T.; Zhou C.; Chohan M. O.; Gu X.; Wegiel J.; Zhou J.; Hwang Y.-W.; Iqbal K.; Grundke-Iqbal I.; Gong C.-X.; Liu F. Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J. Biol. Chem. 2008, 283, 28660–28669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W.; Liang H.; Shi J.; Jin N.; Grundke-Iqbal I.; Iqbal K.; Gong C.-X.; Liu F. Regulation of the alternative splicing of tau exon 10 by SC35 and Dyrk1A. Nucleic Acids Res. 2011, 39, 6161–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.; Jin N.; Gu J.; Shi J.; Zhou J.; Gong C.-X.; Iqbal K.; Grundke-Iqbal I.; Liu F. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J. Biol. Chem. 2012, 287, 30497–30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A.; Rujescu D.; Giannakouros T.; Nikolakaki E.; Goedert M.; Mandelkow E. M.; Gao Q. S.; Andreadis A.; Stamm S. Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Mol. Cell Neurosci 2001, 18, 80–90. [DOI] [PubMed] [Google Scholar]
- Jain P.; Karthikeyan C.; Moorthy N. S. H. N.; Waiker D. K.; Jain A. K.; Trivedi P. Human CDC2-like kinase 1 (CLK1): A novel target for Alzheimer’s disease. Curr. Drug Targets 2014, 15, 539–550. [DOI] [PubMed] [Google Scholar]
- Zaharieva E.; Chipman J. K.; Soller M. Alternative splicing interference by xenobiotics. Toxicology 2012, 296, 1–12. [DOI] [PubMed] [Google Scholar]
- Muraki M.; Ohkawara B.; Hosoya T.; Onogi H.; Koizumi J.; Koizumi T.; Sumi K.; Yomoda J.; Murray M. V.; Kimura H.; Furuichi K.; Shibuya H.; Krainer A. R.; Suzuki M.; Hagiwara M. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 2004, 279, 24246–24254. [DOI] [PubMed] [Google Scholar]
- Yomoda J.; Muraki M.; Kataoka N.; Hosoya T.; Suzuki M.; Hagiwara M.; Kimura H. Combination of Clk family kinase and SRp75 modulates alternative splicing of Adenovirus E1A. Genes Cells 2008, 13, 233–244. [DOI] [PubMed] [Google Scholar]
- Fedorov O.; Huber K.; Eisenreich A.; Filippakopoulos P.; King O.; Bullock A. N.; Szklarczyk D.; Jensen L. J.; Fabbro D.; Trappe J.; Rauch U.; Bracher F.; Knapp S. Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing. Chem. Biol. 2011, 18, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch B.; Allemand E.; Facompré M.; Facompre M.; Bailly C.; Riou J.; Soret J.; Tazi J. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001, 61, 6876–6884. [PubMed] [Google Scholar]
- Ogawa Y.; Nonaka Y.; Goto T.; Ohnishi E.; Hiramatsu T.; Kii I.; Yoshida M.; Ikura T.; Onogi H.; Shibuya H.; Hosoya T.; Ito N.; Hagiwara M. Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A. Nat. Commn. 2010, 1, 86. [DOI] [PubMed] [Google Scholar]
- Taira N.; Mimoto R.; Kurata M.; Yamaguchi T.; Kitagawa M.; Miki Y.; Yoshida K. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J. Clin. Invest. 2012, 122, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Davies S. P.; Augustin M.; Woodward A.; Patel U. A.; Kovelman R.; Harvey K. J. A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochem. J. 2013, 451, 313–328. [DOI] [PubMed] [Google Scholar]
- Spadaro A.; Frotscher M.; Hartmann R. W. Optimization of hydroxybenzothiazoles as novel potent and selective inhibitors of 17β-HSD1. J. Med. Chem. 2012, 55, 2469–2473. [DOI] [PubMed] [Google Scholar]
- Duncan P. I.; Stojdl D. F.; Marius R. M.; Bell J. C. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol. Cell. Biol. 1997, 17, 5996–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A.; Gattoni R.; Dooghe Y.; Stévenin J.; Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001, 20, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K.; Patel N. A.; Watson J. E.; Apostolatos H.; Kleiman E.; Hanson O.; Hagiwara M.; Cooper D. R. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCβII messenger ribonucleic acid. Endocrinology 2009, 150, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida A.; Kataoka N.; Takeshima Y.; Yagi M.; Awano H.; Ota M.; Itoh K.; Hagiwara M.; Matsuo M. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat. Commun. 2011, 2, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.