Abstract
Math1 is the defining molecule of the cerebellar rhombic lip and Pax6 is downstream in the Math1 pathway. In the present study, we discover that Wntless (Wls) is a novel molecular marker of the cells in the interior face of the rhombic lip throughout normal mouse cerebellar development. Wls expression is found complementary to the expression of Math1 and Pax6, which are localized to the exterior face of the rhombic lip. To determine the interaction between these molecules, we examine the loss-of-Math1 or loss-of-Pax6 in the cerebellum, i.e., the Math1-null and Pax6-null (Sey) mutant cerebella. The presence of Wls-positive cells in the Math1-null rhombic lip indicates that Wls expression is independent of Math1. In the Sey mutant cerebellum, there is an expansion of Wls-expressing cells into regions that are normally colonized by Pax6-expressing cells. The ectopic expression of Wls in the Pax6-null cerebellum suggests a negative interaction between Wls-expressing cells and Pax6-positive cells. These findings suggest that the rhombic lip is dynamically patterned by the expression of Wls, Math1, and Pax6. We also examine five rhombic lip cell markers (Wls, Math1, Pax6, Lmx1a, and Tbr2) to identify four molecularly distinct compartments in the rhombic lip during cerebellar development. The existence of spatial compartmentation in the rhombic lip and the interplay between Wls, Math1, and Pax6 in the rhombic lip provides novel views of early cerebellar development.
Keywords: cerebellar compartments, cerebellum, developmental biology, Pax6, rhombic lip, Wls
Introduction
Tissue patterning by gene expression is an important developmental process in the generation of different structures and specific cell types in multicellular organisms. In the developing vertebrate CNS, the midbrain and cerebellar primordia are specified by the opposing expression of Otx2 and Gbx2, which patterns the developing neural plate and positions the isthmic organizer (ISO) at the midbrain–hindbrain boundary (Broccoli et al., 1999; Millet et al., 1999). Subsequently, expression of Math1 and Ptf1a specify two distinct germinal zones in the cerebellar anlage, the rhombic lip (RL), and the ventricular zone, respectively (Ben-Arie et al., 1997; Hoshino et al., 2005; Pascual et al., 2007; Yamada et al., 2014). The Math1+ rhombic lip gives rise to the glutamatergic lineages including the large cerebellar nuclear (CN) neurons, granule cells, and unipolar brush cells (UBCs); while the Ptf1a+ ventricular zone gives rise to GABAergic lineages such as the GABAergic nuclear neurons, Purkinje cells, and interneurons.
An important molecule in the development of the main glutamatergic cell to emanate from the RL, the cerebellar granule cell, is Pax6, a paired-domain transcription factor (Engelkamp et al., 1999; Swanson et al., 2005). Pax6 expression marks the Math1+ cells that arise from the RL and the granule cell progenitors (Stoykova and Gruss, 1994). The loss of Pax6, as in the Small Eye mutant (Sey) mouse, results in the disruption of cerebellar foliation and the organization of the external germinal layer (EGL; Engelkamp et al., 1999; Swanson et al., 2005; Swanson and Goldowitz, 2011). To identify Pax6-related genes that are presumably causal for the granule cell mutant phenotype, our group studied the transcriptional network regulated by Pax6 where we measured the whole genome transcriptome of Sey mutant and wild-type cerebella at different development stages (Ha et al., 2012). By comparing the transcriptomes between Sey mutant and wild-type cerebella, we identified genes that are differentially regulated in the developing cerebellum in the loss of Pax6.
One of the genes that exhibited an upregulation in the Sey mutant cerebellum is Wntless (Wls, also known as GPR177). Wls is a highly conserved, multipass transmembrane molecule. Studies in Drosophila have indicated a role for Wls in promoting Wnt molecule secretion (Bänziger et al., 2006; Bartscherer et al., 2006). A recent study in the mouse has demonstrated the requirement of Wls in body-axis establishment (Fu et al., 2009). However, the role of Wls in cerebellar development remains unknown. The microarray analysis of transcript expression shows that in the wild-type cerebellum, Wls is highly expressed during early development and diminished over time. However, in the Sey mutant, cerebellar expression of Wls is found to be upregulated at the time when expression normally decreases in the wild-type cerebellum.
In this study, we describe Wls as a novel molecular marker of the RL that joins four other cell markers (Math1, Pax6, Lmx1a, and Tbr2) in identifying four molecularly distinct compartments in the developing RL. Math1-null and Sey mutants are used to test the interaction among Wls, Math1, and Pax6. We find that Wls expression is independent of Math1 influence in the RL, while Wls expression is negatively regulated by Pax6.
Materials and Methods
Mouse strains and husbandry.
ES cells heterozygous for a WlsLacZ reporter allele were obtained from BayGenomics gene trap mutation project (Cell line: RRJ545, RRID:IMSR_MMRRC:003140). This cell line is characterized by a β-geo gene-trap vector integrated in the intron between exon 9 and 10 of the endogenous Wls sequence. The resulting knock-in allele encodes a fusion protein between a truncated Wls and a β-gal reporter protein, and transcription is controlled under the native Wls 5′ region.
To generate the Wls-LacZ reporter animals, ES cells were injected into C57BL/6J blastocysts to create chimeras for germline transmission, and chimeras were bred to C57BL/6J mice to obtain WlsLacZ heterozygotes. Ear notches were collected at weaning and ear DNA was prepared by digestion with Proteinase K in 1X PCR tissue homogenization buffer at 55°C incubation overnight, followed by a Proteinase K inactivation step at 95°C for 10 min. PCR genotyping was performed using forward primer specific to the wild-type Wls sequence (Wls-F1: atgcaccacatacacaactgg) and reverse primers specific to the wild-type Wls sequence (Wls-R1: caggtcatgaggctgtcaat) and to the LacZ insertion sequence (LacZ: ggttgcggtggtgatataaa) that amplifies DNA fragments of 126 and 80 bp for the wild-type and WlsLacZ alleles, respectively. Primer concentrations for multiplex PCR genotyping were 575 (Wls-F1), 288 (Wls-R1), and 575 nm (LacZ). PCRs contained a final concentration of 185 μm dNTPs, 1.8 mm MgCl2, and 1 U of TaqDNA polymerase. Cycling conditions were as follows: first denaturation step at 94°C for 2 min, 35 cycles of denaturation at 94°C for 30 s, hybridization at 60°C for 45 s and elongation at 72°C for 1 min, and end with a final elongation step at 72°C for 6 min. PCR product was applied to TBE agarose gel for analysis.
The Pax6-null mutant strain, Pax6Sey (obtained from Robert Grainger and Marilyn Fisher, University of Virginia), was used in the study of Wls expression. The strain was bred, phenotyped, and genotyped as previously described (Swanson et al., 2005).
The Math1-lacZ reporter strain (obtained from Huda Zoghbi, Baylor College of Medicine) was used in the study of RL marker expression and Math1-KO experiments. The Math1 genotype was determined by PCR according to protocol previously described (Jensen et al., 2002).
Experimental wild-type Wls+/+ and WlsLacZ/+ heterozygous mice were generated by intercrossing WlsLacZ/+ mice or outcrossing carriers to ICR mice. No phenotypic differences were noted between embryos generated by either approach. Mice of wild-type Pax6, Pax6 mutants, wild-type Math1, and Math1LacZ/LacZ mutants were generated by heterozygote matings. The morning of the day that a vaginal plug was detected was designated as E0.5. All studies were conducted according to the protocols approved by Institutional Animal Care and Use Committee and Canadian Council on Animal Care at the University of Tennessee Health Science Center and the University of British Columbia.
BrdU labeling.
To examine cell proliferation in the cerebellar RL, timed pregnant females were injected intraperitoneally with BrdU (Sigma, B5002; 50 μg/g body weight) 1 h before the collection of embryos. Tissue was processed and sectioned as described below. To quantify the number of BrdU+ cells in the cerebellar RL, ∼50 sections that were equally distributed across the full cerebellum, right and left sides inclusive, were analyzed.
Tissue preparation and histology.
Embryos of either sex harvested between E10.5 and E16.5 were fixed by immersion in 4% paraformaldehyde in 0.1 m PB, pH 7.4, for 1 h at 4°C. Embryos harvested at E18.5 were perfused with 4% paraformaldehyde in 0.1 m PB and brain tissues were isolated and further fixed in 4% paraformaldehyde in 0.1 m PB for 1 h at room temperature. For tissues to be sectioned, fixed tissues were rinsed with PBS, followed by cryoprotection with 30% sucrose/PBS overnight at 4°C before embedding in OCT compound. Tissues were sagittally sectioned at 12 μm for immunohistochemistry or 16 μm for in situ hybridization; cryosections were mounted on SuperFrost slides (Fisher), air dried at room temperature, and stored at −80°C until used. For whole-mount β-gal staining, fixed tissues were rinsed twice with PBS and finished with one PBS-T wash (0.1 m PBS with 0.1% Triton X-100) before staining. In all cases, observations were based on a minimum of three embryos to a maximum of eight embryos per experiment.
In situ hybridization.
Sense and antisense riboprobes corresponding to the cDNA fragment of Wls (position 371–1487 in NM026582.3) were generated and labeled with DIG-UTP. Wls cDNA was generated from a cDNA library obtained from E15.5 and P0 mouse brain generated with a cDNA synthesis kit (Invitrogen) using the forward (GCAGTACCCTACACGGCAAT) and reverse (GGCTAGACTGCTTCCCACTG) primers. The resultant Wls cDNA was cloned into the pGEM-T vector, and was used to generate cDNA templates for the sense and antisense riboprobes, with the primers M13F or M13R and the aforementioned forward or reverse primers. Before hybridization, sections were acetylated with acetic anhydride in 0.1 m triethanolamine at pH 8.0 and dehydrated with graded concentrations of ethanol. Sections were incubated with cDNA probe in Ultra-hybridization buffer (Ambion) at 55°C overnight in a humid chamber. After hybridization, the slides were washed and rinsed in descending concentrations of salt: 4X SSC, 2X SSC, 1X SSC, and 0.5X SSC at 55°C, and then incubated with an anti-DIG antibody (Roche) for 2 h at room temperature at a concentration of 1:300. After washing, slides were colorized with NTP/BICP (Roche) and mounted with 1.5% gelatin containing 15% glycerol.
Immunohistochemistry.
Tissue sections were rehydrated to PBS. For bright-field immunohistochemistry, endogenous peroxidase activity was inhibited by treating the sections with 1% H2O2 in PBS followed by a PBS-T rinse. Sections were incubated at room temperature for 20 min with blocking solution (1% BSA and 5% normal serum in PBS-T), and subsequently incubated at room temperature overnight with primary antibodies in a humid chamber. Following PBS-T washes, the sections were incubated with biotinylated secondary antibodies (1:200; Vector Laboratories) and processed for PAP immunohistochemistry using the ABC Kit (Vector Laboratories) according to the manufacturer's protocol. Slides were dehydrated and coverslips were applied. For immunofluorescence, secondary antibodies labeled with fluorochrome were used to recognize the primary antibodies. The slides were counterstained and mounted with Vectashield mounting media with DAPI (Vector Laboratories). Primary antibodies used were as follows: rabbit anti-WlsN (1:500; YenZym Antibodies), rabbit anti-WlsC (1:1000; initially a gift from Richard Lang, University of Cincinnati; then purchased from Seven Hills Bioreagents, WLAB-177), rabbit anti-Pax6 (1:200; Covance Research Products, PRB-278P-100, RRID:AB_10092959), rabbit anti-LMX-1 (1:2000; EMD Millipore, AB10533, RRID:AB_10805970), rabbit anti-Tbr2 (1:600; Millipore, Ab2283, RRID:AB_10806889), mouse anti-BrdU (1:200; Becton Dickinson, 347580, RRID:AB_400326), and chicken anti-β-gal (1:10,000; Abcam, Ab9361, RRID:AB_307210).
Detection of β-gal activity.
To detect β-gal activity in the WlsLacZ/+ reporter mice, 4% paraformaldehyde-fixed embryos or cryosections were incubated in an X-gal reaction buffer (containing 5 mm potassium ferricyanide, 5 mm potassium ferricyanide, 2 mm MgCl2, 0.02% Nonidet P-40, 0.01% sodium deoxycholate in 0.1 m PBS-T, and 1 mg/ml X-gal; Invitrogen, 15520-018; dissolved in DMSO) at 37°C overnight in a humid chamber. After incubation, tissues were rinsed with PBS. Cryosections processed for X-gal activity were counterstained with neutral red.
Microscopy.
Analysis and photomicroscopy were performed with a Zeiss Axiovert 200M microscope with the AxioCam/AxioVision hardware–software components (Carl Zeiss). Confocal microscopy was performed using an Olympus FV500 confocal laser scanning microscope and the FluoView image-capture and analysis software.
Results
Expression profile of Wls in the embryonic cerebellum
To elucidate the role Wls plays in cerebellar development, we first determined the expression profile of Wls in cerebellum during embryonic development. In situ hybridization was performed on sectioned brain tissues collected from E13.5, E15.5, and E18.5 wild-type embryos. Expression of Wls transcript is localized predominately to the cerebellar RL throughout these embryonic stages (Fig. 1A–C, arrows). Contiguous with the cerebellar RL, the choroid plexus epithelium of the fourth ventricle and the lower RL of the hindbrain are also found to express Wls. At the midbrain–hindbrain boundary, the ISO region, Wls transcript is also detected at all embryonic stages examined (Fig. 1A, asterisk; data not shown). Cells in the ventricular zone are also positive, particularly at E15.5. Staining is less evident in the ventricular zone at E13.5 and E18.5. Wls transcript is found to be lightly expressed in the meninges and roof plate over the cerebellum.
Figure 1.
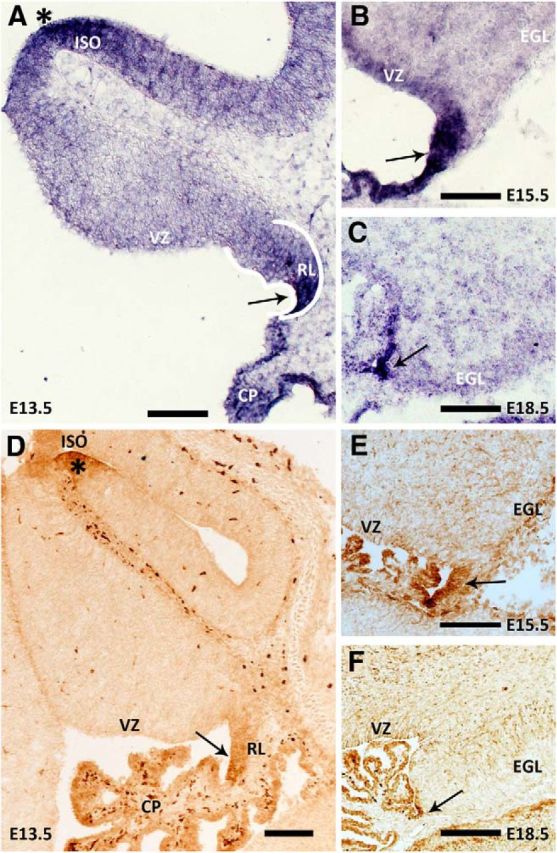
Wls expression is localized to the cerebellar RL during embryonic development. Expression of Wls transcript and protein in the cerebellum, across embryonic development, is revealed by in situ hybridization (A–C) and immunohistochemistry (D–F) analyses on wild-type brain tissues collected at E13.5, E15.5, and E18.5. Wls expression is localized predominately to the cerebellar RL at all ages examined (arrows). Wls is also expressed at the ISO region (asterisks in A and D), in the cells of the choroid plexus of the fourth ventricle, and in the meninges over the cerebellum and roof plate. CP, choroid plexus; EGL, external germinal layer; VZ, ventricular zone. Scale bars: 100 μm.
To further characterize Wls in the developing cerebellum, we examined the expression of Wls protein using immunohistochemistry with two antibodies that recognize either the N-terminal or the C-terminal amino acid of the Wls peptide. Wls protein expression is detected in the cerebellum at all developmental stages examined (E13.5, E15.5, and E18.5; Fig. 1D–F). Expression is localized to the cerebellar RL, choroid plexus, lower RL, and ISO region as well as the meninges and roof plate over the cerebellum. The expression patterns are identical between the two antibodies used for Wls protein detection (data not shown). This pattern of immunostaining is identical to the expression profile of the Wls transcript. These results demonstrate that Wls has a very restricted expression domain in the developing cerebellum. In the adult, the only expression of Wls, which is light, is found in the Purkinje cells (data not shown).
Expression of the β-gal reporter in WlsLacZ/+ mice
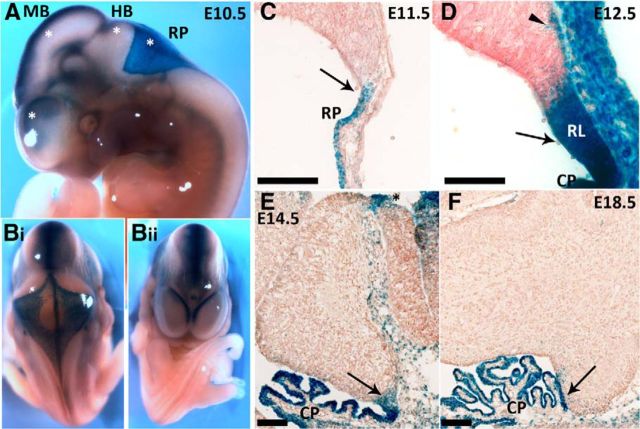
To further elucidate the expression pattern of Wls, we used a β-gal reporter mouse strain (see Materials and Methods section). Mice heterozygous for the knock-in allele appear normal and are fertile, homozygous WlsLacZ/LacZ, however, is early embryonic lethal. Embryos ranging from E10.5 to E18.5 were harvested from heterozygote × wild-type matings. Tissues from heterozygote and wild-type embryos were processed for X-gal staining. The presence of β-gal reporter activity is readily detected with X-gal staining in whole-mount and sectioned tissues of heterozygous embryos, while staining is absent, as would be expected, from wild-type tissues (Fig. 2; data not shown). At the early embryonic stage, E10.5, a strong dorsal midline expression of the reporter protein is detected in the developing brain, including the telencephalon, diencephalon, mesencephalon, and the rhombencephalon (Fig. 2A,B). Also noticeable at this early stage is a broad expression at the hindbrain roof plate that gives rise to the future choroid plexus (Fig. 2A). Examination at the cellular level reveals that in the early developing cerebellum, expression of the reporter protein is seen in the cells of the roof plate and the developing choroid plexus (E11.5; Fig. 2C). At later stages of embryonic development, X-gal staining is largely restricted to cells in the cerebellar RL and the choroid plexus (Fig. 2D–F), the ISO region (Fig. 2E, black asterisks), and the meninges. The embryonic expression pattern of the reporter protein recapitulates endogenous Wls expression in the cerebellum (Fig. 1). There are a few Wls-immunonegative but β-gal-positive cells in the subpial stream from E11.5 to E14.5 suggesting that these cells are from the Wls lineage that has downregulated Wls expression (Fig. 2D, arrowhead).
Figure 2.
Expression of the β-gal reporter protein in WlsLacZ/+ mice recapitulates the expression of endogenous Wls during cerebellar development. The WlsLacZ/+ reporter strain carries a transgene that expresses the β-gal protein under the control of the endogenous Wls 5′ region. Sections of WlsLacZ/+ embryos and brain tissues were stained for β-gal activity. A, Expression of the β-gal protein is observed in the CNS as early as E10.5, at the developing forebrain, midbrain, hindbrain, and the roof plate (asterisks). B, A strong midline expression is observed in these developing brain regions. The embryos are positioned to provide a view from the posterior (looking at the roof of the fourth ventricle) in Bi and from the anterior (looking at the telencephalic vesicles) in Bii. C–F, Expression of the β-gal reporter protein in the developing cerebellum is localized to the upper RL (arrows in C–F), ISO region (black asterisk in E), choroid plexus, and the meninges over the cerebellum and roof plate. A cohort of cells positive for β-gal but Wls immunonegative is observed at the cerebellar surface subpial stream (arrowhead in D). CP, choroid plexus; HB, hindbrain; MB, midbrain; RP, roof plate. Scale bars: 100 μm.
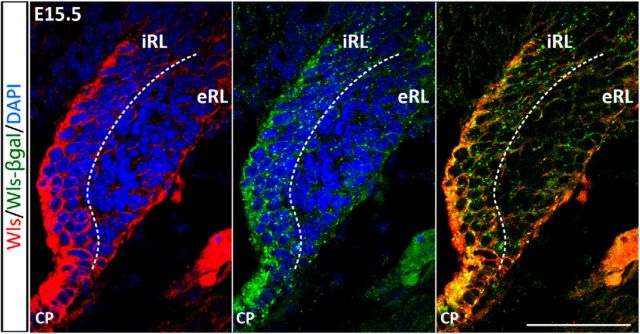
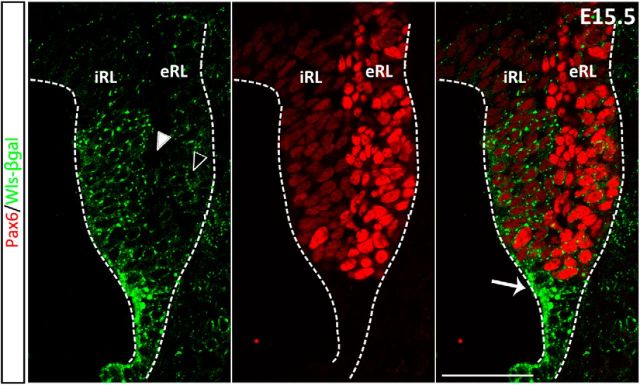
At the cellular level, Wls immunostaining indicates that the protein is localized in the cytoplasm (Fig. 3, left). Staining for β-gal reporter protein with β-gal antibody also shows a cytoplasmic localization and staining appears punctate (Fig. 3, middle). The double-labeling for β-gal and Wls protein (using the antibody against the C terminus) in heterozygous embryos illustrates colocalization in the cells of the cerebellar RL (Fig. 3, right). When labeled with the Wls antibody that recognizes the N terminus of the Wls protein, the staining of Wls appears punctate and is perfectly colocalized with the β-gal staining (data not shown). These findings corroborate the notion that each of these three visualization techniques provides a faithful rendition of Wls expression in the developing cerebellum. Most importantly, these three approaches identify the Wls-positive cells as largely localized to the interior face of the RL (iRL), while cells of the exterior face of the RL (eRL) are Wls negative (Fig. 3, regions separated by dashed line). Furthermore, these findings validate the use of this transgene reporter model to study Wls expression.
Figure 3.
Wls and β-gal reporter proteins are colocalized in the RL. Immunohistochemistry was performed on brain tissues of E15.5 WlsLacZ/+ embryos using anti-β-gal antibody and an antibody that targets the C terminus of Wls (absent in the truncated reporter protein). Expression of endogenous Wls is localized predominately to the iRL (red fluorescence, left), whereas the eRL is Wls negative. The β-gal reporter is expressed at the same region in the iRL (green punctate fluorescence, middle). Double labeling shows that the expression of Wls and β-gal proteins is colocalized in the cytoplasm of the iRL cells (merged image, right). CP, choroid plexus. Scale bar, 50 μm.
Expression of Wls is complementary to, and independent of, Math1 expression in the RL
Math1 is the master control gene that defines the cerebellar RL and is critical for the generation of RL derivatives. The novel finding that Wls is expressed in the RL prompted us to ask whether Wls is a member of the Math1 molecular cascade. As a first step, we compared the anatomical expression of Wls with Math1 in the wild-type RL. Math1 is expressed in the RL as early as E9.5 and in the RL derivatives during embryonic development (Machold and Fishell, 2005; Wang et al., 2005). By using the Math1LacZ/+ reporter mouse (β-gal reporter protein expression driven under Math1 locus; Ben-Arie et al., 2000), we found that during early development (E11.5), Wls expression in the RL is largely complementary to the Math1-expressing cells that are located at the surface of the emerging cerebellar anlagen (Fig. 4A). In the E13.5 cerebellum, Wls is expressed throughout the RL (Fig. 5A) and the overlap of expression of Math1 (β-gal positive; Fig. 5B) and Wls occurs in columns oriented in the anterior-to-posterior domain (Fig. 5C, bounded by the dotted line). In the E15.5 cerebellum, Math1 and Wls expression domains further segregate, where the expression of Wls is localized predominately to the iRL (Figure 5D) and Math1 expression to the eRL (Fig. 5E), with only a few cells coexpressing Wls and Math1 in the RL (Fig. 5F, arrow). At the E15.5 time point the Wls-positive cells end abruptly at the presumed dorsal border of the RL (Fig. 5D) while Math1-positive cells are seen to be continuous with the forming EGL (Fig. 5F). By E18.5, Wls expression is restricted to the diminishing RL and completely segregated from the Math1-expressing cells in the EGL (Fig. 5G–I). Thus, Wls is largely expressed in different cells than those that express Math1.
Figure 4.
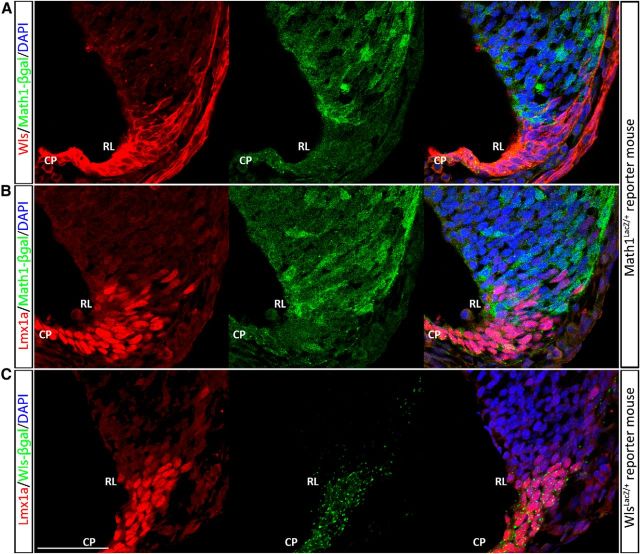
The cerebellar RL is comprised of two molecular populations (Wls+/Lmx1a+/Math1− and Wls−/Lmx1a+/Math1+) during early development at E11.5. Immunocytochemical demonstration of expression of Wls (A, red fluorescence, left) and Lmx1a (B and C, red fluorescence, left) and Math1 (A and B, green fluorescence, as seen in Math1LacZ/+ mice). Math1 is expressed in cells that are at the surface of the cerebellar anlagen and nonoverlapping with Wls (A) expression in the RL. There is intermixing of Wls+ Math1−cells and Wls−/Math1+ cells at the boundary of the two expression domains (A, right); a minimal number of cells shows coexpression of Lmx1a and Math1 (B, right). C, Examination of Lmx1a-positive (red fluorescence, left) and Wls-positive cells (green fluorescence, middle) reveals that Wls and Lmx1a are largely coexpressed in the cells of RL and choroid plexus (CP) at early cerebellar development. Scale bar, 50 μm.
Figure 5.
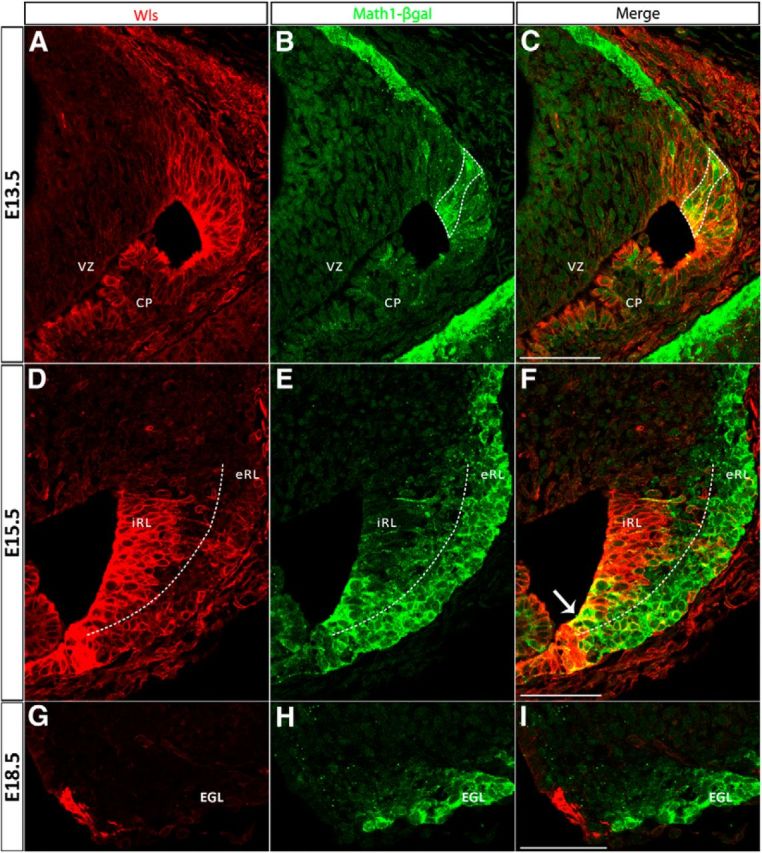
The cerebellar RL progressively develops molecularly distinct populations during embryonic development identified by Wls and Math1 expression. The expression of Wls and Math1 was studied in Math1LacZ/+ mice, which express a β-gal reporter protein under control of the Math1 locus. Immunolabeling for Wls (red fluorescence, left) and β-gal (green fluorescence, middle) proteins is shown at E13.5 (A–C, top), E15.5 (D–F, middle), and E18.5 (G–I, bottom). At E13.5, Wls is expressed throughout the rhombic lip (A) and Math1 is strongly expressed in the emerging EGL and exhibits columns of expression in the RL (B). The overlap of Wls and Math1 expression domains in the cerebellar RL is shown in C (bounded by dotted line) where a subset of Math1-positive cells are also Wls positive at this early stage. Later at E15.5, Wls expression is localized predominately to the iRL (D), whereas Math1 expression is localized to the eRL and EGL (E). Expression at this time is largely nonoverlapping with only a few cells (arrow in F) coexpressing Wls and Math1 at the RL. At E18.5, the Wls-positive population found in the RL (G) is completely segregated from the Math1-positive population in the EGL (H). CP, choroid plexus; EGL, external germinal layer; VZ, ventricular zone. Scale bars: 50 μm.
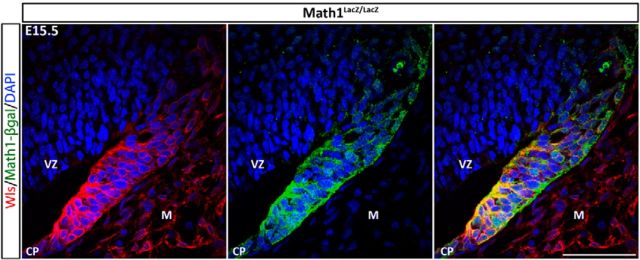
Given the differential cellular localization of Math1 and Wls, we hypothesized that these two molecules play independent roles in the life of the RL. To test this hypothesis we examined the Math1-null mutant to determine whether the fate of Wls-expressing cells was altered in the KO. At E15.5, the EGL is absent in the Math1-null as previously reported (Ben-Arie et al., 2000; Jensen et al., 2004; Wang et al., 2005), and the cerebellar RL appears much smaller in the mutant cerebellum compared with that of the wild-type. Surprisingly, Wls expression appears unaltered in the RL, choroid plexus, and ISO region of the Math1-null cerebellum (Fig. 6; data not shown). This Wls-positive domain in the Math1-null mutant maps onto the cytoarchitectonics of the cells of the iRL [i.e., in columnar arrays as noted by Jensen et al., (2004)]. Interestingly, there is now coexpression of Wls and Math1 in many cells of the iRL. This coexpression is only seen in a limited number of cells in the wild-type RL (compare Figs 5F, Fig. 6, right). The finding of a lack of alterations of Wls immunoreactivity in the Math1-null is consistent with the transcript level of Wls in the Math1-null cerebellum (Ha et al., 2012). These results demonstrate that the Wls-expressing domain in the RL is independent of the regulation of Math1.
Figure 6.
Wls expression is unchanged in the Math1-null RL. The expressions of Wls (red fluorescence, left) and the β-gal reporter protein of Math1 (green fluorescence, middle) are examined in the Math1LacZ/LacZ-null mutant cerebellum at E15.5. Math1-null mutant cerebella lack EGL and RL derivatives. Nevertheless, Wls expression remains unaffected in the Math1-null cerebellum and is detected in the remaining cells of RL, which are arranged in columnar arrays that cytoarchitecturally resemble the cells of the iRL. The merging of images of Wls and Math1 expression demonstrates, on the right, the coexpression of Wls and Math1 in many cells of the iRL. This coexpression is only seen in a limited number of cells in the wild-type RL (Fig. 5F). CP, choroid plexus; M, meninges; VZ, ventricular zone. Scale bar, 50 μm.
Expression of Wls is complementary to, and negatively regulated by, Pax6 in the RL
Pax6 expression is eliminated from the Math1-null cerebellum and therefore considered to be downstream Math1 in the RL (Wang et al., 2005; Englund et al., 2006; Fink et al., 2006). Given that Wls expression is independent of Math1, as shown above, we hypothesized that Pax6 and Wls are also independently expressed. However, our previous Pax6 transcriptome analysis suggests the hypothesis that Wls expression is regulated by Pax6 as we observed an upregulation of Wls transcript in the Pax6-null cerebellum (Ha et al., 2012). Initially, to examine these hypotheses, we compared the anatomical expression of Wls with Pax6 in the wild-type RL. Immunohistochemical analysis revealed three configurations of Pax6-positive and Wls-positive cells in the E15.5 RL (Fig. 7). First, in the eRL, as with Math1, there are strongly positive Pax6 cells that do not express Wls. A second population is found of low-expressing Pax6 cells in the iRL, which strongly express Wls. Finally, there is a set of cells in the most distal part of the RL that does not express Pax6, but strongly expresses Wls (Fig. 7, arrow). At E18.5, the cells in the iRL now coexpress Wls and Pax6, in contrast to the exclusionary relationship between Wls and Math1at the same time point (Fig. 5I; data not shown).
Figure 7.
The E15.5 RL is comprised of three domains as shown by Wls- and Pax6-expressing cells. The E15.5 RL was immunostained for Pax6 (red fluorescence, middle) and Wls (green fluorescence, left) in E15.5 WlsLacZ/+ embryos. Staining reveals that the iRL contains cells that robustly express Wls but are also lightly positive for Pax6. The eRL contains cells that robustly express Pax6 but are also lightly positive for Wls. There is also a population of cells that exclusively expresses Wls at the RL (white arrow, right). At the boundary of eRL and iRL, a population of cells is found negative for Wls and Wls reporter expression (compare white arrowhead to black arrowhead). Scale bar, 50 μm.
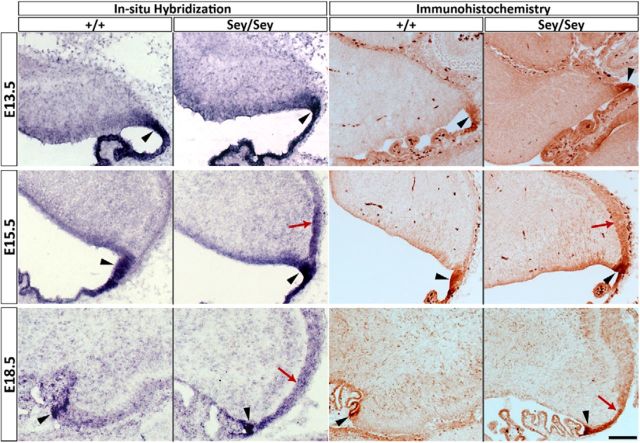
A more direct test of the above hypotheses would be to examine the expression of Wls in the Sey cerebellum. We find that Wls transcript expression is upregulated as early as E15.5 by 1.35-fold (p = 0.002) and 2.15-fold (p = 3.21 × 10−5) at E18.5 (Ha et al., 2012). There are two possibilities that could explain these molecular findings. (1) There is an upregulation of Wls in Wls-positive cells in the mutant cerebellum, which would imply that Pax6 normally exerts control (i.e., suppression) on the expression level of Wls in the iRL, and the absence of Pax6 releases the suppression and results in a higher expression of Wls in the same number of cells. (2) A second possibility is that Wls expression expands into domains not normally occupied by Wls-expressing cells, suggesting that Pax6 suppresses Wls expression in the Pax6-expressing cells. To assess these nonexclusionary possibilities, we examined the expression of Wls in the Sey mutant cerebellum by in situ hybridization and immunohistochemistry. At E13.5 there are no apparent differences in the expression of Wls in wild-type and Sey mutant cerebella (Fig. 8). However, at E15.5 we observed a striking expansion of the Wls expression domain beyond the cerebellar RL into the nascent EGL in the Sey mutant cerebellum (Fig. 8, E15.5). This exuberant expression of Wls is maintained at E18.5 in the Sey mutant cerebellum, at a time when wild-type Wls is on the wane (Fig. 8). Furthermore, the expanded expression of Wls in the Sey mutant is specific to the EGL as the isthmus expression is unaltered. In wild-type and heterozygous Sey embryos, cerebellar morphology and Wls expression patterns are indistinguishable (data not shown). The study of the Sey mutant cerebellum indicates that Wls expression is normally negatively regulated by the expression of Pax6 in the cells of the EGL.
Figure 8.
Expansion of Wls expression domain in the Sey mutant cerebellum. Expression of Wls was examined with in situ hybridization and immunohistochemistry in Sey mutant and wild-type cerebellum across embryonic development. At E13.5 (top), Wls expression is restricted to the RL (black arrowheads) in wild-type and Sey cerebella. A striking expansion of the Wls expression domain in the Sey mutant cerebellum is found at E15.5 and E18.5. At E15.5 (middle), expression of Wls has expanded beyond the RL (black arrowheads) into the nascent EGL in the Sey mutant (red arrows). This upregulation of Wls expression is maintained at E18.5 (bottom) in the Sey cerebellum (red arrows), when Wls expression has markedly decreased in the wild-type RL (the RL is identified by black arrowheads in wild-type and mutant cerebella). Scale bar, 100 μm.
Expression of Wls in relation to markers of the RL
In this study, we find evidence that the RL is actively patterned by gene expression. We show that Wls is expressed in the iRL and complementary to Math1 and Pax6 expression in the eRL. Furthermore, we demonstrate that the Wls-expression domain arises independent of Math1, but Wls is restricted to the iRL by the negative regulation of Pax6 in the eRL. To further characterize the different molecular domains in the RL, we examined the expression of Wls in relation to two additional markers of the cells in RL, Lmx1a, and Tbr2.
Lmx1a is a roof plate marker expressed in the RL and choroid plexus in the developing hindbrain (Chizhikov et al., 2006). Expression of Lmx1a is observed in the fourth ventricle choroid plexus throughout embryonic development. During cerebellar development Lmx1a marks the CN neuron precursors at E13.5 that migrate to the nuclear transitory zone (NTZ; Chizhikov et al., 2010). Lmx1a is also expressed in the UBCs that are found in the RL during E15.5 to E18.5 (Chizhikov et al., 2010). The relationship between the Lmx1a and Wls expression domains is dynamic during cerebellar development. Early at E11.5, expression of Wls and Lmx1a overlap in the roof plate (Fig. 4C). Lmx1a expression is largely complementary to that of Math; with an intermixing of Lmx1a-positive and Math1-positive cells at the border of the expression domain of each molecule, the number of cells coexpressed Math1 and Lmx1a is minimal (Fig. 4B). At E15.5, Lmx1a is weakly expressed in the iRL that contains Wls+ cells. However, a stream of cells with strong Lmx1a expression is detected at the interface of iRL and eRL (Fig. 9A, bounded by the yellow dotted line), and these cells are Wls negative. It is worth noting that these cells are devoid of β-gal expression (Figs. 7, 9, white arrowheads) in contrast to the Pax6-positive cells in the eRL, which still maintain a weak expression of β-gal (Figs. 7, 9, black arrowheads). At E18.5, expression of Lmx1a is excluded from the Wls-positive cells in the iRL (data not shown).
Figure 9.
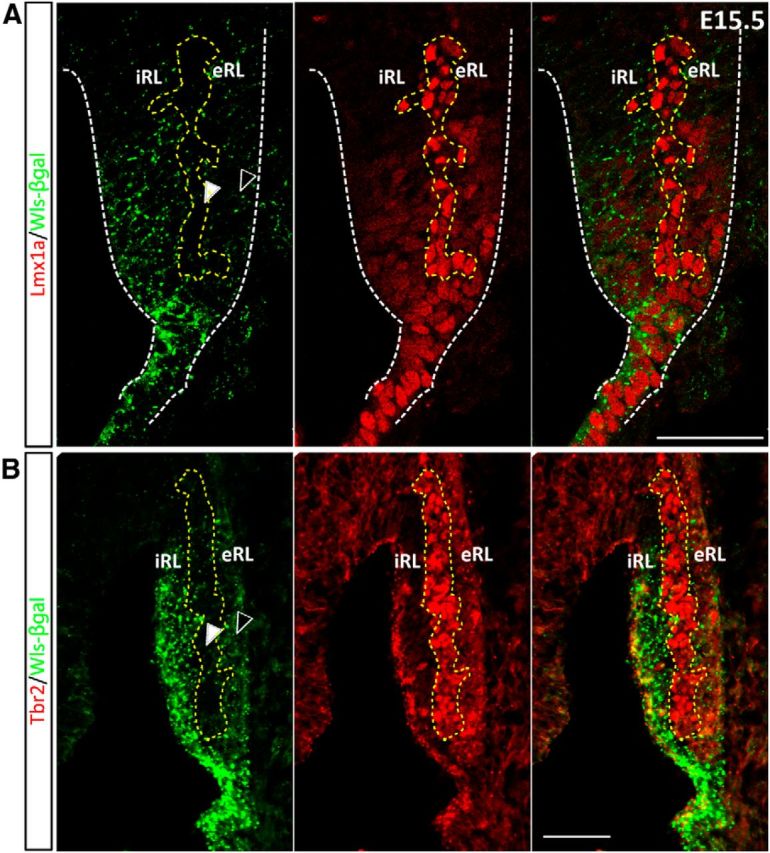
The expression of Wls, Lmx1a, and Tbr2 defines a distinct molecular domain within the E15.5 RL. A, The E15.5 RL was immunostained for Lmx1a (red fluorescence, middle) and Wls (green fluorescence, left) in E15.5 WlsLacZ/+ embryos. Staining reveals that there is a cohort of Lmx1a-positive cells that exist between the Wls and Pax6 populations of cells. These cells are not only immunonegative for Wls, but also very low in Wls reporter protein expression (bounded by yellow dotted line and white arrowhead, left) as compared with the Wls-negative cells in the adjacent eRL (black arrowhead, left). B, The E15.5 RL was immunostained for Tbr2 (red fluorescence, middle) and Wls (green fluorescence, left) in E15.5 WlsLacZ/+ embryos. Cells positive for Tbr2 (bounded by yellow dotted line) are negative for Wls and Wls reporter expression (compare white arrowhead to black arrowhead). Scale bar, 50 μm.
Tbr2 has been demonstrated to be a marker of UBCs and NTZ cells in the developing cerebellum (Englund et al., 2006; Fink et al., 2006). Tbr2 is not expressed in cells of the RL until E15.5. At this time Tbr2 is strongly expressed in a subset of cells localized to the interface between the iRL and eRL (Fig. 9B, bounded by the yellow dotted line). At E15.5, these Tbr2+ cells are negative for Wls expression, and interestingly, these cells are also devoid of β-gal expression (Fig. 9B, white arrowhead), similar to the observation of Lmx1a expression at the same age (compare Fig. 9A, white arrowhead). Expression of Tbr2 remained complementary to Wls+ iRL at E18.5 (data not shown).
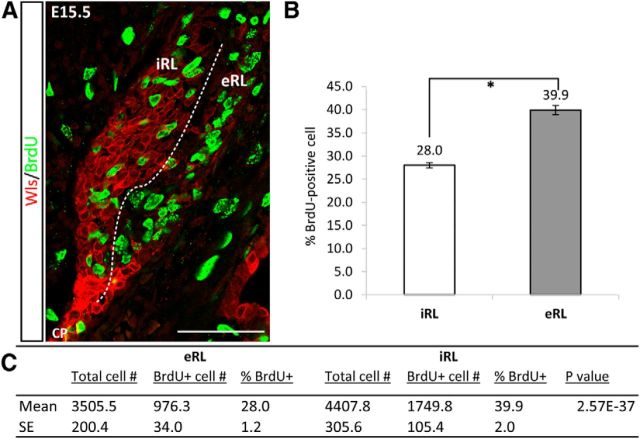
With the identification of cell populations with different molecular identities in the RL, i.e., Wls+ cells in the iRL and Math1+ and Pax6+ in the eRL, we wondered if there were any cellular phenotypic differences that distinguished these cells. We examined cell death and cell proliferation in these two populations. Cell death is limited in the RL and displayed no obvious differences (data not shown). To assess cell proliferation, we pulse labeled embryos with BrdU at E15.5. The proliferative population of cells, as demonstrated with BrdU immunohistochemistry, appeared to be fewer in the iRL compared with eRL (Fig. 10A). To quantify whether the proliferative activity of Wls-positive cells in the iRL was reduced when compared with eRL cells, a labeling index was calculated for the iRL and eRL cell populations. In the E15.5 cerebella, the proliferative index is significantly higher in the eRL (39.9%) compared with that in the iRL (28.0%; Fig. 10B,C). These results suggest that the molecularly distinct cell populations in the iRL and eRL are also phenotypically different.
Figure 10.
The iRL and eRL have differential-proliferative activity. Cell-proliferative activity in the iRL and eRL was studied with pulse-BrdU labeling and quantitative assessment at E15.5. A, The E15.5 RL is double labeled for Wls (red fluorescence) and BrdU (green fluorescence), and proliferative cells were observed in both the Wls+ iRL and Wls− eRL. B, Proliferative indices are calculated for the iRL and eRL cell populations (n = 4 embryos), and proliferation activity is significantly different between the two populations of cells. Error bars represent SE; *p < 0.0001. C, Cell counts of total cells and BrdU-positive cells in iRL and eRL (n = 4 embryos). Scale bar, 50 μm.
Discussion
The RL refers to the proliferative neuroepithelium along the dorsal alar plate (His, 1891). The RL can be further separated into two domains: the cerebellar RL that arises from rhombomere 1 and gives rise to cells in the cerebellum, and the lower RL that arises from rhombomere 2 to 8 and generates the precerebellar nuclei of the hindbrain (Rodriguez and Dymecki, 2000; Ray and Dymecki, 2009). Studies have shown that precursor cells emerge from the cerebellar RL to form the adjacent EGL, and classically believed to be specified exclusively to a granule cell lineage (Hatten and Heintz, 1995; Alder et al., 1996). More recently, genetic lineage tracing of RL cells marked by Math1 and Wnt1 gene expression identified that the CN neurons and UBCs also arise from the RL (Machold and Fishell, 2005; Wang et al., 2005; Englund et al., 2006; Hagan and Zervas, 2012). Interestingly, Altman and Bayer (Altman and Bayer, 1997) made the observation that the RL is comprised of two cytologically distinct epithelial faces, providing the possible morphogenetic basis for multiple cell types arising from the RL.
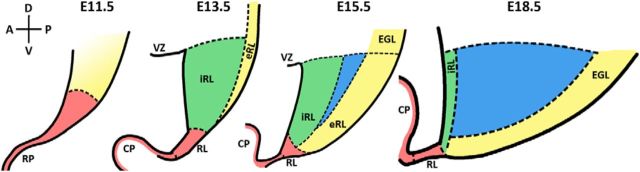
In the present studies, we describe a novel RL marker, Wls, which marks a cell population in the iRL that is found to be molecularly and cellularly distinct from the complementary Math1-positive eRL cell populations. Our finding of the Wls+/Math1− population indicates molecular heterogeneity in the RL. Recent studies have also noted molecularly heterogeneous populations of cells in the RL, defined by Lmx1a+/Math1− and Wnt1+/Math1− expression (Chizhikov et al., 2010; Cheng et al., 2012; Hagan and Zervas, 2012). To further examine the ideas of RL compartmentation, we performed the expression of a panel of five RL markers (Wls, Math1, Pax6, Lmx1a, and Tbr2) at early and later developmental time points. Our results reveal a more complex compartmentation in the RL compared with previous studies (Chizhikov et al., 2010; Cheng et al., 2012; Hagan and Zervas, 2012) and define four molecularly distinct compartments in the RL (see summary schematic; Fig. 11). These compartments are defined by (1) Wls-positive cells in the roof plate and the distal tip of the RL from E11 to E18 (Fig. 11, pink region). These cells only express low levels of Math1 and Lmx1a and are negative for Pax6 and Tbr2. (2) Strong expression of Wls in cells of the interior face of the RL, a cytoarchitecturally distinct region characterized by a columnar arrangement of cells (Altman and Bayer, 1997; Fig. 11, green region). This compartment becomes apparent at approximately E13.5 with a strong Wls expression and an intermixing of Math1+ cells and a low level of Pax6 and Lmx1a expression. At E15.5, these cells are found to be largely Math1 negative and are Math1 independent (Jensen et al., 2004 and the current paper). As development progresses (e.g., by E18.5), these Wls+ cells are completely segregated from the Math1-expressing cells. (3) Strong expression of Math1 and Pax6 is seen in cells at the exterior face of the RL (Fig. 11, yellow region) as early as E13.5. These molecules are also expressed in cells that continue into the EGL. Expression of Wls is downregulated in this compartment at all developmental times, although the cells in this compartment are likely of Wls lineage as suggested by the presence of Wls-reporter protein noted in the Results section. (4) Cells with the molecular signature of Tbr2+/Lmx1a+/Pax6+/Wls− are found between the iRL and eRL regions at E15.5 and later (Fig. 11, blue region). It is noteworthy that cells in this compartment do not show any Wls-reporter expression (compared with the low level of reporter expression in cells of the adjacent eRL). Cells in this compartment may arise from a non-Wls lineage or from Wls-expressing cells that have extinguished previous Wls expression.
Figure 11.
Summary schematic of molecularly distinct developmental compartments in the cerebellar RL. Four compartments marked by differential molecular expression in the RL evolve over developmental time in the mouse cerebellum. The pink region represents the population of Wls-positive cells in the roof plate and the distal tip of the RL found in the E11–E18 cerebellum. This region is devoid of Pax6 and Tbr2 expression. The yellow region is defined by strong expression of Math1. At E11, this region is largely Math1 positive with a few cells that coexpress Lmx1a. By E13 and onward, this region is marked by strong Math1 and Pax6 expression. Wls is downregulated in this region but Wls reporter expression is detected. The green region is characterized by strong Wls expression and weak expression of Math1 and Pax6. This region becomes apparent as early as E13, and clearly segregates from the yellow region by E15. The blue region defines a population of cells marked by the molecular signature of Tbr2+/Lmx1a+/Pax6+/Wls−, which is found in the RL from E15 and later. This region is devoid of Wls-reporter expression. A, anterior; CP, choroid plexus; D, dorsal; EGL, external germinal layer; P, posterior; RP, roof plate; V, ventral; VZ, ventricular zone.
In the RL, how do the compartments we define in this study relate to the generation of specific cell types from the RL? It has been demonstrated that CN neurons arise from the Math1+ RL at E10.5–E12.5 (Machold and Fishell, 2005). At this time the RL is demarcated by Wls and Math1 into two compartments, Wls+/Math1− and Wls−/Math1+ (Fig. 11, yellow and pink zones). A third molecule, Lmx1a, is largely expressed in Wls+ cells and a few Math1+ cells at the compartment boundary. It is found that Lmx1a lineage does not contribute to CN neurons (Chizhikov et al., 2010). Therefore, the domain that gives rise to CN neurons maps to the compartment identified in this study by Math1+/Wls−/Lmx1a− at E11.5 (Fig. 11, yellow compartment). Granule cell progenitors arise later from the RL starting at E12.5 (Machold and Fishell, 2005) and have strong expression of Math1 (Machold and Fishell, 2005) and Pax6 (Engelkamp et al., 1999) but not Lmx1a (Chizhikov et al., 2010) in the EGL, which aligns with the eRL in Figure 11 at E13.5–E15.5. This is supported by the observation that in the Math1-null RL, the yellow compartment is absent and so is granule cell production (Wang et al., 2005 and current findings). The origins of UBCs coincide with the blue region in Figure 11 defined by Tbr2+/Lmx1a+/Pax6+/Wls− at E15.5–E18.5 (Englund et al., 2006; Chizhikov et al., 2010). Thus, our gene mapping in combination with mutant analyses defines four distinct, molecularly defined compartments in the developing RL.
In this work we examine the interaction between Math1 and Wls in the RL using the Math1-null mutant, and find the presence of Wls-expressing cells in the Math1-null RL, which indicates that the Wls+ domain is independent of Math1 regulation. Given that Math1 is required and sufficient in generating cerebellar glutamatergic neurons from the neuroepithelium (Wang et al., 2005; Yamada et al., 2014) and that all glutamatergic RL derivatives are lost in the absence of Math1: Does our finding imply that Wls+ domain is dispensable in the generation of cerebellar cell types? Interestingly, we observed a weak expression of the Wls-reporter protein in some cells in the adjacent Math1+ eRL, the subpial stream, and the EGL, which may indicate that the Math1+ RL cells are of Wls lineage. A genetic fate mapping study has shown that RL cells that express Math1 rapidly migrate out of the RL (Machold and Fishell, 2005), which opens the question of the source of progenitors that feed into the Math1+ population. Our observation could suggest that Wls+ cells give rise to the Math1+ RL cells and replenish the cells that exit the RL once Math1 expression is switched on. Thus, the Wls+ domain in the iRL may serve as the reservoir of precursors for the Math1+ RL cells (Fig. 12A). In line with this, a recent study has found that β-catenin, the key mediator of Wnt signaling and downstream of Wls, activates in vitro expression of Math1 in neural progenitor cells (Shi et al., 2010). By a similar mechanism, Wls may induce Math1 expression in the cells of the RL through the activation of Wnt signaling. Thus, the in vitro observation and our present data raise the possibility that Wls is upstream of Math1 and that the Wls+ domain provides both the cells and the signal for the Math1-progenitor population. Further support for this idea comes from our observation that Math1-reporter expression is found in Wls-expressing cells in the iRL of the Math1-null cerebellum. A possible explanation for this observation is that Wls in the iRL induces Math1 reporter expression in the iRL cells, but without actual Math1 expression, the cells are not instructed to migrate from the iRL.
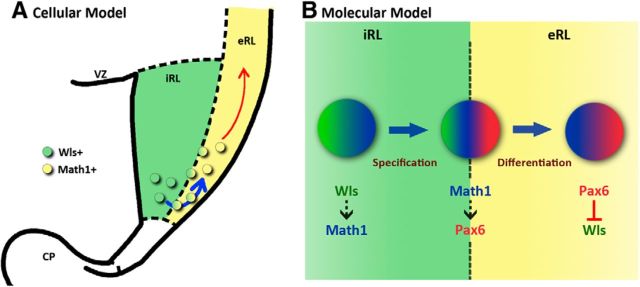
Figure 12.
A cellular and molecular model of the role of the Wls-positive domain in the cerebellar RL development. A, Cellular, The Wls+ iRL serves as a reservoir for RL progenitors that will migrate out of the RL through the eRL when these cells turn on Math1 expression. B, Molecular, The molecular interaction between Wls (green), Math1 (blue), and Pax6 (red) in the RL progenitors is proposed based upon experimental results in this paper and other studies. Cells in the iRL express Wls, which activates the expression of Math1 through the action of β-catenin. Expression of Math1 in these cells specifies an RL cell fate and instructs the cells to migrate to the adjacent eRL. In the eRL, these Math1-positive RL progenitors express Pax6. In turn, Pax6 represses the expression of Wls in these RL progenitor cells, providing a negative regulation on Wls and promoting Math1 expression in order for the progenitor cells to differentiate appropriately. CP, choroid plexus; VZ, ventricular zone.
We also examined the interaction between Pax6 and Wls in the RL using the Sey mutant, and found evidence that Pax6 negatively regulates Wls. In the Sey mutant we observed an expansion of Wls expression into the eRL and the EGL, areas that normally express Pax6 and are devoid of Wls-positive cells, suggesting that Wls is normally restricted to the iRL by the suppressive action of Pax6. This idea is supported by our Pax6 transcriptome analysis (www.cbgrits.org) where we found a downregulation of Wnt antagonists such as Dkk3 and Sfrp2 in the absence of Pax6, indicating that Pax6 exerts suppression on Wnt signaling through the action of Wnt inhibitors. One possible function for Pax6 suppression of Wls is to control the expression level of Wls downstream genes. As discussed above, Wls may activate Math1 expression in the RL. In the loss of Pax6 suppression ectopic Wls expression in the eRL and EGL is found, and we would expect an upregulation of Math1 expression as a result. This is consistent with our Pax6 transcriptome analysis (www.cbgrits.org) in which we observed a significant increase of Math1 expression at E18.5. Furthermore, it has been demonstrated that a balanced level of Math1 expression is required for proper granule cell differentiation (Helms et al., 2001), thus Pax6 can provide a negative feedback control on the Math1 expression through Wls, which is required for the proper development of granule cells (Fig. 12B).
In conclusion, our current work identifies the existence of molecular heterogeneity in the RL and uncovers dynamic interactions between the novel RL molecule, Wls, and Math1 and Pax6. These interactions may serve to establish a molecular cascade that controls the specification of RL derivatives. We also describe four molecularly distinct compartments that evolve during embryonic development, and each may represent the developmental domain that gives rise to different RL derivatives in the cerebellum.
Footnotes
This work was supported by National Institutes of Health Grant R01 HD52472, NeuroDevNet, and CMMT/CFRI start-up funds. We thank Dr. Richard Lang for generously providing us with antibody. We also thank Derek Rains and Fernando Lucero Villegas for their expert technical assistance.
The authors declare no competing financial interests.
References
- Alder J, Cho NK, Hatten ME. Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron. 1996;17:389–399. doi: 10.1016/S0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the cerebellar system: in relation to its evolution, structure, and functions. Boca Raton, FL: CRC; 1997. [Google Scholar]
- Bänziger C, Soldini D, Schütt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of wnt proteins from signaling cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Broccoli V, Boncinelli E, Wurst W. The caudal limit of Otx2 expression positions the isthmic organizer. Nature. 1999;401:164–168. doi: 10.1038/43670. [DOI] [PubMed] [Google Scholar]
- Cheng FY, Huang X, Sarangi A, Ketova T, Cooper MK, Litingtung Y, Chiang C. Widespread contribution of Gdf7 lineage to cerebellar cell types and implications for hedgehog-driven medulloblastoma formation. PLoS One. 2012;7:e35541. doi: 10.1371/journal.pone.0035541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Mishima Y, Roberts RW, Aldinger KA, Miesegaes GR, Currle DS, Monuki ES, Millen KJ. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc Natl Acad Sci U S A. 2010;107:10725–10730. doi: 10.1073/pnas.0910786107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development. 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Jiang M, Mirando AJ, Yu HM, Hsu W. Reciprocal regulation of wnt and Gpr177/mouse wntless is required for embryonic axis formation. Proc Natl Acad Sci U S A. 2009;106:18598–18603. doi: 10.1073/pnas.0904894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha TJ, Swanson DJ, Kirova R, Yeung J, Choi K, Tong Y, Chesler EJ, Goldowitz D. Genome-wide microarray comparison reveals downstream genes of Pax6 in the developing mouse cerebellum. Eur J Neurosci. 2012;36:2888–2898. doi: 10.1111/j.1460-9568.2012.08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan N, Zervas M. Wnt1 expression temporally allocates upper rhombic lip progenitors and defines their terminal cell fate in the cerebellum. Mol Cell Neurosci. 2012;49:217–229. doi: 10.1016/j.mcn.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Helms AW, Gowan K, Abney A, Savage T, Johnson JE. Overexpression of MATH1 disrupts the coordination of neural differentiation in cerebellum development. Mol Cell Neurosci. 2001;17:671–682. doi: 10.1006/mcne.2000.0969. [DOI] [PubMed] [Google Scholar]
- His W. Die entwicklung des menschlichen rautenhirns vom ende des ersten bis zum beginn des dritten monats. I. Verlangertes Mark. Bhandlungen der koniglicher sachsischen Gesellschaft dser Wissenschaften, Mathematische-physikalische Klasse. 1891;29:1–74. [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jensen P, Zoghbi HY, Goldowitz D. Dissection of the cellular and molecular events that position cerebellar Purkinje cells: a study of the math1 null-mutant mouse. J Neurosci. 2002;22:8110–8116. doi: 10.1523/JNEUROSCI.22-18-08110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P, Smeyne R, Goldowitz D. Analysis of cerebellar development in math1 null embryos and chimeras. J Neurosci. 2004;24:2202–2211. doi: 10.1523/JNEUROSCI.3427-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Millet S, Campbell K, Epstein DJ, Losos K, Harris E, Joyner AL. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature. 1999;401:161–164. doi: 10.1038/43664. [DOI] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martínez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RS, Dymecki SM. Rautenlippe redux– toward a unified view of the precerebellar rhombic lip. Curr Opin Cell Biol. 2009;21:741–747. doi: 10.1016/j.ceb.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Dymecki SM. Origin of the precerebellar system. Neuron. 2000;27:475–486. doi: 10.1016/S0896-6273(00)00059-3. [DOI] [PubMed] [Google Scholar]
- Shi F, Cheng YF, Wang XL, Edge AS. Β-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3′ enhancer. J Biol Chem. 2010;285:392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A, Gruss P. Roles of pax-genes in developing and adult brain as suggested by expression patterns. J Neurosci. 1994;14:1395–1412. doi: 10.1523/JNEUROSCI.14-03-01395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson DJ, Goldowitz D. Experimental Sey mouse chimeras reveal the developmental deficiencies of Pax6-null granule cells in the postnatal cerebellum. Dev Biol. 2011;351:1–12. doi: 10.1016/j.ydbio.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Swanson DJ, Tong Y, Goldowitz D. Disruption of cerebellar granule cell development in the Pax6 mutant, sey mouse. Brain Res Dev Brain Res. 2005;160:176–193. doi: 10.1016/j.devbrainres.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Yamada M, Seto Y, Taya S, Owa T, Inoue YU, Inoue T, Kawaguchi Y, Nabeshima Y, Hoshino M. Specification of spatial identities of cerebellar neuron progenitors by Ptf1a and Atoh1 for proper production of GABAergic and glutamatergic neurons. J Neurosci. 2014;34:4786–4800. doi: 10.1523/JNEUROSCI.2722-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]