Abstract
Importance
One approach to understanding the genetic complexity of schizophrenia is to study associated behavioral and biological phenotypes that may be more directly linked to genetic variation.
Objective
To identify single nucleotide polymorphisms associated with general cognitive ability (“g”) in people with schizophrenia and controls.
Design
Genome-wide association study (GWAS), followed by analyses in unaffected siblings and independent schizophrenia samples, functional magnetic resonance imaging studies of brain physiology in vivo, and RNA sequencing in post-mortem brain samples.
Setting
The discovery cohort and unaffected siblings were participants in the NIMH Clinical Brain Disorders Branch schizophrenia genetics studies. Additional schizophrenia cohorts were from psychiatric treatment settings in the United States, Japan, and Germany.
Participants
The discovery cohort comprised 339 with schizophrenia and 363 community controls. Follow-up analyses studied 147 unaffected siblings of the schizophrenia cases, and independent schizophrenia samples of 279, 95 and 294 participants. Imaging analyses included 87 schizophrenia cases and 397 controls. Brain tissue samples were available for 64 cases and 61 controls.
Main Outcome Measures
We studied genome-wide association with g, by group, in the discovery cohort. We used selected genotypes to test specific associations in unaffected siblings and independent schizophrenia samples. Imaging analyses focused on activation in prefrontal cortex during working memory. Brain tissue studies yielded mRNA expression levels for RefSeq transcripts.
Results
The schizophrenia discovery cohort showed GWAS-significant association of g with polymorphisms in sodium channel gene SCN2A, accounting for 10.4% of g variance (rs10174400, P=9.27×10−10). Controls showed a trend for g/genotype association with reversed allelic directionality. The genotype-by-group interaction was also GWAS-significant (P=1.75×10−9). Siblings showed a genotype association with g parallel to the schizophrenia group, and the same interaction pattern. Parallel, but weaker, associations with cognition were found in independent schizophrenia samples. Imaging analyses showed a similar pattern of genotype associations by group and genotype-by-group interaction. RNA sequencing revealed reduced expression in 2 of 3 SCN2A alternative transcripts in the patient group, with genotype-by-group interaction, that again paralleled the cognition effects.
Conclusions
The findings implicate SCN2A and sodium channel biology in cognitive impairment in schizophrenia cases and unaffected relatives, and may facilitate development of cognition-enhancing treatments.
Schizophrenia is a heritable neurodevelopmental disorder characterized by disturbed patterns of behavior and abnormalities of brain function.1,2 Genome-wide association studies (GWAS) are beginning to yield insights into the genetic architecture of schizophrenia, although effect sizes for individual genes are modest.3–5 However, few GWAS have examined behavioral or biological traits associated with the disorder, which may reflect more penetrant effects of common genetic variation.
Broad cognitive impairment is common in schizophrenia.6–8 Subtle cognitive differences are often measurable years before psychotic symptoms or exposure to medications,9–13 and impairment is seen in attenuated form in unaffected relatives,6,7,14–16 suggesting that impaired cognition is an intermediate phenotype related to genetic risk for schizophrenia.17 Studies in non-clinical groups,18–20 and in patients with schizophrenia,6,21,22 indicate that cognitive data are characterized by a hierarchical structure, in which individual measures group into domain-specific cognitive factors (e.g., “working memory”), which underlie a higher-order construct referred to as general cognitive ability or “ g.” g is reliably indexed with standard measurement tools,23 stable over time,24,25 and associated with life outcomes from academic and vocational success26–30 to health and mortality.31,32 Physiologically, g is closely related to the efficiency of the prefrontal cortex (PFC),33,34 an important focus of schizophrenia research.35
The heritability of g has been estimated at between 40% and 80%,25,36–38 but genetic associations with cognitive performance in non-clinical samples have been difficult to find and replicate,27,39 likely due to the interaction of multiple genetic and environmental influences on brain development and function. Gene-cognition associations within clinical groups present additional complexities because of the potential role of illness epiphenomena (e.g., medication), but may be enriched for illness-specific mechanisms of cognitive impairment (e.g., APOE4 in Alzheimer’s samples). A fast-emerging but inconsistent literature has explored the association of cognitive performance with suspected genetic markers of schizophrenia.40–46 One twin study suggested significant overlap in the genes that contribute to cognition and schizophrenia,47 whereas another concluded that overlap was more limited.48 Thus, it remains unclear to what degree the set of genes that gives rise to schizophrenia risk also impact brain systems that underlie cognitive performance.
Here, we report a GWAS of cognition in Americans of European ancestry with DSM-IV schizophrenia and community controls from the CBDB/NIMH Study of Schizophrenia Genetics (DRW, PI). In the sodium channel gene, SCN2A (Gene ID: 6326) – previously associated with seizure disorders, intellectual disability, and autism49–53 – we have identified single-nucleotide polymorphisms (SNPs rs10174400 and rs10182570) that show GWAS-significant association with general cognitive ability in schizophrenia. We found consistent evidence in a sample of the unaffected siblings of these probands and in independent schizophrenia samples. Further support comes from analyses of blood oxygen level-dependent functional magnetic resonance imaging (BOLD fMRI) during working memory and of RNA sequencing in post-mortem prefrontal cortex (PFC) tissue samples.
METHODS
SUBJECTS IN THE CBDB/NIMH SAMPLE
The GWAS discovery sample included 363 community controls and 339 people with DSM IV schizophrenia,54,55 after exclusions and genotyping QC (Table 1). Main findings were tested further in a sample of full siblings of 147 of these probands (eTable 1, see Supplement for details regarding inclusion and exclusion of participants). All research participants were competent adults and provided written informed consent pursuant to IRB reviewed and approved protocols.
Table 1.
Descriptive statistics for discovery sample
| Variable or Composite | Group | N | Mean | SD | Minimum | Maximum | Statistic | df | P value |
|---|---|---|---|---|---|---|---|---|---|
| Age | Schizophrenia | 339 | 34.95 | 9.84 | 18 | 60 | |||
| Control | 363 | 31.16 | 9.88 | 18 | 60 | t = 5.09 | 701 | 4.68×10−7 | |
| Years of Education | Schizophrenia | 339 | 14.13 | 2.11 | 8 | 23 | |||
| Control | 362 | 16.73 | 2.45 | 12 | 25 | t = −14.96 | 700 | 4.26×10−44 | |
| Sex -- Males, % Male | Schizophrenia | 339 | 262 | 77.3% | |||||
| Control | 363 | 172 | 47.4% | χ2 = 66.41 | 701 | 3.21×10−16 | |||
| Duration of Illness | Schizophrenia | 320 | 13.809 | 9.526 | 0 | 41 | |||
| GAF | Schizophrenia | 319 | 46.339 | 13.182 | 20 | 90 | |||
| Positive Symptoms (1–7) | Schizophrenia | 276 | 2.678 | 1.675 | 1.00 | 7.00 | |||
| Negative Symptoms (1–7) | Schizophrenia | 282 | 2.930 | 1.634 | 1.00 | 7.00 | |||
| Disorganized Symptoms (1–7) | Schizophrenia | 278 | 2.941 | 1.713 | 1.00 | 7.00 | |||
| Estimated WAIS Full Scale IQ | Schizophrenia | 339 | 93.068 | 11.271 | 70.00 | 129.00 | |||
| Control | 362 | 108.569 | 8.955 | 86.00 | 130.00 | t = −20.22 | 700 | 5.88×10−72 | |
| g | Schizophrenia | 339 | −1.208 | 0.738 | −3.33 | .50 | |||
| Control | 363 | −0.009 | 0.418 | −1.17 | 1.00 | t = −26.68 | 701 | 9.38×10−109 | |
N, Number. SD, standard deviation. df, degrees of freedom. GAF, Global Assessment of Functioning. IQ, intelligence quotient. g, general cognitive ability composite.
COGNITIVE PHENOTYPES FOR CBDB/NIMH SAMPLE
Cognitive phenotypes were composites of individual measures constructed to represent verbal memory, visual memory, N-back, processing speed, card sorting, and working memory span, and g (eTable 2, see Supplement). All composites were unweighted and were calculated in exactly the same way for probands, controls and unaffected siblings.6
GENOTYPING AND QUALITY CONTROL FOR CBDB/NIMH SAMPLE
DNA samples were genotyped using Illumina HumanHap550K/610Quad Bead Chips (San Diego, CA) according to the manufacturer’s protocol (see Supplement). After QC procedures (see Supplement), 495,089 high quality autosomal SNPs were available for analysis. QC of individual genotyping results (see Supplement) left a total of 933 individuals with good genotype information. Of these, 339 probands and 363 controls had cognitive test data and were retained for discovery analyses (g could not be calculated for 5 probands because of missing data).
For siblings, SCN2A rs10174400 genotypes were determined using the 5’ exonuclease TaqMan assay. SNP probe and primer sets were acquired from Applied Biosystems (Carlsbad, CA). Genotype accuracy was assessed by re-genotyping within a subsample, and reproducibility was routinely greater than 99%.
STATISTICAL ANALYSIS FOR THE CBDB/NIMH SAMPLE
We performed multi-dimensional scaling (MDS) on the matrix of genome-wide IBS pairwise distances using PLINK (v1.07)56 and, to control for population stratification, included the first four MDS axes as covariates in GWAS analyses. Analyses of the associations of 495,089 SNPs with 7 cognitive variables were performed in PLINK, assuming an additive genetic model and also controlling for age and sex. We did not control for education as it is confounded with illness and with g.57 Analyses in unaffected siblings were conducted using PASW Statistics 18.0 (IBM, Armonk, NY).
ADDITIONAL SAMPLES AND COGNITIVE VARIABLES
Study design details for the multisite CATIE schizophrenia antipsychotic effectiveness trial have been published, including details of cognitive assessments, genotyping, and genotype QC methods.58–60 (For details related to the current comparison sample, see Supplement.) Details of data collection for the Japanese sample have been previously published.61 The cognitive battery was comparable to the CBDB/NIMH battery. Genotyping and QC are described in the Supplement. Genetic and cognitive data were available for 95 people (eTables 1 and 2). The German sample consisted of 294 clinically stable individuals of European ancestry with DSM-IV schizophrenia, as described previously (see Supplement for details).62
STATISTICAL ANALYSIS FOR ADDITIONAL SAMPLES
Genotype-cognition association analyses in independent schizophrenia samples were conducted using PASW Statistics 18.0. We performed unidirectional tests (i.e., one-tailed), assuming a minor allele disadvantage in schizophrenia, and using an additive genetic model, controlling for age and sex. For meta-analysis of effect sizes across schizophrenia samples, we calculated sample-weighted effect sizes with a bias correction for the small number of samples combined.
BOLD fMRI ANALYSES
To test the relationship between SCN2A rs10174400 and cognition-related activation patterns as measured by BOLD fMRI, we studied 397 controls and 87 schizophrenia cases from the CBDB/NIMH sibling study who were genotyped and completed the N-back working memory task while scanned at 3T (details in Supplement). After quality screening and correction for covariates of no interest (e.g., head motion), we used ANCOVAs controlling for age and sex to test SCN2A genotype within each diagnostic group and the interaction of diagnosis-bygenotype. Genotype groups within diagnoses did not differ in terms of demographic and performance variables. Thus, differences in activation are thought to reflect neural efficiency (i.e., less activation at similar performance implying greater efficiency) – such differences representing a familial and heritable phenotype.63–66 Given our interest in prefrontal information processing efficiency, we used a prefrontal region of interest with small volume statistical correction (family wise error or FWE).
RNA SEQUENCING IN INDPENDENT POST-MORTEM BRAIN SAMPLES
RNA sequencing data was performed on post-mortem PFC grey matter from 61 adult, controls (51 males; age: 44±14.6) and 64 adult, probands (51 males; age: 44.3±14.8), all of European ancestry. Detailed brain tissue collection methods used by the Lieber Institute and CBDB/NIMH have been published67 and details of RNA sequencing are described in the Supplement. The relative abundances of the three common SCN2A RefSeq transcripts, NM_21007, NM_001040142, and NM_001040143, were estimated by Cufflinks v2.0.2 and compared to Illumina iGenome gene annotation. The three transcripts can be differentiated based on 5’ exons, thus allowing a reliable estimation of relative abundance of each transcript. We used ANCOVAs, with age and sex covariates, to investigate main effects and interactions among diagnosis, SCN2A rs10174400 genotype, and SCN2A transcript levels for the three transcripts. Analyses were also corrected for post-mortem interval and RNA integrity number. We calculated Cohen’s d effect sizes. With the low number of rs10174400 minor allele homozygotes (8 probands, 7 controls), we combined heterozygotes with minor allele homozygotes (T carriers) for analyses based on genotype.
SUPPLEMENTARY ANALYSES
The Supplement describes covariate sensitivity analyses (medication, chronicity, age of onset, family socioeconomic status), analysis of the potential role in current findings of low frequency exonic SNPs, and tests of the association of g with SNP sets representing the whole SCN2A gene, other sodium channel genes, and the whole sodium and calcium channel gene families.
RESULTS
CBDB/NIMH DISCOVERY SAMPLE
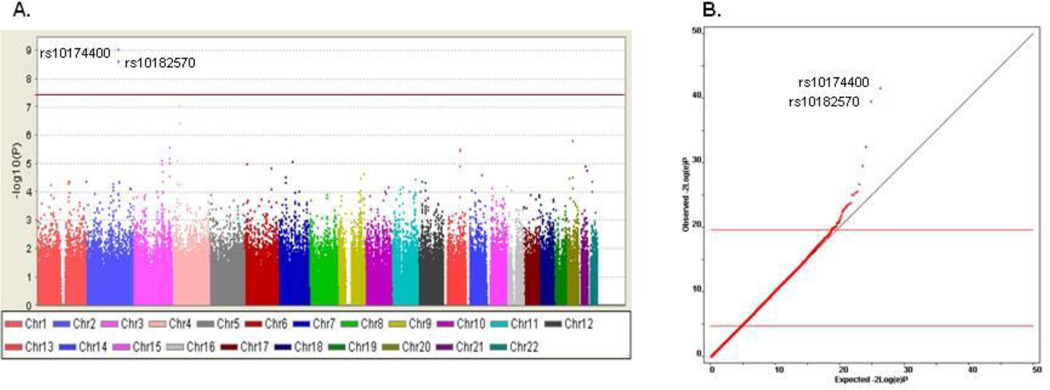
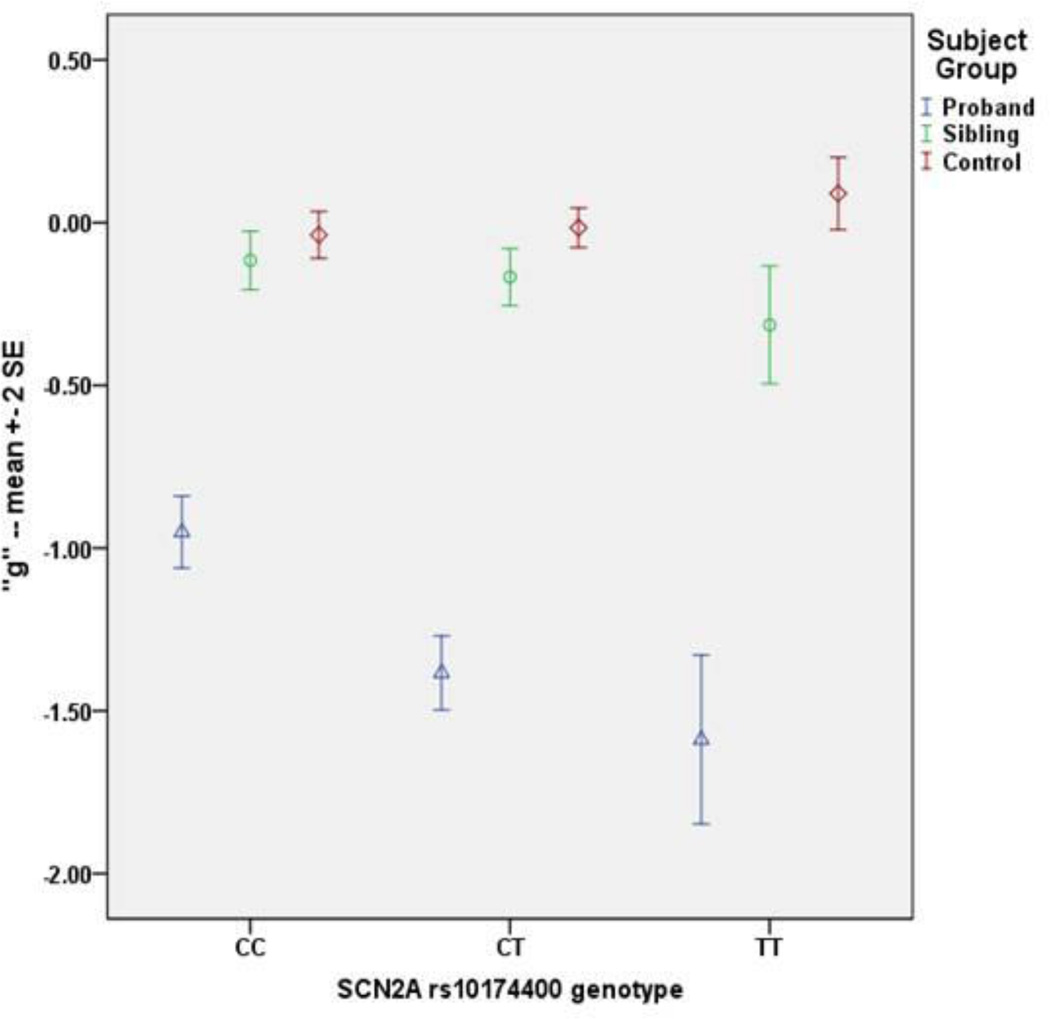
The GWAS in the schizophrenia sample identified a strong association signal (Figure 1a). Two linked, intronic SNPs in SCN2A surpassed GWAS significance (i.e., P=5.0×10−8) for association with g (rs10174400, P=9.27×10−10; rs10182570, P=2.56×10−9; Table 2; eFigure 1) – accounting for 10.4% of g variance – with no evidence of inflation of test statistics due to population effects (λgenomic control = 1; Figure 1b, Supplement, and eTables 3–6). Performance was least impaired in subjects homozygous for the major C allele, intermediate in heterozygotes, and most impaired in subjects homozygous for the T allele (Figure 2). In non-independent analyses, SCN2A rs10174400 genotype was also associated with the six cognitive domain variables in schizophrenia (Table 2). Each of the domains showed directionally consistent and at least nominally significant association with rs10174400 genotype, but none met the GWAS threshold.
Figure 1. Manhattan plot (A.) and QQ plot (B.) for SCN2A GWAS findings in 334 people with schizophrenia.
A: Manhattan plot of GWAS results from 495,089 SNPs tested for association with g in 334 individuals with schizophrenia. On the y-axis is −log10(P). The red line denotes the p-value of 5.0×10−8. B: Quantile–quantile (QQ) plots of actual versus expected −2log(e)P for g in cases and controls. −2log(e)P follows a χ2 distribution with 2 degrees of freedom and can be used for statistical inference. Points above the horizontal line indicate an enrichment of low p-values beyond what would be expected by chance.
Table 2.
Association results in discovery sample and additional cohorts for analyses of SCN2A rs10174400 and proxies
| Cohort | Group(s) | N | Target Variable | Reference SNP ID |
Position | MA | MAF % |
Statistical Model | Statistic | df* | P Value | % Var |
Direction of Effect |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. GWAS-significant findings for SCN2A SNPs in discovery cohort | |||||||||||||
| CBDB/NIMH | Schizophrenia | 334 | g | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −6.33 | 327 | 9.27×10−10 | 10.4 | CC>CT>TT |
| CBDB/NIMH | Schizophrenia | 334 | g | rs10182570 | 166109634 | C | 33.7 | Additive model | t = −6.18 | 327 | 2.56×10−9 | 10.0 | AA>AC>CC |
| 2. Post hoc analyses of SCN2A rs10174400 in discovery cohort | |||||||||||||
| CBDB/NIMH | Controls | 363 | g | rs10174400 | 166125219 | T | 33.7 | Additive model | t = 1.64 | 356 | 0.102 | <0.1 | TT>CT>CC |
| CBDB/NIMH | Schizophrenia/ Controls | 697 | g | rs10174400 | 166125219 | T | 33.8 | Group status by genotype interaction | F = 20.77 | 2, 685 | 1.75×10−9 | 5.7 | Opposite |
| CBDB/NIMH | Schizophrenia/ Controls | 697 | Case/Control status | rs10174400 | 166125219 | T | 33.8 | Logistic regression | Wald = 1.34 | 688 | 0.248 | n/a | n/a |
| CBDB/NIMH | Schizophrenia | 337 | Span | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −5.10 | 330 | 7.61×10−7 | 7.1 | CC>CT>TT |
| CBDB/NIMH | Schizophrenia | 320 | Card Sorting | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −4.55 | 313 | 8.24×10−6 | 6.0 | CC>CT>TT |
| CBDB/NIMH | Schizophrenia | 339 | Processing Speed | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −4.06 | 332 | 6.60×10−5 | 4.6 | CC>CT>TT |
| CBDB/NIMH | Schizophrenia | 339 | Verbal Memory | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −3.38 | 332 | 0.0008 | 3.2 | CC>CT>TT |
| CBDB/NIMH | Schizophrenia | 332 | Visual Memory | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −3.25 | 325 | 0.0012 | 2.8 | CC>CT>TT |
| CBDB/NIMH | Schizophrenia | 236 | Nback | rs10174400 | 166125219 | T | 34.0 | Additive model | t = −2.26 | 229 | 0.0179 | 2.0 | CC>CT>TT |
| 3. Analyses of SCN2A rs10174400 (or proxy) in additional cohorts | |||||||||||||
| CBDB/NIMH | Unaffected Siblings | 147 | g | rs10174400 | 166125219 | T | 28.6 | Additive model | t = −2.24 | 144 | 0.026 | 3.4 | CC>CT>TT |
| CBDB/NIMH | Siblings/Controls | 510 | g | rs10174400 | 166125219 | T | 32.2 | Group status by genotype interaction | F = 20.77 | 2, 502 | 0.026 | 1.5 | Opposite |
| CATIE | Schizophrenia | 279 | g | rs10192208 | 166117399 | G | 37.3 | Additive (one-tailed) | t = −2.12 | 276 | 0.017 | 1.5 | AA>AG>GG |
| CATIE | Schizophrenia | 279 | Processing Speed | rs10192208 | 166117399 | G | 37.3 | Additive (one-tailed) | t = −2.11 | 276 | 0.018 | 1.4 | AA>AG>GG |
| CATIE | Schizophrenia | 279 | Working Memory | rs10192208 | 166117399 | G | 37.3 | Additive (one-tailed) | t = −2.03 | 276 | 0.022 | 1.4 | AA>AG>GG |
| Japanese | Schizophrenia | 95 | g | rs10192208 | 166117399 | G | 39.0 | Additive (one-tailed) | t = −1.86 | 91 | 0.034 | 3.4 | AA>AG>GG |
| Japanese | Schizophrenia | 95 | g | rs10192208 | 166117399 | G | 39.0 | Recessive (one-tailed) | t = −2.22 | 91 | 0.015 | 4.8 | AA>G carriers |
| Japanese | Schizophrenia | 95 | Verbal Memory | rs10192208 | 166117399 | G | 39.0 | Additive (one-tailed) | t = −1.78 | 91 | 0.040 | 3.2 | AA>AG>GG |
| Japanese | Schizophrenia | 95 | Verbal Memory | rs10192208 | 166117399 | G | 39.0 | Recessive (one-tailed) | t = −2.82 | 91 | 0.006 | 7.7 | AA>G carriers |
| German | Schizophrenia | 278 | g | rs10174400 | 166125219 | T | 33.5 | Additive (one-tailed) | t = −0.91 | 275 | 0.254 | n/a | n/a |
| German | Schizophrenia | 294 | Span | rs10174400 | 166125219 | T | 33.5 | Additive (one-tailed) | t = −1.99 | 291 | 0.024 | 1.3 | CC>CT>TT |
N, Number. MA, minor allele. MAF %, observed minor allele frequency percent. df, degrees of freedom (*for current effect, after accounting for covariates). % Var, percentage of variance explained. GWAS, genome-wide association study. g, general cognitive ability composite.
Figure 2. Effect of SCN2A rs10174400 genotype on g composite performance, by group, in 334 probands, 147 siblings, and 363 controls.
On the y-axis are values of g. The blue triangles (schizophrenia), green circles (siblings), and red diamonds (controls) represent mean g values by genotype subgroups. The error bars are ± 2 standard errors.
For controls, no SNP association approached GWAS significance (see Supplement, eFigure 2, eTable7) and SCN2A rs10174400 genotype was not a predictor of case/control status (Table 2). Unexpectedly, the allelic trend for the control association with g was in the direction opposite the schizophrenia association (Figure 2), and an analysis of the interaction of rs10174400 genotype-by group was also GWAS-significant (P=1.75×10−9; Table 2).
UNAFFECTED SIBLINGS
Although not independent of proband results, the sibling analyses addressed the concern that the proband association might be primarily related to illness characteristics (e.g., ongoing symptoms) or medications. In unaffected siblings, there was a robust, directionally parallel association between rs10174400 genotype and g, accounting for 3.4% of performance variation, and a significant genotype-by-group interaction (Table 2).
In healthy populations, g has been shown to predict educational attainment,18,27 so a genotype that predicts g might be associated with education. In 147 unaffected siblings, rs10174400 genotype accounted for 5.7% of the variance in years of education completed (P=.003), with T allele carriers showing clearly reduced educational attainment compared with C allele homozygotes (eFigure 3). This association was not present in the full schizophrenia sample (P=.384), likely because of the confounding effect of illness on educational attainment.57
ADDITIONAL SAMPLES
In 279 schizophrenia cases from the CATIE trial, regression analysis confirmed the association of an rs10174400 proxy to the CATIE “neurocognitive composite,”68 a general cognitive ability index similar to g, again showing directionality parallel to the discovery analyses (Table 2). Genotype associations to subsidiary composites for processing speed and working memory were also significant and parallel. In 95 Japanese schizophrenia cases, regression analysis yielded a directionally consistent significant association of the same proxy SNP with g,accounting for 3.4% of the variance in performance (Table 2). Post hoc analysis using a recessive model showed an even more pronounced effect and we observed a similar pattern for a verbal memory composite. Finally, we examined gene/cognition associations in 295 Germans with schizophrenia. Regression analyses failed to replicate the association of rs10174400 with g in schizophrenia in this sample (Table 2). However, there was a parallel genotype association with the working memory span composite in the German cases, which was the strongest domain-specific effect in the discovery sample. Together, the three replication samples included 649 people with schizophrenia. Across the three groups, rs10174400 genotype accounted for 1.0% of the variance in g (sample-weighted mean effect size). Including the discovery sample, as well, with the replication samples (total N=983), genotype accounted for 3.0% of g variance in schizophrenia, on average.
BOLD fMRI ANALYSES
Looking beyond performance, we tested for genotype effects at the level of brain physiology, using an N-back working memory paradigm that robustly engages prefrontal cortical circuitry. rs10174400 was differentially associated with PFC efficiency in cases and controls, analogously to the cognitive results pattern. Among controls, CC homozygotes were most efficient, among schizophrenia cases they were least efficient, and the interaction effect was significant (MNI coordinates: −36 27 33, FWE-corrected P=0.02; eFigure 4). There were also main effects of SCN2A genotype in both diagnostic groups consistent with the direction of this interaction and with the cognitive associations (see Supplement).
RNA SEQUENCING ANALYSES
Analysis of RNA sequencing data from post-mortem PFC grey matter tissue samples showed significantly reduced expression of SCN2A mRNA in the schizophrenia sample relative to controls for two of three RefSeq transcripts and significant genotype effects and interactions for these two transcripts (Table 3 and eFigure 5). Effect sizes for significant findings were small to medium in magnitude. The directions of genotype effects were opposite for the two groups and the diagnosis-by-genotype interactions were significant – patterns remarkably similar to those in the cognitive and imaging data.
Table 3.
Results for analyses of mRNA expression of SCN2A alternative transcripts in PFC tissue samples from schizophrenia cases and controls
| Group(s) | N | Target for Expression Analysis |
Statistical Analysis | F Value | df* | P Value | Cohen’s d | Direction of Effect |
|---|---|---|---|---|---|---|---|---|
| Schizophrenia | 64 | NM_021007 | rs10174400 genotype main effect (recessive model) | 5.6 | 1, 60 | 0.021 | 0.61 | CC>T carriers |
| Control | 61 | NM_021007 | rs10174400 genotype main effect (recessive model) | 2.8 | 1, 57 | 0.100 | 0.44 | T carriers>CC |
| Schizophrenia/Controls | 125 | NM_021007 | Diagnosis main effect | 11.0 | 1. 121 | 0.001 | 0.60 | Control>Schizophrenia |
| Schizophrenia/Controls | 125 | NM_021007 | Diagnosis by rs10174400 genotype interaction | 7.8 | 1, 119 | 0.006 | 0.38 | Opposite |
| Schizophrenia | 64 | NM_001040142 | rs10174400 genotype main effect (recessive model) | 5.0 | 1, 60 | 0.029 | 0.58 | CC>T carriers |
| Control | 61 | NM_001040142 | rs10174400 genotype main effect (recessive model) | 0.4 | 1, 57 | 0.549 | n/a | n/a |
| Schizophrenia/Controls | 125 | NM_001040142 | Diagnosis main effect | 10.7 | 1. 121 | 0.001 | 0.59 | Control>Schizophrenia |
| Schizophrenia/Controls | 125 | NM_001040142 | Diagnosis by rs10174400 genotype interaction | 3.8 | 1, 119 | 0.054 | 0.19 | Opposite |
| Schizophrenia | 64 | NM_001040143 | rs10174400 genotype main effect (recessive model) | 1.5 | 1, 60 | 0.223 | n/a | n/a |
| Control | 61 | NM_001040143 | rs10174400 genotype main effect (recessive model) | 2.0 | 1, 57 | 0.160 | n/a | n/a |
| Schizophrenia/Controls | 125 | NM_001040143 | Diagnosis main effect | 0.1 | 1. 121 | 0.750 | n/a | n/a |
| Schizophrenia/Controls | 125 | NM_001040143 | Diagnosis by rs10174400 genotype interaction | 0.1 | 1, 119 | 0.949 | n/a | n/a |
N, Number. df, degrees of freedom (* for current effect, after accounting for covariates). Cohen’s d, Standardized mean difference.
SUPPLEMENTARY ANALYSES
Our main findings showed little change in analyses with additional covariates (medication, age of prodrome onset, chronicity, positive and negative symptoms, or family SES). Analysis of low frequency exonic SNPs was inconclusive. Tests of the association of g with SNP sets were an initial step in determining whether the association of sodium channel biology with general cognitive performance extended beyond the influence of the two GWAS-significant SNPs (all in Supplement and eTables 8–12).
COMMENT
In our GWAS analyses of general cognitive ability in patients with schizophrenia, two LD-linked SNPs in SCN2A showed GWAS-significant association. The effect accounted for 10.4% of the variance in overall cognitive performance. A parallel association of rs10174400 genotype with g in 147 unaffected siblings indicated that the schizophrenia association cannot be attributed solely to illness epiphenomena (e.g. medication). Notably, in the siblings, educational attainment also varied with rs10174400 genotype, accounting for 5.7% of sibling education variance. We found evidence for weaker, but parallel, genotype/cognition associations in independent schizophrenia samples. Across these three replication samples, totaling 649 probands, genotype accounted for 1.0% of g variance. Controls showed a trend for genotype association with allelic directionality opposite to the schizophrenia association, and the rs10174400 genotype-by-group interaction was also GWAS-significant.
Neuroimaging findings and RNA sequencing data from postmortem PFC samples provided a measure of biological validation for the behavioral association findings. Analyses of prefrontal information processing efficiency during working memory revealed a genotype-by-diagnosis interaction. The rs10174400 minor (T) allele conferred efficiency advantages for controls but maximal inefficiency in schizophrenia. In postmortem RNA sequencing experiments, the schizophrenia sample showed reduced expression of mRNA for two of three common alternative transcripts, and genotype-by-diagnosis interactions analogous to the imaging results. Thus, the pattern of differential rs10174400 genotype associations for cases versus controls that was hinted at in the behavioral data (i.e., a clear allele dose-dependent effect on cognitive performance in schizophrenia, and a weak opposite trend in controls), came more clearly into focus in biological analyses. In sum, congruent evidence spanning behavior, physiology, and mRNA expression suggests an interaction between SCN2A genetic markers and schizophrenia-associated phenomena.
Our discovery sample effect was dramatic and likely reflects the “winner’s curse” seen in other some other genetic association studies of relatively small samples. Evidence from independent schizophrenia samples suggested that the SCN2A effect on cognition may generally be smaller – on average genotype accounted for 1.0% of variance in our replication samples, though in two of these three samples the effect was in the range of 1.5–3.4%. While smaller, these effects in independent samples were directionally consistent with discovery sample effects – notwithstanding considerable differences in ascertainment, genotyping and phenotyping. Additionally, the magnitude of the main schizophrenia finding may have reflected enrichment of the CBDB/NIMH sample for a particular form of schizophrenia risk-associated cognitive impairment, due to uniform, restrictive inclusion criteria (e.g., IQ>70, no substance abuse). Across the discovery and replication samples (N=983), the mean sample-weighted association effect size was 3.0% of g variance. Neuroimaging findings, and mRNA expression findings in wholly independent post-mortem brain tissue samples, offered further, directionally-consistent support for the main finding. At the same time, the parallel findings in siblings, although non-independent, suggested that the schizophrenia findings were not determined by illness epiphenomena. Altogether, the data alleviate concerns that these are not true genotype effects on SCN2A biology. A better understanding of the magnitude of these effects will require further analyses in other samples.
The findings are also plausible, both biologically and in terms of known clinical associations. SCN2A encodes the α2 subunit of a voltage-gated sodium ion channel that is widely expressed in the brain and contributes to the initiation and propagation of action potentials.69,70 Na(v)1.2 (the protein encoded by SCN2A) is abundant in parvalbumin-positive GABAergic inhibitory interneurons, at least in hippocampus and temporal lobe.70 GABA system abnormalities have been a particular focus of cognitive impairment research in schizophrenia.71,72 Multiple mutations in SCN2A have been associated with childhood epilepsies, sometimes combined with intellectual disability and/or autism-like symptoms,69,73 and antiepileptic medications that block sodium channels (e.g., topiramate) have adverse cognitive effects.74 Notably, each of three recent whole-exome sequencing studies focused on nonsyndromic intellectual disability found de novo coding mutations in SCN2A (3/55 sequenced individuals in one study,52 1/12 in the second,51 and 1/100 in the third49). Results from a large exome sequencing study of autism recently identified 279 independent de novo mutations, and highlighted SCN2A as the single gene disrupted by two of these.53
The hypothesis that cognition is an intermediate phenotype for schizophrenia implies that rs10174400 should discriminate cases from controls, at least to some degree.17 No case/control signal was observed in the discovery sample. Our results therefore suggest that a strong, directionally-specific SCN2A association with impaired cognition may emerge in the context of the complex genetic risk architecture of schizophrenia, which is shared by patients and family members, even though there is little or no association of SCN2A with cognitive performance in the general population. We have very limited evidence as to possible mechanisms, but the involvement of sodium channel biology – and its apparent effect at the most general level of cognitive performance – suggests mediation through low-level and widely-acting neural systems. Dysfunction in GABAergic inhibitory systems could fit this description, although there are many possibilities. The findings in unaffected siblings may be quite important in further refining hypotheses about mechanisms. The sibling results clarified that the genotype association to cognition was not driven mainly by illness-specific phenomena. The association was not unique to family members with a schizophrenia diagnosis and was not tightly linked either to positive or negative symptoms, illness chronicity, or antipsychotic medication (see eTable 8). Although impaired cognition and psychotic symptoms are defining characteristics of the schizophrenia syndrome, the sibling results reported here frame the question whether these characteristics may be related to distinct genetic components. At the same time, the current study was dramatically smaller than case/control samples that have shown high P-value SNP associations with diagnosis. It may be that, with sufficient samples sizes, associations of SCN2A SNPs with the schizophrenia diagnosis will emerge. In the latest published Psychiatric Genomics Consortium analysis of over 20,000 subjects (>9,000 cases),5 several SNPs in SCN2A show association with schizophrenia at P~5.0×10−3 (searched using Ricopili tool, Broad Institute).
Despite ample evidence of heritability for widely used cognitive measures,24 in controls no common variant reached genome-wide significance or approached the magnitude of the rs10174400 effect in schizophrenia. Our results echo findings in earlier, larger cognition GWAS.75,76 Perhaps especially for traits as conserved and fundamental as non-disordered cognition, the causal effects of individual, common genetic markers cannot be detected at present amid the complex interaction of genetic, environmental, and random influences that affect individuals over decades of development.77
In sum, we have identified common variants in SCN2A that, in the context of schizophrenia and risk for schizophrenia, show substantial and consistent associations with broad cognitive performance, brain physiology, and mRNA expression in the brain. These findings intersect with prominent lines of schizophrenia research and suggest testable hypotheses about the biological roots of cognitive impairment in schizophrenia and avenues for new treatment development.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Division of Intramural Research Programs, National Institute of Mental Health, National Institutes of Health, Bethesda, Maryland, USA, as direct funding of the Clinical Brain Disorders Branch (DRW PI), and the Lieber Institute for Brain Development, Baltimore, Maryland, USA. Some of the authors work or worked within the institutions, and DRW directs the Lieber Institute, but beyond the contributions of the various authors and support staff to this work, the institutions did not participate in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Drs. Dickinson and Weinberger had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. The authors declare no conflict of interest.
REFERENCES
- 1.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44(7):660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 2.Weinberger DR, Levitt P. Neurodevelopmental origins of schizophrenia. In: Weinberger DR, Harrison P, editors. Schizophrenia. 3rd ed. Vol. 165. Oxford: Wiley-Blackwell; 2011. pp. 579–587. [Google Scholar]
- 3.Chen X, Lee G, Maher BS, Fanous AH, Chen J, Zhao Z, et al. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol Psychiatry. 2011;16(11):1117–1129. doi: 10.1038/mp.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 5.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickinson D, Goldberg TE, Gold JM, Elvevag B, Weinberger DR. Cognitive factor structure and invariance in people with schizophrenia, their unaffected siblings, and controls. Schizophr Bull. 2011;37(6):1157–1167. doi: 10.1093/schbul/sbq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickinson D, Ramsey M, Gold JM. Overlooking the obvious: A meta-analytic comparison of digit symbol coding tasks and other cognitive measures in schizophrenia. Arch Gen Psychiatry. 2007;64:532–542. doi: 10.1001/archpsyc.64.5.532. [DOI] [PubMed] [Google Scholar]
- 8.Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology. 1998;12(3):426–445. doi: 10.1037//0894-4105.12.3.426. [DOI] [PubMed] [Google Scholar]
- 9.Cannon TD, Bearden CE, Hollister JM, Rosso IM, Sanchez LE, Hadley T. Childhood cognitive functioning in schizophrenia patients and their unaffected siblings: a prospective cohort study. Schizophr Bull. 2000;26(2):379–393. doi: 10.1093/oxfordjournals.schbul.a033460. [DOI] [PubMed] [Google Scholar]
- 10.Cannon M, Caspi A, Moffitt TE, Harrington H, Taylor A, Murray RM, et al. Evidence for early-childhood, pan-developmental impairment specific to schizophreniform disorder: results from a longitudinal birth cohort. Arch Gen Psychiatry. 2002;59(5):449–456. doi: 10.1001/archpsyc.59.5.449. [DOI] [PubMed] [Google Scholar]
- 11.Lewandowski KE, Cohen BM, Ongur D. Evolution of neuropsychological dysfunction during the course of schizophrenia and bipolar disorder. Psychol Med. 2011;41(2):225–241. doi: 10.1017/S0033291710001042. [DOI] [PubMed] [Google Scholar]
- 12.Niendam TA, Bearden CE, Rosso IM, Sanchez LE, Hadley T, Nuechterlein KH, et al. A prospective study of childhood neurocognitive functioning in schizophrenic patients and their siblings. Am J Psychiatry. 2003;160(11):2060–2062. doi: 10.1176/appi.ajp.160.11.2060. [DOI] [PubMed] [Google Scholar]
- 13.Reichenberg A, Caspi A, Harrington H, Houts R, Keefe RS, Murray RM, et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am J Psychiatry. 2010;167(2):160–169. doi: 10.1176/appi.ajp.2009.09040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egan MF, Goldberg TE, Gscheidle T, Weirich M, Rawlings R, Hyde TM, et al. Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry. 2001;50(2):98–107. doi: 10.1016/s0006-3223(01)01133-7. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg TE, Torrey EF, Gold JM, Bigelow LB, Ragland RD, Taylor E, et al. Genetic risk of neuropsychological impairment in schizophrenia: a study of monozygotic twins discordant and concordant for the disorder. Schizophr Res. 1995;17(1):77–84. doi: 10.1016/0920-9964(95)00032-h. [DOI] [PubMed] [Google Scholar]
- 16.Snitz BE, Macdonald AW, 3rd, Carter CS. Cognitive deficits in unaffected first-degree relatives of schizophrenia patients: a meta-analytic review of putative endophenotypes. Schizophr Bull. 2006;32(1):179–194. doi: 10.1093/schbul/sbi048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7(10):818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 18.Carroll JB. Human Cognitive Abilities: A Survey of Factor-Analytic Studies. New York: Cambridge University Press; 1993. [Google Scholar]
- 19.Deary IJ. Human intelligence differences: a recent history. Trends Cogn Sci. 2001;5(3):127–130. doi: 10.1016/s1364-6613(00)01621-1. [DOI] [PubMed] [Google Scholar]
- 20.Jensen AR. The g Factor: The Science of Mental Ability. Westport, CN: Praeger; 1998. [Google Scholar]
- 21.Dickinson D, Ragland JD, Calkins ME, Gold JM, Gur RC. A comparison of cognitive structure in schizophrenia patients and healthy controls using confirmatory factor analysis. Schizophr Res. 2006;85(1–3):20–29. doi: 10.1016/j.schres.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladsjo JA, McAdams LA, Palmer BW, Moore DJ, Jeste DV, Heaton R. A six-factor model of cognition in schizophrenia and related psychotic disorders: relationships with clinical symptoms and functional capacity. Schizophr Bull. 2004;30(4):739–754. doi: 10.1093/oxfordjournals.schbul.a007127. [DOI] [PubMed] [Google Scholar]
- 23.Johnson W, Nijenhuis Jt, Bouchard TJ., Jr Still just 1 g: Consistent results from five test batteries. Intelligence. 2008;36(1):81–95. [Google Scholar]
- 24.Deary IJ, Johnson W, Houlihan LM. Genetic foundations of human intelligence. Hum Genet. 2009;126(1):215–232. doi: 10.1007/s00439-009-0655-4. [DOI] [PubMed] [Google Scholar]
- 25.Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482(7384):212–215. doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- 26.Bowie CR, Reichenberg A, Patterson TL, Heaton RK, Harvey PD. Determinants of real-world functional performance in schizophrenia subjects: correlations with cognition, functional capacity, and symptoms. Am J Psychiatry. 2006;163(3):418–425. doi: 10.1176/appi.ajp.163.3.418. [DOI] [PubMed] [Google Scholar]
- 27.Deary IJ. Intelligence. Annu Rev Psychol. 2012;63:453–482. doi: 10.1146/annurev-psych-120710-100353. [DOI] [PubMed] [Google Scholar]
- 28.Gottfredson LS. What do we know about intelligence? The American Scholar. 1996 Winter;:15–30. [Google Scholar]
- 29.Green MF. What are the functional consequences of neurocognitive deficits in schizophrenia? Am J Psychiatry. 1996;153(3):321–330. doi: 10.1176/ajp.153.3.321. [DOI] [PubMed] [Google Scholar]
- 30.Green MF, Kern RS, Heaton RK. Longitudinal studies of cognition and functional outcome in schizophrenia: implications for MATRICS. Schizophr Res. 2004;72(1):41–51. doi: 10.1016/j.schres.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Batty GD, Deary IJ. Early life intelligence and adult health. BMJ. 2004;329(7466):585–586. doi: 10.1136/bmj.329.7466.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batty GD, Deary IJ, Gottfredson LS. Premorbid (early life) IQ and later mortality risk: systematic review. Ann Epidemiol. 2007;17(4):278–288. doi: 10.1016/j.annepidem.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Duncan J, Owen AM. Common regions of the human frontal lobe recruited by diverse cognitive demands. Trends Neurosci. 2000;23(10):475–483. doi: 10.1016/s0166-2236(00)01633-7. [DOI] [PubMed] [Google Scholar]
- 34.Duncan J, Seitz RJ, Kolodny J, Bor D, Herzog H, Ahmed A, et al. A neural basis for general intelligence. Science. 2000;289(5478):457–460. doi: 10.1126/science.289.5478.457. [DOI] [PubMed] [Google Scholar]
- 35.Callicott JH, Bertolino A, Mattay VS, Langheim FJ, Duyn J, Coppola R, et al. Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cereb Cortex. 2000;10(11):1078–1092. doi: 10.1093/cercor/10.11.1078. [DOI] [PubMed] [Google Scholar]
- 36.Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16(10):996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devlin B, Daniels M, Roeder K. The heritability of IQ. Nature. 1997;388(6641):468–471. doi: 10.1038/41319. [DOI] [PubMed] [Google Scholar]
- 38.Plomin R, Spinath FM. Intelligence: genetics, genes, and genomics. J Pers Soc Psychol. 2004;86(1):112–129. doi: 10.1037/0022-3514.86.1.112. [DOI] [PubMed] [Google Scholar]
- 39.Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging. 2011;32(1):63–74. doi: 10.1016/j.neurobiolaging.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Burdick KE, Goldberg TE, Funke B, Bates JA, Lencz T, Kucherlapati R, et al. DTNBP1 genotype influences cognitive decline in schizophrenia. Schizophr Res. 2007;89(1–3):169–172. doi: 10.1016/j.schres.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98(12):6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004;101(34):12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldberg TE, Straub RE, Callicott JH, Hariri A, Mattay VS, Bigelow L, et al. The G72/G30 gene complex and cognitive abnormalities in schizophrenia. Neuropsychopharmacology. 2006;31(9):2022–2032. doi: 10.1038/sj.npp.1301049. [DOI] [PubMed] [Google Scholar]
- 44.Greenwood TA, Lazzeroni LC, Murray SS, Cadenhead KS, Calkins ME, Dobie DJ, et al. Analysis of 94 candidate genes and 12 endophenotypes for schizophrenia from the Consortium on the Genetics of Schizophrenia. Am J Psychiatry. 2011;168(9):930–946. doi: 10.1176/appi.ajp.2011.10050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porteous DJ, Thomson P, Brandon NJ, Millar JK. The genetics and biology of DISC1--an emerging role in psychosis and cognition. Biol Psychiatry. 2006;60(2):123–131. doi: 10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Walters JT, Corvin A, Owen MJ, Williams H, Dragovic M, Quinn EM, et al. Psychosis susceptibility gene ZNF804A and cognitive performance in schizophrenia. Arch Gen Psychiatry. 2010;67(7):692–700. doi: 10.1001/archgenpsychiatry.2010.81. [DOI] [PubMed] [Google Scholar]
- 47.Toulopoulou T, Picchioni M, Rijsdijk F, Hua-Hall M, Ettinger U, Sham P, et al. Substantial genetic overlap between neurocognition and schizophrenia: genetic modeling in twin samples. Arch Gen Psychiatry. 2007;64(12):1348–1355. doi: 10.1001/archpsyc.64.12.1348. [DOI] [PubMed] [Google Scholar]
- 48.Fowler T, Zammit S, Owen MJ, Rasmussen F. A population-based study of shared genetic variation between premorbid IQ and psychosis among male twin pairs and sibling pairs from Sweden. Arch Gen Psychiatry. 2012;69(5):460–466. doi: 10.1001/archgenpsychiatry.2011.1370. [DOI] [PubMed] [Google Scholar]
- 49.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N Engl J Med. 2012 doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 50.Meisler MH, O'Brien JE, Sharkey LM. Sodium channel gene family: epilepsy mutations, gene interactions and modifier effects. J Physiol. 2010;588(Pt 11):1841–1848. doi: 10.1113/jphysiol.2010.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49(6):353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 53.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for Axis I DSM-IV. New York: Biometrics Research Dept, New State Psychiatric Institute; 1994. [Google Scholar]
- 55.Spitzer R, Williams J, First M. Structured Clinical Interview for DSM-IV, Patient Version. Washington: American Psychiatric Press; 1996. [Google Scholar]
- 56.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Resnick SM. Matching for education in studies of schizophrenia. Arch Gen Psychiatry. 1992;49(3):246. doi: 10.1001/archpsyc.1992.01820030078011. [DOI] [PubMed] [Google Scholar]
- 58.Keefe RS, Mohs RC, Bilder RM, Harvey PD, Green MF, Meltzer HY, et al. Neurocognitive assessment in the Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project schizophrenia trial: development, methodology, and rationale. Schizophr Bull. 2003;29(1):45–55. doi: 10.1093/oxfordjournals.schbul.a006990. [DOI] [PubMed] [Google Scholar]
- 59.Stroup TS, McEvoy JP, Swartz MS, Byerly MJ, Glick ID, Canive JM, et al. The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull. 2003;29(1):15–31. doi: 10.1093/oxfordjournals.schbul.a006986. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13(6):570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hashimoto R, Ohi K, Yasuda Y, Fukumoto M, Iwase M, Iike N, et al. The impact of a genome-wide supported psychosis variant in the ZNF804A gene on memory function in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(8):1459–1464. doi: 10.1002/ajmg.b.31123. [DOI] [PubMed] [Google Scholar]
- 62.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blokland GA, McMahon KL, Hoffman J, Zhu G, Meredith M, Martin NG, et al. Quantifying the heritability of task-related brain activation and performance during the N-back working memory task: a twin fMRI study. Biol Psychol. 2008;79(1):70–79. doi: 10.1016/j.biopsycho.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blokland GA, McMahon KL, Thompson PM, Martin NG, de Zubicaray GI, Wright MJ. Heritability of working memory brain activation. J Neurosci. 2011;31(30):10882–10890. doi: 10.1523/JNEUROSCI.5334-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callicott JH, Weinberger DR. Brain imaging as an approach to phenotype characterization for genetic studies of schizophrenia. Methods Mol Med. 2003;77:227–247. doi: 10.1385/1-59259-348-8:227. [DOI] [PubMed] [Google Scholar]
- 66.Koten JW, Jr, Wood G, Hagoort P, Goebel R, Propping P, Willmes K, et al. Genetic contribution to variation in cognitive function: an FMRI study in twins. Science. 2009;323(5922):1737–1740. doi: 10.1126/science.1167371. [DOI] [PubMed] [Google Scholar]
- 67.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keefe RS, Bilder RM, Davis SM, Harvey PD, Palmer BW, Gold JM, et al. Neurocognitive effects of antipsychotic medications in patients with chronic schizophrenia in the CATIE Trial. Arch Gen Psychiatry. 2007;64(6):633–647. doi: 10.1001/archpsyc.64.6.633. [DOI] [PubMed] [Google Scholar]
- 69.Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Invest. 2005;115(8):2010–2017. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang W, Takashima S, Segawa Y, Itoh M, Shi X, Hwang SK, et al. The developmental changes of Na(v)1.1 and Na(v)1.2 expression in the human hippocampus and temporal lobe. Brain Res. 2011;1389:61–70. doi: 10.1016/j.brainres.2011.02.083. [DOI] [PubMed] [Google Scholar]
- 71.Coyle JT. The GABA-glutamate connection in schizophrenia: which is the proximate cause? Biochem Pharmacol. 2004;68(8):1507–1514. doi: 10.1016/j.bcp.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez-Burgos G, Lewis DA. NMDA Receptor Hypofunction, Parvalbumin-Positive Neurons and Cortical Gamma Oscillations in Schizophrenia. Schizophr Bull. 2012 doi: 10.1093/schbul/sbs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Catterall WA, Dib-Hajj S, Meisler MH, Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J Neurosci. 2008;28(46):11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salinsky MC, Storzbach D, Spencer DC, Oken BS, Landry T, Dodrill CB. Effects of topiramate and gabapentin on cognitive abilities in healthy volunteers. Neurology. 2005;64(5):792–798. doi: 10.1212/01.WNL.0000152877.08088.87. [DOI] [PubMed] [Google Scholar]
- 75.Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, et al. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18(7):815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis OS, Butcher LM, Docherty SJ, Meaburn EL, Curtis CJ, Simpson MA, et al. A three-stage genome-wide association study of general cognitive ability: hunting the small effects. Behav Genet. 2010;40(6):759–767. doi: 10.1007/s10519-010-9350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turkheimer E. Commentary: Variation and Causation in the Environment and Genome. International Journal of Epidemiology. 2011;40(3):598–601. doi: 10.1093/ije/dyq147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.