Abstract
DNA glycosylases that remove alkylated and deaminated purine nucleobases are essential DNA repair enzymes that protect the genome, and at the same time confound cancer alkylation therapy, by excising cytotoxic N3-methyladenine bases formed by DNA targeting anticancer compounds. The basis for glycosylase specificity toward N3- and N7-alkylpurines is believed to result from intrinsic instability of the modified bases and not from direct enzyme functional group chemistry. Here, we present crystal structures of the recently discovered Bacillus cereus AlkD glycosylase in complex with DNAs containing alkylated, mismatched, and abasic nucleotides. Unlike other glycosylases, AlkD captures the extrahelical lesion in a solvent-exposed orientation, providing the first illustration for how hydrolysis of N3- and N7-alkylated bases may be facilitated by increased lifetime out of the DNA helix. The structures and supporting biochemical analysis of base flipping and catalysis reveal how AlkD’s HEAT-repeats distort the DNA backbone to detect non-Watson-Crick base pairs without duplex intercalation.
Alkylation of DNA by endogenous methyl donors, environmental toxins, and chemotherapeutic agents produces a diverse spectrum of cytotoxic and mutagenic lesions, including N3-methyladenine (3mA), N7-methylguanine (7mG), and 1,N6-ethenodenine (εA), that threaten the survival of all organisms 1–5. 3mA is highly toxic owing to its inhibition of DNA polymerases during replication 6,7, and production of such lesions is the rationale behind the use of alkylating agents in chemotherapy. N7-substituted guanines are the most prevalent alkylation lesions and display a wide range of toxic and mutagenic biological properties 8. By virtue of their positive charges at physiological pH, 3mA and 7mG are especially susceptible to spontaneous depurination, which generates abasic sites in DNA that can ultimately lead to single- and double-strand breaks.
DNA glycosylases initiate base excision repair of N3- and N7-methylpurines from the genome by catalyzing hydrolysis of the N-glycosidic bond (Fig. 1a,b). Despite their structural diversity, all DNA glycosylases studied to date utilize a common base flipping mechanism to access damaged DNA and orient the substrate for catalysis by rotating the target nucleotide 180° around the phosphoribose backbone into a complementary shaped active site pocket 9,10. The resulting distortion to the DNA is stabilized by an intercalating side chain “plug” that fills the void created by the extrahelical nucleotide. Glycosylases typically excise their target nucleobases by using a carboxylate side chain as a general base to activate a water nucleophile or to stabilize the carbocation transition state during base dissociation 11. Mutation of this residue, however, does not abolish catalytic activity in all cases, leading to a model in which conformational strain in the DNA arising from extensive binding energy helps to drive the reaction forward 12,13. The lack of a residue capable of performing general base catalysis in 3mA-specific DNA glycosylases (e.g., E. coli TAG) 14–16 is consistent with the idea that excision of positively charged 3mA and 7mG does not require the same level of catalytic assistance as more stable ethenoadducts, although direct evidence for this has not been reported.
Figure 1.
Base excision repair of alkylated DNA by AlkD. a, AlkD catalyzes the hydrolysis of the N-glycosidic bond to liberate an abasic site and free nucleobase. The enzyme is specific for positively charged N3-methyladenine (a) and N7-methylguanine (b). c,d, Structures of 3-deaza-3-methyladenosine (c) and tetrahydrofuran (d) used to trap AlkD in complex with alkylated and abasic DNA. e, Crystal structure of AlkD bound to 3d3mA-DNA. Each of the 6 HEAT-repeats are colored red-to-violet. The DNA is colored silver with the 3d3mA nucleotide colored magenta.
AlkC and AlkD proteins, recently discovered in Bacillus cereus and subsequently identified in all three kingdoms of life (Fig. S1), have emerged as a unique DNA glycosylase superfamily specific for N3- and N7-alkylpurines 17,18. AlkD accelerates the rate of 7mG hydrolysis from DNA 100-fold over the spontaneous rate of 7mG depurination 19, prompting us to investigate the mechanism by which AlkD excises destabilized alkylated bases. Here, we present crystal structures of B. cereus AlkD in complex with DNA damage resembling the substrate and product of the glycosylase reaction. These structures, together with supporting biochemistry of base flipping and 7mG depurination activities, demonstrate how AlkD utilizes an unprecedented strategy to trap non-canonical base pairs that allows for specific hydrolysis of destabilized N-glycosidic bonds without direct chemical attack from the enzyme.
A new architecture for binding nucleic acids
We previously determined the crystal structure of B. cereus AlkD and identified residues important for DNA binding and catalysis 19. AlkD is comprised entirely of HEAT repeats – tandem α-helical pairs that generate extended, non-enzymatic scaffolds that typically mediate protein, but not nucleic acid interactions within their concave surfaces 20–22. AlkD’s concave surface contains highly conserved residues important for 7mG excision and DNA binding activities and protection against bacterial sensitivity to alkylating agents 17–19.
To investigate the mechanisms by which this novel enzyme binds DNA and catalyzes base excision, we determined crystal structures of B. cereus AlkD in complex with DNAs resembling the substrate and product of 3mA excision (Fig. 1a). Trapping an alkylpurine DNA glycosylase onto a 3mA-containing substrate has presented a formidable challenge owing to the inherent instability of the N-glycosidic bond. To overcome this obstacle, we crystallized AlkD in complex with DNA containing 3-deaza-3-methyladenine (3d3mA), a structural 3mA mimetic in which the N3 nitrogen is replaced with carbon (Fig. 1c). The 3d3mA base is refractory to spontaneous depurination or excision by AlkD or human alkyladenine DNA glycosylase (AAG) 7, presumably because the 3d3mA purine ring lacks the formal positive charge associated with 3mA. Importantly, the N3 → C3 substitution does not affect duplex stability (Supplementary Information) 23. We also crystallized AlkD in complex with DNA containing a tetrahydrofuran (THF) moiety (Fig. 1d), which resembles the abasic site product. The AlkD/3d3mA-DNA and AlkD/THF-DNA structures were determined by molecular replacement and refined to 1.6 Å (R/Rfree=15.9/18.3%) and 1.75 Å (R/Rfree = 18.5%/22.5%), respectively (Table S1 and Fig. S2).
Both 3d3mA and THF complexes show the same general mode of nucleic acid binding despite their unique DNA sequences and crystal packing arrangements (Fig. 2). The DNA is positioned along AlkD’s concave surface, which is lined with positively charged residues from the C-terminal α-helix of each HEAT repeat (Fig. 1e and Fig. S3). The C-shaped protein wraps halfway around the DNA helix with a footprint of ~10 base-pairs. The contact surface is dominated by electrostatic interactions between side chains at the protein mid-region and the phosphoribose backbone of the DNA strand opposite the lesion. In contrast, contacts to the lesioned strand are limited to base pairs further removed from the lesion and the protein termini (Fig. 2). The DNA axes are bent 30° away from AlkD’s N-terminus as a result of helix αB (the only non-HEAT repeat in AlkD) projecting into the minor groove (Fig 2). A 2-Å shift in helix αB is the only noticeable movement in the protein upon DNA binding (Fig. S4).
Figure 2.
Crystal structures of AlkD in complex with 3d3mA-DNA (a) and THF-DNA (b). The top of each panel shows orthogonal views of the AlkD protein (green) wrapping around the DNA duplex (gold). The modified 3d3mA and THF nucleotides are colored blue, and the opposing thymine is magenta. At the bottom, a side view of the atomic model and corresponding schematic illustrates the interactions between the modified base pairs and the protein. Dashed lines represent hydrogen bonds and wavy lines represent van der Waals interactions.
A novel lesion capture mechanism
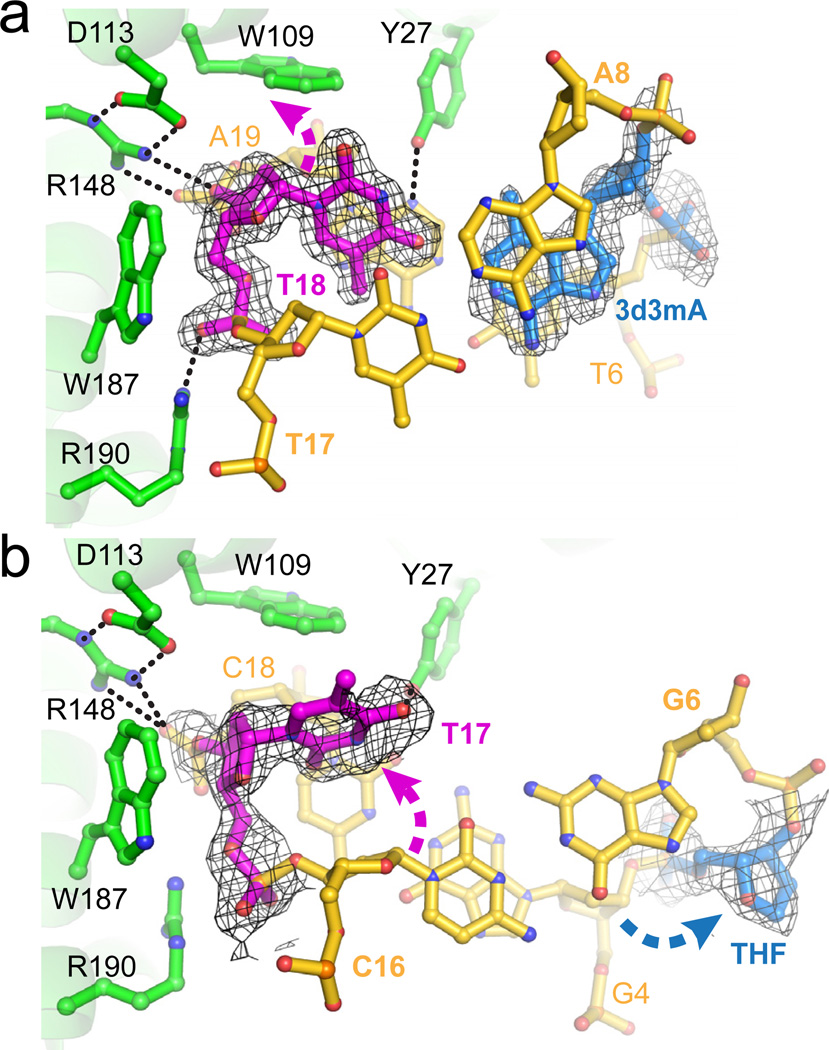
The most striking feature of the AlkD/DNA complexes is that both 3d3mA and THF reside on the face of the DNA duplex not in contact with the protein, whereas the base opposite the lesion is nestled into a cleft on the protein’s concave surface (Figs. 2 and 3). The 3d3mA•T unpredictably forms a highly sheared base pair in which 3d3mA remains stacked between T6 and A8, while the opposite thymine (T18) is displaced into the minor groove with no hydrogen bonds to 3d3mA (Fig. 3a). There are no protein contacts to the T18 base. Rather, it is held in this position by distortion of the T18/A19 backbone as a result of a hydrogen bond network among Asp113-Arg148 and Arg190. The protein-DNA interface is further strengthened by van der Waals interactions between tryptophans 109 and 187 and the phosphoribose backbone flanking the damaged base pair.
Figure 3.
Recognition of DNA damage by AlkD. a, 3d3mA-DNA (substrate) complex; b, THF-DNA (product) complex. Composite omit electron density (contoured to 1σ) for the modified base pairs is superimposed against the crystallographic models. Dashed arrows denote displacement of THF and opposing thymine from their positions in B-DNA. Hydrogen bonds are shown as dashed lines. Views are down the DNA helix axis.
In the product complex, the abasic site is rotated ~90° around the phosphoribose backbone into the major groove, and is fully solvent exposed (Figs. 2b and 3b). Interestingly, the opposite thymine is slipped completely out of the base stack and into the minor groove of the DNA, and is rotated (χ = 58°) so that the plane of the pyrimidine ring is virtually parallel with the helical axis. Unlike other DNA glycosylases, there is no intercalating side chain plugging the gap left by the flipped base. As a consequence, the duplex has collapsed in order to maintain base stacking interactions. Guanine G4, immediately 5′ to the THF, is now stacked with cytosine C16 on the opposite strand (Fig. 3b). Importantly, the DNA backbone is highly distorted as a result of the large slide (4.4 Å) and twist (58°) between G4•C18 and G6•C16 base pairs (Figs. 2b, 3b, S12). A hydrogen bond between Tyr27 at the C-terminal end of helix αB and the base 3′ to the tipped thymine is the only specific AlkD-nucleobase contact (Fig. 3). Thus, AlkD stabilizes the distortions in both substrate and product DNA—a sheared 3d3mA•T base pair and a single base THF•T bubble—through interactions with the phosphoribose backbone of the non-lesioned strand.
The solvent-exposed capture of DNA damage in the AlkD/DNA structures is both unexpected and unprecedented for a DNA glycosylase, and raises the possibilities that either AlkD utilizes a different mechanism to catalyze base excision or that the crystal structures represent non-specific, catalytically incompetent protein/DNA complexes. Indeed, the aromatic region at the center of the concave cleft loosely resembles nucleobase binding pockets of other alkylpurine DNA glycosylases 18,19. However, several important differences argue against a traditional lesion binding pocket in AlkD. First, AlkD lacks the plug residue universally used by DNA glycosylases to prevent the flipped substrate base from re-entering the DNA base stack. Second, an extrahelical nucleobase would be sterically prohibited from full 180° rotation into this shallow cleft (Fig. 3). Third, high concentrations of free nucleobases do not inhibit base excision activity by AlkD as observed in other alkylpurine glycosylases (Fig. S5) 15. Fourth, the electrostatic interaction between Asp113 and Arg148 reduces the likelihood that Asp113 acts as a general base in catalysis. Fifth, mutation of a putative base binding cleft directly adjacent to the catalytic Asp113 and Arg148 did not affect 7mG excision activity (Fig. S6). Finally, whereas alkylpurine DNA glycosylases normally exhibit enhanced excision activity for mispaired alkylbases, presumably because of their greater propensity to base-flip 15,24,25 AlkD does not discriminate against the base opposite the lesion (Table S2; Fig. S7).
To determine the orientation of DNA relative to the central cleft during catalysis, we measured the rate of 7mG excision opposite a bulky nucleotide. A pyrene nucleotide wedge across from uracil has been shown to enhance base excision by uracil DNA glycosylase (UDG) and rescue the loss of activity of UDG mutants that lack the Leu191 plug side chain 26. In contrast, placing pyrene across from 7mG reduced AlkD’s activity 10-fold relative to a 7mG•C pair (Fig. S7). Superposition of the pyrene onto the opposite thymine in the AlkD/DNA crystal structures showed that this bulky group would be hindered from rotating into this tipped position. Thus, the consistency between the crystal structures and partial inhibition of 7mG activity by an opposing pyrene argues strongly that the crystal structures represent a catalytically competent orientation of DNA.
In a converse experiment, we tested AlkD’s ability to excise bulky pyridyloxobutyl (POB) base adducts (Fig. 4a), which arise in DNA upon exposure to cigarette smoke-derived nitrosamine carcinogens 27. The expectation was that AlkD should excise POB-bases from DNA, whereas the tightly constrained nucleobase binding pocket of human AAG would discriminate against bulky alkyl adducts 28. Indeed, AlkD liberated positively charged 7-POB-Gua and O2-POB-Cyt adducts from DNA, whereas neither of these modified bases was detected after treatment with AAG or in a mock reaction containing no enzyme (Fig. 4b,c). Neutral adducts O6-POB-Gua and O2-POB-Thy present in the DNA were not detected in the supernatant upon reaction with AlkD, consistent with the specificity of AlkD for positively charged lesions. This result indicates that AlkD need not flip the substrate base into an active site cavity in order to excise N3- or N7-alkylpurines from DNA.
Figure 4.
Excision of N7- and O2- pyridyloxobutyl (POB) base adducts by AlkD. a, Chemical structures of N7-POB-deoxyguanine and O2-POB-deoxycytosine. b,c, Time courses for the release of N7-POB-dG (black squares) and O2-POB-dC (red circles) in the presence (closed symbols, solid lines) and absence (open symbols, dashed lines) of Bacillus cereus AlkD (b) or human AAG (c). Error bars represent the standard deviation from three independent measurements.
AlkD traps and restructures destabilized base pairs
Recent work suggests that DNA glycosylases and oxidative demethylases detect damage by using side chains to probe for free energy differences between normal and modified base pairs 29–32. The lack of lesion-specific and DNA intercalating interactions in the AlkD/DNA complexes implies that AlkD detects damage solely on the basis of DNA duplex destabilization resulting from altered stacking or pairing of non-canonical base pairs. In support of this, we crystallized the protein in complex with DNA containing a G•T mismatch (Fig. 5a and Table S1), for which AlkD has no activity, but were unable to trap the protein onto the same oligonucleotide containing a G•C or A•T base pair at this same position. The resulting 1.5-Å AlkD/G•T-DNA structure is virtually identical to the 3d3mA•T complex (Table S1, Fig. S4). The similarity in these structures, together with thermodynamic differences between modified and unmodified nucleobases, suggests that AlkD detects these energetic differences as opposed to specifically recognizing the N3- or N7-methyl groups (see Supplementary Information) 23,33–36.
Figure 5.
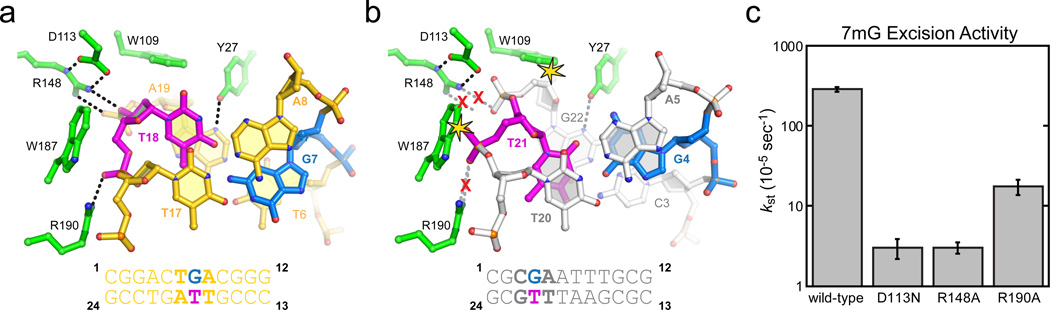
Remodeling of a G•T wobble base pair by AlkD. a, AlkD/G•T-DNA complex viewed down the helical axis. b, The structure of a G•T wobble base pair in DNA alone (PDB 113D) is superimposed onto the AlkD/G•T complex. Steric clashes between the protein and DNA are highlighted by yellow stars, and disrupted hydrogen bonds (dashed lines) are shown by a red X. c, Relative single-turnover rates (kst) of 7mG excision from a 25mer oligonucleotide duplex by wild-type and mutants of AlkD. Wild-type, D113N, and R148A data from ref. 19. Error bars represent the standard deviation from three independent measurements.
Comparison of the G•T mismatch bound by AlkD and in the context of DNA alone provides a basis for DNA damage recognition by AlkD (Fig. 5). In DNA, G•T wobble mismatches form two Watson-Crick hydrogen bonds and are well stacked within the duplex 37 (Fig. S10). AlkD restructures the G•T wobble so that the two bases protrude into opposite DNA grooves, disrupting base stacking and leaving only a single hydrogen bond between guanine N2 and thymine O4 (Fig. 5a). Superposition of a canonical G•T wobble onto the AlkD structure revealed that the protein stabilizes this conformation by inducing a specific distortion to the DNA backbone in order to alleviate steric clashes (Fig. 5b) and to create optimal hydrogen bonding and van der Waals interactions at the DNA capture site (Fig. S10). Thus, the enzyme detects non-Watson-Crick base pairs by resculpting the DNA backbone to create an optimized protein-DNA binding surface. In both 3d3mA•T and G•T complexes, specific protein-DNA contacts are mediated by Arg148-Asp113 and Arg190. Substitution of any of these highly conserved residues reduces single-turnover rates of 7mG excision by an order of magnitude (Fig. 5c), highlighting the importance of these interactions to catalysis.
Base excision by solvent exposure
The specific structure of the DNA trapped in the AlkD complexes provides a rationale for the enzyme’s specificity toward bases with a high propensity for depurination. We propose the lesion capture mechanism facilitates base hydrolysis by increasing the lifetime that the N-glycosidic bond is exposed to solvent, consistent with spontaneous depurination rates of 7mG in different DNA secondary structural contexts (see Supplementary Information). However, the 100-fold rate enhancement of 7mG hydrolysis from duplex DNA by AlkD cannot be explained on the basis of solvent exposure alone. Close inspection of the highly distorted DNA backbone in the flipped abasic structure revealed that the deoxyribose ring is positioned directly above a neighboring phosphate and that several water molecules bridge this phosphate and the extrahelical deoxyribose C1′ carbon (Fig. S12a), raising the possibility that the phosphate groups participate in catalysis. DNA-mediated water positioning to facilitate hydrolysis is a plausible catalytic mechanism given the lack of a requirement for a general base in these likely highly dissociative reactions. Alternatively, electrostatic stabilization of an oxocarbenium intermediate by nearby phosphates, which has been reported for uracil DNA glycosylase 38,39, offers a second possible mechanism for DNA-stimulated catalysis.
Discussion
AlkD represents a novel glycosylase found in bacteria, archaea, plants and lower eukaryotes (Fig. S1) 17. To our knowledge, most if not all of these organisms contain at least one other alkylpurine DNA glycosylase, raising the question as to why an alternate mechanism has evolved to eliminate genomic alkylation damage. The redundancy of alkylation repair may provide enhanced protection to organisms faced with an onslaught of methylating agents. Alternatively, AlkD may be a general DNA binding protein that coincidentally accelerates hydrolysis of unstable N-glycosidic bonds, or, as speculated below, may play a supporting role in general lesion detection.
AlkD’s activity toward bulky POB-DNA adducts normally associated with nucleotide excision repair (NER) 40,41 may be indicative of a more generalized function of AlkD in genome maintenance. AlkD’s lesion capture strategy is reminiscent of Rad4/XPC, which recognizes cyclopyrimidine dimers by binding to the opposing nucleotides 42. Exposure of the lesion away from the protein has the biological advantage of damage accessibility by the rest of the NER machinery. The AlkD-product complex may provide a platform for recruitment of a protein against the extrahelical abasic site, as seen in human APE1-DNA complexes 43. It is intriguing to speculate that AlkD may participate in alternative repair pathways by virtue of its ability to expose DNA damage. Indeed, non-enzymatic alkyltransferase-like proteins were recently found to trigger NER of O6-alkylguanines by inducing a specific protein-DNA complex, as illustrated by the crystal structure of ATL bound to DNA containing O6-POB-dG 44.
The AlkD-DNA structures illustrate how HEAT repeats engage nucleic acids and, to our knowledge, provide the first structural example of a HEAT motif with enzymatic activity. Comparison with nuclear import factor importin β, which uses HEAT repeats to bind a highly charged region of importin α 21 and Ran GTPase 22, demonstrates that the concave surface of the HEAT domain is a generalized macromolecular binding platform. HEAT repeats have been identified in chromatin-remodeling factors, including condensins, cohesins, and some SWI2/SNF2 proteins 45, as well as DNA-damage response protein kinases ATM, ATR, and DNA-PK 46. Recently, HEAT domains were visualized in EM and crystal structures of the catalytic subunit of DNA-PK 47,48, raising the possibility that other structurally uncharacterized DNA processing enzymes utilize HEAT domains to bind DNA in a manner similar to AlkD.
Methods
AlkD purification and crystallization
AlkD proteins were purified as described previously 19. Briefly, Bacillus cereus AlkD was overexpressed as an N-terminal His6-SUMO-AlkD fusion protein in E. coli HMS174 cells for 3 h at 37°C. AlkD was isolated using Ni-NTA (Qiagen) affinity chromatography, followed by cleavage of the His6-SUMO tag and further purification by heparin affinity and gel filtration chromatography. Protein was concentrated to 12.5 mg/mL and stored in 20 mM Bis-Tris Propane, 100 mM NaCl, 2 mM DTT and 0.1 mM EDTA. Site-directed mutagenesis of the wild-type AlkD vector was performed using a Quik-Change Kit (Stratagene), Mutant proteins were overexpressed and purified identically to wild-type AlkD, and their structure verified by circular dichroism spectroscopy.
AlkD/DNA complexes were assembled by incubating 0.45 mM protein and 0.54 mM oligonucleotide for 15 min at 4°C. Oligonucleotide sequences used were d(TGGG(THF)GGCTT)/d(AAAGCCYCCC), in which Y = thymine or cytosine, and d(CGGACTXACGGG)/d(CCCGTTTCCG), in which X was either 3d3mA or G. AlkD/THF-DNA crystals were grown at 16°C by mixing 2 µL protein/DNA complex with 2 µL reservoir solution containing 0.1 M Bis-Tris pH 6.5, 0.1 mM NaCl, and 19% PEG 3350 and 2% glycerol. Crystals were soaked in 30% glycerol/reservoir solution for 1 min and flash frozen in a liquid nitrogen stream. Crystals of 3d3mA-DNA and G•T-DNA complexes were grown from reservoir solutions containing 85 mM NaAcetate pH 4.6, 170 mM ammonium acetate, 25.5% PEG 4000, and 15% glycerol at 21°C, and were flash frozen in liquid nitrogen directly from this solution.
X-ray Data Collection, Phasing, and Structure Refinement
X-ray data (Table S1) were collected at a wavelength of 0.97850 Å and 110 K at the Advanced Photon Source beamlines 21-ID-D and 21-ID-G (LS-CAT) and processed with HKL2000 50. Molecular replacement using unliganded AlkD (PDB ID 3BVS) as a search model in Phaser 51 gave a clear solution for each structure. Following one round of simulated annealing refinement in CNS 52, the entire DNA molecules could be discerned and were built into 2Fo-Fc and Fo-Fc electron density using XtalView 53. Atomic coordinates and B-factors for the protein/DNA models were refined in Phenix 54. TLS refinement with protein and each DNA chain defined as three separate TLS groups was carried out for each model except the GT-complex. Individual anisotropic B-factors were derived from the refined TLS parameters and held fixed during subsequent rounds of refinement, which significantly decreased the crystallographic residuals and improved the electron density maps. Instead of TLS refinement, individual anisotropic B-factors were explicitly refined for the G•T-complex. Adjustments to the model, including addition of solvent molecules, using Coot 55 were guided by manual inspection of 2Fo-Fc and Fo-Fc electron density maps and were judged successful by a decrease in Rfree during refinement.
Protein and DNA models were validated using PROCHECK 56 and DNA parameters were quantified using CURVES 5.2 57. All but one out of the total 223–231 protein residues resided in the most favored (191–198 residues) or allowed (14–15 residues) regions of the Ramachandran plot. As in the unliganded structure 19, Thr54 in all four DNA complex structures remained in the disallowed region despite an excellent fit to 2Fo-Fc electron density maps.
Enzyme Activity
Excision of 7mG by AlkD was measured by incubating the enzyme with a 25mer oligonucleotide containing a centrally located 7mG and following the appearance of abasic DNA product after alkaline cleavage. 7mG was enzymatically incorporated into DNA duplexes using the previously described method 58, in which an oligonucleotide primer [d(GACCACTACACC)] was 32P-labeled at the 5′-end, annealed to a 3-fold excess of the complementary strand [d(GTTGTTAGGAAACGGTGTAGTGGTC)] and extended using DNA polymerase I Klenow fragment (New England Biolabs) in the presence of 2′-deoxy-7-methylguanosine 5′-triphosphate (Sigma), dCTP, dTTP, and dATP. To create 7mG mispairs, 100-fold excess of complementary strand with T, G, A or pyrene in place of C at position 13 was reannealed to the 7mG containing oligonucleotide. Single-stranded 7mG containing strands were obtained by reannealing to 100-fold excess of unlabeled lesion strand with G in place of 7mG [d(GACCACTACACCGTTTCCTAACAAC)].
In a 10 µL glycosylase reaction, 100 nM [32P]-DNA duplex was incubated with 0–20 µM AlkD in 50 mM HEPES pH 7.5, 100 mM KCl, 10 mM DTT, and 2 mM EDTA. The reaction was quenched at various times by addition of 0.2 N NaOH and heated at 70°C for 2 min. Substrate 25mer and product 12mer DNA strands were separated by denaturing 20% polyacrylamide gel electrophoresis in 7 M urea and quantitated by autoradiography. Kinetic data were analyzed by standard single-turnover techniques 59, which have been extensively used for DNA glycosylases 60–65. Enzymatic rate constants (k) were obtained from single-exponential fits to the data (fP = 1 – e−kt, in which fP is the fraction of product). For determination of the single-turnover rate constant, kst, AlkD was at least 5-fold in excess over the K½ for a particular labeled DNA substrate (e.g., 5 µM for 7mG•C). For K½ determinations, the 7mG excision assay was performed over a range of enzyme concentrations and K½ obtained by fitting the Michaelis-Menten plot with the equation, kobs = Vmax[AlkD] / (K½ + [AlkD]). We note that our K½ for maximal activity may differ from the Km value for multiple turnover because the Km can be affected by product release. Stoichiometric 7mG excision was performed in the presence of 10 µM unlabeled 25mer DNA duplex (K½ for this DNA was determined to be 0.9 ± 0.1 µM). Spontaneous rates of 7mG hydrolysis were determined using the sequence d(GACCACTACACC(7mG)ATTCCTTACAAC) that had been re-annealed to 100-fold excess complementary strand d(GTTGTAAGGAAT(C/T)GGTGTAGTGGTC).
POB Adduct Excision
Materials
DNA (cat. no. D1501), alkaline phosphatase (P8361), esterase (E2884) micrococcal nuclease (N3755), and phosphodiesterase II (P9041) were purchased from Sigma. The tetra-deuterated standards were provided by Dr Stephen S. Hecht of the University of Minnesota. NNKOAc was synthesized by Dhimant Desai of the Organic Synthesis facility of the Penn State college of Medicine.
NNKOAc-damaged DNA
Five milliliters DNA (2 mg/mL) dissolved in 100 mM sodium phosphate (pH 7.0), 1 mM EDTA and 50 mM NaCl was reacted with 1 mM NNKOAc and esterase (200 units) at 37°C for 2 h. The reaction was diluted to 10 mL with H2O and extracted with 10 mL CHCl3/iso-amyl alcohol (24/1) to remove the protein and 10 mL ethyl acetate to remove any unreacted NNKOAc. The DNA was precipitated by the addition of 40 mL ethanol and washed twice 70% ethanol. Residual amounts of ethanol was removed by rotary evaporation and the DNA was dissolved in H2O, aliquoted and stored at −80°C prior to use.
Glycosylase reactions
The damaged DNA (1 mg/mL) was incubated with 1 µM glycosylase in 400 µL buffer (50 mM HEPES (pH 7.5), 1 mM EDTA, 100 mM KCl, 1 mM DTT) at 37°C. Aliquots (100 µL) were quenched at various times by the addition of 5 µL 3 M sodium acetate (pH 5.2) and 200 µL ice cold ethanol. The mixture was centrifuged for 10 min and the supernatant decanted and saved for analysis.
HPLC-MS/MS
Deuterated standards (100 fmol each of O2-POB-C-d4 and 7-POB-G-d4) were added to the ethanol supernatant and the solvent evaporated. The sample was dissolved in 50 µL methanol for MS analysis. The samples were analyzed with a MDS/Sciex 4000 QTrap instrument with electrospray ionization (ESI) coupled to an Agilent 1100 HPLC system. Samples (20 µL) were loaded onto a column (Luna C18(2) 150×2 mm, 3 micron) which was eluted with 10 mM ammonium formate at 0.1 mL/min. The POB−DNA adducts, along with their deuterated standards, were monitored by selected reaction monitoring (SRM). The ion transitions were as follows 7-POB-Gua, m/z 299.1 [M + 1] + to m/z 148.1 [POB]+; [pyridine-D4]7-POB-Gua, m/z 303.1 [M + 1] + to m/z 152.1 ([pyridine-D4]POB])+ and [Gua + H]+; O2-POB-Cyt, m/z 259.1 [M + 1] + to m/z 148.1 [POB]+; [pyridine-D4]O2-POB-Cyt, m/z 263.1 [M + 1]+ to m/z 152.1 ([pyridine-D4]POB])+. Prior to HPLC-ESI-MS/MS analysis of the samples the MS parameters were optimized for each deuterated POB−DNA adduct standard. For analysis, the MS parameters were set as follows: Curtain gas, 40 psi; Ion spray voltage, 4 kV; source temperature, 650°C; nebulizer gas (GS1), 70 psi; heater gas (GS2), 70 psi; and collision gas, 12 psi. The fragmentation potentials were optimized for each ion: 299.1 and 303.1, declustering potential (DP), 65 V, entrance potential (EP), 10 V; collision energy (CE), 20 V; collision cell exit potential (CXP), 12 V and for 259.1 and 263.1: DP, 40V; EP, 8V; EP, 15V; and CE, 6V. The amount of each POB−DNA adduct was determined by comparing the MS peak area ratio of each adduct to its deuterated standard with a calibration curve. Calibration standards were prepared by spiking different amounts of each adduct with a constant amount of the corresponding internal standard in H2O and then analyzed by LC-MS/MS without undergoing the sample preparation procedure described above. The calibration curves were constructed by plotting concentration ratio vs MS peak area ratios of each adduct to its deuterated standard.
DNA Binding
DNA binding was monitored by a change in fluorescence anisotropy as increasing concentrations of protein were added to an oligonucleotide duplex that contained a THF abasic modification in the middle of one strand [d(TGACTACTACAT(THF)GTTGCCTACCAT)] and a 6-carboxyfluorescein (FAM) on the 3′-end of the complementary strand [d(ATGGTAGGCAACTATGTAGTAGTCA)-FAM]. For stoichiometric binding measurements, increasing concentrations of protein (0–200 µM) were added to a solution containing 50 nM FAM-DNA and 20 µM unlabeled 25mer DNA (Kd = 3.1 ± 0.3 µM) in 20 mM Bis-Tris propane pH 6.5, 100 mM NaCl, 2 mM DTT, and 0.1 mM EDTA. Polarized fluorescence intensities using excitation and emission wavelengths of 485 and 538 were measured at ambient temperature using a SpectraMax M5 microplate reader (Molecular Devices). Dissociation constants were derived by fitting a two-state binding model to data from three independent experiments.
Supplementary Material
Acknowledgements
We thank James Stivers for providing the pyrene phosphoramidite, Zdzislaw Warzak and LS-CAT beamline staff at the Advanced Photon Source (APS) for assistance with X-ray data collection, and Tom Ellenberger, James Stivers, and Patrick O’Brien for helpful comments on the manuscript. Use of the APS was supported by the U.S. Department of Energy Office of Basic Energy Sciences. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. This research was supported by a grant from the American Cancer Society (to B.F.E.). E.H.R. was supported in part by the Vanderbilt Training Program in Molecular Toxicology. Additional support for local crystallography facilities was provided by the Vanderbilt Center in Molecular Toxicology and the Vanderbilt-Ingram Cancer Center.
Footnotes
Author Contributions
E.H.R. purified and crystallized AlkD, determined crystal structures, and performed 7mG activity assays; B.G. synthesized 3d3mA oligonucleotides; A.S.P.G. and T.E.S. performed POB activity assays; B.F.E. designed the project and analyzed data; B.F.E. and E.H.R. wrote the paper. All authors discussed the results and commented on the manuscript.
Author Information
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3JX7 (3d3mA•T), 3JXY (G•T), 3JXZ (THF•T), and 3JY1 (THF•C). Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Friedberg EC, et al. DNA repair: from molecular mechanism to human disease. DNA repair. 2006;5(8):986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens: Intrinsic Properties of Nucleic Acids. Plenum Press: New York; 1983. [Google Scholar]
- 3.Holt S, Yen TY, Sangaiah R, Swenberg JA. Detection of 1,N6-ethenoadenine in rat urine after chloroethylene oxide exposure. Carcinogenesis. 1998;19(10):1763–1769. doi: 10.1093/carcin/19.10.1763. [DOI] [PubMed] [Google Scholar]
- 4.Shuker DE, Bailey E, Parry A, Lamb J, Farmer PB. The determination of urinary 3-methyladenine in humans as a potential monitor of exposure to methylating agents. Carcinogenesis. 1987;8(7):959–962. doi: 10.1093/carcin/8.7.959. [DOI] [PubMed] [Google Scholar]
- 5.Shuker DE, Farmer PB. Relevance of urinary DNA adducts as markers of carcinogen exposure. Chem Res Toxicol. 1992;5(4):450–460. doi: 10.1021/tx00028a001. [DOI] [PubMed] [Google Scholar]
- 6.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutation research. 1985;150(1–2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 7.Plosky BS, et al. Eukaryotic Y-family polymerases bypass a 3-methyl-2'-deoxyadenosine analog in vitro and methyl methanesulfonate-induced DNA damage in vivo. Nucleic acids research. 2008;36(7):2152–2162. doi: 10.1093/nar/gkn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7- alkylguanine residues in DNA. Chem Res Toxicol. 2004;17(7):839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 9.Stivers JT. Site-specific DNA damage recognition by enzyme-induced base flipping. Prog Nucleic Acid Res Mol Biol. 2004;77:37–65. doi: 10.1016/S0079-6603(04)77002-6. [DOI] [PubMed] [Google Scholar]
- 10.Stivers JT. Extrahelical damaged base recognition by DNA glycosylase enzymes. Chemistry. 2008;14(3):786–793. doi: 10.1002/chem.200701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chemical reviews. 2003;103(7):2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 12.Parikh SS, et al. Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci U S A. 2000;97(10):5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mol CD, Arvai AS, Begley TJ, Cunningham RP, Tainer JA. Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases. Journal of molecular biology. 2002;315(3):373–384. doi: 10.1006/jmbi.2001.5264. [DOI] [PubMed] [Google Scholar]
- 14.Drohat AC, Kwon K, Krosky DJ, Stivers JT. 3-Methyladenine DNA glycosylase I is an unexpected helix-hairpin-helix superfamily member. Nat Struct Biol. 2002;9(9):659–664. doi: 10.1038/nsb829. [DOI] [PubMed] [Google Scholar]
- 15.Eichman BF, O'Rourke EJ, Radicella JP, Ellenberger T. Crystal structures of 3- methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. The EMBO journal. 2003;22(19):4898–4909. doi: 10.1093/emboj/cdg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metz AH, Hollis T, Eichman BF. DNA damage recognition and repair by 3- methyladenine DNA glycosylase I (TAG) The EMBO journal. 2007;26(9):2411–2420. doi: 10.1038/sj.emboj.7601649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alseth I, et al. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Molecular microbiology. 2006;59(5):1602–1609. doi: 10.1111/j.1365-2958.2006.05044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalhus B, et al. Structural insight into repair of alkylated DNA by a new superfamily of DNA glycosylases comprising HEAT-like repeats. Nucleic acids research. 2007;35(7):2451–2459. doi: 10.1093/nar/gkm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubinson EH, Metz AH, O'Quin J, Eichman BF. A new protein architecture for processing alkylation damaged DNA: the crystal structure of DNA glycosylase AlkD. Journal of molecular biology. 2008;381(1):13–23. doi: 10.1016/j.jmb.2008.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11(2):115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 21.Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399(6733):221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 22.Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97(5):635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 23.Ganguly M, Wang R-W, Marky LA, Gold B. Thermodynamic characterization of DNA with 3-deazaadenine and 3-methyl-3-deazaadenine substitutions. J. Phys. Chem. B. 2010 doi: 10.1021/jp101004k. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. The Journal of biological chemistry. 2004;279(11):9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. The Journal of biological chemistry. 2004;279(26):26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
- 26.Jiang YL, Kwon K, Stivers JT. Turning On uracil-DNA glycosylase using a pyrene nucleotide switch. The Journal of biological chemistry. 2001;276(45):42347–42354. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 27.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutation research. 1999;424(1–2):127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 28.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95(2):249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311(5764):1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434(7033):612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 31.Yang CG, Garcia K, He C. Damage detection and base flipping in direct DNA alkylation repair. Chembiochem. 2009;10(3):417–423. doi: 10.1002/cbic.200800580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CG, et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452(7190):961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboul-ela F, Koh D, Tinoco I, Jr, Martin FH. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T) Nucleic acids research. 1985;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ezaz-Nikpay K, Verdine GL. Aberrantly methylated DNA: site-specific introduction of N-7-methyl-2'-deoxyguanosine into the Dickerson/Drew dodecamer. J. Am. Chem. Soc. 1992;114(16):6562–6563. [Google Scholar]
- 35.Ezaz-Nikpay K, Verdine GL. The effects of N7-methylguanine on duplex DNA structure. Chem Biol. 1994;1(4):235–240. doi: 10.1016/1074-5521(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 36.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J Am Chem Soc. 2008;130(35):11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter WN, et al. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-A resolution. The Journal of biological chemistry. 1987;262(21):9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 38.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413(6857):752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 39.Jiang YL, Ichikawa Y, Song F, Stivers JT. Powering DNA repair through substrate electrostatic interactions. Biochemistry. 2003;42(7):1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 40.Brown PJ, Bedard LL, Massey TE. Repair of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced DNA pyridyloxobutylation by nucleotide excision repair. Cancer Lett. 2008;260(1–2):48–55. doi: 10.1016/j.canlet.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Li L, et al. The Influence of Repair Pathways on the Cytotoxicity and Mutagenicity Induced by the Pyridyloxobutylation Pathway of Tobacco-Specific Nitrosamines. Chem Res Toxicol. 2009 doi: 10.1021/tx9001572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449(7162):570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 43.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403(6768):451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 44.Tubbs JL, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459(7248):808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10(10):1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112(2):151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 47.Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16(3):468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463(7277):118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to Methods
- 49.Irani RJ, SantaLucia J. Jr. The synthesis of anti-fixed 3-methyl-3-deaza-2'- deoxyadenosine and other 3H-imidazo[4,5-c]pyridine analogs. Nucleosides Nucleotides Nucleic Acids. 2002;21:737–751. doi: 10.1081/NCN-120016477. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 52.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 53.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. Journal of structural biology. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, et al. In: Evolving Methods for Macromolecular Crystallography. Read RJ, Sussman JL, editors. Springer; 2007. pp. 101–109. [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 57.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 58.Asaeda A, et al. Substrate specificity of human methylpurine DNA N-glycosylase. Biochemistry. 2000;39:1959–1965. doi: 10.1021/bi9917075. [DOI] [PubMed] [Google Scholar]
- 59.Jones BN, Quang-Dang DU, Oku Y, Gross JD. A kinetic assay to monitor RNA decapping under single- turnover conditions. Methods Enzymol. 2008;448:23–40. doi: 10.1016/S0076-6879(08)02602-5. [DOI] [PubMed] [Google Scholar]
- 60.Baldwin MR, O'Brien PJ. Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyons DM, O'Brien PJ. Efficient recognition of an unpaired lesion by a DNA repair glycosylase. J Am Chem Soc. 2009;131:17742–17743. doi: 10.1021/ja908378y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maher RL, Bloom LB. Pre-steady-state kinetic characterization of the AP endonuclease activity of human AP endonuclease 1. The Journal of biological chemistry. 2007;282:30577–30585. doi: 10.1074/jbc.M704341200. [DOI] [PubMed] [Google Scholar]
- 63.Maher RL, Vallur AC, Feller JA, Bloom LB. Slow base excision by human alkyladenine DNA glycosylase limits the rate of formation of AP sites and AP endonuclease-1 does not stimulate base excision. DNA repair. 2007;6:71–81. doi: 10.1016/j.dnarep.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Maiti A, Morgan MT, Drohat AC. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. The Journal of biological chemistry. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennett MT, et al. Specificity of human thymine DNA glycosylase depends on Nglycosidic bond stability. J Am Chem Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.