SUMMARY
The response of animals to hypoxia is mediated by the hypoxia-inducible transcription factor (HIF). Human HIF is regulated by four Fe(II) and 2-oxoglutarate (2OG) dependent oxygenases: Prolyl hydroxylase domain enzymes (PHDs or EGLNs) 1–3 catalyse hydroxylation of two prolyl-residues in HIF, triggering its degradation by the proteasome. Factor inhibiting HIF (FIH) catalyses hydroxylation of an asparagine-residue in HIF, inhibiting its transcriptional activity. Collectively, the HIF hydroxylases negatively regulate HIF in response to increasing oxygen concentration. Prolyl hydroxylase domain 2 (PHD2) is the most important oxygen sensor in human cells; however the underlying kinetic basis of the oxygen sensing function of PHD2 is unclear. We report analyses of the reaction of PHD2 with oxygen. Chemical quench/mass spectrometry experiments showed that reaction of a complex of PHD2, Fe(II), 2OG and the C-terminal oxygen-dependent degradation domain of HIF-α (CODD) with oxygen to form hydroxylated CODD and succinate is much slower (~100 fold) than for other similarly studied 2OG oxygenases. Stopped flow/UV-visible spectroscopy experiments showed that the reaction produces a relatively stable species absorbing at 320nm; Mössbauer spectroscopic experiments implied that this species is likely not a Fe(IV)=O intermediate, as observed for other 2OG oxygenases. Overall the results suggest that, at least compared to other studied 2OG oxygenases, PHD2 reacts relatively slowly with oxygen, a property that may be associated with its function as an oxygen sensor.
Keywords: Hypoxia Inducible Factor, 2-Oxoglutarate, Oxygen, Oxygenase, Prolyl Hydroxylase, Spectroscopy
INTRODUCTION
The cellular response of animals to hypoxia is mediated by a heterodimeric transcription factor, hypoxia-inducible factor (HIF) [1]. Under hypoxic conditions, HIF upregulates an array of genes including those encoding vascular endothelial growth factor and erythropoietin (for review see [2–4]) that work to counteract the effects of hypoxia. Levels of HIF-α, but not HIF-β, are regulated directly by oxygen availability [5]. Prolyl-4-hydroxylation in either of the N- or C-terminal oxygen-dependent degradation domains (Pro402 of 'NODD' and Pro564 of 'CODD') of HIF-α increases its binding to the von Hippel Lindau protein elongin C/B complex (VCB), which targets HIF-α for proteasomal degradation [6, 7]. In a separate oxygen-dependent mechanism of HIF regulation, asparaginyl hydroxylation (Asn803) in the HIF-α C-terminal transactivation domain (C-TAD) reduces the interaction of HIF with transcriptional coactivators [8]. HIF hydroxylation is catalysed by Fe(II)- and 2-oxoglutarate (2OG) dependent oxygenases. In human cells, there are three prolyl hydroxylase domain enzymes 1–3 (PHDs 1–3), which have significant homology in their catalytic domains [9–11], and one asparaginyl hydroxylase, termed Factor Inhibiting HIF (FIH) [12, 13]. Based on evidence from cell cultures and mouse models [14, 15], PHD2 has been identified as the most important of the HIF hydroxylases for the hypoxic response in normal human tissues.
Fe(II)/2OG-dependent oxygenases constitute a ubiquitous family of enzymes that perform a range of biologically important oxidation reactions [16]. It is proposed that most 2OG-dependent oxygenases employ a conserved reaction mechanism [17–20] (Figure 1), which has been adapted from that proposed for the collagen-modifying prolyl-4-hydroxylase [21]. Evidence for this mechanism stems from detailed crystallographic and spectroscopic analyses of the stable Fe(II)-containing intermediates, and the characterization of reaction intermediates, including the Fe(IV)=O complex and the Fe(II) product complex, by a combination of rapid kinetic and spectroscopic methods [22]. The Fe(II) centre is normally coordinated by three protein-derived ligands, that form a ‘facial His2-(Glu/Asp)1 triad’ (for reviews see [23, 24]). 2OG binds to the Fe(II) in a bidentate manner [25, 26], which gives rise to a metal-to-ligand charge transfer band at ~520 nm [27]. Substrate binding adjacent to the Fe(II) is proposed to weaken binding of the remaining coordinated water, so enabling binding of oxygen [28, 29]. Oxidative decarboxylation of 2OG then produces succinate (into which one of the dioxygen atoms is incorporated [30]), carbon dioxide and a reactive Fe(IV)=O (ferryl) intermediate. The ferryl intermediate has been detected for two 2OG-dioxygenases: taurine 2OG dioxygenase (TauD) [31] and Paramecium bursaria Chlorella virus 1 prolyl-4-hydroxylase (PBCV-1 P4H) [32]. The Fe(IV)=O intermediate can cleave the target substrate C-H bond by H-abstraction[33]; Rebound of the substrate radical with a hydroxyl radical equivalent derived from the ensuing Fe(III)-OH complex [34] then leads to a Fe(II)-product complex [32, 35]. Product dissociation completes the catalytic cycle.
Figure 1.
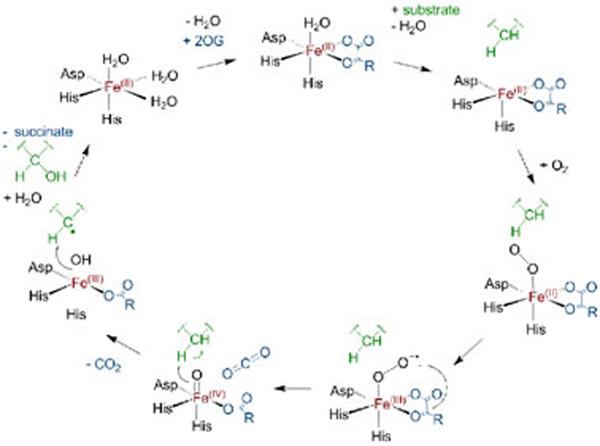
Proposed general catalytic mechanism for the Fe(II)/2OG oxygenases.
Crystal structures of PHD2 [36, 37] have revealed the double stranded beta helix core fold that is characteristic of the 2OG oxygenases, and that the Fe(II) is bound by a His2-(Glu/Asp)1 triad. Evidence has been reported that PHD2 has, possibly unusually, tight binding constants for Fe(II) and 2OG and that the Fe(II)and Fe(II).2OG complexes of PHD2 are unusually stable [38–40]. Kinetic studies on PHD2 have focused on steady state analyses, and have monitored activity by a range of methods including oxygen consumption and production of 14CO2. kcat values for PHD2 have been reported to range from 0.004s−1 using PHD21–426 expressed in insect cells and biotinylated HIF-1α566–574 substrate [40], through ~0.03s−1 using PHD2181–426 expressed in Escherichia coli with a HIF-1α566–574 (CODD) substrate [41, 42], to 0.3s−1 using cell extracts of endogenous PHD21–426 with biotinylated HIF-1α566–574 substrate [43] (kinetic data obtained in different conditions are more fully compared in [41]). Some of these differences likely reflect, at least in part, variations in assay compositions.
Although all 2OG oxygenases necessarily react with oxygen, an important question with respect to PHD2 is whether its kinetic properties are consistent with its role as the most important of the identified human oxygen sensors. Preliminary analyses with crude extracts and isolated enzymes have led to reported apparent Km values for oxygen for PHD2 and FIH of 65–240µM [41, 44, 45]; one study has even estimated a Km value for oxygen for PHD2 of 1.7mM [40]. However, there is little information on the kinetic details of individual steps in catalysis by these enzymes. In particular there is no reported direct information on whether the rate of reaction of PHD2 with oxygen is actually limiting; in order for PHD2 to act as an oxygen sensor, as proposed, in cells its hydroxylation of HIF must be limited by oxygen availability. Here we report combined kinetic analyses on purified PHD2 and its preferred CODD substrate, focussing on single catalytic turnover events, employing mass spectrometry (MS) and nuclear magnetic resonance (NMR), UV-visible absorption, and Mössbauer spectroscopies. The results provide evidence that at least under the studied conditions, the rate of reaction of the PHD2:Fe(II):2OG:HIF-α complex with oxygen is very much slower than for other studied 2OG oxygenases.
RESULTS
The catalytically productive single turnover reaction of the PHD2:Fe(II):2OG:CODD complex with oxygen is slow
To investigate the kinetics of the reaction of a PHD2:Fe(II):2OG:CODD complex with oxygen in a single turnover, an anoxic solution of 0.8 mM PHD2 (catalytic domain, residues 181–426), 0.7 mM Fe(II), 0.5 mM 2OG and 1 mM CODD was rapidly mixed with O2-saturated buffer (in the presence of ascorbate, which stimulates PHD2 activity [46, 47]). Following quenching of the reaction with 0.1 M HCl, liquid chromatography-MS analyses were used to monitor the conversion of 2OG to succinate: 2OG and succinate levels were measured over time intervals ranging from 34 ms to 50 min. The results (Figure 2) demonstrate that full conversion of 2OG to succinate takes ~220 s and occurs with an apparent first-order rate constant of 0.018 (±0.0014) s−1 (Supplementary Figure S1). When the same reaction was monitored in the absence of CODD, conversion of 2OG to succinate was still observed. However, the apparent first-order rate constant for this reaction (0.0006 (±0.0000) s−1, Supplementary Figure S1) was much less (~30 fold) than in the presence of CODD. These observations are consistent with the known ability of 2OG oxygenases to catalyze 2OG turnover in the absence of their prime substrate [18]; it is notable that the rate of this ‘uncoupled’ turnover is particularly slow for PHD2.
Figure 2.
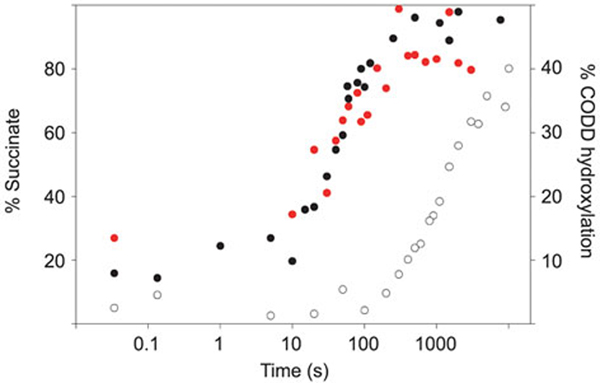
PHD2:Fe(II):2OG:CODD reacts slowly with oxygen in vitro. In the presence of CODD peptide substrate, 2OG decarboxylation to succinate (black circles) and CODD hydroxylation (red circles) occur at similar rates of 0.018s−1 and 0.013s−1 respectively, as determined by LC/MS and MALDI MS respectively. In the absence of CODD peptide substrate, 2OG decarboxylation to succinate (white circles) is 30-fold slower, at 0.0006s−1. Data are shown against time on a logarithmic scale. Concentrations before mixing were PHD2 (0.8 mM), Fe (0.7 mM), 2OG (0.5 mM), ascorbate (5 mM), CODD peptide (1 mM if present) and oxygen (1.9 mM). All reactions were carried out at 5 °C.
Analogous experiments were then performed to monitor the extent of CODD hydroxylation by matrix assisted laser desorption/ionisation mass spectrometry (MALDI MS) analyses (Figure 2). As for 2OG turnover, the CODD hydroxylation reaction appeared to be complete after ~220s, and occurred at a rate of 0.013 (±0.003) s−1 (Supplementary Figure S1). Figure 2 shows that in the presence of CODD, consumption of 2OG, formation of succinate, and formation of the hydroxylated-CODD product are near contemporaneous, and sufficiently rapid to account for the steady-state turnover rate of 0.03s−1 [41, 42] under similar conditions [differences are likely due to the significantly lower temperature at which the single turnover experiments took place (5°C) compared to that at which steady state experiments are usually conducted (37°C)]. Given that 2OG consumption and succinate formation are considerably slower in the absence of the CODD substrate, binding of the CODD substrate appears to stimulate reaction of the PHD2 complex with O2; however, at least under our current conditions, the extent of this stimulation is substantially less than for other similarly studied 2OG oxygenases, e.g. TauD has a substrate triggering effect of 1000-fold [48, 49]. This sluggish reaction of the PHD2 complexes with O2 may be related to the role of PHD2 as an O2 sensor.
Prime substrate hydroxylation and 2OG decarboxylation are fully coupled under steady state conditions
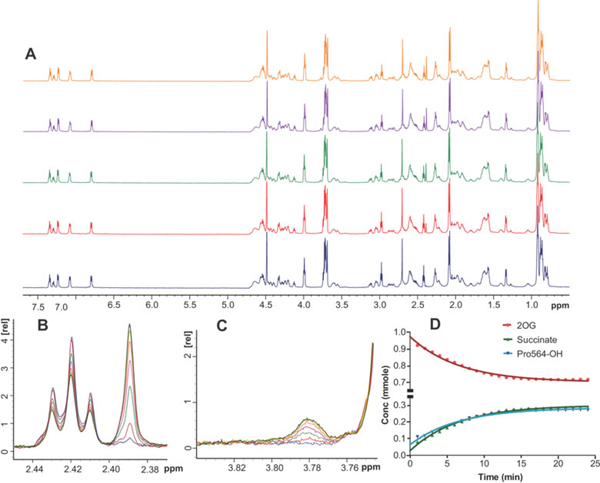
To determine the extent to which 2OG decarboxylation is coupled to CODD hydroxylation under our typical steady-state turnover conditions, we used 1H NMR spectroscopy (700MHz) to simultaneously monitor both events (Figure 3). 2OG turnover was quantified by integration of the 2OG (δH ~ 2.42) and succinate (δH ~ 2.38) methylene protons; CODD hydroxylation was monitored by integration of the intensity associated with the C5-bonded hydrogen of hydroxyproline at δH ~ 3.78 (Figure 3B and 3C). The results clearly demonstrate that 2OG depletion occurs concomitantly with both succinate production and CODD hydroxylation (Figure 3D), showing complete, or almost complete, coupling of these two events under steady-state turnover conditions.
Figure 3.
1H-Nuclear magnetic resonance time course demonstrating that 2-oxoglutarate decarboxylation is coupled to conversion to succinate and CODD peptide hydroxylation during reaction of PHD2. A, Full spectra of assay mixtures (see Experimental Details) as measured at 0, 5, 10, 15 and 20 minutes (blue, red, green, violet and yellow respectively). B, Conversion of 2OG to succinate: 2OG was monitored by the triplet at 2.42ppm, and succinate by the singlet at 2.39ppm. C, An increase in the intensity of the 1H-NMR signal at 3.78ppm, previously assigned as the δ proton of Pro-564 [39]. For clarity in B and C, only spectra recorded every 225s are shown. D, Integrated 1H-NMR signal intensities for 2OG, succinate and hydroxylated CODD (n=3), showing that 2OG decarboxylation and CODD hydroxylation rates are coupled in steady state turnover experiments. Data were fitted by the equation, y=(y0 − plateau)*exp(−K*X) + plateau, using Prism 5™.
Stopped-flow absorption and Mössbauer spectroscopic studies of the reaction of the enzyme complex with oxygen
Stopped-flow absorption spectroscopy was then used to monitor reaction of the enzyme:Fe(II):2OG complex with oxygen, with the aim of detecting intermediate complexes. Anoxic PHD2:Fe(II):2OG and PHD2:Fe(II):2OG:CODD complexes demonstrated absorption features at 530 nm and 520 nm, respectively (Supplementary Figure S2), consistent with the values reported for analogous complexes with previously studied 2OG-dependent oxygenases [31, 32, 50, 51]. However, upon mixing of the complexes with oxygen-saturated buffer, the hallmarks of rapid O2 activation observed in previous studies on TauD [31], PBCV-1 P4H [32], and the related halogenases, CytC3 [52] and SyrB2 [53], are not observed with PHD2, at least under our current assay conditions. Because the substrate affinity of PHD2 can increase with peptide length (the Km,app,sub for CODD (HIF-1α556–574) is 22 µM compared to a Km,app,sub for the longer HIF-1α(530–698) protein substrate of ~ 2 µM [41, 54]), we repeated the stopped-flow absorption analyses in the presence of a HIF-1α(530–698) protein substrate (Supplementary Figure S3). Significantly, development of absorption was no more rapid in the presence of the longer protein substrate than with CODD peptide, indicating that inefficient substrate binding is probably not the cause of the slow reaction with oxygen.
In each of the other similarly studied 2OG-dependent oxygenases [31, 32, 52, 53] [55] rapid development of an absorption feature at ~ 320 nm reflects accumulation of the Fe(IV)=O intermediate, and decay of this feature reflects abstraction of hydrogen from the substrate followed by rapid radical recombination to form the hydroxylated or halogenated product. In the reaction of PHD2 with O2, absorption develops only slowly and all across the UV/visible regime, both in the absence and in the presence of the CODD prime substrate (Figure 4, panels A and B, respectively). Moreover, the developing absorption decays even more slowly. The number and identities of the species responsible for the UV/visible absorption features cannot readily be determined, because there are no obvious correlations among these features and any new features detected in freeze-quench Mössbauer experiments (see below); it is therefore not possible to correlate particular spectral features with intermediates. Nevertheless, the stopped-flow absorption data do reveal an effect of the prime substrate - the decay phase is significantly faster in its presence. This effect supports the results observed in the chemical quenched-flow experiments (see above).
Figure 4.
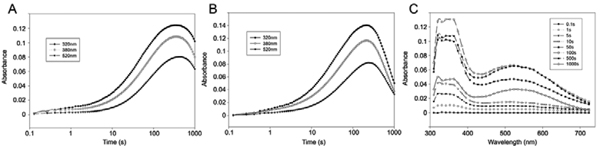
UV-visible absorption spectra on reaction of PHD2:Fe(II):2OG with an equal volume of an oxygen-saturated buffer, with and without CODD peptide substrate. A, Formation of species absorbing at 320, 380 and 520nm with time in the absence of substrate. B, Formation of species absorbing at 320, 380 and 520 over time in the presence of CODD. C, Broad spectral features observed at a range of time points in the presence of CODD. Concentrations before mixing were PHD2 (0.8 mM), Fe (0.7mM), 2OG (10mM), ascorbate (5mM), CODD peptide (1.0mM) and oxygen (1.9mM). Reactions were carried out at 5°C.
Mössbauer experiments were carried out with the intention of further characterizing the PHD2 reaction. The 4.2-K/zero-field Mössbauer spectrum of a sample of the PHD2:Fe(II):2OG:CODD complex (Figure 5A) exhibits several (at least two, perhaps even more) overlapping quadrupole doublet features with parameters typical of high-spin Fe(II) [δ1 (isomer shift) = 1.24 mm/s and ΔEQ,1 (quadrupole splitting parameter) = 2.04 mm/s (69%, red line) and δ2 = 1.25 mm/s and ΔEQ,2 = 3.16 mm/s (31%, blue line)]. The presence of multiple species suggests conformational heterogeneity of the PHD2:Fe(II):2OG:CODD complex. When this state is reacted with O2 for 200 s (the time at which A320 is maximal in the stopped-flow absorption experiments), the Mössbauer spectrum changes (Figure 5B). The first quadrupole doublet partially decays, and several poorly defined features develop. First, features attributable to another high-spin Fe(II) species with smaller quadrupole splitting parameter develop, as evidenced by the shoulders at the inside of the prominent lines at 0.2 mm/s and 2 mm/s (indicated by the black arrows). These differences can also be seen in the difference spectrum (Figure 5C). Second, a broad absorption at ~0.8 mm/s also develops (indicated by the red arrows). Although this second feature is at the correct position to arise from a high-spin Fe(IV)=O intermediate, the cognate complexes in several other non-heme Fe(II) enzymes have exhibited sharp quadrupole doublets in the 4.2K/zero-field Mössbauer spectra, due to their integer spin (S = 2) ground states [32, 52, 53, 56, 57]. At most ~6% of the absorption intensity in the spectrum of Figure 5B can be attributed to such a quadrupole doublet, implying that an Fe(IV)=O species accumulates to a minor extent, if at all, in the reaction of the PHD2:Fe(II):2OG:CODD complex with O2.
Figure 5.
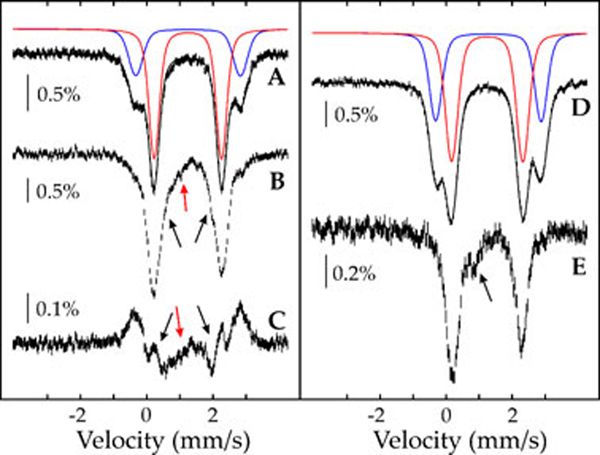
4.2-K/zero-field Mössbauer spectra of the PHD2:Fe(II):2OG:CODD complex before (A) and after reaction with an O2–saturated buffer solution for 200 s (B), and of the PHD2:Fe(II):2OG: complex before (D) and after reaction with an equal volume of an O2–saturated buffer solution for 200 s (E). Reaction conditions are given in Materials and Methods. (C) is the difference spectrum (B) – (A). The solid lines in (A) and (D) are quadrupole doublet simulations using the parameters described in the text.
The spectrum of the PHD2:Fe(II):2OG complex in the absence of CODD and O2 also reveals two quadrupole doublets with parameters nearly identical to those arising from the PHD2:Fe(II):2OG:CODD complex (Figure 5D) [δ1 = 1.25 mm/s and ΔEQ,1 = 2.16 mm/s (60%, red line) and δ2 = 1.28 mm/s and ΔEQ,2 = 3.20 mm/s (blue line, 40%)]. Upon reaction of this complex with O2 for 200 s (Figure 5E), the features of the first quadrupole doublet decay, and features with parameters similar to those of the second quadrupole doublet develop. A broad feature at 0.9 mm/s (indicated by the arrow) also develops. The nature of this species is unknown, but it is similar to features we have observed in the reaction of other Fe(II)- and 2OG-dependent oxygenases with O2 in the absence of their prime substrates [52].
DISCUSSION
In normoxia, i.e. when oxygen availability is not limiting, HIF-α hydroxylation (catalysed by the HIF hydroxylases) and degradation occur very efficiently, thus levels of HIF-α are very low in most normal healthy cells (reviewed in [58, 59]). When oxygen availability is below a threshold level, HIF hydroxylase activity reduces and HIF-α levels rise; it can then form a heterodimeric complex with HIF-β and initiate transcription of the array of genes involved in the hypoxic response. The oxygen-dependent role of the HIF hydroxylases in regulating HIF-α levels is supported by, or consistent with, an extensive body of evidence, involving work with isolated proteins, cellular analyses, and animal work (including work on clinically observed mutations) [2]. The oxygenase activity of the HIF hydroxylases therefore provides a direct mechanism that connects oxygen levels and transcriptional activity.
Other than oxygen availability, many factors may affect, and sometimes directly limit, PHD2 activity; these include the rate of HIF/PHD production (likely to be an important parameter within cells), the availability of iron, 2OG, and/or ascorbate, mutations, redox stress, and inhibitors (for review see [60]). In certain cases, e.g. in some types of tumor cell, it is likely that these, or other factors slow HIF-α degradation and result in its accumulation, even under aerobic conditions [61]. However, in normal cells, although many factors may regulate the rate of HIF hydroxylation, in order for the PHDs to act in their proposed role as oxygen sensors, their catalytic activity must be dependent on oxygen availability within physiologically relevant limits.
Previous studies have shown that PHD2, the most important of the human PHDs in oxygen sensing, forms relatively stable complexes with Fe(II) and 2OG (Kd values for both ≤2µM [39]). Moreover, and unusually, the PHD2:Fe(II):2OG complex appears to be quite stable in vitro, even in the presence of oxygen (i.e. uncoupled turnover of 2OG is slow [39]). The spectroscopic and other analyses reported here support these proposals. Our observation that binding of 2OG to the PHD2.Fe complex gives rise to absorption bands with maxima at ~530nm and ~520nm, in the absence and presence (respectively) of CODD, suggests that the 2OG binds to the iron in a bidentate manner as for other 2OG oxygenases, and as proposed for PHD2 on the basis of crystallographic analyses using 2OG analogues [36, 37]. The shift in λmax from 527nm to 521nm is consistent with a shift from a 6-coordinate to a 5-coordinate Fe-centre, facilitating binding of oxygen [28, 29]. PHD2 catalysis therefore likely proceeds via an ordered sequential mechanism as observed for other 2OG oxygenases [32].
Interestingly, however, we have observed that, although binding of CODD stimulates reaction of the PHD2:Fe(II):2OG complex with oxygen relative to that in the absence of CODD (by ~30-fold), the reaction of the PHD2:Fe(II):2OG:CODD complex with oxygen is still very much (~100-fold) slower than the reactions of analogous complexes of other 2OG oxygenases [31, 32]. We are aware of the dangers of correlating individual kinetic parameters determined in vitro with the in vivo situation, including the use of modified enzymes and peptide substrates. Nonetheless, given the assigned oxygen sensing role of the PHDs, the slow reaction of PHD2 with oxygen, as monitored by mass-spectrometric analyses of chemically-quenched samples taken over the time course of a single turnover reaction, is striking. In other studied 2OG oxygenases, upon reaction with oxygen, a transient species absorbing at 320nm has been observed and characterized as an Fe(IV)=O intermediate [31, 32]. For PHD2, broadly absorbing spectral features were observed by stopped-flow UV-visible spectroscopy. However, it was not possible on the basis of these and Mössbauer-spectroscopic analyses, to assign these features to catalytic intermediates, and further analyses are necessary.
Crystallographic analyses suggest that upon binding of CODD (and by implication, NODD) to PHD2, substantial conformational changes may occur, including away from the metal centre [36, 37] and that PHD2 may have an unusually narrow entrance to its active site. However, it seems unlikely that these factors alone can account completely for the slow reaction of PHD2 with oxygen. In terms of its immediate iron coordination by the facial triad of side chains, PHD2 appears similar to other 2OG oxygenases. However, crystallographic studies on a complex of PHD2:CODD with Mn(II) and N-oxalylglycine substituting for Fe(II) and 2OG, respectively, suggests that the coordinated water which must be displaced in order for oxygen to bind [28, 29] is stabilized by hydrogen bonding to the protein, It is possible that this interaction accounts, at least in part, for the slow reaction of PHD2 with oxygen. Because the 2OG 1-carboxylate has been observed to adopt different coordination positions relative to the other Fe ligands [62] in 2OG oxygenase crystal studies, it is also possible that a metal centered rearrangement contributes to the rate limiting nature of the reaction of PHD2 with oxygen.
Our observation of an unusually slow reaction of PHD2:Fe(II):2OG:CODD with oxygen is interesting with respect to its proposed role. We cannot rule out the possibility that our assay conditions are non-optimal / do not reflect cellular conditions; however based on the current evidence we propose that PHD2 has evolved to be tailored to its role as an oxygen sensor: a ‘stable’ PHD2:Fe(II):2OG complex in cells can readily bind HIF-α, as tightly as possible within the context of catalysis (Kd, CODD = 14µM [63]), and is then ‘primed’ to react with oxygen. In this way, PHD2 may have evolved to have its activity regulated by oxygen availability.
EXPERIMENTAL DETAILS
Materials
The HIF-1α556–574 peptide sequence (DLDLEMLAPYIPMDDDFQL), hereafter referred to as CODD, was from Peptide Protein Research Ltd., Fareham, U.K. DNA encoding PHD2181–426 (PHD2 hereafter) has previously been ligated into the pET-24a vector (Novagen) [41] Recombinant PHD2 was produced in Escherichia coli BL21(DE3) cells and purified by cation exchange and size exclusion chromatography, as described [39]. Protein purity was > 90% as assessed by SDS-PAGE and electrospray ionization (ESI) mass spectrometry. Apo-PHD2 was prepared by incubation in 0.2 mM EDTA/15 mM ammonium acetate pH 7.0 overnight at 4 °C (at < 1 mg/mL) followed by size exclusion chromatography (Superdex75 300 mL column, GE Healthcare).
Rapid Chemical Quench Mass Spectrometric Experiments
Deoxygenated solutions of, typically, PHD2, 2OG, ascorbate, CODD and (NH4)2Fe(SO4)2 (used as a Fe(II) source throughout) were mixed in an anaerobic glove box (Belle Laboratories, U.K., < 2 ppm O2). The resulting solution was rapidly mixed (at 5 °C) in a 1:1 ratio with a buffered solution that had been saturated with O2 [31]. Rapid chemical quench experiments were performed as described [31], quenching with either 0.2 M HCl (2OG/succinate measurements) or 0.1% HCOOH (CODD hydroxylation measurements). Ratios of 2OG and succinate were determined by separation on a Hamilton PRP-X300 anion exclusion column followed by mass spectrometry (MS) analyses using a Waters Micromass 2000 Mass Spectrometer. Ratios of hydroxylated and unhydroxylated CODD were determined by MALDI MS. Briefly, recrystallised CHCA (α-cyano-4-hydroxycinnamic acid) MALDI matrix (1µL) and the quenched assay mix (1 µL) were spotted onto a MALDI sample plate, and analysed using a Waters Micromass™ MALDI microMX™ mass spectrometer in negative ion mode [42]. Ion counts for hydroxylated and unhydroxylated CODD as a fraction of the total CODD ion count were used to calculate hydroxylation ratios.
Nuclear Magnetic Resonance Experiments
Reaction components (20 µM apo-PHD2, 50 µM (NH4)2Fe(SO4)2, 1 mM HIF-1α556–574, (the solubility limit in our assay conditions),1 mM 2OG, and 4 mM ascorbate) were prepared in deuterated Tris buffer (pD 7.5, 50 mM in D2O, D = 2H). The reaction was carried out at 310 K in a 2 mm diameter NMR tube, and initiated by addition of PHD2. 1H-NMR spectra were recorded using a Bruker AVIII 700 machine (with inverse cryoprobe optimised for 1H observation and running TOPSPIN 2 software) and reported in ppm relative to D2O (δH 4.72). The deuterium signal was also used as an internal lock signal and the solvent signal was suppressed by presaturating its resonance. Spectra were obtained at 75 s intervals and integrated using absolute intensity scaling to monitor changes in the intensity of signals of interest. Synthetic hydroxylated CODD is identical to the enzymatically produced hydroxylated CODD [39].
Stopped-Flow Absorption Spectroscopic and Mössbauer-Spectroscopic Experiments
Deoxygenated reaction solutions were prepared and mixed with an O2 saturated solution, as described above. Subsequent analysis used an SX20 stopped flow spectrometer (Applied Photophysics), as reported [31]. Mössbauer samples were prepared and spectroscopic measurements were carried out as described [31].
Supplementary Material
Acknowledgments
We thank Mr E. Barr for assistance with the rapid chemical quench assays, and Dr T. D. W. Claridge for assistance with the NMR assay design and data analysis. These studies were supported by the Engineering and Physical Sciences Research Council [EP/DO48559/1], the National Institutes of Health (NIH GM-69657 to JMB and CK) the National Science Foundations (NSF MCB-642058 and NSF CHE-724084 to JMB and CK) and the Pennsylvania Department of Health Tobacco Settlement Funds (to JMB and CK).
Abbreviations
- HIF
Hypoxia-Inducible Factor
- 2OG
2-Oxoglutarate
- PHD2
Prolyl Hydroxylase Domain 2
- NODD
N-terminal Oxygen Dependent Degradation Domain
- CODD
C-terminal Oxygen Dependent Degradation Domain
- VCB
von Hippel Lindau protein elongin C/B complex
- TauD
Taurine 2OG Dioxygenase
- PBCV-1 P4H
Paramecium bursaria Chlorella virus 1 Prolyl-4-Hydroxylase
- FIH
Factor Inhibiting HIF
- MS
Mass Spectrometry
- MALDI
Matrix-Assisted Laser Desorption/Ionization
- ESI
Electrospray Ionization
- NMR
Nuclear Magnetic Resonance
Footnotes
Conflict of Interest: Prof. C.J. Schofield is a co-founder of ReOx Ltd., a company working on the exploitation of the hypoxic response.
REFERENCES
- 1.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via denovo protein- synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 5.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor-1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 6.Ivan M, Kondo K, Yang HF, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG. HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 7.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 8.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain: A hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 9.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 10.Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C-elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 11.Taylor MS. Characterization and comparative analysis of the EGLN gene family. Gene. 2001;275:125–132. doi: 10.1016/s0378-1119(01)00633-3. [DOI] [PubMed] [Google Scholar]
- 12.Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RWD, Elkins JM, Oldham NJ, Battacharya S, Gleadle J, Ratcliffe PJ, Pugh CW, Schofield CJ. Hypoxia inducible factor (HIF) asparagine hydroxylase is identical to Factor Inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 13.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, Cowan A, Fong GH. Essential role for prolyl hydroxylase domain protein 2 in oxygen homeostasis of the adult vascular system. Circulation. 2007;116:774–781. doi: 10.1161/CIRCULATIONAHA.107.701516. [DOI] [PubMed] [Google Scholar]
- 15.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 17.Costas M, Mehn MP, Jensen MP, Que L. Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 18.Hausinger RP. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 19.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev. 2000;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 20.Krebs C, Galonić Fujimori D, Walsh CT, Bollinger JM., Jr Non-heme Fe(IV)-oxo intermediates. Acc Chem Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanauske-Abel HM, Günzler V. A stereochemical concept for the catalytic mechanism of prolyl hydroxylase: applicability to classification and design of inhibitors. J Theor Biol. 1982;94:421–455. doi: 10.1016/0022-5193(82)90320-4. [DOI] [PubMed] [Google Scholar]
- 22.Bollinger JM, Jr, Price JC, Hoffart LM, Barr EW, Krebs C. Mechanism of oxygen activation by taurine: α-ketoglutarate dioxygenase (TauD) from Escherichia coli. Eur J Inorg Chem. 2005;2005:4245–4254. [Google Scholar]
- 23.Koehntop KD, Emerson JP, Que L., Jr The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 24.Que L. One motif - many different reactions. Nat Struct Biol. 2000;7:182–184. doi: 10.1038/73270. [DOI] [PubMed] [Google Scholar]
- 25.Valegård K, van Scheltinga ACT, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, Thompson A, Lee HJ, Baldwin JE, Schofield CJ, Hajdu J, Andersson I. Structure of a cephalosporin synthase. Nature. 1998;394:805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Ren J, Stammers DK, Baldwin JE, Harlos K, Schofield CJ. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat Struct Biol. 2000;7:127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- 27.Pavel EG, Zhou J, Busby RW, Gunsior M, Townsend CA, Solomon EI. Circular dichroism and magnetic circular dichroism spectroscopic studies of the non-heme ferrous active site in clavaminate synthase and its interaction with alpha-ketoglutarate cosubstrate. J Am Chem Soc. 1998;120:743–753. [Google Scholar]
- 28.Zhou J, Gunsior M, Bachmann BO, Townsend CA, Solomon EI. Substrate binding to the α-ketoglutarate-dependent non-heme iron enzyme clavaminate synthase 2: Coupling mechanism of oxidative decarboxylation and hydroxylation. J Am Chem Soc. 1998;120:13539–13540. [Google Scholar]
- 29.Whiting AK, Que L, Saari RE, Hausinger RP, Fredrick MA, McCracken J. Metal coordination environment of a Cu(II)-substituted α-keto acid-dependent dioxygenase that degrades the herbicide 2,4-D. J Am Chem Soc. 1997;119:3413–3414. [Google Scholar]
- 30.Welford RW, Kirkpatrick JM, McNeill LA, Puri M, Oldham NJ, Schofield CJ. Incorporation of oxygen into the succinate co-product of iron(II) and 2-oxoglutarate dependent oxygenases from bacteria, plants and humans. FEBS Lett. 2005;579:5170–5174. doi: 10.1016/j.febslet.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 31.Price JC, Barr EW, Tirupati B, Bollinger JM, Jr, Krebs C. The first direct characterization of a high-valent iron intermediate in the reaction of an alpha-ketoglutarate-dependent dioxygenase: a high-spin FeIV complex in taurine/alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 32.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci U S A. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price JC, Barr EW, Glass TE, Krebs C, Bollinger JM., Jr Evidence for hydrogen abstraction from C1 of taurine by the high-spin Fe(IV) intermediate detected during oxygen activation by taurine:alpha-ketoglutarate dioxygenase (TauD) J Am Chem Soc. 2003;125:13008–13009. doi: 10.1021/ja037400h. [DOI] [PubMed] [Google Scholar]
- 34.Groves JT. Key elements of the chemistry of cytochrome P-450. J Chem Educ. 1985;62:928–931. [Google Scholar]
- 35.Price JC, Barr EW, Hoffart LM, Krebs C, Bollinger JM., Jr Kinetic dissection of the catalytic mechanism of taurine:alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry. 2005;44:8138–8147. doi: 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- 36.McDonough MA, Li V, Flashman E, Chowdhury R, Mohr C, Lienard BM, Zondlo J, Oldham NJ, Clifton IJ, Lewis J, McNeill LA, Kurzeja RJ, Hewitson KS, Yang E, Jordan S, Syed RS, Schofield CJ. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2) Proc Natl Acad Sci U S A. 2006;103:9814–9819. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhury R, McDonough MA, Mecinovic J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ. Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure. 2009;17:981–989. doi: 10.1016/j.str.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Bleijlevens B, Shivarattan T, Flashman E, Yang Y, Simpson PJ, Koivisto P, Sedgwick B, Schofield CJ, Matthews SJ. Dynamic states of the DNA repair enzyme AlkB regulate product release. EMBO Rep. 2008;9:872–877. doi: 10.1038/embor.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McNeill LA, Flashman E, Buck MR, Hewitson KS, Clifton IJ, Jeschke G, Claridge TD, Ehrismann D, Oldham NJ, Schofield CJ. Hypoxia-inducible factor prolyl hydroxylase 2 has a high affinity for ferrous iron and 2-oxoglutarate. Mol Biosyst. 2005;1:321–324. doi: 10.1039/b511249b. [DOI] [PubMed] [Google Scholar]
- 40.Dao JH, Kurzeja RJ, Morachis JM, Veith H, Lewis J, Yu V, Tegley CM, Tagari P. Kinetic characterization and identification of a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase 2 using a time-resolved fluorescence resonance energy transfer-based assay technology. Anal Biochem. 2009;384:213–223. doi: 10.1016/j.ab.2008.09.052. [DOI] [PubMed] [Google Scholar]
- 41.Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, Schofield CJ. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J. 2007;401:227–234. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flashman E, Bagg EAL, Chowdhury R, Mecinovic J, Loenarz C, McDonough MA, Hewitson KS, Schofield CJ. Kinetic Rationale for Selectivity toward N- and C-terminal Oxygen-dependent Degradation Domain Substrates Mediated by a Loop Region of Hypoxia-Inducible Factor Prolyl Hydroxylases. J Biol Chem. 2008;283:3808–3815. doi: 10.1074/jbc.M707411200. [DOI] [PubMed] [Google Scholar]
- 43.Tuckerman JR, Zhao Y, Hewitson KS, Tian YM, Pugh CW, Ratcliffe PJ, Mole DR. Determination and comparison of specific activity of the HIF-prolyl hydroxylases. FEBS Lett. 2004;576:145–150. doi: 10.1016/j.febslet.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 45.Koivunen P, Hirsilä M, Günzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 46.Flashman E, Davies SL, Yeoh KK, Schofield CJ. Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem J. 2010;427:135–142. doi: 10.1042/BJ20091609. [DOI] [PubMed] [Google Scholar]
- 47.Knowles HJ, Raval RR, Harris AL, Ratcliffe PJ. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 2003;63:1764–1768. [PubMed] [Google Scholar]
- 48.Grzyska PK, Ryle MJ, Monterosso GR, Liu J, Ballou DP, Hausinger RP. Steady-state and transient kinetic analyses of taurine/alpha-ketoglutarate dioxygenase: effects of oxygen concentration, alternative sulfonates, and active-site variants on the FeIV-oxo intermediate. Biochemistry. 2005;44:3845–3855. doi: 10.1021/bi048746n. [DOI] [PubMed] [Google Scholar]
- 49.Bollinger JM, Jr, Krebs C. Stalking intermediates in oxygen activation by iron enzymes: motivation and method. J Inorg Biochem. 2006;100:586–605. doi: 10.1016/j.jinorgbio.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 50.Ryle MJ, Padmakumar R, Hausinger RP. Stopped-flow kinetic analysis of Escherichia coli taurine/alpha-ketoglutarate dioxygenase: interactions with alpha-ketoglutarate, taurine, and oxygen. Biochemistry. 1999;38:15278–15286. doi: 10.1021/bi9912746. [DOI] [PubMed] [Google Scholar]
- 51.Neidig ML, Brown CD, Light KM, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM, Jr, Krebs C, Walsh CT, Solomon EI. CD and MCD of CytC3 and taurine dioxygenase: role of the facial triad in alpha-KG-dependent oxygenases. J Am Chem Soc. 2007;129:14224–14231. doi: 10.1021/ja074557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galonić DP, Barr EW, Walsh CT, Bollinger JM, Jr, Krebs C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat Chem Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- 53.Matthews ML, Krest CM, Barr EW, Vaillancourt FH, Walsh CT, Green MT, Krebs C, Bollinger JM. Substrate-triggered formation and remarkable stability of the C-H bond-cleaving chloroferryl intermediate in the aliphatic halogenase, SyrB2. Biochemistry. 2009;48:4331–4343. doi: 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koivunen P, Hirsilä M, Kivirikko KI, Myllyharju J. The length of peptide substrates has a marked effect on hydroxylation by the hypoxia-inducible factor prolyl 4-hydroxylases. J Biol Chem. 2006;281:28712–28720. doi: 10.1074/jbc.M604628200. [DOI] [PubMed] [Google Scholar]
- 55.Matthews ML, Neumann CS, Miles LA, Grove TL, Booker SJ, Krebs C, Walsh CT, Bollinger JM., Jr Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc Natl Acad Sci U S A. 2009;106:17723–17728. doi: 10.1073/pnas.0909649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eser BE, Barr EW, Frantom PA, Saleh L, Bollinger JM, Jr, Krebs C, Fitzpatrick PF. Direct spectroscopic evidence for a high-spin Fe(IV) intermediate in tyrosine hydroxylase. J Am Chem Soc. 2007;129:11334–11335. doi: 10.1021/ja074446s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krebs C, Price JC, Baldwin J, Saleh L, Green MT, Bollinger JM., Jr Rapid freeze-quench 57Fe Mössbauer spectroscopy: monitoring changes of an iron-containing active site during a biochemical reaction. Inorg Chem. 2005;44:742–757. doi: 10.1021/ic048523l. [DOI] [PubMed] [Google Scholar]
- 58.Chowdhury R, Hardy A, Schofield CJ. The human oxygen sensing machinery and its manipulation. Chem Soc Rev. 2008;37:1308–1319. doi: 10.1039/b701676j. [DOI] [PubMed] [Google Scholar]
- 59.Hirota K, Semenza GL. Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem Biophys Res Commun. 2005;338:610–616. doi: 10.1016/j.bbrc.2005.08.193. [DOI] [PubMed] [Google Scholar]
- 60.Schofield CJ, Ratcliffe PJ. Signalling hypoxia by HIF hydroxylases. Biochemical and Biophysical Research Communications. 2005;338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 61.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z, Ren J, Harlos K, McKinnon CH, Clifton IJ, Schofield CJ. Crystal structure of a clavaminate synthase.Fe(II).2-oxoglutarate.substrate.NO complex: Evidence for metal centered rearrangements. FEBS Letters. 2002;517:7–12. doi: 10.1016/s0014-5793(02)02520-6. [DOI] [PubMed] [Google Scholar]
- 63.Leung IKH, Flashman E, Yeoh KK, Schofield CJ, Claridge TD. Using NMR solvent water relaxation to investigate metalloenzyme-ligand binding interactions. J Med Chem. 2010;53:867–875. doi: 10.1021/jm901537q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.