Abstract
Genome editing with site-specific nucleases, such as zinc-finger nucleases or transcription activator-like effector nucleases (TALENs), and RNA-guided nucleases, such as the CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated) system, is becoming the new standard for targeted genome modification in various organisms. Application of these techniques to the manufacture of knockout mice would be greatly aided by simple and easy methods for genotyping of mutant and wild-type pups among litters. However, there are no detailed or comparative reports concerning the identification of mutant mice generated using genome editing technologies. Here, we genotyped TALEN-derived enhanced green fluorescent protein (eGFP) knockout mice using a combination of approaches, including fluorescence observation, heteroduplex mobility assay, restriction fragment length polymorphism analysis and DNA sequencing. The detection sensitivities for TALEN-induced mutations differed among these methods, and we therefore concluded that combinatorial testing is necessary for the screening and determination of mutant genotypes. Since the analytical methods tested can be carried out without specialized equipment, costly reagents and/or sophisticated protocols, our report should be of interest to a broad range of researchers who are considering the application of genome editing technologies in various organisms.
Keywords: genome editing, knockout mouse, TALEN, targeted mutagenesis
Introduction
Transcription activator-like effector (TALE) nuclease (TALEN)-mediated gene knockout technology is now applicable to a wide variety of cells and organisms [5]. Each TALEN comprises a TALE domain that binds to a specified DNA sequence and a nuclease domain derived from the FokI restriction endonuclease. When a pair of TALENs designed for a specific genomic locus is introduced into embryos, a DNA double-strand break (DSB) occurs at the target site. DSBs are mainly repaired by error-prone non-homologous end-joining (NHEJ), resulting in randomly induced insertions and deletions that cause disruption of gene functions [7].
Conventionally, knockout mice have been created using an embryonic stem (ES) cell-mediated strategy based on spontaneous homologous recombination between genomic DNA and a targeting construct [2]. This method is time-consuming and requires several laborious processes, such as construction of a gene targeting vector, isolation of targeted ES cell clones, production of chimeras, test breeding for germline transmission and, in some cases, backcrossing to another inbred background. However, the use of TALENs for gene targeting enables knockout mice to be produced in a short time because the TALEN mRNAs are simply injected into the embryos of any intended inbred strain. Recently, several groups reported that knockout mice and rats could be efficiently created by TALEN-mediated gene targeting [4, 6, 10, 14, 16]. The convenience of this technique means that TALEN-mediated gene knockout will become a major method for the production of genetically modified rodents in the near future. However, while ES cell-mediated chimeric mice can easily be determined by their coat color, with TALEN-mediated gene targeting it is difficult to distinguish genetically modified mice from wild-type pups unless a phenotype is apparent.
Here, we injected TALEN mRNAs targeting the enhanced green fluorescent protein (eGFP) gene into fertilized mouse eggs expressing eGFP ubiquitously under control of the CAG promoter (pCAG). Pups were analyzed by observation of green fluorescence, heteroduplex mobility assay (HMA), restriction fragment length polymorphism (RFLP) analysis and DNA sequencing to consolidate the method for detecting pups with TALEN-induced mutations.
Materials and Methods
TALEN plasmid construction and mRNA preparation
Synthesized TALE repeats were cloned into pBluescript SK and assembled using the Golden Gate cloning method [12]. The N- and C-terminal domains of TALE and the FokI nuclease domain were taken from pTALEN_v2 (Addgene, Cambridge, MA, USA) [13]. The eGFP TALEN target sequence was described previously [11] and is indicated in Fig. 1. TALEN mRNAs were synthesized from plasmids linearized by SmaI digestion using an mMessage mMachine T7 Ultra Kit (Life Technologies, Carlsbad, CA, USA) and purified with an RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturers’ instructions and as previously described [11].
Fig. 1.
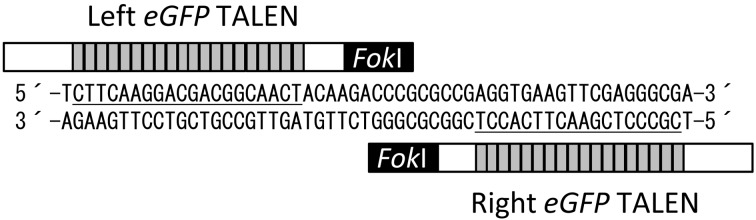
Diagram depicting engineered TALENs binding to the eGFP gene. The TALENs comprise DNA-binding repeats (gray boxes), the N- and C-terminal domains of TALE (white boxes) and a FokI nuclease domain (black box). The left and right TALEN target sequences are underlined.
pCAG-eGFP mouse embryos
The parental pCAG-eGFP mouse strain has been deposited in the Center for Animal Resources and Development (CARD), Kumamoto University (B6;D2-Tg (CAG-EGFP) 49SImeg; CARD ID: 267; http://cardb.cc.kumamoto-u.ac.jp/transgenic/strainsDetailAction.do?strainId=267). The background strain is C57BL/6. The pCAG-eGFP gene, illustrated in Supplementary Fig. 1A, was confirmed to be one copy by Southern blot analysis (Supplementary Fig. 1B). Expression of the eGFP gene is detected throughout the whole body.
To obtain mouse embryos for TALEN injections, C57BL/6N female mice were induced to super-ovulate using pregnant mare serum gonadotropin (PMSG; Serotropin; ASKA Pharmaceutical, Tokyo, Japan) and human chorionic gonadotropin (hCG; Veterinary Puberogen; Novartis Animal Health, Tokyo, Japan) at 5 weeks of age, and then mated with male pCAG-eGFP mice described above. Fertilized eggs were collected from females displaying vaginal plugs.
Microinjection of TALEN mRNAs
TALEN mRNAs were diluted in RNase-free PBS at 100 or 150 ng/µl for injection of each TALEN into the pronuclei or cytoplasm of zygotes. Approximately 2–3 pL of capped mRNAs was injected into the zygotes. The injected embryos were cultured in potassium simplex optimized medium with amino acids (KSOM-AA) at 37°C in 5% CO2 and 95% humidified air for 1 h. Surviving embryos were transferred to the oviducts of pseudopregnant ICR female mice.
Genomic PCR for HMA and DNA sequencing
Genomic DNA was extracted from the tail of each pup using a DNeasy Blood & Tissue Kit (Qiagen). Genomic PCR was performed using LA Taq DNA polymerase (TAKARA Biotechnology, Shiga, Japan) under the following conditions: 94°C for 2 min; followed by 94°C for 30 s, 64°C for 30 s, and 72°C for 20 s for 38 cycles. The PCR primers were as follows: 5′-CCTCGTGACCACCCTGACCTAC-3′ and 5′-CTGTTGTAGTTGTACTCCAGCTTGTGC-3′. The PCR products were subjected to agarose gel electrophoresis and ethidium bromide staining for the HMA.
For DNA sequence analysis, the PCR products were subcloned into pGEM-T Easy (Promega, Madison, WI, USA). The plasmids were extracted and sequenced using a T7 (5′-TAATACGACTCACTATAGGG-3′) or SP6 (5′-CATACGATTTAGGTGACACTATAG-3′) primer with a BigDye Terminator Cycle Sequencing Kit (Life Technologies), and then analyzed using an ABI PRISM 3130 Genetic Analyzer (Life Technologies).
RFLP analysis
The PCR products were purified using a Wizard SV Gel and PCR Clean-Up System (Promega). The purified products were digested with 3 units of AccII (TAKARA Biotechnology), and then subjected to agarose gel electrophoresis and ethidium bromide staining.
Results and Discussion
As a model for TALEN-mediated knockout, we used fertilized eggs from mice ubiquitously expressing pCAG-eGFP. All of the fertilized eggs were heterozygous for the pCAG-eGFP gene. We selected the same eGFP TALEN target sequence as described previously [11] and newly constructed TALEN expression vectors as described in the Materials and Methods.
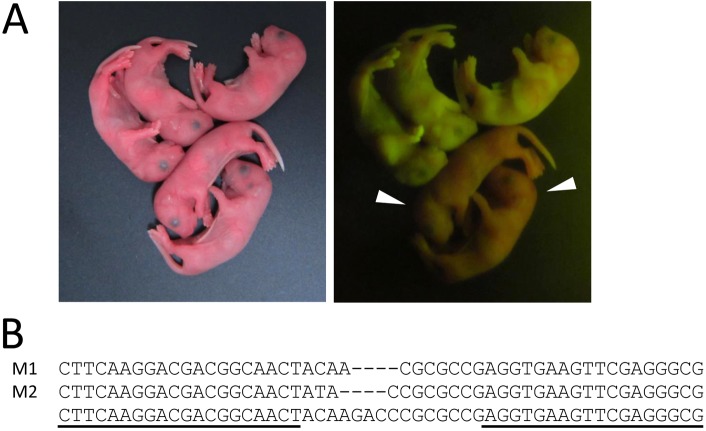
First, to judge the efficiency and toxicity between pronuclear and cytoplasmic injections, we microinjected eGFP TALEN mRNAs at 100 or 150 ng/µl into the pronuclei of zygotes. After the microinjection, 92.2% (130/141) and 90.6% (116/128) of the TALEN-injected embryos survived, respectively (Table 1). Following transfer of the surviving embryos to pseudopregnant females, 51 pups were born from 130 transferred embryos (39.2%) at 100 ng/µl and 32 pups were born from 116 transferred embryos (27.6%) at 150 ng/µl. Observation of the pups for green fluorescence under ultraviolet light on the day of birth revealed that eGFP fluorescence was completely abolished in three pups (5.9%) at 100 ng/µl and in four pups (12.5%) at 150 ng/µl (Table 1). The fluorescence images and DNA sequences of two of the three eGFP-disrupted pups at 100 ng/µl are shown in Fig. 2A and 2B, respectively. In addition, we obtained some pups with mosaic disruption of eGFP fluorescence at 150 ng/µl (Supplementary Fig. 2).
Table 1. TALEN-mediated eGFP gene disruption in mice.
| Route | Dose (ng/µl) |
Injected | Transferred | Newborns | eGFP disap- peared pups* |
Analyzed pups |
HMA | RFLP | Mutants |
|---|---|---|---|---|---|---|---|---|---|
| pronucleus | 100 | 141 | 130 | 51 (39.2%) | 3 (5.9%) | NT | NT | NT | NT |
| pronucleus | 150 | 128 | 116 | 32 (27.6%) | 4 (12.5%) | NT | NT | NT | NT |
| cytoplasm | 150 | 80 | 69 | 33 (47.8%) | 4 (12.1%) | 33 | 12 (36.4%) | 11 (33.3%) | 17 (51.5%) |
| no injection | – | – | 21 | 8 (38.1%) | 0 (0%) | NT | NT | NT | NT |
*Not including mosaic pups. HMA, heteroduplex mobility assay. RFLP, restriction fragment length polymorphism. NT, not tested.
Fig. 2.
TALEN-mediated disruption of the eGFP gene in mice. (A) Bright-field (left panel) and fluorescence microscopy (right panel) images of newborn mice. eGFP TALEN mRNAs were injected into fertilized eggs heterozygous for eGFP. Embryos were transferred to pseudopregnant females. The arrowheads indicate pups in which eGFP is disrupted. (B) eGFP sequences in pups displaying a disrupted eGFP gene (M1 and M2). The original sequence is shown at the bottom with the TALEN target sequences (underlined). Deletions are indicated by dashes.
Next, eGFP TALEN mRNAs were injected at 150 ng/µl into the cytoplasm of zygotes. After the microinjection, 86.3% (69/80) of TALEN-injected embryos survived (Table 1) and 33 pups were born from 69 transferred embryos (47.8%). On the other hand, after transfer of pCAG-eGFP mouse embryos that had not been injected with TALEN mRNAs, eight pups were born from 21 embryos (38.1%). Thus, toxicity was not observed after the microinjection of TALEN mRNAs into the cytoplasm. When we observed the pups for green fluorescence under ultraviolet light, we found four pups in which fluorescence was completely absent and four mosaic pups (12.1% each) (Tables 1 and 2). Although the percentage of the green fluorescence-disappeared pups differed little from pronuclear injection, the birth rate of the pups with cytoplasmic injection was much higher than that with pronuclear injection (Table 1). Therefore, we concluded that TALEN mRNAs should be injected into the cytoplasm rather than the pronuclei. We then examined several analytical methods using the 33 pups described above.
Table 2. Summary of the analyses for mutant screening.
| Founders | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| eGFP disruption | mosaic | ND | ND | ND | ND | ND | mosaic | ND | ND | ND | ND |
| HMA | + | ND | ND | ND | + | + | ND | ND | ND | ND | + |
| RFLP | + | ND | ND | ND | + | ND | ND | ND | ND | ND | + |
| Sequence mutation | NT | NT | NT | NT | NT | + | + | NT | NT | NT | NT |
| Genotype | MT | MT | MT | MT | MT | ||||||
| Founders | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 |
| eGFP disruption | mosaic | ND | ND | ND | ND | ND | ND | ND | + | + | mosaic |
| HMA | + | ND | ND | ND | ND | + | ND | ND | + | ND | ND |
| RFLP | + | ND | +/− | ND | ND | + | ND | ND | ND | + | ND |
| Sequence mutation | NT | NT | + | NT | NT | NT | NT | NT | NT | + | + |
| Genotype | MT | MT | MT | MT | MT | MT | |||||
| Founders | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 |
| eGFP disruption | ND | ND | + | + | ND | ND | ND | ND | ND | ND | ND |
| HMA | + | + | ND | + | ND | ND | + | ND | + | ND | ND |
| RFLP | + | + | ND | ND | ND | ND | +/− | ND | + | ND | ND |
| Sequence mutation | NT | NT | + | NT | ND | NT | NT | NT | NT | NT | NT |
| Genotype | MT | MT | MT | MT | MT | MT | |||||
HMA, heteroduplex mobility assay. RFLP, restriction fragment length polymorphism. ND, not detected. NT, not tested. MT, mutant.
Genomic DNA was extracted from all pups, and subjected to genomic PCR. The amplified product, including the TALEN target site, was 264 bp if no mutation was present. The individual products were subjected to agarose gel electrophoresis for the HMA. A recent study demonstrated that PCR products including TALEN-induced mutations could be detected by the HMA [9]. The HMA is the easiest method for the detection of mutations, because it only requires the performance of agarose gel electrophoresis after the genomic PCR. If mutations are introduced in the target DNA fragments, the shifted bands appear in proportion to the mutation rate based on the formation of heteroduplexes between the mutated alleles and non-mutated alleles. We confirmed shifted bands in 12 pups (#1, 5, 6, 11, 12, 17, 20, 23, 24, 26, 29 and 31) (Table 2 and Supplementary Fig. 3). In two pups (#21 and 29), we detected PCR products of 264 bp and about 200 bp. These smaller bands suggested that extensive deletions were induced in these pups.
RFLP analysis is also often used for the detection of mutant alleles [1, 8, 15]. If there is a unique restriction site in the center of the TALEN spacer sequence, the mutated alleles obtain resistance to the restriction enzyme. The PCR products were purified from all individuals, and examined for whether they could be digested by AccII. The original 264-bp PCR product is cleaved into two DNA fragments of 150 bp and 114 bp by AccII digestion (Supplementary Fig. 4A). The PCR products of 11 pups (#1, 5, 11, 12, 14, 17, 21, 23, 24, 29 and 31) showed some resistance to AccII, including less obvious ones such as #14 and 29 (Table 2 and Supplementary Fig. 4B). Although these results were similar to the HMA results (Supplementary Fig. 3), there were still some individuals that we could not confirm as mutants. Therefore, we determined the genomic eGFP sequences and expected amino acid sequences of such individuals (Supplementary Figs. 5A and 5B). Finally identified 17 pups as TALEN-mediated mutants (#1, 5, 6, 7, 11, 12, 14, 17, 20, 21, 22, 23, 24, 25, 26, 29 and 31; 51.5%) (Table 2).
The different results for the HMA and RFLP analysis were thought to be caused by the difference in the principles between these methods. If mutations are present that do not cause disruption of the recognition sequence of the restriction enzyme, they cannot be identified as RFLP-positive. On the other hand, a single base substitution or an extremely small insertion or deletion might not be detected by the HMA because of the minor effects on the heteroduplex mobility. In addition, some mice that were identified as mutants by HMA and/or RFLP analysis did not show the eGFP disruption (Table 2). There are some possible reasons of inconsistent results between green fluorescence observation and mutation analyses. One possibility is a failure of disrupting protein functions. If in-frame mutations or base substitutions are induced, green fluorescence could be remained. Another possibility is the timing of mutagenesis. If mutations are induced at late developmental stages, disruption of green fluorescence could be undetectable.
In summary, we assessed multiple analytical methods, comprising eGFP observation, HMA, RFLP analysis and DNA sequencing, for the detection of TALEN-induced mutants. Although DNA sequencing is generally the best way to definitely confirm mutations, it is very laborious work to subclone and sequence multiple clones for all pups. Although direct sequencing of genomic PCR products is easier than the above-described clone sequencing method, it can only detect mutations occurring at very early developmental stages. Considering all the various factors together, we think that there is a need to narrow down the candidates for mutants using the HMA and/or RFLP analysis before sequencing, even though it may lead to some mutant pups being overlooked, such as #25.
In contrast to other methods for analyzing TALEN-induced mutations that require specialized and costly equipment, such as high-resolution melting analysis [3] and surveyor nuclease assay [16], the methods that we tested require only commonly used reagents and equipment. We hope that this report will become a good model for the genotyping of TALEN-mediated knockout mutations, in mice and other organisms.
Supplementary
Acknowledgments
We thank Ms. Kazuko Kuroda and Ms. Reika Yoshimatsu for excellent technical assistance. Male pCAG-eGFP mice were kindly provided by Dr. Ken-ichi Yamamura at Kumamoto University. This study was supported by KAKENHI (24591016) to M. Ohmuraya from the Japan Society for the Promotion of Science.
References
- 1.Ansai S., Sakuma T., Yamamoto T., Ariga H., Uemura N., Takahashi R., Kinoshita M.2013. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 193: 739–749. doi: 10.1534/genetics.112.147645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capecchi M.R.2005. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6: 507–512. doi: 10.1038/nrg1619 [DOI] [PubMed] [Google Scholar]
- 3.Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F., Grunwald D.J.2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8: e1002861. doi: 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies B., Davies G., Preece C., Puliyadi R., Szumska D., Bhattacharya S.2013. Site Specific Mutation of the Zic2 Locus by Microinjection of TALEN mRNA in Mouse CD1, C3H and C57BL/6J Oocytes. PLoS ONE 8: e60216. doi: 10.1371/journal.pone.0060216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joung J.K., Sander J.D.2013. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14: 49–55. doi: 10.1038/nrm3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mashimo T., Kaneko T., Sakuma T., Kobayashi J., Kunihiro Y., Voigt B., Yamamoto T., Serikawa T.2013. Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci. Rep. 3: 1253. doi: 10.1038/srep01253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mussolino C., Cathomen T.2012. TALE nucleases: tailored genome engineering made easy. Curr. Opin. Biotechnol. 23: 644–650. doi: 10.1016/j.copbio.2012.01.013 [DOI] [PubMed] [Google Scholar]
- 8.Ochiai H., Fujita K., Suzuki K., Nishikawa M., Shibata T., Sakamoto N., Yamamoto T.2010. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells. 15: 875–885. [DOI] [PubMed] [Google Scholar]
- 9.Ota S., Hisano Y., Muraki M., Hoshijima K., Dahlem T.J., Grunwald D.J., Okada Y., Kawahara A.2013. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells. 18: 450–458. doi: 10.1111/gtc.12050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Z., Liu M., Chen Z., Shao Y., Pan H., Wei G., Yu C., Zhang L., Li X., Wang P., Fan H.Y., Du B., Liu B., Liu M., Li D.2013. High-efficiency and heritable gene targeting in mouse by transcription activator-like effector nucleases. Nucleic. Acids Res. 41: e120. doi: 10.1093/nar/gkt258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakuma T., Hosoi S., Woltjen K., Suzuki K., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y., Matsuura S., Okada Y., Kawahara A., Hayashi S., Yamamoto T.2013. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 18: 315–326. doi: 10.1111/gtc.12037 [DOI] [PubMed] [Google Scholar]
- 12.Ochiai H., Kaneko T., Mashimo T., Tokumasu D., Sakane Y., Suzuki K., Miyamoto T., Sakamoto N., Matsuura S., Yamamoto T.2013. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 3: 3379. doi: 10.1038/srep03379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjana N.E., Cong L., Zhou Y., Cunniff M.M., Feng G., Zhang F.2012. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7: 171–192. doi: 10.1038/nprot.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung Y.H., Baek I.J., Kim D.H., Jeon J., Lee J., Lee K., Jeong D., Kim J.S., Lee H.W.2013. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 31: 23–24. doi: 10.1038/nbt.2477 [DOI] [PubMed] [Google Scholar]
- 15.Suzuki K.T., Isoyama Y., Kashiwagi K., Sakuma T., Ochiai H., Sakamoto N., Furuno N., Kashiwagi A., Yamamoto T.2013. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol. Open. 2: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesson L., Usal C., Ménoret S., Leung E., Niles B.J., Remy S., Santiago Y., Vincent A.I., Meng X., Zhang L., Gregory P.D., Anegon I., Cost G.J.2011. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 29: 695–696. doi: 10.1038/nbt.1940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.