Abstract
We have developed an immortalized oral epithelial cell line, ROE2, from fetal transgenic rats harboring temperature-sensitive simian virus 40 large T-antigen gene. The cells grew continuously at either a permissive temperature of 33°C or an intermediate temperature of 37°C. At the nonpermissive temperature of 39°C, on the other hand, growth decreased significantly, and the Sub-G1 phase of the cell cycle increased, indicating that the cells undergo apoptosis at a nonpermissive temperature. Histological and immunocytochemical analyses revealed that ROE2 cells at 37°C had a stratified epithelial-like morphology and expressed cytokeratins Krt4 and Krt13, marker proteins for oral nonkeratinized epithelial cells. Global-scale comprehensive microarray analysis, coupled with bioinformatics tools, demonstrated a significant gene network that was obtained from the upregulated genes. The gene network contained 16 genes, including Cdkn1a, Fos, Krt13, and Prdm1, and was associated mainly with the biological process of skin development in the category of biological functions, organ development. These four genes were validated by quantitative real-time polymerase chain reaction, and the results were nearly consistent with the microarray data. It is therefore anticipated that this cell line will be useful as an in vitro model for studies such as physiological functions, as well as for gene expression in oral epithelial cells.
Keywords: differentiation, gene network, oral epithelial cell, temperature-sensitive simian virus 40 large T-antigen
Introduction
Oral epithelial tissues form an important barrier between the tissue and the environment, and they protect against exogenous substances and pathogens. The oral cavity contains two types of stratified epithelia: keratinized epithelia located in the gingival and hard palate tissues, and nonkeratinized epithelia in buccal tissues, the underside of the tongue, and soft palate tissues. To maintain these epithelial barriers, epithelial tissues undergo constant renewal. Oral keratinized and nonkeratinized epithelia are composed of cells that show various differentiated phenotypes, and the progenitor cells in the basal layer give rise to all of the epithelial cell types in the oral epithelia [32, 38]. Cell junctions including desmosomes function as points in cell-cell and cell-basement membranes, and connect to the keratin cytoskeleton to the cell surface. It is also well known that cytokeratins, important components of cytoskeletons, are reliable indicators of oral epithelial cell differentiation [32, 36,37,38]. Cytokeratins keratin 5 (Krt5) and Krt14 are expressed in basal cells in both oral keratinized and nonkeratinized epithelia, whereas the keratin pairs Krt1/Krt10 and Krt4/Krt13 are found in keratinized and nonkeratinized epithelia, respectively.
The use of in vitro cell culture systems has been of central importance for research into the physiology, pharmacology, and toxicology of cell lines; and such systems function at both the cellular and molecular levels. Primary cell culture methods have been developed for the study of oral epithelial cells [17, 46]; however, these primary cultures contain multiple cell types with different developmental stages and are frequently invaded by fibroblastic cells. Moreover, the proliferation activity of primary culture cells is limited, and some variability among cultured cells from individual sources has been observed between experiments. On the other hand, it has also been thought that cell lines are a good tool for molecular biology, especially recombinant DNA experiments, because they are a continuous source of readily available cells. Many oral epithelial cell lines have been established from normal cell cultures [18] or carcinoma cells [19, 25]. However, the genetic backgrounds of these cell lines are undefined and unstable, and usually are lacking some of their normal properties. Immortalization of primary cultures can be achieved directly with viral oncogenes. Either simian virus 40 (SV40) large T-antigen or its mutant temperature-sensitive simian virus 40 (tsSV40) large T-antigen can establish continuous proliferation without a transformed phenotype in primary culture cells [15, 16]. Previous reports demonstrated that the SV40 large T-antigen can stabilize cell-type-specific functions in oral epithelial immortalized cell lines [9, 20]. Moreover, transgenic (TG) mice harboring tsSV40 large T-antigen have been very useful for establishing immortalized cell lines from many kinds of tissues including epithelia [27, 28]. Using TG mice, several kinds of epithelial cell lines with specific functions have been developed from kidney distal tubule [45, 54], gastric fundic mucosa [39], gingival epithelium [12], tracheal epithelium [40], intestinal epithelium [42, 52], and epididymal epithelium [3]. Recently, some groups, including ours, have reported that TG rats bearing the tsSV40 large T-antigen [47] are a good source of conditionally immortalized epithelial cell lines, including gastric fundic mucosal [43], small intestinal [13], and tracheal epithelial cells [44].
The present study was undertaken to establish an oral epithelial cell line to constitute a continuous source of cells readily available for oral epithelial investigations using TG rats harboring the tsSV40 large T-antigen and to characterize these cells’ biological functions, including epithelial functions and gene expressions.
Materials and Methods
Establishment of an oral epithelial cell line and cell culture
TG rats (Wistar strain) which have a tsSV40 large T-antigen gene [pSVtsA58ori (−)-2] [47] were obtained from PhoenixBio Co., Ltd. (Hiroshima, Japan). The experiments were performed according to guidelines presented by the Animal Care and Use Committee of University of Toyama.
Tongues were dissected out from fetal rats (18-day-old). The tissues from underside of tongues were rinsed with phosphate-buffered saline (PBS), and were minced finely with scissors. The minced tissues were incubated in Dulbecco’s modified Eagle medium/Ham F-12 (1:1) (DMEM/F12) medium containing 0.1% collagenase for 30 min at 37°C, and epithelial tissues were separated from mesenchyme with the aid of forceps under a dissecting microscope, and cultured in DMEM/F12 medium supplemented with 2% fetal bovine serum (FBS), 1% ITES (2.0 mg/l insulin, 2.0 mg/l transferrin, 0.122 mg/l ethanolamine and 9.14 µg/l sodium selenite), 10 ng/ml epidermal growth factor (EGF), 100 unit/ml penicillin, 100 µg/ml streptomycin and 2.5 µg/ml amphotericin B in a collagen type I-precoated culture dish for 24 h at 37°C. After 24 h, culture temperature was lowered at a permissive temperature of 33°C, and cells attached to the dish were cultured under the same culture condition. To remove contaminated fibroblastic cells from the culture, dispase (25 U/ml) was added to the culture medium for 24 h. Then the cells with epithelial cell morphology were cloned by colony formation. This procedure was repeated twice [40]. The cells were routinely maintained in DMEM/F12 medium supplemented with 2% FBS, 1% ITES and 10 ng/ml EGF in a collagen type I-precoated culture vessel at 33°C.
Anchorage-independent growth
For cell culture in soft agar, the cells were suspended in agar solution which consisted of DMEM/F12 medium supplemented with 0.3% agar, 1% ITES, 10 ng/ml EGF, and 2% FBS, layered on a semisolid feeder layer, which differed from the agar solution in having a high agar percentage (0.5%), and incubated at 33°C. Two weeks later, the number of colonies was counted under a microscope [42].
Measurements of cell growth and cell cycle
The number of cells was counted by using a hematocytometer. Cell cycle was determined by flow cytometry as described previously [7]. In short, cells were fixed with 70% ice-cold ethanol, and subsequently treated with 0.25 mg/ml RNase A and 50 µg/ml propidium iodide (PI). The samples were run on an Epics XL flow cytometer (Beckman Coulter, Fullerton, CA, USA).
Immunocytochemistry
The cells were grown in a collagen type I-precoated eight-well chamber slide. The cells were washed with PBS, fixed in methanol for 7 min at 25°C. The cells were incubated with the primary antibody for 18 h at 4°C, and placed in the ChromeoTM 488-labeled secondary antibody (Active Motif, Carlsbad, CA, USA) for 1 h at 25°C. The fluorescence was examined under a fluorescence microscope. Primary antibodies used were as follows: mouse monoclonal anti-SV40 large T-antigen antibody (ab16879, Abcam, Cambridge, MA, USA); mouse monoclonal anti-Pcna (proliferating cell nuclear antigen) antibody (sc-56, Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit polyclonal anti-cytokeratin antibody (PAN, Zymed Laboratories, Inc., San Francisco, CA, USA); rabbit polyclonal anti-actin antibody (65–096, ICN ImmunoBiologicals, Lisle, IL, USA); rabbit polyclonal anti-laminin antibody (PS040, Sanbio, AM Uden, The Netherlands); rabbit polyclonal anti-collagen type IV antibody (AB756, Millipore Co., Temecula, CA, USA); mouse monoclonal anti-E-cadherin antibody (610182, BD Biosciences, San Jose, CA, USA); rabbit polyclonal anti-connexin 43 antibody (#3512, Cell Signaling Technology, Inc., Danvers, MA, USA); normal mouse IgG (PP54, Chemicon International, Inc., Temecula, CA, USA); normal rabbit IgG (02–6102, Zymed Laboratories, Inc.).
Transmission electron microscopy
For transmission electron microscopic study, cells were fixed with 3% glutaraldehyde in PBS and osmicated in 1% osmium tetroxide. After dehydrated in graded aceton solution, they were embedded in Epon 812. Ultrathin sections were stained with lead citrate and uranyl acetate and examined with an H-7100 electron microscope (Hitachi Ltd., Tokyo, Japan) at 75 kV.
SDS-polyacrylamide gel electrophoresis and Western blotting
The cells were washed once with PBS and scraped using a plastic policeman. Cellular material was placed into 50 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl and 1% NP-40 and homogenized by an ultrasonic disruptor. SDS-polyacrylamide gel electrophoresis and Western blotting were carried out as described elsewhere [21, 49]. The polyvinylidene difluoride membranes were incubated with the primary antibody at 4°C for 18 h, and exposed to the peroxidase-conjugated secondary antibody at room temperature for 1 h. Immunoreactive proteins were visualized by a luminescent image analyzer using a chemiluminescence detection system. Primary antibodies used were as follows: rabbit polyclonal anti-Krt4 antibody (ab51600, Abcam); mouse monoclonal anti-Krt10 antibody (ab9025, Abcam); mouse monoclonal anti-Krt13 antibody (1C7, MUbio Products BV, Oxfordlaan, Maastricht, The Netherlands); mouse monoclonal anti-Krt14 antibody (Millipore Co.); rabbit monoclonal anti-Krt17/19 antibody (#3984, Cell Signaling Technology, Inc.); mouse monoclonal anti-P63 antibody (sc-8431, Santa Cruz Biotechnology); mouse monoclonal anti-involucrin antibody (ab80530, Abcam); mouse monoclonal anti-Gapdh (glyceraldehyde 3-phosphate dehydrogenase) antibody (MAB374, Millipore Co.).
RNA isolation
Total RNA was extracted from cells using an RNeasy Total RNA Extraction kit (Qiagen, Valencia, CA, USA) along with on-column DNase I treatment. RNA quality was analyzed using a Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). RNA samples that had RIN (RNA integrity number) values above 9.5 were considered acceptable.
Microarray and computational gene expression analyses
Microarray analysis was carried out using a GeneChip® system with a Rat Genome 230 2.0 array, which was spotted with 31,099 probe sets (Affymetrix, Inc., Santa Clara, CA, USA) according to the manufacturer’s instructions. In brief, 500 ng of total RNA was used to synthesize cRNA with a GeneChip® 3′ IVT Express Kit (Affymetrix, Inc.). After fragmentation, biotin-labeled cRNA was hybridized to the array at 45°C for 16 h. The arrays were washed, stained with streptavidin-phycoerythrin, and scanned using a probe array scanner. The obtained hybridization intensity data were analyzed using GeneSpring® GX (Agilent Technologies, Inc.) to extract the significant genes. To examine gene ontology, including biological processes, cellular components, molecular functions, and gene networks, the obtained data were analyzed using Ingenuity® Pathway Analysis tools (Ingenuity Systems, Inc., Mountain View, CA, USA), a web-delivered application that enables the identification, visualization, and exploration of molecular interaction networks in gene expression data [1, 6].
Real-time quantitative polymerase chain reaction (PCR) assay
Real-time quantitative PCR was performed on an Mx3005P real-time PCR system (Agilent Technologies, Inc.) using SYBR PreMix ExTaq (Takara Bio Inc., Shiga, Japan) or Premix Ex Taq (for the use of TaqMan probes; Takara Bio Inc.) according to the manufacturer’s protocols. The reverse transcriptase reaction was carried out with total RNA by using a random 6 mers and an oligo dT primer. PCR primers were designed as based on the databases; Fos (FBJ osteosarcoma oncogene; GenBank Accession No.: NM_022197, sense primer position: 529–550, and antisense primer position: 692–673), Gapdh (NM_017008, 775–794, and 1062–1043), Krt13 (NM_001004021, 323–342, and 464–445) and Prdm1(PR domain containing 1, with ZNF domain; NM_001107639, 341–360, and 586–567). Based on the database, the specific primers and a probe were designed; Cdkn1a(cyclin-dependent kinase inhibitor 1A; GenBank Accession No.: U24174, sense primer position: 109–128, antisense primer position: 176–157, and probe position: 153–174). Each mRNA expression level was normalized with respect to the mRNA expression of Gapdh.
Statistical analysis
Data are presented as the means ± SDs. Differences between groups were analyzed by ANOVA, and correction for multiple comparisons was made using Dunnett’s multiple comparison test. Statistically significant differences were assumed at P<0.05.
Results
Establishment of an oral epithelial cell line from TG rats
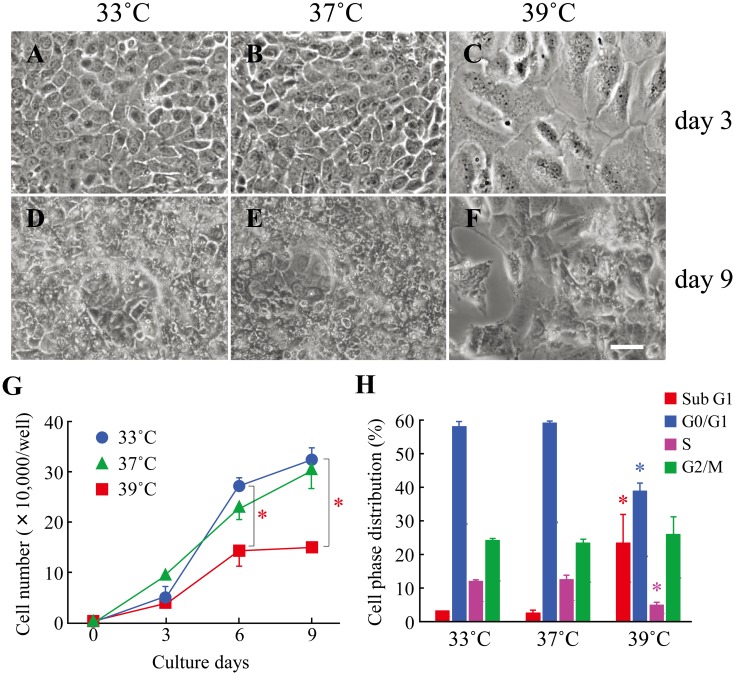
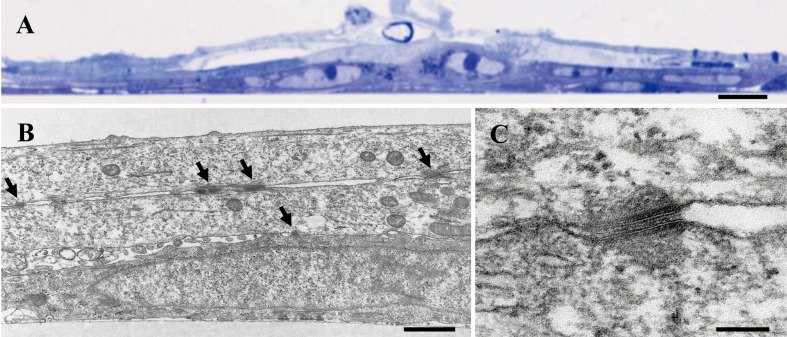
In our series of two attempts, several cell lines were developed from primary cultures of underside epithelial tissues of tongues from feral transgenic rats harboring the mutant oncogene. We chose a cell line, designated ROE2, on the basis of its ability to exhibit an epithelial morphology and protein expression of epithelial cell markers. ROE2 cells had a stable homogeneous epithelial-like morphology and maintained tight connections with neighboring cells at either a permissive temperature of 33°C or an intermediate temperature of 37°C (Figs. 1A and B). When the culture temperature was shifted to a nonpermissive temperature of 39°C, the cells were significantly larger than they were at the two lower temperatures (Fig. 1C), as was also true in our previous studies [43, 44]. A multilayered cell sheet was observed in ROE2 cells cultured for 9 days at both 33°C and 37°C but not at 39°C (Figs. 1D–F). Electron microscopic observation confirmed that the cells cultured at 37°C for 9 days formed a multilayered structure (Figs. 3A and B).
Fig. 1.
Effects of culture temperatures on the cell morphology, cell growth and cell cycle in ROE2 cells. The cells were cultured at a permissive temperature (33°C) followed by culturing at an intermediate temperature (37°C) and a nonpermissive temperature (39°C) for 0–9 days. (A–F) The cultures were examined microscopically after 3 days (A–C) and 9 days (D–F) at different temperature conditions (33°C: A, D; 37°C: B, E; 39°C: C, F). Bar, 50 µm. (G) The number of cells was counted using a hematocytometer. Closed circles, 33°C; closed triangles, 37°C; closed squares, 39°C. *: P<0.05 vs. culture at 33°C (Dunnett’s multiple comparison test). (H) The cells were cultured at different temperatures for 3 days, and then cell phase distribution in the cell cycle was monitored by using a flow cytometer. Red columns, sub G1 phase; blue columns, G0/G1 phase; red-purple columns, S phase; green columns, G2/M phase. *: P<0.05 vs. each control (culture at 33°C) (Dunnett’s multiple comparison test).
Fig. 3.
Transmission electron micrographs of ROE2 cells. The cells were cultured at 37°C for 9 days. (A) A low-magnification image of the semi-thin section stained with toluidine blue. (B) Section cut perpendicular to the plane of the dish. Arrows indicate desmosomes. (C) Higher magnification of desmosome. Bar in A, 10 µm; bar in B, 1 µm; bar in C, 0.1 µm.
Cell growth characteristics
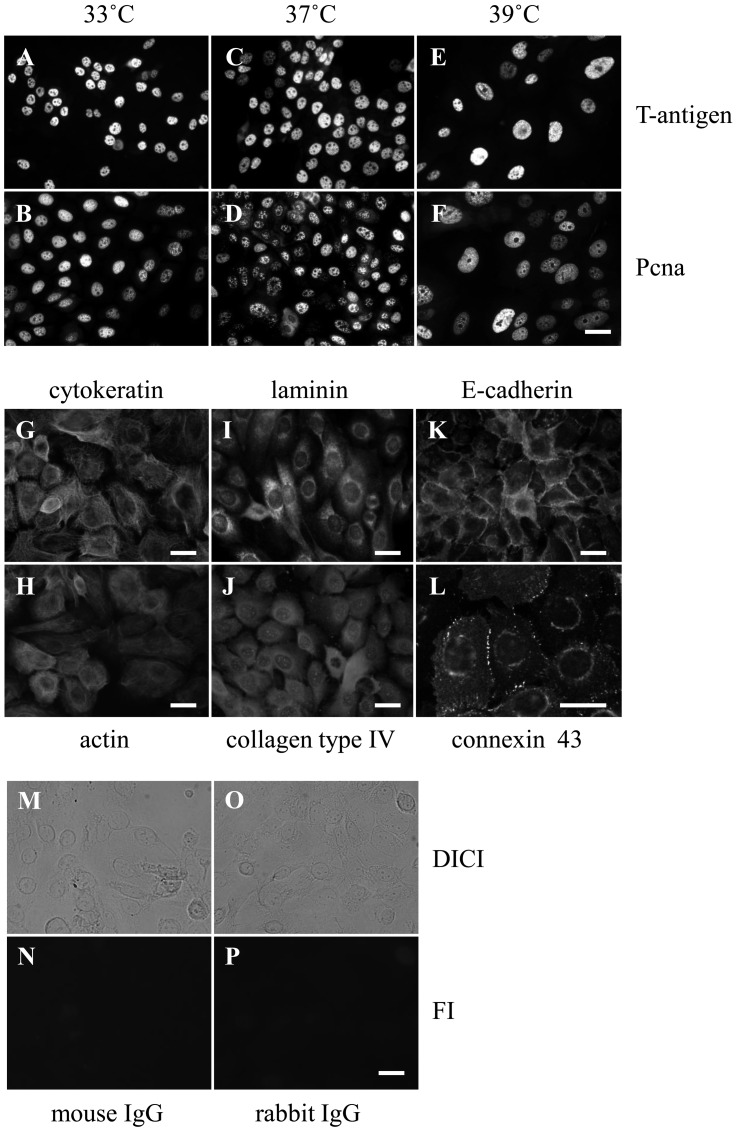
ROE2 cells proliferated constantly at both 33°C and 37°C, whereas growth decreased significantly at 39°C (Fig. 1G), indicating that cell growth is sensitive to temperature. On the other hand, large T-antigen and Pcna were expressed in the nuclei at all temperatures tested (Figs. 2A–F). However, under the negative control conditions, no fluorescence signals were detected in the cells treated with mouse and rabbit normal IgG at 33°C (Figs. 2M–P), and almost no signals were detected at the higher temperatures of 37°C and 39°C (data not shown). Next, the cell-cycle distribution was monitored by PI staining with flow cytometry. The percentages of ROE2 cells at 33°C in the sub-G1, G0/G1, S, and G2/M phases were 3.2 ± 0.2%, 58.0 ± 1.4%, 12.0 ± 0.7%, and 24.2 ± 0.7% (mean ± SD), respectively. No changes in the distribution of cell-cycle phases were observed at 37°C. Cells cultured at 39°C showed significantly lower percentages of G0/G1 and S phases (Fig. 1H). In contrast, those cultured at 39°C showed marked elevation of the sub-G1 phase of the cell cycle, a marker for apoptosis, compared to those at 33°C and 37°C; the percentage of sub-G1 in the cell cycle at 39°C was 23.4 ± 8.4% (mean ± SD; 3.2 ± 0.2% at 33°C). These data indicate that the nonpermissive temperature induces apoptosis.
Fig. 2.
Expressions of nuclear, cytoskeletal, basement, and junctional proteins in ROE2 cells. (A–F) The cells were cultured at different temperatures for 3 days. Immunocytochemical analyses of nuclear proteins, large T-antigen (A, C, E) and Pcna (B, D, F) at 33°C (A, B), 37°C (C, D) and 39°C (E, F). (G–L) The cells were cultured at 33°C for 3 days. Immunocytochemical analyses of cytoskeletal proteins, cytokeratin (G) and actin (H), basement proteins, laminin (I) and collagen type IV (J), and junctional proteins, E-cadherin (K) and connexin 43 (L). (M–P) The cells were cultured at 33°C for 3 days. Immnocytochemical analyses of normal mouse IgG (M, N) and normal rabbit IgG (O, P). Differential interference contrast images (DICI) (M, O) and fluorescence images (FI) (N, P) are shown. Bar, 25 µm.
ROE2 cells have been maintained in culture for approximately one year at 33°C, with stable morphological phenotype. Moreover, when the cells were cultured in a soft agar gel at 33°C, the cells did not show any colony-forming activity (data not shown). These data thus demonstrated that ROE2 cells are only immortalized and not transformed. Although immortalized cell lines by infection with the SV40 large T-antigen occasionally undergo malignant transformation [14], no transformed cell lines developed from the TG mice [40, 42] and rats [43, 44] that harbor tsSV40 large T-antigen.
Expressions of marker proteins for epithelial cells
We investigated the epithelial phenotype of an established cell line. Immunocytochemical investigations demonstrated that the cytoskeletal proteins cytokeratin and actin; basement membrane proteins laminin and collagen type IV; and junctional complex proteins E-cadherin and connexin 43 were detected in the ROE2 cells at 33°C (Figs. 2G–L). In addition, under electron microscopic observations, when the cells were cut parallel to the plane of the dish, several desmosomes were observed between the cells in the multilayered sheet of ROE2 cells (Figs. 3B, C). These results provide strong evidence that this cell line preserves some characteristics of oral epithelial cells, as in the cases of mouse gingival epithelial cell lines from TG mice bearing the tsSV40 large T-antigen [12], human buccal epithelial cell line SVpgC2a transformed by SV40 large T-antigen [11], and human oral primary epithelial cell cultures [2, 11].
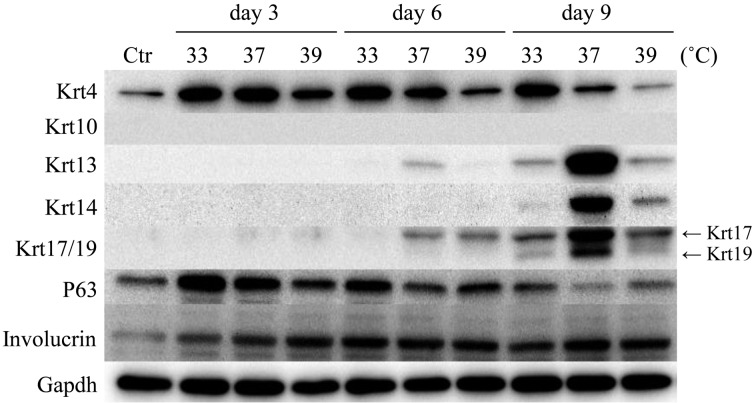
The assessment of the expression of different cytokeratin subtypes is very useful for identifying cell types in oral epithelia [36, 37]. Cells of the basal layer in both oral keratinized and nonkeratinized epithelia express Krt14, whereas Krt10 or Krt4 and Krt13 are found in cells of the spinous layer in keratinized epithelia or the intermediate layer in nonkeratinized epithelia, respectively. In addition, Krt19 [24] and P63 [31] are reported to be good candidates for epidermal stem cell markers. Involucrin is used as a marker of oral epithelial cell differentiation [32, 38]. Figure 4 shows the protein levels of cytokeratins, P63, and involucrin, as demonstrated by Western blotting under different temperature conditions. Krt4 was expressed at all temperature conditions, and the level was higher at 33°C than at 37°C or 39°C. The expression levels of Krt13, Krt14, Krt17, and Krt19 were below the detection limit under control conditions. Culturing for 9 days induced significant expression levels of Krt13, Krt14, Krt17, and Krt19 at 33°C, 37°C, and 39°C, and remarkable elevations of these four cytokeratins were observed in ROE2 cells at 37°C. On the other hand, Krt10 was not expressed under any experimental conditions. The peak expression of P63 was detected at 3 days of culture, and the level gradually and time-dependently decreased under all temperature conditions. A constant expression of involucrin was maintained throughout the experiment under all conditions employed (Fig. 4).
Fig. 4.
Effects of culture periods and culture temperatures on protein expressions for cytokeratins, P63 and involucrin in ROE2 cells. The cells were cultured at different temperatures for 3, 6, and 9 days. Western blot analyses were performed with specific primary antibodies for Krt4, Krt10, Krt13, Krt14, Krt17/19, P63, involucrin, and Gapdh. Gapdh served as a loading control. Ctr, control (day 0).
Global-scale gene expression and functional category analyses
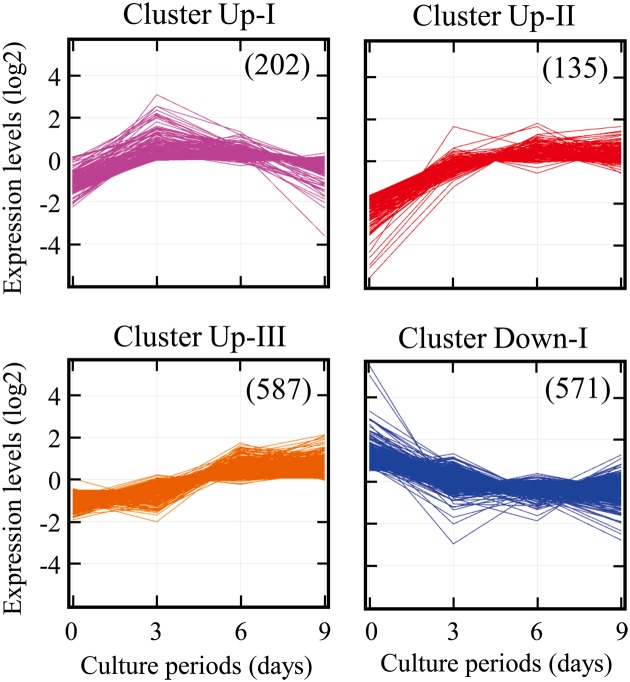
To identify genes involved in ROE2 cell differentiation, global-scale gene expression analysis was carried out using the GeneChip® system. We detected 11,264 probe sets that were expressed in ROE2 cells cultured at 37°C for 3–9 days. The complete lists of genes from ROE2 cell samples have been deposited in the Gene Expression Omnibus (GEO), a public database (an accession number: GSE44560). Expression analysis using GeneSpring® software revealed many probe sets that were differentially regulated by a factor of 2.0 or greater. We found a total of 1,495 probe sets; 924 upregulated and 571 downregulated. Moreover, K-means clustering, a nonhierarchical gene clustering algorithm, was performed to generate the major patterns of gene expression during cell differentiation. Differentially expressed probe sets were grouped in four clusters with distinct expression profiles, designated Up-I, Up-II, Up-III, and Down-I, which included 202, 135, 587, and 571 probe sets, respectively. The Up-I cluster contained gradually increased probe sets whose peak expressions occurred at day 3; the Up-II cluster contained increased probe sets whose basal expression levels at day 0 were very low. The expression levels of probe sets in the Up-III and Down-I clusters gradually increased and decreased, respectively (Fig. 5).
Fig. 5.
Gene expression analysis. ROE2 cells were cultured at 37°C for 0–9 days. K-means clustering of the probe sets that were differentially expressed by a factor of 2.0 or greater was performed using bioinformatics analysis tools. Figures in the parentheses indicate probe sets numbers.
Functional category analyses of differentially expressed probe sets in each cluster were conducted by using the Ingenuity® Pathways Knowledge Base. The top biological function and associated genes in each cluster are summarized in Table 1. The top biological function in the upregulated gene cluster Up-I was amino acid metabolism, which was functionally annotated with the synthesis of amino acids and included 7 genes. In the downregulated gene cluster Down-I, the top biological function was cellular movement, which was functionally annotated with the invasion of cells and included 37 genes. Furthermore, the top biological function, organ development, which was functionally annotated with skin development, was observed in both upregulated clusters Up-II and Up-III. Nine genes, including Krt13, Lor (loricrin), and Prdm1; and 23 genes, including Cdkn1a, Egfr (epidermal growth factor receptor), and Tgm (transglutaminase)1, were associated with skin development in clusters Up-II and Up-III, respectively (Table 1).
Table 1. The top biological function and associated genes in each cluster.
| Cluster | Category | Functional annotation (P-value) | Molecules (numbers) |
|---|---|---|---|
| Up-I | Amino acid metabolism | Synthesis of amino acids (7.05E-06) | Asns, Cth, Egr1, Gch1, Phgdh, Pycr1, Slc1a3 (7) |
| Up-II | Organ development | Skin development (1.34E-05) | Abhd5, Asprv1, Elovl4, Grhl3, Krt1, Krt13, Krt15, Lor, Prdm1 (9) |
| Up-III | Organ development | Skin development (1.05E-06) | Bmp4, Calml5, Ccnd2, Cdkn1a, Celsr1, Egfr, Eif2c2, Errfi1, Evpl, Foxe1, Gba, Krtdap, Nab1, Prss8, Rbp2, Scel, Spink5, Tacstd2, Tfap2a, Tgfb2, Tgm1, Tgm3, Ugcg (23) |
| Down-I | Cellular movement | Invasion of cells (1.22E-04) | Acsl4, Apex1, Areg/Aregb, B4galt5, Capn2, Casp3, Cd14, Csf3, Dab2, Dock1, F2r, Fgf13, Fhl2, Fhod1, Fn1, Frk, Fstl1, Gna12, Gzmb, Id2, Itgb6, Kitlg, Lama5, Mmp3, Ppic, Ptgs2, Pxn, Rras2, Ruvbl1, Sdc2, Serpine2, Slc12a2, Sod2, Spry2, Timp1, Traf3, Vim (37) |
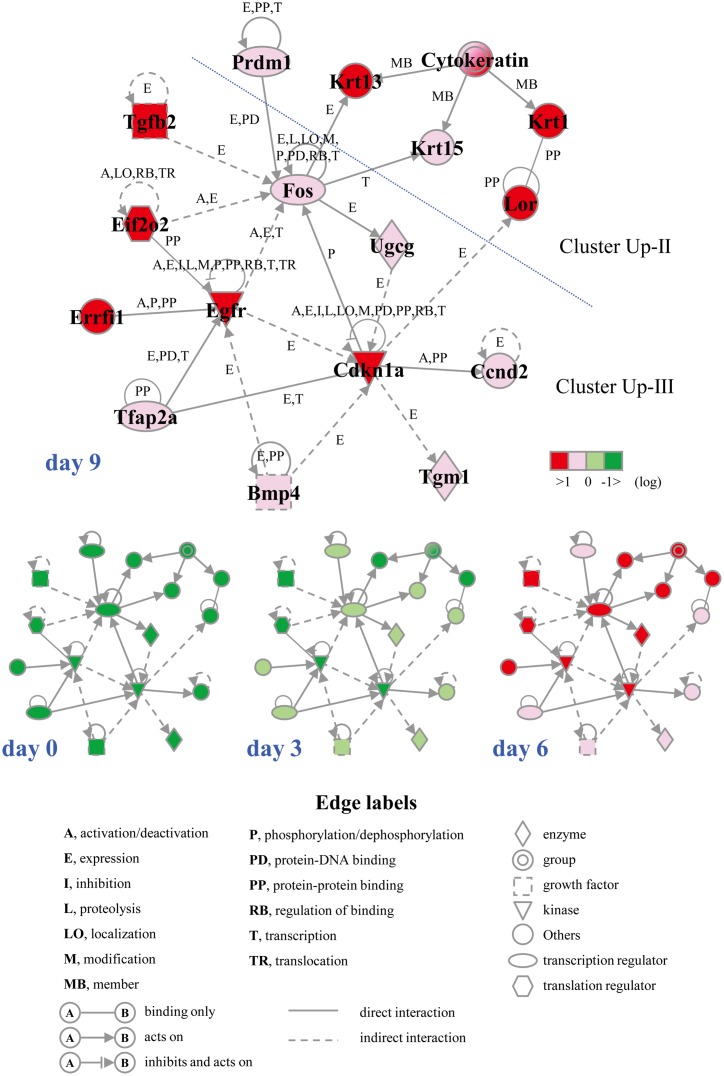
Gene network analysis
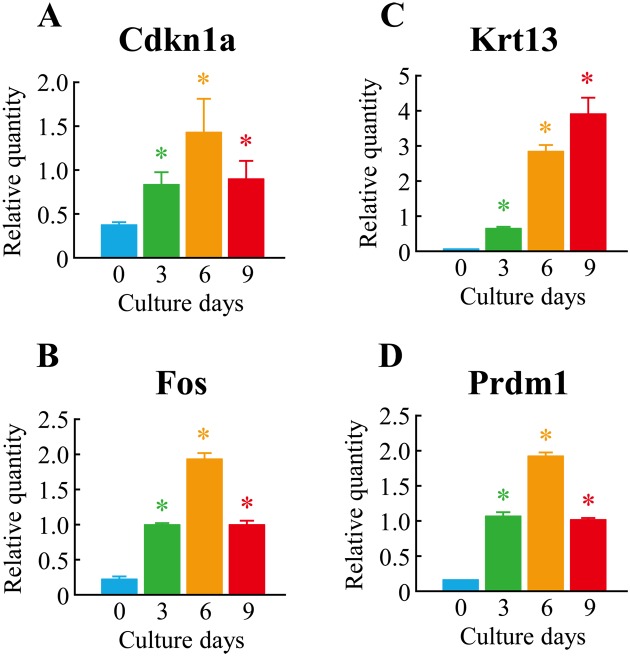
To define the molecular mechanism underlying cell differentiation induced by culturing at 37°C, we performed gene network analysis on upregulated genes belonging to clusters Up-II and Up-III using the Ingenuity Pathways Knowledge Base. We found a significant gene network, D, which included Cdkn1a, Egfr, Fos, Krt13, and Prdm1(a total of 16 genes). Interestingly, almost all genes in the gene network D were associated with skin development (functions annotation) in the category of biological functions (organ development) (Fig. 6). To verify the results of the microarrays, we performed a real-time quantitative PCR assay. Four genes–Cdkn1a, Fos, Krt13, and Prdm1–were selected from the genes that belonged to the gene network and were upregulated by culturing at 37°C for 3–9 days. As shown in Fig. 7, the expression levels of the four genes were significantly upregulated in the case of cell culturing at 37°C. This was almost comparable to the results of microarray analysis.
Fig. 6.
The gene network D. ROE2 cells were cultured at 37°C for 0–9 days. Probe sets that were up-regulated in the clusters Up-II and Up-III were analyzed by the Ingenuity Pathway analysis software. The network is displayed graphically as nodes (genes) and edges (the biological relationships between the nodes). Nodes and edges are displayed various shapes and labels that present the functional class of genes and the nature of the relationship between the nodes, respectively.
Fig. 7.
Verification of microarray results by real-time quantitative PCR. The cells were cultured at 37°C for 0–9 days. Real-time quantitative PCR was performed. Each expression level was normalized by Gapdh. (A)Cdkn1a; (B)Fos; (C)Krt13; (D)Prdm1. The data represent means ± SDs from 4 different experiments. *P<0.05 vs. control (day 0) (Dunnett’s multiple comparison test).
Discussion
TG mice harboring tsSV40 large T-antigen have been found to be a good source for establishing immortalized cell lines from many kinds of tissues including epithelia [27, 28]. The aim of this study, the development of an oral epithelial cell line with differentiation potential, was achieved by using TG rats. Interestingly, rat oral epithelial cell line ROE2, which was established here, retained some functions of stratified oral epithelial cells and differentiated into nonkeratinized epithelial cells under standard culture conditions. To our knowledge, this is the first report regarding the establishment of an oral epithelial cell line having differentiation potential from TG rats harboring the mutant oncogene.
It has been well known that wild-type SV40 large T-antigen induces immortalization by inactivating functions of several tumor suppressor molecules, such as p53 and retinoblastoma binding protein (pRb) [41]. In contrast, mutant tsSV40 large T-antigen can form complexes with p53 and pRb at permissive temperatures but at nonpermissive temperatures is inactivated and degraded and releases p53 or pRb from the complexes [16, 55]. A variety of the cell lines developed from TG animals harboring the mutant oncogene exhibited temperature-dependent growth characteristics, and nonpermissive temperatures induced cell differentiation or cell growth arrest accompanying apoptosis [28]. In the present study, although ROE2 cells showed temperature-sensitive growth, the expression levels of large T-antigen and Pcna, a valuable marker for S-phase cells, did not decrease in the nonpermissive temperature of 39°C. However, cell-cycle analysis demonstrated that the culture at 39°C significantly increased in the sub-G1 phase, a marker for apoptosis, but decreased in the S-phase in the cell cycle. We thought the immortalization and growth of ROE2 cells were due to the expression of tsSV40 large T-antigen. However, further studies are needed to investigate the molecular mechanism underling nonpermissive temperature-induced apoptosis in ROE2 cells.
In cell lines established from TG animals harboring the tsSV40 large T-antigen, their differentiated phenotypes were usually observed under nonpermissive temperature conditions at 39°C [28]. However, when apoptosis is induced by the culture at 39°C, differentiated phenotypes are reported to be detected even at the intermediate temperatures of 37°C or the permissive temperatures of 33°C [12, 48]. In line with these findings, the most differentiated phenotype of ROE2 cells was found to be long-term culture at 37°C. Under the differentiated conditions, ROE2 cells expressed not only superficial and intermediate layer cell markers, Krt4, Krt13, and involucrin, but also basal layer cell markers, Krt14, Krt19, and P63 [31, 32, 38], indicating that the culture may include several transit cells that show distinct types of cell differentiation.
To clarify the molecular mechanism underling the differentiation of ROE2 cells at 37°C, we further performed global-scale comprehensive microarray and computational gene expression analyses. The results showed changes in gene expression and gene network D involved in the cell differentiation of ROE2 cells. Very interestingly, the top biological function in both upregulated gene clusters Up-II and Up-III was the biological process of skin development. Among a total of 32 genes identified (9 + 23; Table 1), 28 genes (the exceptions being Prdm1, Calml5(calmodulin-like 5), Foxe1(forkhead box E1), and Tfap2a (transcription factor AP-2 alpha)) were also found to be expressed in normal differentiated keratinocytes in mouse oral mucosa [53], the gene expression data of which were obtained from GEO, an accession number of GSE28035. In these genes, it has been reported that Evpl (envoplakin) [35], Scel (sciellin) [4], Tgm1 [8], and Tgm3 [56] are involved in the development of epidermis. In this study, of particular interest was our identification of the skin development-associated gene network D, whose core contains Cdkn1a, Egfr, and Fos derived from upregulated genes in cluster Up-II or Up-III. Krt13 is a marker protein for oral nonkeratinized and differentiated epithelial cells [32, 38], and is shown to play an important role in epidermal development. Richard et al. [33] presented evidence to support the notion that morphological changes, including epithelial thickening, parakeratosis, and extensive vacuolization of the suprabasal keratinocytes in the upper spinous layers occur in epidermal disorders due to Krt13 defects. Previous findings also demonstrated that Prdm1 [23] and Cdkn1a [30] are involved in the terminal differentiation of epidermis. Moreover, as regards the gene network D, relationships have been reported between Fos and Krt13, and between both of these and Cdkn1a and Prdm1; namely, the downregulation of Krt13 was observed in skin keratinocytes of Fos-deficient mice [5], Cdkn1a increased the phosphorylation of Fos [26], and Fos elevated the expression level of Prdm1 [29]. According to previous findings, Lor [22], Tfap2a [50], and Ugcg (UDP-glucose ceramide glucosyltransferase) [51] are involved in keratinocyte differentiation. Bmp4 (bone morphogenetic protein 4) [10] and Ccnd2 (cyclin D2) [34] are also reported to positively regulate epidermis morphology in experimental animal models. The differentially expressed genes and/or acted within the gene network D identified here are likely to be involved in the differentiation of ROE2 cells. However, the interaction between gene expression and cell differentiation is as yet not fully understood. Important questions need to be elucidated in further investigations.
Recently, Alaminos et al. [2] reported that the expression of 1,085 genes was upregulated at least 10-fold in stratified differentiated cells in comparison to monolayer undifferentiated cells formed using human normal oral epithelial cells on fibrin-agarose scaffolds. For the expression profiles of these 1,085 genes, we identified 44 genes involved in skin development by using computational gene expression analysis tools. In 44 genes, only 6 genes–Calml5, Celsr1 (cadherin, EGF LAG seven-pass G-type receptor 1), Evpl, Krtdap (keratinocyte differentiation associated protein), Scel, and Spink5 (serine peptidase inhibitor, Kazal type 5)–were upregulated under differentiated conditions in both ROE2 cells and the human oral culture system [2]. This discrepancy may be due to cell-type differences and different experimental conditions, such as culture conditions, or to differences in the analytical methods used.
In conclusion, rat oral epithelial cell line ROE2 having specific functions would be useful as an in vitro model for studies of physiological functions and of gene expression in oral epithelial cells. In addition, the established cell line may offer an alternative to experiments using living animals, thereby providing a solution to ethical issues.
Acknowledgments
This study was supported in part by a Grant-in-Aid for Challenging Exploratory Research (23650303) and a Grant-in-Aid for Scientific Research B (24310046) from Japan Society for the Promotion of Science, and by research grants from the University of Toyama.
References
- 1.Ahmed K., Furusawa Y., Tabuchi Y., Emam H.F., Piao J.L., Hassan M.A., Yamamoto T., Kondo T., Kadowaki M.2012. Chemical inducers of heat shock proteins derived from medicinal plants and cytoprotective genes response. Int. J. Hyperthermia 28: 1–8. doi: 10.3109/02656736.2011.627408 [DOI] [PubMed] [Google Scholar]
- 2.Alaminos M., Garzón I., Sánchez-Quevedo M.C., Moreu G., González-Andrades M., Fernández-Montoya A., Campos A.2007. Time-course study of histological and genetic patterns of differentiation in human engineered oral mucosa. J. Tissue Eng. Regen. Med. 1: 350–359. doi: 10.1002/term.38 [DOI] [PubMed] [Google Scholar]
- 3.Araki Y., Suzuki K., Matusik R.J., Obinata M., Orgebin-Crist M.C.2002. Immortalized epididymal cell lines from transgenic mice overexpressing temperature-sensitive simian virus 40 large T-antigen gene. J. Androl. 23: 854–869. [PubMed] [Google Scholar]
- 4.Champliaud M.F., Baden H.P., Koch M., Jin W., Burgeson R.E., Viel A.2000. Gene characterization of sciellin (SCEL) and protein localization in vertebrate epithelia displaying barrier properties. Genomics 70: 264–268. doi: 10.1006/geno.2000.6390 [DOI] [PubMed] [Google Scholar]
- 5.Durchdewald M., Guinea-Viniegra J., Haag D., Riehl A., Lichter P., Hahn M., Wagner E.F., Angel P., Hess J.2008. Podoplanin is a novel fos target gene in skin carcinogenesis. Cancer Res. 68: 6877–6883. doi: 10.1158/0008-5472.CAN-08-0299 [DOI] [PubMed] [Google Scholar]
- 6.Furusawa Y., Tabuchi Y., Wada S., Takasaki I., Ohtsuka K., Kondo T.2011. Identification of biological functions and gene networks regulated by heat stress in U937 human lymphoma cells. Int. J. Mol. Med. 28: 143–151. [DOI] [PubMed] [Google Scholar]
- 7.Furusawa Y., Iizumi T., Fujiwara Y., Zhao Q.L., Tabuchi Y., Nomura T., Kondo T.2012. Inhibition of checkpoint kinase 1 abrogates G2/M checkpoint activation and promotes apoptosis under heat stress. Apoptosis 17: 102–112. doi: 10.1007/s10495-011-0660-7 [DOI] [PubMed] [Google Scholar]
- 8.Gentile V., Cooper A.J.2004. Transglutaminases − possible drug targets in human diseases. Curr. Drug Targets CNS Neurol. Disord. 3: 99–104. doi: 10.2174/1568007043482552 [DOI] [PubMed] [Google Scholar]
- 9.Gilchrist E.P., Moyer M.P., Shillitoe E.J., Clare N., Murrah V.A.2000. Establishment of a human polyclonal oral epithelial cell line. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 90: 340–347. doi: 10.1067/moe.2000.107360 [DOI] [PubMed] [Google Scholar]
- 10.Goswami M., Uzgare A.R., Sater A.K.2001. Regulation of MAP kinase by the BMP-4/TAK1 pathway in Xenopus ectoderm. Dev. Biol. 236: 259–270. doi: 10.1006/dbio.2001.0338 [DOI] [PubMed] [Google Scholar]
- 11.Hansson A., Bloor B.K., Sarang Z., Haig Y., Morgan P.R., Stark H.J., Fusenig N.E., Ekstrand J., Grafström R.C.2003. Analysis of proliferation, apoptosis and keratin expression in cultured normal and immortalized human buccal keratinocytes. Eur. J. Oral Sci. 111: 34–41. doi: 10.1034/j.1600-0722.2003.00010.x [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama S., Ohara-Nemoto Y., Yanai N., Obinata M., Hayashi S., Satoh M.2001. Establishment of gingival epithelial cell lines from transgenic mice harboring temperature sensitive simian virus 40 large T-antigen gene. J. Oral Pathol. Med. 30: 296–304. doi: 10.1034/j.1600-0714.2001.300507.x [DOI] [PubMed] [Google Scholar]
- 13.Hosoya K., Tomi M., Takayama M., Komokata Y., Nakai D., Tokui T., Nishimura K., Ueda M., Obinata M., Hori S., Ohtsuki S., Amidon G.L., Terasaki T.2004. Transporter mRNA expression in a conditionally immortalized rat small intestine epithelial cell line (TR-SIE). Drug Metab. Pharmacokinet. 19: 264–269. doi: 10.2133/dmpk.19.264 [DOI] [PubMed] [Google Scholar]
- 14.Isom H.C., Tevethia M.J., Kreider J.W.1981. Tumorigenicity of simian virus 40-transformed rat hepatocytes. Cancer Res. 41: 2126–2134. [PubMed] [Google Scholar]
- 15.Jat P.S., Sharp P.A.1986. Large T antigens of simian virus 40 and polyomavirus efficiently establish primary fibroblasts. J. Virol. 59: 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jat P.S., Sharp P.A.1989. Cell lines established by a temperature-sensitive simian virus 40 large-T-antigen gene are growth restricted at the nonpermissive temperature. Mol. Cell Biol. 9: 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jepsen A.1974. An in vitro model of an oral keratinizing squamous epithelium. Scand. J. Dent. Res. 82: 144–146. [DOI] [PubMed] [Google Scholar]
- 18.Jepsen A., MacCallum D.K., Lillie J.H.1980. Fine structure of subcultivated stratified squamous epithelium. Exp. Cell Res. 125: 141–152. doi: 10.1016/0014-4827(80)90198-6 [DOI] [PubMed] [Google Scholar]
- 19.Krause C.J., Carey T.E., Ott R.W., Hurbis C., McClatchey K.D., Regezi J.A.1981. Human squamous cell carcinoma. Establishment and characterization of new permanent cell lines. Arch. Otolaryngol. 107: 703–710. doi: 10.1001/archotol.1981.00790470051012 [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni P.S., Sundqvist K., Betsholtz C., Höglund P., Wiman K.G., Zhivotovsky B., Bertolero F., Liu Y., Grafström R.C.1995. Characterization of human buccal epithelial cells transfected with the simian virus 40 T-antigen gene. Carcinogenesis 16: 2515–2521. doi: 10.1093/carcin/16.10.2515 [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U.K.1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. doi: 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 22.Lee C.H., Marekov L.N., Kim S., Brahim J.S., Park M.H., Steinert P.M.2000. Small proline-rich protein 1 is the major component of the cell envelope of normal human oral keratinocytes. FEBS Lett. 477: 268–272. doi: 10.1016/S0014-5793(00)01806-8 [DOI] [PubMed] [Google Scholar]
- 23.Magnúsdóttir E., Kalachikov S., Mizukoshi K., Savitsky D., Ishida-Yamamoto A., Panteleyev A.A., Calame K.2007. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc. Natl. Acad. Sci. USA. 104: 14988–14993. doi: 10.1073/pnas.0707323104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel M., Török N., Godbout M.J., Lussier M., Gaudreau P., Royal A., Germain L.1996. Keratin 19 as a biochemical marker of skin stem cells in vivo and in vitro: keratin 19 expressing cells are differentially localized in function of anatomic sites, and their number varies with donor age and culture stage. J. Cell Sci. 109: 1017–1028. [DOI] [PubMed] [Google Scholar]
- 25.Momose F., Araida T., Negishi A., Ichijo H., Shioda S., Sasaki S.1989. Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J. Oral Pathol. Med. 18: 391–395. doi: 10.1111/j.1600-0714.1989.tb01570.x [DOI] [PubMed] [Google Scholar]
- 26.Neise D., Sohn D., Budach W., Jänicke R.U.2010. Evidence for a differential modulation of p53-phosphorylating kinases by the cyclin-dependent kinase inhibitor p21WAF1/CIP1. Cell Cycle 9: 3575–3583. doi: 10.4161/cc.9.17.12799 [DOI] [PubMed] [Google Scholar]
- 27.Noble M., Groves A.K., Ataliotis P., Ikram Z., Jat P.S.1995. The H-2KbtsA58 transgenic mouse: a new tool for the rapid generation of novel cell lines. Transgenic Res. 4: 215–225. doi: 10.1007/BF01969114 [DOI] [PubMed] [Google Scholar]
- 28.Obinata M.2007. The immortalized cell lines with differentiation potentials: their establishment and possible application. Cancer Sci. 98: 275–283. doi: 10.1111/j.1349-7006.2007.00399.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohkubo Y., Arima M., Arguni E., Okada S., Yamashita K., Asari S., Obata S., Sakamoto A., Hatano M., O-Wang J., Ebara M., Saisho H., Tokuhisa T.2005. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J. Immunol. 174: 7703–7710. [DOI] [PubMed] [Google Scholar]
- 30.Paramio J.M., Segrelles C., Ruiz S., Martin-Caballer J., Page A., Martinez J., Serrano M., Jorcano J.L.2001. The ink4a/arf tumor suppressors cooperate with p21cip1/waf in the processes of mouse epidermal differentiation, senescence, and carcinogenesis. J. Biol. Chem. 276: 44203–44211. doi: 10.1074/jbc.M105650200 [DOI] [PubMed] [Google Scholar]
- 31.Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M.2001. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA. 98: 3156–3161. doi: 10.1073/pnas.061032098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Presland R.B., Jurevic R.J.2002. Making sense of the epithelial barrier: what molecular biology and genetics tell us about the functions of oral mucosal and epidermal tissues. J. Dent. Educ. 66: 564–574. [PubMed] [Google Scholar]
- 33.Richard G., De Laurenzi V., Didona B., Bale S.J., Compton J.G.1995. Keratin 13 point mutation underlies the hereditary mucosal epithelial disorder white sponge nevus. Nat. Genet. 11: 453–455. doi: 10.1038/ng1295-453 [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Puebla M.L., LaCava M., Miliani De Marval P.L., Jorcano J.L., Richie E.R., Conti C.J.2000. Cyclin D2 overexpression in transgenic mice induces thymic and epidermal hyperplasia whereas cyclin D3 expression results only in epidermal hyperplasia. Am. J. Pathol. 157: 1039–1050. doi: 10.1016/S0002-9440(10)64616-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruhrberg C., Hajibagheri M.A., Simon M., Dooley T.P., Watt F.M.1996. Envoplakin, a novel precursor of the cornified envelope that has homology to desmoplakin. J. Cell Biol. 134: 715–729. doi: 10.1083/jcb.134.3.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakamoto K., Aragaki T., Morita K., Kawachi H., Kayamori K., Nakanishi S., Omura K., Miki Y., Okada N., Katsube K., Takizawa T., Yamaguchi A.2011. Down-regulation of keratin 4 and keratin 13 expression in oral squamous cell carcinoma and epithelial dysplasia: a clue for histopathogenesis. Histopathology 58: 531–542. doi: 10.1111/j.1365-2559.2011.03759.x [DOI] [PubMed] [Google Scholar]
- 37.Sawaf M.H., Ouhayoun J.P., Forest N.1991. Cytokeratin profiles in oral epithelial: a review and a new classification. J. Biol. Buccale. 19: 187–198. [PubMed] [Google Scholar]
- 38.Squier C.A., Kremer M.J.2001. Biology of oral mucosa and esophagus. J. Natl. Cancer Inst. Monogr. 29: 7–15. doi: 10.1093/oxfordjournals.jncimonographs.a003443 [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama N., Tabuchi Y., Horiuchi T., Obinata M., Furusawa M.1993. Establishment of gastric surface mucous cell lines from transgenic mice harboring temperature-sensitive simian virus 40 large T-antigen gene. Exp. Cell Res. 209: 382–387. doi: 10.1006/excr.1993.1324 [DOI] [PubMed] [Google Scholar]
- 40.Sugiyama N., Tabuchi Y., Numata F., Uchida Y., Horiuchi T., Ishibashi K., Ono S., Obinata M., Furusawa M.1998. Establishment and characterization of tracheal epithelial cell lines, TM01 and TM02-3, from transgenic mice bearing temperature-sensitive simian virus 40 large T-antigen gene. Cell Struct. Funct. 23: 119–127. doi: 10.1247/csf.23.119 [DOI] [PubMed] [Google Scholar]
- 41.Sullivan C.S., Pipas J.M.2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66: 179–202. doi: 10.1128/MMBR.66.2.179-202.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabuchi Y., Ohta S., Arai Y., Kawahara M., Ishibashi K., Sugiyama N., Horiuchi T., Furusawa M., Obinata M., Fuse H., Takeguchi N., Asano S.2000. Establishment and characterization of a colonic epithelial cell line MCE301 from transgenic mice harboring temperature-sensitive simian virus 40 large T-antigen gene. Cell Struct. Funct. 25: 297–307. doi: 10.1247/csf.25.297 [DOI] [PubMed] [Google Scholar]
- 43.Tabuchi Y., Arai Y., Ohta S., Shioya H., Takahashi R., Ueda M., Takeguchi N., Asano S., Obinata M.2002. Development and characterization of conditionally immortalized gastric epithelial cell lines from transgenic rats harboring temperature-sensitive simian virus 40 large T-antigen gene. Cell Struct. Funct. 27: 71–79. doi: 10.1247/csf.27.71 [DOI] [PubMed] [Google Scholar]
- 44.Tabuchi Y., Doi T., Takasaki I., Takahashi R., Ueda M., Suzuki Y., Obinata M.2008. Establishment and functional characterization of a tracheal epithelial cell line RTEC11 from transgenic rats harboring temperature-sensitive simian virus 40 large T-antigen. Cell Biol. Int. 32: 1344–1352. doi: 10.1016/j.cellbi.2008.07.025 [DOI] [PubMed] [Google Scholar]
- 45.Tai G., Hohenstein P., Davies J.A.2012. Making immortalized cell lines from embryonic mouse kidney. Methods Mol. Biol. 886: 165–171. doi: 10.1007/978-1-61779-851-1_15 [DOI] [PubMed] [Google Scholar]
- 46.Taichman L., Reilly S., Garant P.R.1979. In-vitro cultivation of human oral keratinocytes. Arch. Oral Biol. 24: 335–341. doi: 10.1016/0003-9969(79)90099-2 [DOI] [PubMed] [Google Scholar]
- 47.Takahashi R., Hirabayashi M., Yanai N., Obinata M., Ueda M.1999. Establishment of SV40-tsA58 transgenic rats as a source of conditionally immortalized cell lines. Exp. Anim. 48: 255–261. doi: 10.1538/expanim.48.255 [DOI] [PubMed] [Google Scholar]
- 48.Tavelin S., Milovic V., Ocklind G., Olsson S., Artursson P.1999. A conditionally immortalized epithelial cell line for studies of intestinal drug transport. J. Pharmacol. Exp. Ther. 290: 1212–1221. [PubMed] [Google Scholar]
- 49.Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 76: 4350–4354. doi: 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X., Pasolli H.A., Williams T., Fuchs E.2008. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J. Cell Biol. 183: 37–48. doi: 10.1083/jcb.200804030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe R., Wu K., Paul P., Marks D.L., Kobayashi T., Pittelkow M.R., Pagano R.E.1998. Up-regulation of glucosylceramide synthase expression and activity during human keratinocyte differentiation. J. Biol. Chem. 273: 9651–9655. doi: 10.1074/jbc.273.16.9651 [DOI] [PubMed] [Google Scholar]
- 52.Whitehead R.H., Robinson P.S.2009. Establishment of conditionally immortalized epithelial cell lines from the intestinal tissue of adult normal and transgenic mice. Am. J. Physiol. Gastrointest. Liver Physiol. 296: G455–G460. doi: 10.1152/ajpgi.90381.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu T., Jia L., Du R., Tao X., Chen J., Cheng B.2011. Genome-wide analysis reveals the active roles of keratinocytes in oral mucosal adaptive immune response. Exp. Biol. Med. (Maywood). 236: 832–843. doi: 10.1258/ebm.2011.010307 [DOI] [PubMed] [Google Scholar]
- 54.Yanai N., Satoh T., Kyo S., Abe K., Suzuki M., Obinata M.1991. A tubule cell line established from transgenic mice harboring temperature-sensitive simian virus 40 large T-antigen gene. Jpn. J. Cancer Res. 82: 1344–1348. doi: 10.1111/j.1349-7006.1991.tb01803.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanai N., Obinata M.1994. Apoptosis is induced at nonpermissive temperature by a transient increase in p53 in cell lines immortalized with temperature-sensitive SV40 large T-antigen gene. Exp. Cell Res. 211: 296–300. doi: 10.1006/excr.1994.1090 [DOI] [PubMed] [Google Scholar]
- 56.Zhang J., Zhi H.Y., Ding F., Luo A.P., Liu Z.H.2005. Transglutaminase 3 expression in C57BL/6J mouse embryo epidermis and the correlation with its differentiation. Cell Res. 15: 105–110. doi: 10.1038/sj.cr.7290274 [DOI] [PubMed] [Google Scholar]