Abstract
The tremor rat is an autosomal recessive mutant exhibiting sterility with gonadal hypoplasia in both sexes. The causative mutation tremor (tm) is known as a genomic deletion spanning >200 kb in Chr 10q24. Spermatogenesis associated 22 (Spata22) has been shown to be a vertebrate-specific gene essential for the progression of meiosis through prophase I and completion of chromosome synapsis and meiotic recombination using a mouse repro42 mutant carrying an N-ethyl-N-nitrosourea (ENU)-induced nonsense mutation in Spata22. In this study, we show that Spata22 was identified as the gene responsible for the failure of gametogenesis to progress beyond meiosis I in tm homozygous rats by a transgenic rescue experiment. Meiosis was arrested during prophase I in the mutant testis. Precise mapping of the breakage point revealed that the deleted genomic region spanned approximately 240 kb and comprised at least 13 genes, including Spata22. Rat Spata22 was predominantly expressed in the testis, and its transcription increased with the first wave of spermatogenesis, as seen in the mouse ortholog. These results suggest that Spata22 may play an important role in meiotic prophase I in rats, as seen in mice, and that the tm homozygous rat may be useful for investigating the physiological function of Spata22, as an experimental system for clarifying the effect of a null mutation, and may be an animal model for studying the pathogenesis and treatment of infertility caused by impaired meiosis.
Keywords: gametogenesis, gonadal development, meiotic failure, spontaneous mutation, sterile rat
Introduction
Human infertility extends to 15% of couples, and 60% of idiopathic infertility cases in males have a recessive autosomal cause [9, 16]. Genetically well-defined animal models are considered to be useful not only for understanding the pathogenesis of infertility but also for the development of treatments for it. At present, mice are widely used as animal models for human diseases. However, considering the effect of species variation of suitability for animal experiments and phenotypic differences due to species differences, it is desirable to prepare animal models of various species.
Meiosis is a developmentally programmed complex process that produces haploid germ cells from diploid parental cells in reproducing eukaryotes [20]. In vivo experimental systems are indispensable for investigating molecular mechanisms of vertebrate meiosis because of the lack of reproducible in vitro experimental systems. Animals exhibiting meiotic defects caused by gene mutations may be important tools not only for investigating molecular mechanisms underlying meiosis but also for studying human infertility.
Spermatogenesis associated 22 (SPATA22), formally known as NYD-SP20, was originally cloned from human testes by comprehensive expression analysis [24], and later, novel alternative spliced forms were reported [11]. Recently, SPATA22 has been shown to be a candidate gene for human oligospermia or aspermia [1] and identified as the causative gene for an N-ethyl-N-nitrosourea (ENU)-induced autosomal recessive mutation, repro42, in mice. This mouse model exhibits arrest of meiosis at prophase I with abnormal chromosome synapsis and double-stranded DNA break (DSB) repair in both sexes [15]. SPATA22 and the orthologs in mouse and rat encode 363-, 358- and 361-amino acid proteins, respectively. Confirmed or predicted SPATA22 protein sequences from various vertebrates, including the human, chimpanzee, mouse, rat, dog, cattle, chicken, and zebrafish, display evolutionarily conserved regions in their N- and C-termini [15]. SPATA22 comprises numerous putative functional sites across the protein sequence, including phosphopeptide motifs interacting with the BRCT domain of breast cancer 1, early onset (BRCA1), Polo-like kinase phosphorylation sites, and phosphoinositide-3-OH kinase-related kinase (PIKK) phosphorylation motifs; however, it has no significant homology with any other previously described proteins or functional domains [4, 15]. Although the essential role of SPATA22 in the progression of meiosis I has been revealed, its molecular function remains unknown.
The mouse repro42 mutation is a single nucleotide substitution, resulting in the appearance of a premature stop codon in the C-terminal conserved domain in the deduced amino acid sequence of Spata22 [15]. The deduced protein lacks the C-terminal 275−358-amino acid region. In testes of homozygous mutant mice, the expression level of Spata22 mRNA is decreased, and the Spata22 protein cannot be detected [15], suggesting that repro42 is a loss-of-function mutation and that the resultant phenotype is caused by the low level of expression and/or a structural defect in Spata22.
The tremor (tm) mutation arose spontaneously in a colony of Kyoto:Wistar rats in 1980 and was initially characterized for its recessive body tremor, hair and whisker abnormalities, and infertility with dysgenesis of gonads in both sexes [29]. A subsequent report indicated that tm mutant males and females have recessive and semidominant absence-like seizure and recessive spongiform degeneration in the central nervous system (CNS) [23]. The tremor rat had been established as a segregating inbred strain by brother−sister inbreeding of tm heterozygous rats. The tm mutation has been mapped to rat chromosome 10q24 and has been identified as a deletion spanning a >200-kb genomic region containing aspartoacylase (Aspa), transient receptor potential vanilloid 1 (Trpv1), and several genes encoding olfactory receptors (ORs) [13]. According to the rat genomic sequence (RGSC v3.4), the region deleted in the tm locus is considered to comprise Spata22. In the present study, we found that gametogenesis failed to progress beyond meiosis I in the tm homozygous rat, and that meiosis was arrested during prophase I in the tm homozygous male testis. We revealed that the deleted genomic region in the tm locus comprised nearly 240 kb and harbored at least 13 genes, including Spata22. We identified Spata22 as the causative gene of infertility and abnormal meiosis in homozygous mutants by a transgenic rescue experiment. This is the first description of meiotic failure caused by a null mutation of Spata22, and the first report of infertility associated with meiotic failure caused by a naturally or spontaneously occurring mutation of Spata22, including human infertility.
Materials and Methods
Animals
The tremor rat TRM/Kyo was supplied by the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University (Kyoto, Japan), and maintained in the Genome Dynamics Research Center at Hokkaido University. Heterozygous (tm/+) females were mated with heterozygous (tm/+) males to obtain the affected (tm/tm) and control (+/+) rats used in this study. All the rats were maintained in a room with controlled temperature and a 12-h light–dark cycle. Food and water were provided ad libitum. All the animal experiments in this study were approved by Institutional Animal Care and Use Committee, Hokkaido University.
Histological analysis
Testes were collected from three each of tm/tm and wild-type rats and tm homozygous mutants carrying Spata22 transgenes at 3 months after birth. Ovaries were collected from two each of tm/tm and wild-type rats at 7 days after birth. They were fixed using Bouin’s fixative solution, embedded in paraffin, cut into 5-µm-thick sections, and then stained with hematoxylin and eosin (HE).
Chromosome analysis
The preparation of nuclei and chromosomes for light microscopy of spermatogenic cells from two each of tm/tm and wild-type rats at 30 days after birth and two tm homozygous mutants carrying Spata22 transgenes at 3 months after birth was conducted using the air-drying method [12], with the omission of colchicine treatment. The preparations were stained with 4% Giemsa in phosphate buffer (pH 6.8).
Physical mapping
We conducted a homology search of the sequence of a breakage point (GenBank entry: AB023434) in the mutant genome using the BLAST program (http://blast.ncbi.nlm.nih.gov). PCR amplifications were performed with primer pairs for DNA fragments localized around both ends of the deleted region. Sequences of six primer sets used in sequence-tagged site (STS) analysis and map positions of their target DNA sequences on rat chromosome 10 (RGSC v3.4) are as follows: brk1, CCCATCTCACCATCACCCCAACAGT and ACACCCCACTCTTGGAGCTGTTGAA (60,098,543–60,098,723 bp); brk2, TTGTGCTGGGCTGGGTGCTATCTCT and TGCCGTGGGTATTAAATGAGAAGGA (60,119,939–60,120,055 bp); brk3, GCTCTCACACTACAACAAG and GTAGTTGGAAAGGTCTCCC (60,324,857–60,324,992 bp); brk4, TTGTAATAACAGGGATTGC and GCCCATTTACAAACCTCAG (60,333,641–60,333,799 bp); brk5, ATTGGATAACCCAACCTGG and GGTCGTGGTCACCTAAACT (60,345,158–60,345,336 bp); and brk6, GTTCCCACCATTTGCCTACGAATTT and GCTCCACTATGTTCATCGCAGCCT (60,346,178–60,346,348 bp).
RT-PCR
Total RNA was extracted from various organs using TRIzol reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA), and first-strand cDNA was synthesized with oligo-dT primers. The following primer sequences were used for PCR amplification: Spo11, CCGAGGCCTCGTTCTTCGAC and TGTCCACCGCAGCCTGGTTC; Spata22, TCAACTCGAAGTACAGCAGGCTGTT and GCCACCTTGGCTTCTTTTAGCGTTT; Msh4, GCTGTGTACCATCTGGCTACA and CTCCTCAGTCTTCTCTGGAAGG; and Gapdh, ACCACAGTCCATGCCATCAC and TCCACCACCCTGTTGCTGTA.
Generation of transgenic rats
Transgenic rats for rescue experiments were generated using the pCAGGS vector (provided by J. Miyazaki). Full-length Spata22 cDNA was obtained by screening a rat testis cDNA library (Takara Bio, Otsu, Shiga, Japan). Full-length Spata22 cDNA was inserted into the EcoRI site of the vector. The vector plasmid was digested with SalI and HindIII, electrophoresed, and then purified from agarose gel. Transgenic rats were generated by microinjection of the transgenes into fertilized eggs from Wistar females mated with heterozygous tm males. Two lines of transgenic rats (line 2, line 4) were obtained. These transgenic lines were backcrossed to TRM/Kyo. Gross and histological examinations of gonadal development were conducted mainly using line 2. Two each of 7-month-old homozygous mutant males carrying the transgenes (tm/tm Tg/+) and 21-day-old homozygous mutant females carrying the transgenes (tm/tm Tg/+) were used for gross examinations. Their homozygous (tm/tm) and heterozygous (tm/+) non-transgenic littermates were used as controls.
Accession number
Sequence data of a full-length rat Spata22 cDNA has been deposited in DDBJ/EMBL/GenBank databases under accession number AB236891.
Results
Mutant gonads displayed arrest of meiosis I
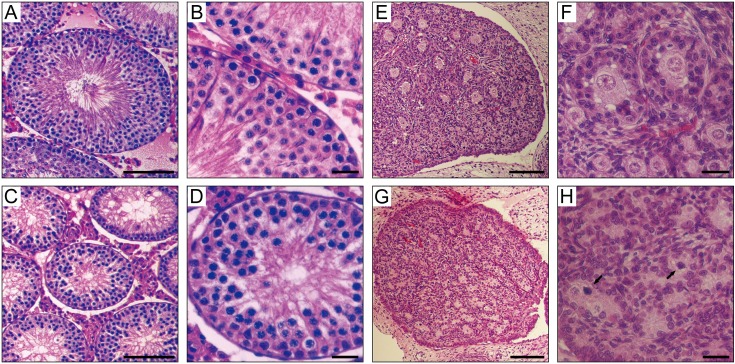
Testis weights of mutants were markedly reduced; average testis weights (mean ± SD) were 391 ± 44 mg in three tm/tm adult males, 1,228 ± 254 mg in three wild type adult males, and 1,395 ± 198 mg in three tm/+ adult males. The average testis weight of the tm homozygous mutants was significantly less than those of the wild types (t-test, P<0.001) and the heterozygotes (t-test, P<0.001). Average testis weights were not significantly different between the wild types and the heterozygotes (t-test, P>0.05). Average ratios (mean ± SD) of testis weight (mg) to body weight (g) were 1.21 ± 0.15 in the homozygous mutants, 4.23 ± 1.08 in the wild-types, and 3.84 ± 0.39 in the heterozygotes. Pathological examination of the testes from adult mutant males demonstrated the accumulation of spermatocytes and depletion of spermatids and spermatozoa, as shown in Figs. 1A, 1B, 1C, and 1D. The ovaries from juvenile homozygous mutant females (tm/tm) had depleted primordial follicles, with many degenerated oocyte nuclei (Figs. 1E, 1F, 1G, and 1H). Adult mutant females lacked discernible ovarian structures (data not shown). These observations indicated the arrest of meiosis I during gametogenesis in mutant gonads. Subsequent cytogenetic examination of mutant meiosis was carried out using spermatogenic cells.
Fig. 1.
Abnormal gametogenesis in mutant rats. Upper panels, wild-type. Lower panels, homozygous mutant. Hematoxylin and eosin (HE)-stained sections of seminiferous tubules at 3 months after birth (A–D) and ovaries at 7 days after birth (E–H). Accumulation of spermatocytes and depletion of spermatids and spermatozoa were observed in adult homozygous mutants (C and D). Oocyte was depleted in juvenile homozygous mutants (G and H). Arrows indicate degenerated nuclei. Scale bar: 100 µm in A, C, E, and G and 25 µm in B, D, F, and H.
Mutant male meiosis failed to progress beyond early prophase I
To examine meiotic progression in mutants, we observed spreads of nuclei and chromosomes of spermatocytes. In 30-day-old control testes, zygotene, late zygotene or early pachytene, midpachytene, and metaphase I spermatocyte nuclei were observed (Fig. 2A). In 30-day-old homozygous mutant testes (tm/tm), no normal nuclei that had progressed into midpachytene were observed, whereas nuclei that had nucleus sizes and chromosome morphologies similar to those of control spermatocytes at zygotene and late zygotene or early pachytene stages and many degenerated nuclei were observed (Fig. 2B), indicating that meiosis progressed into the zygotene stage and was arrested at the zygotene or pachytene stage in mutants. This spermatogenesis phenotype, known as the “meiotic arrest” phenotype, is similar to that of the repro42 homozygous mouse, which carries a nonsense mutation in Spata22 [15], and has been previously found in many types of sterile mutant mice and rats for genes related to synaptonemal complex formation, meiotic homologous recombination repair, and regulation of DNA damage checkpoint [2, 8, 10, 21].
Fig. 2.
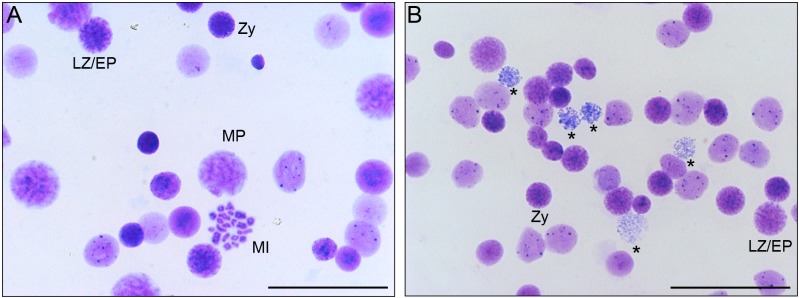
Failure of meiotic progression during prophase I in the mutant testis. A: Giemsa-stained preparation of testicular cell nuclei or chromosomes from 30-day-old wild-type rats indicating the presence of spermatocytes at zygotene (Zy), late zygotene or early pachytene (LZ/EP), midpachytene (MP), and metaphase I (MI) stages. B: Giemsa-stained preparation of testicular cell nuclei or chromosomes from 30-day-old homozygous mutant rats showing the presence of spermatocyte nuclei similar to control spermatocyte nuclei at zygotene (Zy) and late zygotene or early pachytene (LZ/EP) stages. Many degenerated nuclei were observed (asterisks), whereas no normal midpachytene nuclei were present. Scale bar: 50 µm.
The mutant genome lacked a nearly 240-kb genomic region that comprises at least 13 genes, including Spata22
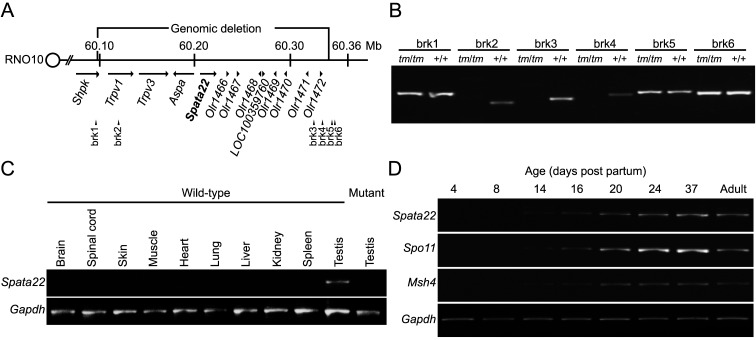
To identify the gene responsible for the abnormal gametogenesis, we determined the precise location of ends of the deleted region on rat chromosome 10 by searching for the DNA fragment (AB023434) containing the tm breakage point using the BLAST program. We verified these results by PCR analysis using STS markers around both ends of the deleted region (Figs. 3A and 3B). The centromere-proximal end of the deleted region was mapped inside the 7th exon of sedoheptulokinase (Shpk), and the centromere-distal end was located within a LINE1 element downstream of Olr1472. Within the determined 240-kb deleted region (60,098,741−60,339,238 bp, RGSC v3.4), entire transcribed regions of 12 genes besides part of the 7th exon of Shpk were localized: transient receptor potential vanilloid 1 and 3 (Trpv1/3); Aspa; the Spata22 ortholog; seven OR genes, Olr1466, Olr1467, Olr1468, Olr1469, Olr1470, Olr1471, and Olr1472; and one hypothetical gene, LOC100359760 (Fig. 3A).
Fig. 3.
Positions of ends of the genomic deletion in the tm locus and expression analysis of Spata22. A: Schematic diagram of the tm critical region of rat chromosome 10 (RNO10) indicating positions of the genomic deletion, Spata22, and other genes within the deleted region. The breakage sites are mapped to sedoheptulokinase (Shpk) and an L1 retrotransposon element in the large cluster of olfactory receptor genes, respectively. The distance between the breakage sites is approximately 240 kb. Arrows above gene names represent approximate gene size and indicate the direction of transcription. Positions of STS markers (brk1–brk6) used in PCR analysis are shown below the chromosome. Map positions in Mb are shown above the chromosome. B: Comparison of PCR amplification of sequence-tagged site (STS) markers between genomic DNA of control and homozygous mutant rats demonstrating loss of PCR amplification of STS makers (brk2–brk4) within the predicted deleted region in mutants. C: mRNA expression of Spata22 and Gapdh in various organs of adult controls and testes of adult homozygous mutants showing enhanced expression of Spata22 in control testes. Weak expression was detected in other organs when the number of PCR cycles was increased. D: mRNA expression of Spata22, Spo11, Msh4, and Gapdh in testes of wild-type rats between the ages of 4 and 37 days and in testes of adult wild-type rats. Spata22 expression begins to increase between the ages of 8 days and 14 days, as shown for Spo11 and Msh4.
High expression of Spata22 that increased with the first meiotic wave
As shown in a previous gene expression analysis [15], RT-PCR analysis revealed that the Spata22 ortholog was predominantly expressed in the testis tissue (Fig. 3C). The expression levels of mRNA and protein of Spata22 in mouse testes began to increase with the first wave of spermatogenesis [15]. In laboratory rats, the first meiotic wave begins at 13–14 days after birth [18]. RT-PCR analysis indicated that ortholog expression began to increase between 8 and 14 days after birth, as seen in two other meiosis-related genes, Spo11 [3, 22] and Msh4 [14] (Fig. 3D). These results suggest that rat Spata22 may be involved in the regulation of meiosis I and that the function of this gene may be conserved between the two rodent species: the rat and mouse.
Spata22 overexpression recovered the reproductive phenotype in homozygous mutants
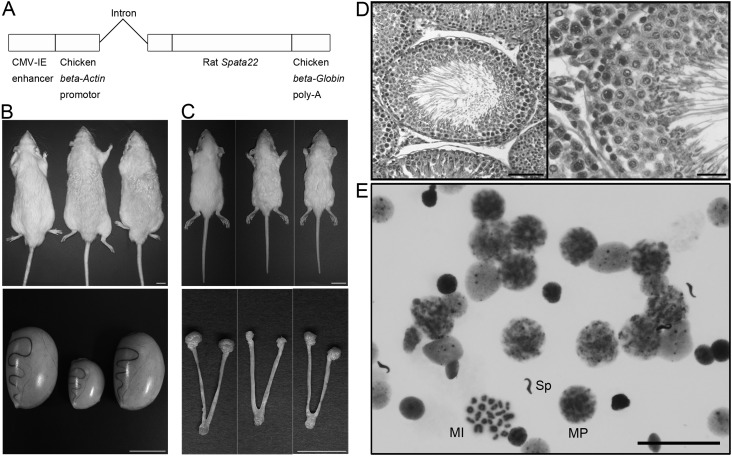
To verify that Spata22 underlies the mutant reproductive phenotype, we performed transgenic rescue experiments with the minigene (Fig. 4A). All homozygous mutant rats (tm/tm) carrying the transgenes at the fifth generation of backcross (=N5) were rescued from sterility: one male and two females in one of the two transgenic lines (line 2) and one male in the other transgenic line (line 4), although they were not rescued from the anomalies in hair, whiskers (Figs. 4B and 4C), and the CNS or from body tremor (data not shown). Spata22 overexpression induced no apparent changes in developmental, morphological, and behavioral phenotypes in homozygous mutants, except for those in the reproductive phenotype. Testes and ovaries of the transgenic rats showed normal development upon gross examination (Figs. 4B and 4C). Histological analysis consistently showed that spermatogenesis proceeded normally in the rescued transgenic rats (Fig. 4D). Cytogenetic examination of meiotic progression also confirmed that meiosis was recovered in homozygous mutant males carrying the transgenes (Fig. 4E). These results indicate that Spata22 is the causative gene for infertility and abnormal gametogenesis in the homozygous mutant rat.
Fig. 4.
Recovery of spermatogenesis by transgenic rescue. A: Transgenic vector constructed by insertion of full-length rat Spata22 cDNA into pCAGGS. B and C: Gross examination of 7-month-old testis (B) and 21-day-old ovary (C) in mutants carrying the transgenes. Upper panels: dorsal views of animals. Lower panels: gross morphologies of gonads, oviducts, and uteri. In each panel, the genotypes of the animals and the organs are as follows: tm/+ (left), tm/tm (middle), and tm/tm Tg/+ (right). The homozygous mutant testis was small, and the homozygous mutant ovary could not be found despite the presence of oviducts and uteri. The testis and the ovary of the tm/tm Tg/+ rats appeared to develop normally, whereas the wavy coat phenotype of the tm/tm Tg/+ rats was similar to that of the tm/tm littermates in both sexes. Scale bar: 2 cm in upper panels, 1 cm in lower panels. D: Hematoxylin and eosin (HE)-stained section of testes from adult homozygous mutants that carry the transgenes, showing normal morphology of the seminiferous epithelium. Scale bar: 100 µm and 25 µm for the low- and high-magnification images, respectively. E: Giemsa-stained preparation of testicular cells, cell nuclei, or chromosomes from adult homozygous mutants carrying the transgenes, showing normal progression of meiosis. Spermatocyte nuclei at midpachytene and metaphase I stages and a spermatozoon head are indicated by MP, MI, and Sp, respectively. Scale bar: 50 µm.
Discussion
In the present study, we have shown that infertility with abnormal gonadal development was associated with the arrest of meiosis I in the tm homozygous rat. Meiotic cell division was blocked during the zygotene or pachytene stage of prophase I in the tm homozygous male rat, similar to the findings in mutant mice and rats for genes related to chromosome synapsis, meiotic recombination, and cell cycle control [2, 8, 10, 21] as well as the repro42 homozygous mouse, which carries a nonsense mutation in Spata22 [15]. Analysis of progression of meiosis in the mutant ovary will be the subject of future study. We revealed that an approximately 240-bp genomic region comprising at least 13 genes, including Spata22, which is known to be a candidate gene for human aspermia or oligospermia and an essential gene for the progression of meiosis I, was lost in the tm genome. Using a transgenic complementation test, we showed that Spata22 is responsible for infertility and abnormal gametogenesis.
A previous study on the repro42 mutant mouse has shown that testicular weights of mutant males are markedly reduced; the average testis weight of repro42 homozygous adults is approximately 20% of that of wild type littermates [15]. Histological analysis of adult repro42 mutant testes showed complete lack of spermatids and spermatozoa despite presence of spermatogonia and spermatocytes [15]. Histological analysis of 10 dpp repro42 mutant ovaries revealed an almost complete depletion of oocytes and presence of degenerating or degenerated oocyte-containing follicles [15]. These findings are quite similar to our present findings concerning the gonadal phenotype of tm mutant rats. In most patients with Canavan disease, point mutations in ASPA are causative [5]. Deletion mutations comprising ASPA and adjacent genes, such as the tm mutation, have been reported in a few cases [5, 27, 30], and gonadal phenotypes of Canavan patients carrying such mutations have not been reported. The similarity in gonadal phenotype between repro42 mutant mice and tm mutant rats suggests that genomic deletion involving lack of SPATA22 in humans may induce gametogenic failure as seen in these mutants.
Concerning the 12 genes other than Spata22 in the deleted region, targeted disruption of Trpv1, Trpv3, and Aspa has no apparent effect on the reproductive system in mice [6, 19, 26], while Trpv3- and Aspa-deficient mice exhibit an abnormal coat and skin [7] and spongiform degeneration in the CNS [26], respectively. Whether the loss of Shpk [28], seven OR genes [25], and a function-unknown gene, LOC100359760, affects the reproductive system has not been known. Although the functions of the seven OR genes are unknown, olfactory receptors are known to be responsible for the recognition and G protein-mediated transduction of odorant signals in cells, such as olfactory receptor neurons and sperms [25]. Therefore, it is not likely that loss of the seven OR genes affects spermatogenesis seriously. In this study, the transgenic rescue experiment showed that deficiencies in Trpv1, Trpv3, and Aspa, partial deletion of Shpk, loss of the other eight genes, and genomic deletion at the tm locus per se had no apparent effect on meiosis. These findings, taken together with a finding that the meiotic phenotype in the tm homozygous rat is similar to a meiotic phenotype caused by only a single nonsense mutation induced in Spata22 in the repro42 homozygous mouse [15], suggest that the deleted genes, except for Spata22, may have no effect or minor effects on the meiotic arrest phenotype caused by the loss of Spata22.
As shown in a previous study on mouse Spata22 [15], the rat Spata22 ortholog was highly expressed in the testis, and its expression level began to be enhanced in conjunction with the initiation of meiosis. Together with the meiotic phenotype in the mutant rat, these results suggested that, like mouse Spata22, rat Spata22 has an important role in the regulation of meiotic prophase I [15]. Despite its importance in the progression of meiosis through prophase I, the molecular function of Spata22 has been poorly understood. The Spata22 allele of the tm genome is a null mutation resulting from complete removal of the gene from the genome. Therefore, the tremor rat (tm/tm) could contribute to further functional analysis of Spata22, as a null mutant, in conjunction with the transgenic rescued mutant.
This is the first report of a deletion mutation of Spata22, which leads to infertility and abnormal gametogenesis. However, in some patients with Canavan disease, which is caused by ASPA deficiency, large genomic deletions involving loss of several genes, including ASPA, were found at Chr 17p13 [5, 27, 30]. In two types of such deletions, approximately 190 kb and 439 kb in size, SPATA22 is located in the deleted regions and seems likely to be removed from the genome with ASPA, although the gonadal phenotypes in the patients were not reported. The sequences around ends of the deleted region in the tm genome have no significant homology with each other, suggesting that the genomic loss may be caused by a nonhomologous recombination event, such as a 190-kb-sized deletion in human Chr 17p13 [27]. The genomic region around the SPATA22 locus comprises no known chromosomal fragile site [17]. Although it is unclear whether these large genomic deletions found at nearly the same map position in Canavan patients and the tremor rat are caused by a common mechanism, similar deletions could also occur in other mammalian species as well as humans and rats. Therefore, the tremor rat could be a useful animal model for studying diseases resulting from such deletions around the Spata22 locus.
Acknowledgments
We thank Ri-ichi Takahashi and Masatsugu Ueda for generation of transgenic rats and Yoshiyuki Deguchi for help with animal breeding. This work was partially supported by a grant from the Uehara Memorial Foundation.
References
- 1.Aston K.I., Krausz C., Laface I., Ruiz-Castané E., Carrell D.T.2010. Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum. Reprod. 25: 1383–1397. doi: 10.1093/humrep/deq081 [DOI] [PubMed] [Google Scholar]
- 2.Bannister L.A., Schimenti J.C.2004. Homologous recombinational repair proteins in mouse meiosis. Cytogenet. Genome Res. 107: 191–200. doi: 10.1159/000080597 [DOI] [PubMed] [Google Scholar]
- 3.Baudat F., Manova K., Yuen J.P., Jasin M., Keeney S.2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6: 989–998. doi: 10.1016/S1097-2765(00)00098-8 [DOI] [PubMed] [Google Scholar]
- 4.Buchold G.M.2012. Meiotic genetics moves forward with SPATA22 (repro42). Biol. Reprod. 86: 42. doi: 10.1095/biolreprod.111.097436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliebe A., Vater I., Plendl H., Gesk S., Siebert R., Cremer F.W., Klein-Hitpass L.2010. A 439 kb-sized homozygous deletion in 17p13.3 leading to biallelic loss of the ASPA as cause of Canavan disease detected by SNP-array analysis. Mol. Genet. Metab. 99: 184–185. doi: 10.1016/j.ymgme.2009.10.011 [DOI] [PubMed] [Google Scholar]
- 6.Caterina M.J., Leffler A., Malmberg A.B., Martin W.J., Trafton J., Petersen-Zeitz K.R., Koltzenburg M., Basbaum A.I., Julius D.2000. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288: 306–313. doi: 10.1126/science.288.5464.306 [DOI] [PubMed] [Google Scholar]
- 7.Cheng X., Jin J., Hu L., Shen D., Dong X.P., Samie M.A., Knoff J., Eisinger B., Liu M.L., Huang S.M., Caterina M.J., Dempsey P., Michael L.E., Dlugosz A.A., Andrews N.C., Clapham D.E., Xu H.2010. TRP channel regulates EGFR signaling in hair morphogenesis and skin barrier formation. Cell 141: 331–343. doi: 10.1016/j.cell.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen P.E., Pollack S.E., Pollard J.W.2006. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr. Rev. 27: 398–426. doi: 10.1210/er.2005-0017 [DOI] [PubMed] [Google Scholar]
- 9.de Kretser D.M., Baker H.W.G.1999. Infertility in men: recent advances and continuing controversies. J. Clin. Endocrinol. Metab. 84: 3443–3450. doi: 10.1210/jc.84.10.3443 [DOI] [PubMed] [Google Scholar]
- 10.de Rooij D.G., de Boer P.2003. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet. Genome Res. 103: 267–276. doi: 10.1159/000076812 [DOI] [PubMed] [Google Scholar]
- 11.Huang X., Li J., Lu L., Xu M., Xiao J., Yin L., Zhu H., Zhou Z., Sha J.2005. Novel development-related alternative splices in human testis identified by cDNA microarrays. J. Androl. 26: 189–196. [DOI] [PubMed] [Google Scholar]
- 12.Imai H.T., Matsuda Y., Shiroishi T., Moriwaki K.1981. High frequency of X-Y chromosome dissociation in primary spermatocytes of F1 hybrids between Japanese wild mice (Mus musculus molossinus) and inbred laboratory mice. Cytogenet. Cell Genet. 29: 166–175. doi: 10.1159/000131565 [DOI] [PubMed] [Google Scholar]
- 13.Kitada K., Akimitsu T., Shigematsu Y., Kondo A., Maihara T., Yokoi N., Kuramoto T., Sasa M., Serikawa T.2000. Accumulation of N-acetyl-L-aspartate in the brain of the tremor rat, a mutant exhibiting absence-like seizure and spongiform degeneration in the central nervous system. J. Neurochem. 74: 2512–2519. doi: 10.1046/j.1471-4159.2000.0742512.x [DOI] [PubMed] [Google Scholar]
- 14.Kneitz B., Cohen P.E., Avdievich E., Zhu L., Kane M.F., Hou H., Jr, Kolodner R.D., Kucherlapati R., Pollard J.W., Edelmann W.2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14: 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 15.La Salle S., Palmer K., O’Brien M., Schimenti J.C., Eppig J., Handel M.A.2012. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 86: 45. doi: 10.1095/biolreprod.111.095752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilford R., Jones A.M., Bishop D.T., Thornton J., Mueller R.1994. Case-control study of whether subfertility in men is familial. BMJ 309: 570–573. doi: 10.1136/bmj.309.6954.570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukusa T., Fryns J.P.2008. Human chromosome fragility. Biochim Biophys Acta 1779: 3–16. doi: 10.1016/j.bbagrm.2007.10.005 [DOI] [PubMed] [Google Scholar]
- 18.Malkov M., Fisher Y., Don J.1998. Developmental schedule of the postnatal rat testis determined by flow cytometry. Biol. Reprod. 59: 84–92. doi: 10.1095/biolreprod59.1.84 [DOI] [PubMed] [Google Scholar]
- 19.Moqrich A., Hwang S.W., Earley T.J., Petrus M.J., Murray A.N., Spencer K.S.R., Andahazy M., Story G.M., Patapoutian A.2005. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science 307: 1468–1472. doi: 10.1126/science.1108609 [DOI] [PubMed] [Google Scholar]
- 20.Roeder G.S.1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. doi: 10.1101/gad.11.20.2600 [DOI] [PubMed] [Google Scholar]
- 21.Roeder G.S., Bailis J.M.2000. The pachytene checkpoint. Trends. Genet. 16: 395–403. doi: 10.1016/S0168-9525(00)02080-1 [DOI] [PubMed] [Google Scholar]
- 22.Romanienko P.J., Camerini-Otero R.D.2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975–987. doi: 10.1016/S1097-2765(00)00097-6 [DOI] [PubMed] [Google Scholar]
- 23.Serikawa T., Ohno Y., Sasa M., Yamada J., Takaori S.1987. A new model of petit mal epilepsy: spontaneous spike and wave discharges in tremor rats. Lab. Anim. 21: 68–71. doi: 10.1258/002367787780740635 [DOI] [PubMed] [Google Scholar]
- 24.Sha J., Zhou Z., Li J., Yin L., Yang H., Hu G., Luo M., Chan H.C., Zhou K.2002. Identification of testis development and spermatogenesis-related genes in human and mouse testes using cDNA arrays. Mol. Hum. Reprod. 8: 511–517. doi: 10.1093/molehr/8.6.511 [DOI] [PubMed] [Google Scholar]
- 25.Spehr M., Munger S.D.2009. Olfactory receptors: G protein-coupled receptors and beyond. J. Neurochem. 109: 1570–1583. doi: 10.1111/j.1471-4159.2009.06085.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surendran S., Szucs S., Tyring S.K., Matalon R.2005. Aspartoacylase gene knockout in the mouse: impact on reproduction. Reprod. Toxicol. 20: 281–283. doi: 10.1016/j.reprotox.2005.02.001 [DOI] [PubMed] [Google Scholar]
- 27.Tahmaz F.E., Sam S., Hoganson G.E., Quan F.2001. A partial deletion of the aspartoacylase gene is the cause of Canavan disease in a family from Mexico. J. Med. Genet. 38: E9. doi: 10.1136/jmg.38.3.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wamelink M.M.C., Struys E.A., Jansen E.E.W., Levtchenko E.N., Zijlstra F.S.M., Engelke U., Blom H.J., Jakobs C., Wevers R.A.2008. Sedoheptulokinase deficiency due to a 57-kb deletion in cystinosis patients causes urinary accumulation of sedoheptulose: elucidation of the CARKL gene. Hum. Mutat. 29: 532–536. doi: 10.1002/humu.20685 [DOI] [PubMed] [Google Scholar]
- 29.Yamada J., Serikawa T., Ishiko J., Inui T., Takada H., Kawai Y., Okaniwa A.1985. Rats with congenital tremor and curled whiskers and hair. Jikken Dobutsu 34: 183–188. [DOI] [PubMed] [Google Scholar]
- 30.Zeng B.J., Wang Z.H., Torres P.A., Pastores G.M., Leone P., Raghavan S.S., Kolodny E.H.2006. Rapid detection of three large novel deletions of the aspartoacylase gene in non-Jewish patients with Canavan disease. Mol. Genet. Metab. 89: 156–163. doi: 10.1016/j.ymgme.2006.05.014 [DOI] [PubMed] [Google Scholar]