Abstract
Purpose
The microRNA-34b/c (miR-34b/c) has been considered a tumor suppressor in different tumor types and it is a known transcriptional target of the tumor suppressor gene TP53. The main objectives of this study were to investigate the clinical implications of miR-34b/c methylation in early stage lung adenocarcinoma (AC) patients and to determine the functional role of miR-34b/c re-expression in lung AC cell lines.
Experimental Design
Aberrant methylation and expression of miR-34b/c were assessed in 15 lung AC cell lines and a cohort of 140 early stage lung AC. Lung AC cell lines were transfected with miR-34b/c and the effects upon cell proliferation, migration, invasion and apoptosis were investigated.
Results
Aberrant methylation of miR-34b/c was detected in 6 (40%) of 15 lung AC cell lines and 64 out of 140 (46%) primary lung adenocarcinomas. Expression of miR-34b/c was significantly reduced in all methylated cell lines and primary tumors, especially in those harboring a TP53 mutation. Patients with high levels of miR-34b/c methylation had significantly shorter disease-free survival and overall survival as compared to patients with unmethylated miR-34b/c or low level of miR-34b/c methylation. Ectopic expression of miR-34b/c in lung AC cell lines decreased cell proliferation, migration and invasion.
Conclusions
Epigenetic inactivation of miR-34b/c by DNA methylation has independent prognostic value in early stage lung AC patients with surgically resected tumors. Re-expression of miR-34b/c leads to a less aggressive phenotype in lung AC cell lines.
Keywords: microRNA, DNA methylation, microRNA-34b/c, lung adenocarcinoma, TP53
Introduction
Lung cancer is the second most common cancer in both men and women and is the leading cause of cancer death for both sexes in industrialized countries (1, 2). Non-small cell lung cancer is a heterogeneous disease, with the most common subtypes being adenocarcinomas and squamous cell carcinomas (3). These histological subtypes not only have diverse clinical outcomes, but also encompass different treatments, revealing heterogeneity in both disease aggressiveness and underlying molecular alterations (4). Significant advances in lung adenocarcinoma (AC) biology have provided biomarkers that predict sensitivity to specific compounds such as erlotinib (EGFR mutations) and crizotinib (ALK rearrangements).
In lung cancer, complete resection gives the highest probability of long-term remission and even cure, but only about 15% of patients are candidates for surgical treatment at the time of diagnosis. Furthermore, even among early stage patients treated by surgery with curative intent, the 5-year survival rate is only 52% (2). Postoperative adjuvant chemotherapy to improve survival has been extensively studied and demonstrated an absolute benefit of 4–5% in 5-year survival rates (5, 6). Adjuvant chemotherapy is the standard of care for patients with resected stage II and III. However it is clear that a subset of stage I patients also have poor prognosis and it would therefore be very relevant to identify these high-risk patients that might benefit from an additional therapeutic intervention.
The application of molecular biomarkers that incorporate with traditional clinicopathological factors might improve the management of patients with NSCLC (7). Genetic alterations such as KRAS and TP53 mutations (8, 9), and gene-expression signatures have been identified for classifying surgically-resected patients with different outcomes (10). Other promising biomarkers include miRs since they are upstream regulators of gene-expression and can play a pathogenic role in the disease process (11). MiRs are small non-coding regulatory RNAs that contribute to cancer development and progression by acting as oncogenes or tumor suppressor genes (12, 13) and might be involved in the regulation of biological processes such as cell proliferation, apoptosis, adhesion, migration, invasion and angiogenesis.
MiR-34b and miR-34c, two members of the miR-34 family, are encoded by a bicistronic transcript from chromosome 11q23 (14). The promoter regions of both miRs contain a palindromic sequence that matches the canonical p53-binding site and its expression may be induced by TP53 in response to DNA damage or cell stress (15). Another important regulatory mechanism of miR-34 expression appears to be aberrant DNA methylation. Indeed, the promoter regions of miR-34b/c contain a CpG island that has been reported hypermethylated in several tumor types and resulted in silencing of miR-34b/c expression (16–21). MiR-34b/c methylation was prognostic in NSCLC stage I patients (22), but this result has not been validated in an independent cohort. In addition, miR-34 family acts as a tumor-suppressor among different tumor types, inducing a less aggressive phenotype (16, 20, 23, 24), however the functional role of miR-34b/c has not been specifically studied in lung AC.
In the present study, we sought to determine the role of miR-34b/c methylation and expression in lung AC cell lines and primary tumors, the relationship to patient prognosis in two independent cohorts of early stage resected lung adenocarcinoma and the functional impact of miR-34b/c ectopic expression on lung AC invasion and proliferation.
Material and Methods
Tissue samples
Frozen primary tumors and corresponding nonmalignant lung tissue samples of 140 patients with stage I-II lung ACs who underwent surgical resection with curative intention were collected at two institutions: the Bellvitge Hospital in Barcelona (2001–2007) and the University of Michigan Health System in Ann Arbor (1991–2007). The informed consent, approved by the respective Institutional Review Board, was obtained and specimens were collected and immediately frozen following resection and stored at −80°C. The percentage of tumor purity in sections adjacent to regions used for DNA and/or RNA isolation was assessed as well as examined for routine histopathological analysis. Regions containing a minimum of 70% of tumor cellularity were used for nucleic acid isolation. None of the patients included in this study received preoperative radiation or chemotherapy. Clinical data was retrospectively collected by checking the medical records and all cases were staged according to the revised 7th TNM classification criteria. Patient clinicopathological characteristics are provided in Supplementary Table S1. The patients operated at the University of Michigan were older and this set included more females and former-smokers than the Bellvitge Hospital set. These differences reflected specific patterns of lung AC according to patient’s site (25). Seventy-five (55%) patients died and sixty-five (47.5%) developed a recurrence at the time of the last follow-up report. The median follow-up time was 6.45 years among the patients that remained alive.
Lung cancer cell lines
Fifteen human lung AC cell lines (SK-LU-1, NCI-H2228, NCI-H1838, NCI-H1563, NCI-H2347, NCI-H1395, Calu-3, A549, NCI-H2087, NCI-H1299, NCI-H838, NCI-H23, NCI-H1792, HCC4006 and HCC827) were purchased from American Type Culture Collection (Manassas, VA). All cells were maintained in RPMI-1640 (except Calu-3 and A549, which were maintained in Eagle’s minimum essential medium and DMEM medium respectively), and supplemented with 10% FBS, 1% Gibco® Antibiotic-Antimycotic (Life Technologies) in a humid atmosphere containing 5% CO2 at 37°C.The mutational status of key genes from the cell lines was obtained from the IARC TP53 database (26) and from Sanger Institute Catalogue of Somatic Mutations In Cancer web site (http://www.sanger.ac.uk/cosmic) (27). Nine cell lines included in this study harbor TP53 mutations or TP53 deletion and are listed in Supplementary Table S2. The presence of loss of heterozygosity (LOH) at the miRNA-34b/c loci was determined by the CONAN copy number analysis (http://www.sanger.ac.uk/cgi-in/genetics/CGP/conan/search.cgi).
Chemicals and demethylation treatment of lung AC cell lines
Original stock solutions of cis-diammine-dichloroplatinum (cisplatin; Sigma-Aldrich), pemetrexed (Lilly), erlotinib hydrochloride (Selleckchem) and 5-aza-2’-desoxycytidine (5-aza-dC, Sigma-Aldrich) at a concentration of 1mM, 1mM, 10nM and 25mM, respectively, were stored at −20°C and freshly dissolved in culture medium before use. SK-LU-1, NCI-H2228, NCI-H1838, NCI-H23 and HCC4006 cells were seeded in 6 well plates, cultured for 24 hours, and then treated with 0.5 µmol/L of 5-aza-dC for 5 days in triplicates, replacing drug-containing medium daily, before DNA and RNA isolation for direct sequencing and miR-34b/c quantification respectively.
Bisulfite genome sequencing (BGS)
DNA was isolated using phenol-chloroform method. Bisulfite conversion was carried out using 1 µg of genomic DNA using an EZ-DNA Methylation Gold kit (Zymo Research). The PCR was performed using Immolase DNA Polymerase (Bioline) and the following oligonucleotides: 5’-GGTTGGGAATTGAAGTTTG-3’ (F) and 5’-TTAATAATTATAACCACCACAATACAA-3’ (R). The reactions were cycled at 95°C for 10 minutes, then 30 cycles of 30 seconds (sec) at 95°C, 30 sec at 58°C and 30 sec at 72°C, including a final extension step for 15 minutes. PCR products were gel purified using QIAquick Gel Extraction kit (Qiagen) and cloned into pCR®4-TOPO® Vector (Invitrogen) using the TOPO® TA Cloning® kit (Invitrogen). Five individual clones were sequenced using M13 primers at the University of Michigan DNA Sequencing Core. The region assessed by BGS included 45 CpG sites from the miR-34b/c promoter and average methylation from individual clones was calculated as a percentage of the number of methylated CpG sites over the number of total CpG sites sequenced.
Real-time PCR temperature dissociation (melting curve analysis, MCA)
Genomic DNA treated with SssI methylase (New England Biolabs) was used as positive control to amplify fully methylated DNA for the dissociation curve. Whole genome amplified DNA obtained by REPLI-G kit (Qiagen) was used as a negative control to amplify fully unmethylated DNA for dissociation curve. Methylation standards (100%, 75%, 50%, 25%, 10%, 5%, 1% and 0%) were prepared by mixing the positive and negative controls accordingly. All primers used for BGS and MCA were designed using the Methyl Primer Express v1.0 and did not target CpG dinucleotides in order to specifically amplify the bisulfite-modified sequences. Melting curve analysis was carried out as described before (28). Changes in cytosine-guanine (CG) content after bisulfite treatment resulted in differences in melting temperature (methylated products have a higher melting temperature). Bisulfite converted DNA was first amplified in a 20 cycle external PCR reaction using the primers and conditions as described for BGS. One µl of amplified DNA was used as a template for a nested PCR using a LightCycler 480 II (Roche Applied Science) in the presence of LightCycler 480 SYBR Green I Master (Roche Applied Science) and the following oligonucleotides: 5’-GTATTTTTGGGGGTTATGG-3’ (F) and 5’-TCAACTAATAACACTACCTACAAACC-3’ (R). The amplified DNA fragment encompassed 37 CpGs which overlap with the 45 CpG sites assessed by BGS. The reactions were cycled for 30 cycles of 10 sec at 95°C, annealing at 67°C for 20 sec and extension at 72°C for 15 sec. After completing the amplification, temperature was gradually increased from 65°C to 95°C to obtain the melting curves. Lightcycler480 software (Roche Applied Science) plotted the melting peaks by calculating the negative derivative of fluorescence over temperature and quantified the area under the curve (AUC) for the melting peak corresponding to the unmethylated and methylated alleles. AUC values for the melting peak corresponding to the methylated alleles were highly correlated with the percentage of methylated DNA of the methylation standards, as it is shown in Supplementary Fig. S1. In addition, the percentage of DNA methylation was estimated based upon the linear regression between the AUC values and the methylation standards. Samples were classified as methylated when the estimated methylation value was higher than 5%, similarly as it has been done when using methods that are able to quantify DNA methylation such as pyrosequencing or DNA methylation arrays.
RNA isolation and miR and mRNA quantification in cell lines and tumors
Total RNA was isolated from tissue samples and cell lines followed by column purification using miRNeasy kit (Qiagen) in accordance with the manufacturer's instructions. Expression of mature miR-34b and miR-34c was assessed by real-time RT-PCR (qRT-PCR) using TaqMan Micro-RNA assays (Applied Biosystems) and the 7900 HT-Fast Real-Time PCR System (Applied Biosystems). Gene expression of reported miR-34b/c targets (AXL, BCL2, HMGA2, MET, NOTCH1 and NOTCH2) was assessed by qRT-PCR in miR-34b/c transfected cells (14, 29). cDNA was prepared from total RNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. The qRT-PCR was carried out using Power SYBR Green (Applied Biosystems) at the 7900 HT-Fast Real-Time PCR System (Applied Biosystems). Primers, designed using Primer-BLAST (30), are shown in Supplementary Table S3. Data were analyzed using the SDS 2.2.2 software (Applied Biosystems) and setting a threshold of 0.2 and a manual baseline from 3 to 18 cycles. Relative quantification was performed by using the 2-ΔΔCt method, using endogenous RNU48 and β-Actin expression as controls for miR and mRNA quantification, respectively.
Plasmid construction and stable transfection
The miR-34b/c was subcloned into pSilencer 4.1-CMV puro Expression Vector (Ambion) containing the flanking regions of the mature miR-34b/c (16). Ten µg of the constructed plasmid and the empty vector were introduced into H1838 and SK-LU-1cells using FUGENE 6 Transfection Reagent (Promega). Forty eight hours post-transfection, cells were cultured in selection media with puromycin (Sigma-Aldrich). Resistant clones were selected for further cell culture and experiments.
Western blot
Cells were grown to 60% confluence and harvested in RIPA buffer supplemented with proteinase inhibitor to extract protein. A total of 10–20 µg of protein were separated by SDS-PAGE and transferred to PVDF membranes. The proteins on membranes were incubated overnight at 4°C with the primary antibodies. We selected MET and BCL2 that had been reported as primary targets of miR-34b/c. The membranes were incubated with the following primary antibodies: anti-MET (25H2, Cell Signaling), anti-PARP antibody (Cell Signaling), anti-BCL2 (clone 10, Millipore) and β-actin (AC-15, Abcam). Next, the membranes were incubated with goat anti-rabbit (Cell signaling) or anti-mouse (Southern Biotech) IgG-conjugated horseradish peroxidase (HRP) and detected by chemiluminescence using Amersham ECL Prime Western Blotting Detection Reagents (GE Healthcare Life Sciences).
Cell proliferation assay
Both empty vector and miR-34b/c stably transfected H1838 and SK-LU-1 cells were plated in 96-well plates for 24 hours. Chemosensitivity was tested by treating miR-34b/c stably transfected cells with cisplatin, pemetrexed and erlotinib for 48 hours or with drug-free medium. The cell proliferation and viability was assessed using 10µl/well of WST-1 reagent (Roche). The absorbance at 450 nm and reference at 630 nm were measured using an automated plate reader (ELx808 Bio-Tek) at different time-points, in accordance with the manufacturer’s protocol. Relative proliferation rates were calculated as a percentage of the initial T0 reading within each sub-cell line.
Apoptosis assay
Both empty vector and miR-34b/c stably transfected SK-LU-1 cells were plated in 60 mm plates and then treated for 48 hours with 5 µmol/L of cisplatin. After drug treatment, cells were harvested and processed for Western Blot.
Wound-healing assay
Empty vector and miR-34b/c stably transfected cells were grown to confluence and a wound was made through the monolayer cells using a p20 tip. Accurate measures of the wounds were taken during the time course to calculate the migration rate according to the equation: percentage wound healing = ((wound length at 0 h) − (wound length at 4, 6, 20 or 27 h)) / (wound length at 0 h)×100. Two independent experiments were performed.
Trans-well invasion assay
Cancer cells were re-suspended in media without growth factors. Tumor cells were then seeded at 25,000 cells per well into matrigel-coated (BD Matrigel), growth-factor-reduced, invasion chambers (8µm pore size, BD Biosciences). The bottom chamber was prepared with 20% FBS media as the chemo-attractant. The cell invasion chambers were incubated overnight in a humidified incubator at 37°C, 5% CO2 atmosphere. The top non-invading cells were removed with a cotton swab moistened with medium and the lower surface of the membrane was stained with Diff-Quick Stain Set (Siemens). The number of cells that had migrated to the basal side of the membrane was visualized with an Olympus microscope at 20× magnification. Pictures of five random fields from replicate wells were obtained and the number of cells stained was quantified relative to the migration of cells through the uncoated membrane.
Statistical analysis
Differences between both cohorts used in the methylation analysis were calculated with Student’s t-test and Fisher’s exact tests. Non-parametric tests (Mann-Whitney and Kruskal-Wallis) were used to identify statistically differences in miR-34b/c expression or methylation between different clinical variables. Survival curves were plot using the Kaplan-Meier method and survival differences were assessed using the log-rank test. The univariate and multivariate Cox proportional hazards model using dichotomous and continuous value of AUC DNA methylation were used to assess survival results. Disease-free survival was measured from the date of surgery to the time of recurrence, death or censoring. Overall survival was measured from date of surgery to the time of death or censoring.
Results
MiR-34b/c methylation and underexpression are a frequent alteration in lung AC cell lines
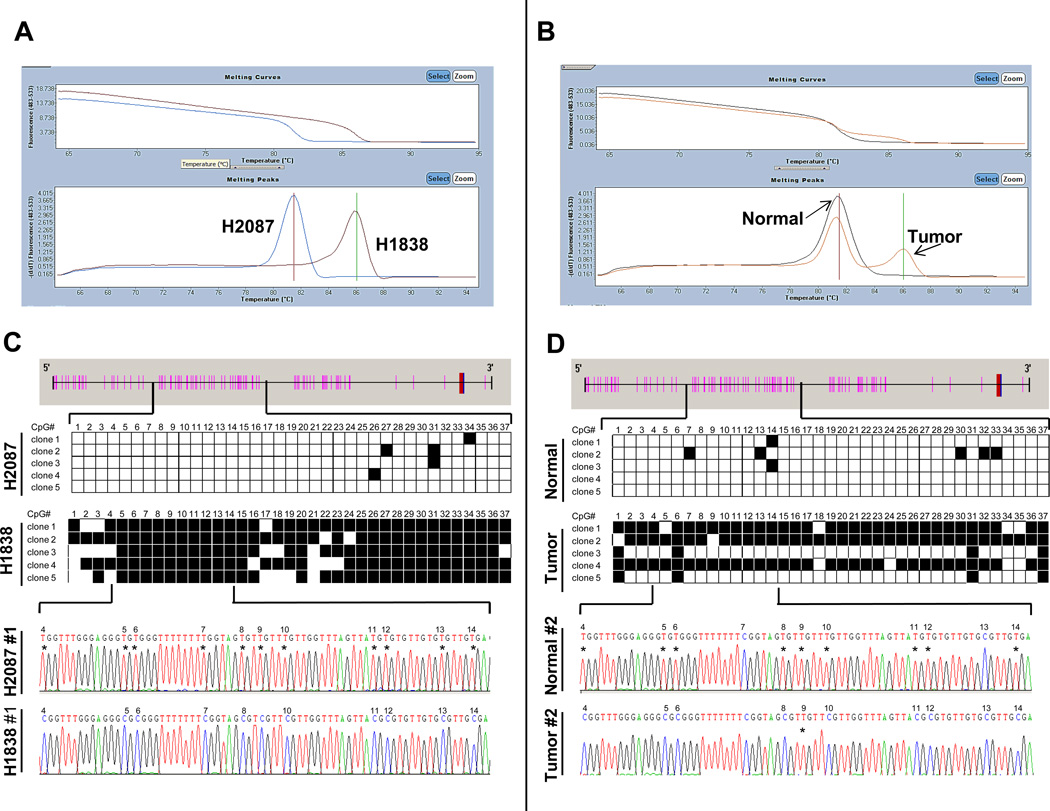
We used MCA to assess the methylation status of miR-34b/c in a panel of lung AC cell lines and BGS was performed to confirm these results in selected cell lines and primary tumors. A description of representative examples of MCA is shown in Fig. 1. An excellent concordance was observed between the results obtained by both methods. Based upon BGS, NCI-H2087 and NCI-H1838 cells had 3% and 78% of 45 CpG sites methylated respectively whereas the estimated percentage of methylation by MCA was 0.5% and 77%. The concordance was also optimal for primary tumors as shown in Fig. 1 a tumor assessed by BGS demonstrated 59% methylation of CpG sites, whereas the estimated percentage of methylation by MCA was 53%. We consider that MCA is a suitable alternative for reliable, efficient and quantitative assessment of DNA from both cell lines and clinical samples.
Figure 1.
Methylation status of miR-34b/c in lung AC cell lines and primary tumors. (A and B) Representative examples of qRT-PCR dissociation analysis (melting curve analysis, MCA) for miR-34b/c. Melting curves and derivative peaks from two cell lines (H2087 and H1838) are shown in the left panel (A) and from a lung AC and its corresponding nonmalignant lung tissue in the right panel (B). Curves were separated according to the level of methylation and an outstanding difference in the melting temperature between unmethylated and methylated samples was observed. The melting peak of lung AC tumor showed a bimodal pattern representing the unmethylated alleles that may correspond to the stroma present in the tumoral sample (<30%). (C and D) Results and representative chromatograms from bisulfite genomic sequencing (BGS) of the miR-34b/c promoter. CpG sites are depicted as pink bars and the transcription start sites as blue bars. Five clones of each sample were sequenced and each row represents one sequenced allele. Black and white squares represent methylated and unmethylated cytosines at CpG sites, respectively.
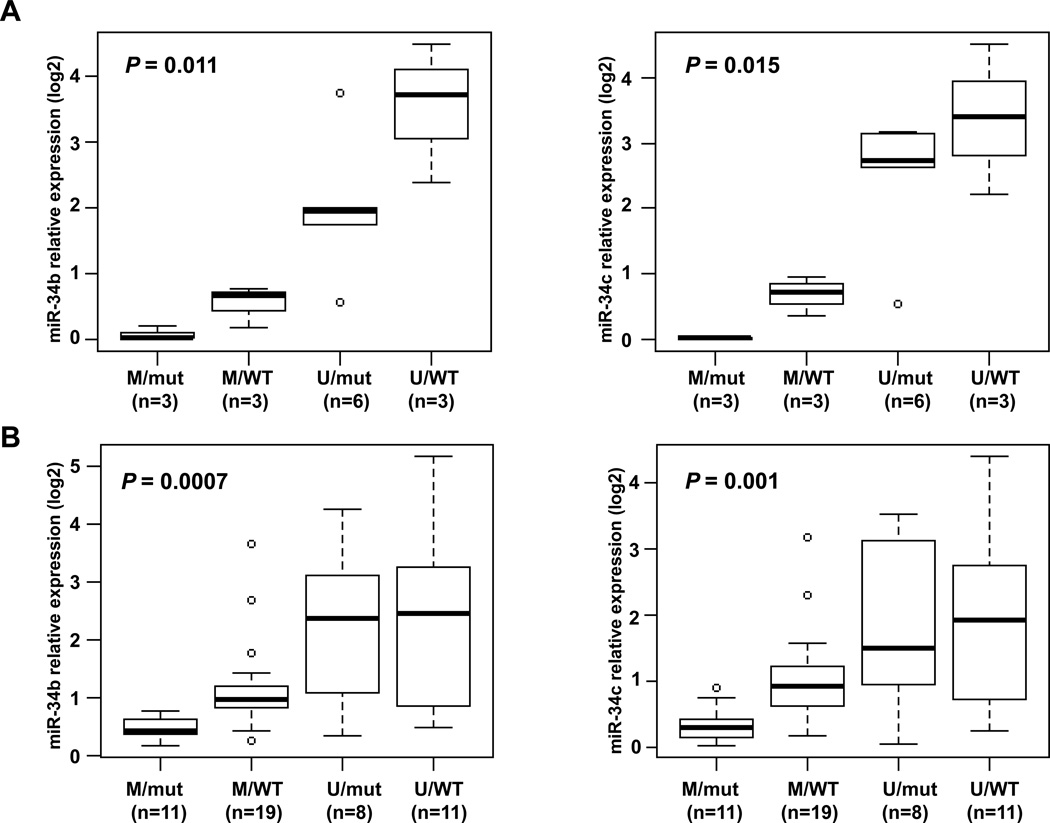
Six of the lung AC cell lines analyzed (40%) had miR-34b/c promoter methylation: H2228, SK-LU-1, H1838, H1563, H2347 and H1395. Expression of miR-34b and miR-34c was lower in cell lines with miR-34b/c methylation as compared with the unmethylated cell lines (P = 0.001 for both miRs). We did not find a significant difference in miR-34b/c expression between cell lines harboring TP53 mutation or those with TP53 wild-type. Interestingly, a significant difference in the expression values for both miRs was detected when the cells were classified based upon the promoter methylation status and their TP53 mutational status (P = 0.011 and 0.015, for miR-34b and miR-34c respectively). As shown in Fig. 2A, the cells with lowest expression demonstrated miR-34b/c promoter methylation and contained a TP53 mutation. LOH at miR-34b/c loci was a frequent event (53% based upon CONAN database) in lung AC cell lines, nevertheless LOH was not associated with lower miR-34b/c expression within this set of cell lines. Remarkably, two cell lines harboring EGFR mutations (HCC827 and HCC4006) had the higher miR-34b and miR-34c expression as compared to EGFR wild-type.
Figure 2.
MiR-34b/c expression in lung AC cell lines (A) and primary tumors (B). The main regulatory mechanisms of miR-34b/c expression appear to be DNA methylation and TP53 status. We observe a strong correlation between DNA methylation, TP53 status and miR-34b/c expression. These boxplots show miR-34b/c expression (log2) in a set of 15 lung AC cell lines (A) and 49 lung AC tumors (B) according to DNA methylation (M, methylated; U, unmethylated) and TP53 status (mut, mutated; WT, wild type). P values correspond to Kruskal-Wallis test among all 4 categories.
The miR-34b/c expression was significantly increased from 29- to >500 fold when cells with hypermethylated miR-34b/c promoter (SK-LU-1, H2228 and H1838) were treated with 5-aza-2’-deoxycytidine (5-aza-dC) for 5 days (Supplementary Fig. S2A). A relative change in the methylation level was detected by direct bisulfite sequencing in cell lines treated with 5-aza-dC (Supplementary Fig S2B). In addition, the miR-34b/c expression was found unchanged when miR-34b/c unmethylated cell lines (H23 and HCC4006) were treated with the DNA demethylating agent.
MiR-34b/c methylation and underexpression are a frequent event in lung AC tumors
MiR-34b/c methylation was assessed by MCA in 140 lung AC tumors and 10 nonmalignant lung tissues. Fifty-nine tumors (42%) showed more than 5% DNA methylation (Supplementary Fig. S3A), whereas only one nonmalignant lung tissue was methylated that was confirmed by bisulfite sequencing. The level of expression of miR-34b and miR-34c were quantified in a subset of 49 early stage lung AC tumors and 10 matched nonmalignant lung samples resected at the University of Michigan. Nonmalignant lung samples had a higher expression of both miRs in non-tumoral samples (median: 3.67 for miR-34b and 2.83 for miR-34c) as compared with lung AC tumors (median: 0.95 for miR-34b and 0.86 for miR-34c; P = 0.002 and 0.003, respectively). Thirty of this subset of tumors (61%) were methylated and has lower expression levels of miR-34b and miR-34c (median: 0.73 and 0.62, respectively) as compared to unmethylated tumors (median: 4.46 and 2.57, P = 0.002 and 0.003, respectively, Supplementary Fig. S3B).
MiR-34b/c methylation/expression and clinicopathological correlations
The level of methylation according to AUC values was correlated with clinical characteristics and survival (Table 1). MiR-34b/c methylation was associated with higher tumor stage among the 140 stage I and II patients examined (P = 0.033), recurrence (P = 0.017) and death (P = 0.019). Tumors from patients with a smoking history had a trend of higher levels of miR-34b/c methylation, but the differences were not statistically significant.
Table 1.
Correlation between miR-34b/c DNA methylation and clinicopathological characteristics (n = 140).
| Clinical covariates | Total | miR-34b/c methylation status | P-value | |
|---|---|---|---|---|
| AUC, mean (±SD) | AUC (mean rank) | |||
| Age | ||||
| <65 years | 60 | 1.82 (±2.72) | 70.69 | ns |
| ≥65 years | 80 | 1.48 (±2.09) | 70.36 | |
| Sex | ||||
| Female | 53 | 1.76 (±2.05) | 76.36 | ns |
| Male | 87 | 1.54 (±2.56) | 66.93 | |
| Smoking history | ||||
| Never | 15 | 1.07 (±1.39) | 64.14 | ns |
| Ever | 123 | 1.65 (±2.41) | 70.15 | |
| Stage | ||||
| Stage I | 96 | 1.35 (±2.21) | 65.73 | 0.033 |
| Stage II | 44 | 2.22 (±2.65) | 80.91 | |
| Recurrence | ||||
| No | 73 | 1.12 (±1.85) | 62.97 | 0.017 |
| Yes | 67 | 2.18 (±2.75) | 78.71 | |
| Recurrence location | ||||
| Loco-regional | 21 | 1.87 (±2.33) | 34.43 | ns |
| Distance | 46 | 2.32 (±2.93) | 33.80 | |
| Life status | ||||
| Alive | 63 | 1.25 (±2.16) | 63.31 | 0.019 |
| Dead | 77 | 2.06 (±2.55) | 78.80 | |
| Event (death or recurrence) | ||||
| No | 68 | 0.93 (±1.64) | 60.17 | 0.002 |
| Yes | 72 | 2.28 (±2.76) | 80.26 | |
AUC: area under the curve.
In the subset of 49 cases where expression levels were analyzed, we determined the correlation between miR-34b/c expression and other clinical and molecular variables (Supplementary Table S4). Interestingly, lower expression of miR-34b was detected in patients who were smokers compared to nonsmokers (P = 0.025). In addition, tumors harboring TP53 mutation had a trend to lower miR-34b and miR-34c expression (P = 0.043 and 0.060, respectively). Consistent with that found in AC cell lines, a significant difference in the expression values of miR-34b/c was observed in lung primary tumors, when tumors were classified based upon the promoter methylation status and the TP53 mutational status (P = 0.0007 and 0.001, respectively, Fig. 2B). MiR-34b/c methylation appears to be the predominant mechanism of regulation of miR-34b/c expression in cooperation with TP53 function. Indeed, tumors with methylated miR-34b/c consistently expressed low levels of miR-34b/c even when TP53 was not mutated. Of note, higher miR-34b/c expression was likewise associated with EGFR mutation (P = 0.007 and 0.013, respectively, Supplementary Table S4). In this set of samples, miR-34b/c expression was not associated with outcome as it was previously reported by others (31).
MiR-34b/c methylation as independent prognostic marker in early stage resected lung AC
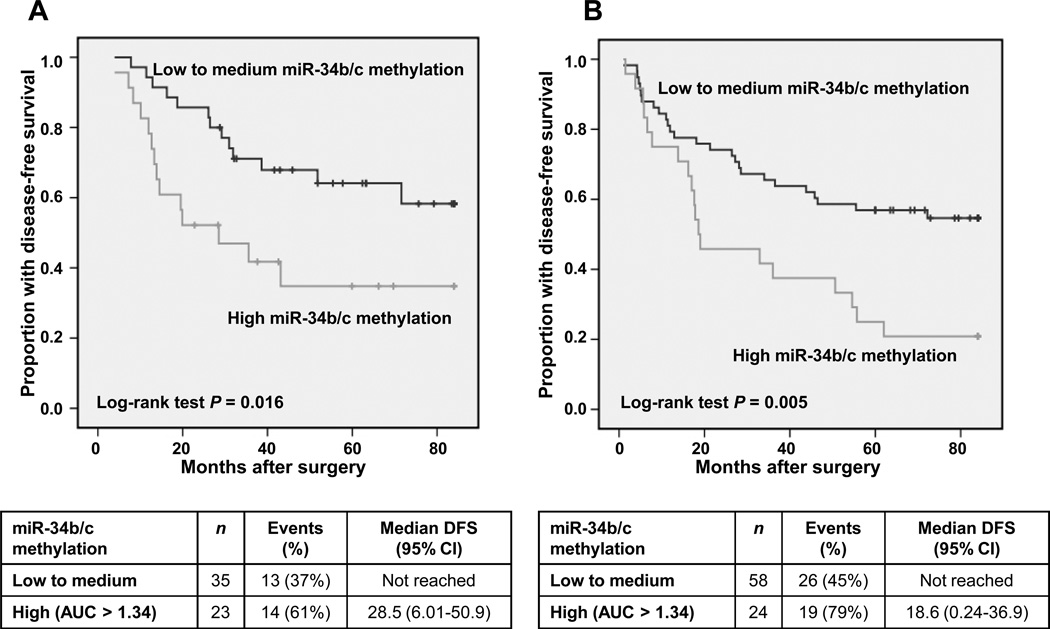
First, we determined the prognostic value of miR-34b/c methylation in the Barcelona set (training set) and we found that patients with higher AUC for miR-34b/c methylation (top third) had a significantly shorter disease-free survival (median: 28.5 months) compared to those patients with low AUC levels (bottom two third) (median: not reached, Log-rank test P = 0.016, Fig. 3A). We used two third cut-off (AUC = 1.34, estimated methylation = 20%) instead of the median, because it provided a higher percentage of events within the group of high levels of DNA methylation. In the multivariate Cox model, higher levels of miR-34b/c methylation (top third) were independently associated with shorter disease-free survival (HR = 3.04, 95% CI 1.26 – 7.03, P = 0.013) as compared to low-medium levels, after adjusting for age, sex and disease stage. Then, we assessed in the University of Michigan set (test set) whether a high AUC miR-34b/c was associated to poorer outcome and we also found that those patients (top third) had a shorter disease-free survival (median: 18.6 months) comparing to patients with low AUC (bottom two third) (median: not reached, Log-rank test P = 0.005, Fig. 3B). In the multivariate analysis, adjusting by the same covariates, high levels of miR-34b/c methylation (top third) were associated with shorter disease-free survival (HR = 2.05, 95% CI 1.09 – 3.84, P = 0.025).
Figure 3.
Prognostic value of miR-34b/c methylation in two independent datasets of early stage lung AC patients. Lung AC tumors were divided according to their source into training (Bellvitge Hospital, A) and test set (University of Michigan, B). Kaplan-Meier plots of DFS according to miR-34b/c methylation level are shown. Patients with tumors showing high methylation of miR-34b/c (top third) had significantly shorter DFS compared with low methylation (bottom two thirds) or unmethylated.
When the survival analysis was carried out using the whole study population (n = 140), miR-34b/c methylation not only was associated with shorter disease-free survival, but also with shorter overall survival (Kaplan-Meier plots are shown in Supplementary Fig. S4). In the multivariate Cox regression analysis, higher levels of miR-34b/c methylation (top third) was independently associated with a shorter disease-free survival (HR = 2.16, 95% CI 1.32 – 3.52, P = 0.002, Table 2) as compared to low to medium levels (bottom two-third), after adjusting for age, sex and disease stage. MiR-34b/c remained as an independent prognostic marker when considered as continuous variable in the Cox regression (Supplementary Table S5). Higher levels of miR-34b/c (top third) were also independently-associated with shorter overall survival (HR = 1.79, 95% CI 1.07 – 3.02, P = 0.027, Supplementary Table S6) as compared to low-medium levels, after adjusting for the same covariates of age, sex and disease stage. Remarkably, methylation of miR-34b/c was also a prognostic marker for stage I patients and those patients with high AUC for miR-34b/c had a shorter disease-free survival (median: 43.1 months) as compared to those with low to medium levels (median: not reached, Log-rank test P = 0.009, Supplementary Fig. S5).
Table 2.
Disease-free survival analysis (Multivariate Cox model) of miR-34b/c methylation in 140 early stage lung AC patients.
| Covariates in the model | Hazard Ratio |
95% confidence interval |
P-value | |
|---|---|---|---|---|
| Age, continuous | 1.02 | 0.994 – 1.05 | ns | |
| Gender | Female | 1.00 | - | |
| Male | 1.17 | 0.72 – 1.92 | ns | |
| Stage | Stage I | 1.00 | - | |
| Stage II | 2.73 | 1.69 – 4.40 | <0.001 | |
| miR-34b/c | Low to medium | 1.00 | - | |
| methylation | High (AUC > 1.34) | 2.16 | 1.32 – 3.52 | 0.002 |
MiR-34b/c ectopic expression reduced cell proliferation, migration and invasion
If epigenetic inactivation of miR-34b/c by DNA methylation has such a strong correlation with clinical outcome, it is logical to anticipate that restoration of miR-34b/c in lung AC cell lines should induce significant changes in the tumor phenotype. Two cell lines that expressed low levels of miR-34b/c (H1838 and SK-LU-1) were transfected with miR-34b/c precursors that mimics endogenous miRs or alternatively with empty vector. The expression values of the miR-34b/c stable transfectants were determined by qRT-PCR and were significantly higher as compared to the empty vector and parental cells (Supplementary Fig. S6A). The gene expression of several genes which have been reported as putative targets of miR-34b/c were examined and a significant reduction of the transcript levels were found for AXL, BCL-2, HMGA2, MET, NOTCH1 and NOTCH2 in both transfected cell lines (Supplementary Fig. S6B). At the protein level significant reduction of expression of MET was observed (Supplementary Fig. S6C).
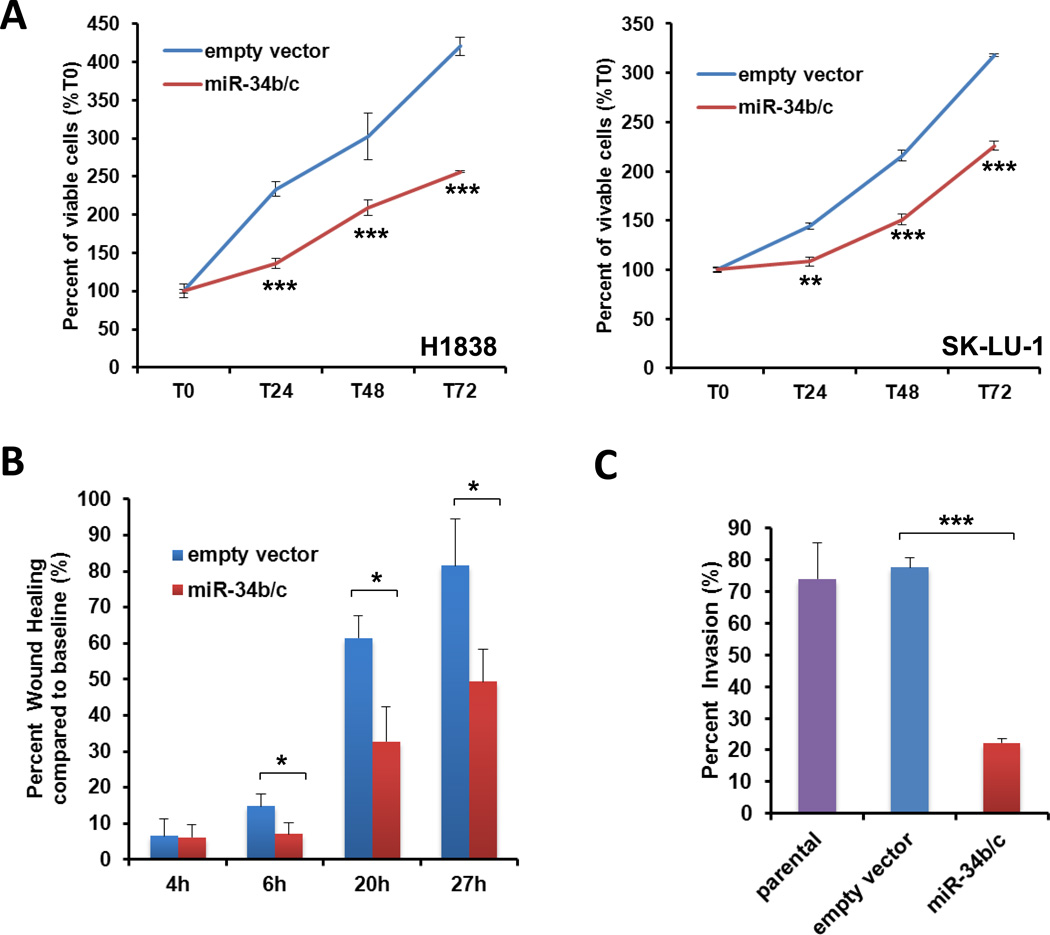
Cells expressing miR-34b/c showed a significantly-lower proliferation rate as compared to empty vector (P < 0.001, Fig. 4A). Accordingly (Supplementary Fig. S7A), stable transfection with miR-34b/c mimics resulted in significantly increased PARP cleavage. We treated the stable cell lines with cisplatin (CDDP), pemetrexed or erlotinib, all of which are currently used for treating lung AC. A modest sensitization effect to CDDP was observed after restoring miR-34b/c expression in SK-LU-1 cells (Supplementary Fig. S7B).
Figure 4.
Impact of miR-34b/c on lung AC cell proliferation, migration and invasion. (A) Cell proliferation of miR-34b/c transfected cells was assessed using WST-1 assay. Values are expressed as the means ± SD of 3 experiments. Cell proliferation was significantly lower in cells expressing miR-34b/c as compared to cells transfected with the empty vector (ev). (* P < 0.05; ** P < 0.01; *** P < 0.001). (B) Wound healing assay in SK-LU-1 cells. The quantitative values indicate the mean distance ± SD between the migration fronts measured at different time-points per 3 wells and are representative of 3 experiments. Cells expressing miR-34b/c closed the induced cell wound significantly slower than cells transfected with ev. (C) Invasion experiments using Boyden chamber in SK-LU-1 cells. Invasive cells stained by Diff-Quick were counted. The quantitative values represent the mean ± SD of 5 microscopic fields per 2 wells and are representative of 2 experiments. Cells expressing miR-34b/c were significantly less invasive as compared to control ev transfected cells.
Migration was assessed using the wound healing assay and invasion was assessed using the Boyden chamber assay. Migration and invasion was significantly suppressed in miR-34b/c stable transfectants of SK-LU-1 (Fig. 4B, 4C and Supplementary Fig. S8) as compared to empty vector or parental cells. However, no differences in migration and invasion were found in transfected H1838 cells. These results might be related to the low invasion ability of parental H1838 cells. We decided to perform transient transfection on H2228 which expresses low levels of miR-34b/c and has a high invasion capacity and a remarkable reduction in cell invasion was found when H2228 were transfected with miR-34b/c (data not shown).
In summary, the restoration of miR-34b/c expression in lung AC lines can reduce cell proliferation, cell migration and invasion, conferring therefore a less aggressive phenotype. Re-expression of these miRs thus might be a novel potential therapeutic strategy in patients expressing low levels of miR-34b/c.
Discussion
MiRs play an important role in tumorigenesis and cancer progression. The discovery of their regulatory function has added a new level of complexity in our understanding of cancer genetics (32). Interestingly, miR expression is often widely down-regulated in cancer cells relative to normal tissues (33), and forced reduction of global miR expression promotes transformation (34). MiR expression is widely deregulated in human cancer, including NSCLC and several regulatory mechanisms have been identified: DNA copy abnormalities (35), mutation (36), failure of post-transcriptional regulation (37), regulation by transcription factors (38), and a defective miR biogenesis pathway (34). Additional epigenetic mechanisms such as methylation of the 5’ regulatory regions associated with specific miR down-regulation in tumors were observed (16, 39, 40).
The miR-34 family consists of miR-34a, the most well studied member located at the chromosome 1, and miR-34b and miR-34c, located at the chromosome 11 as a bicistronic cluster and primarily expressed in non-tumoral lung (23). The putative promoter of miR-34b/c resides 4.5 kb upstream of the miR-coding sequence and includes a dense CpG island and the starting transcription site of BTG4 gene (21). In our study, we found that hypermethylation of miR-34b/c is a frequent event in lung AC tumors and cell lines (40–46%), which is similar to the frequency reported previously by other groups in NSCLCs (22). Interestingly, we observed that patients with higher levels of miR-34b/c methylation had a significantly shorter disease-free survival and overall survival compared to those with lower levels of methylation or unmethylated miR-34b/c. A recent study reported that miR-34b/c methylation might be prognostic for stage I NSCLC patients (22). In this study, DNA methylation was assessed by Methylation-Specific PCR from formalin-fixed paraffin tissue samples. We used frozen samples and DNA methylation was assessed using melting curve analysis, which achieved an excellent analytical sensitivity. We considered that MCA might be a useful technique for detecting and estimating DNA methylation bysimultaneous assessment of multiple CpG residues (37 CpG sites of miR-34b/c promoter), rendering the technique less vulnerable to the behavior of specific residues. When analysing other markers using MCA, we consider relevant to determine the analytical sensitivity for any specific gene to set up a cut-off for classifying the samples as methylated or unmethylatedWhile focusing on lung AC, we validated the prognostic value of miR-34b/c methylation in an independent cohort of this subset of patients.
We also determined the expression of miR-34b and miR-34c in a set of lung AC cell lines and primary tumors. According to previous data (23), global association between TP53 mutational status and miR-34b/c expression was not found in lung AC lines. Indeed, although miR-34b/c are bona fide transcriptional targets of TP53 and their promoters contain TP53-canonical binding sites, miR-34 family appeared to be not necessary for TP53 function using a miR-34 deficient mouse (41). Next, we looked at potential interactions between DNA methylation status and TP53 status and a significant correlation was seen, suggesting that promoter methylation is a relevant mechanism of transcriptional regulation for miR-34b/c. Similarly, a borderline association was detected between TP53 mutational status and miR-34b/c expression in lung AC tumors, but a strong correlation was likewise obtained when TP53 and methylation status were combined. DNA methylation might therefore be the main mechanism of regulation of miR-34b/c expression, as it was previously reported in other tumors. The restoration of miR-34b/c expression with 5-aza-dC suggests that DNA methylation plays a role in the transcriptional regulation of these miRs in lung AC cells.
Interestingly, miR-34b expression was higher in nonsmoking patients and both miRs were significantly overexpressed in EGFR mutant cell lines and primary tumors. Although nonsmoking patients had lower levels of miR-34b/c expression as compared to smokers, we did not find a significant correlation between tobacco use history and DNA methylation. Similarly, during bronchial carcinogenesis of squamous cell carcinomas, expression of miR-34c progressively was reported to decrease from normal epithelium of nonsmokers to invasive bronchial lesions of smokers (42).
The role of miR-34b/c expression as a prognostic marker is controversial. Landi et al (43) found that miR-34b/c expression measured by microarray technology was associated with outcome in surgically-resected lung NSCLC patients. However in a later study (31) miR-34b/c measured by qRT-PCR was not prognostic in a large cohort of early stage NSCLC. In our study, we could not find an association between miR-34b/c expression and outcome potentially due to the limited number of cases analyzed.
In our study, we used stable transfectants concomitantly expressing miR-34b and 34c to study the pathogenic role of these miRs in lung adenocarcinoma lines. Using this in vitro model, we found that the restoration of miR-34b/c expression suppressed cell proliferation, migration and invasiveness. These findings suggest that these miRs might act as a tumor suppressor in lung adenocarcinoma, which is consistent with its role in other human cancers (20, 24, 44, 45).
MiR-34b/c restoration was not able to considerably modify the sensitivity to CDDP, pemetrexed or erlotinib in lung AC lines. Correspondingly, miR-34b/c expression was previously not found to have a predictive effect on survival in lung cancer patients treated with adjuvant chemotherapy after tumor resection (31). On the other hand, miR-34c appears to be significantly overexpressed in erlotinib-sensitive NSCLC cell lines (46) and was associated with a signature predictive for response to erlotinib. However in our study miR-34b/c transfectants did not show a significant difference in their sensitivity to erlotinib (data not shown). This lack of effect can be partially explained since H1838 cells express high levels of MET and SK-LU-1 cells are KRAS mutant.
Several strategies have been proposed to restore the function of miRs with tumor suppressor properties that are down-regulated in cancer (47). One approach potentially useful to restore the expression of genes and miRs regulated by DNA methylation is the use of DNA demethylating agents and histone deacetylase inhibitors. These drugs have shown therapeutic benefit in some hematological diseases and even have demonstrated anti-tumor activity in chemo-refractory NSCLC (48). However we recognize an important limitation of this strategy consists of these agents lack of specificity and that may restore the expression of multiple genes and miRs genome-wide. A more promising strategy may be miR replacement therapy using lipid-based delivery vehicles (49, 50), which are able to restore loss of function activity and to reactivate cellular pathways in cancer that drive a therapeutic response.
In conclusion, our findings show that miR-34b/c is frequently inactivated by promoter DNA methylation in lung AC and restoration of miR-34b/c expression induces a less aggressive and invasive phenotype in this histological subtype of lung tumors. In addition, methylation of miR-34b/c might be an independent prognostic marker in early stage lung AC and is potentially useful for selecting a subset of stage I tumors with higher risk of recurrence or death after lung resection that would benefit from an additional therapeutic intervention. The restoration of miR-34b/c expression was not associated with a differential sensitivity to chemotherapy in vitro and may not be useful as a predictive marker. A miR replacement therapy might be a potential strategy for treating those tumors with hypermethylated miR-34b/c, although further investigation in this area needed.
Supplementary Material
Statement of Translational Relevance.
MiR-34b/c are members of microRNA (miR) 34 family and target relevant genes in lung adenocarcinoma (AC) involved in cell cycle, apoptosis, stem-cell or invasion, such as CCDE2, cMYC, BCL2, NOTCH1 or MET. We analyzed the DNA methylation status of miR-34b/c in both lung AC cell lines and in primary tumors which were then correlated with miR expression. Interestingly, early stage lung AC patients with higher levels of miR-34b/c methylation were found to have significantly shorter disease-free survival and overall survival. Ectopic expression of miR-34b/c in lung AC cell lines decreased cell proliferation, cell migration and cell invasion ability. These results suggest that miR-34b/c methylation is an independent prognostic marker in early stage lung AC patients and might be considered for potential therapeutic targeting.
Acknowledgments
Funds: Supported by the Xarxa de Bancs de Tumors de Catalunya (XBTC) and sponsored by the Pla Director d’Oncologia de Catalunya, The Bonnie J. Addario Lung Cancer Foundation and the University of Michigan Comprehensive Cancer Center. Ernest Nadal was supported by a Rio Hortega Fellowship from the Instituto de Salud Carlos III and by a Spanish Society of Medical Oncology Fellowship.
Footnotes
CIO: The authors have no conflict of interest to disclose.
References
- 1.Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Youlden DR, Cramb SM, Baade PD. The International Epidemiology of Lung Cancer: geographical distribution and secular trends. J Thorac Oncol. 2008;3:819–831. doi: 10.1097/JTO.0b013e31818020eb. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fruh M, Rolland E, Pignon JP, Seymour L, Ding K, Tribodet H, et al. Pooled analysis of the effect of age on adjuvant cisplatin-based chemotherapy for completely resected non-small-cell lung cancer. J Clin Oncol. 2008;26:3573–3581. doi: 10.1200/JCO.2008.16.2727. [DOI] [PubMed] [Google Scholar]
- 6.Group NM-aC. Arriagada R, Auperin A, Burdett S, Higgins JP, Johnson DH, et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet. 2010;375:1267–1277. doi: 10.1016/S0140-6736(10)60059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nature reviews. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 8.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steels E, Paesmans M, Berghmans T, Branle F, Lemaitre F, Mascaux C, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18:705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 10.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 14.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 16.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31:2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- 19.Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 20.Kubo T, Toyooka S, Tsukuda K, Sakaguchi M, Fukazawa T, Soh J, et al. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin Cancer Res. 2011;17:4965–4974. doi: 10.1158/1078-0432.CCR-10-3040. [DOI] [PubMed] [Google Scholar]
- 21.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. 2011;11:490–496. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 23.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka N, Toyooka S, Soh J, Kubo T, Yamamoto H, Maki Y, et al. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer. 2012;76:32–38. doi: 10.1016/j.lungcan.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Curado MP, Edwards B, Shin HR, Storm H, Ferlay J, Heanue M, et al. Cancer Incidence in Five Continents. Report No.: 0300-5085. 2007 [Google Scholar]
- 26.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 27.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 29.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, et al. MicroRNA expression and clinical outcomes in patients treated with adjuvant chemotherapy after complete resection of non-small cell lung carcinoma. Cancer Res. 2010;70:8288–8298. doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 34.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 37.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17–92, E2F, and Myc. Proc Natl Acad Sci U S A. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heller G, Weinzierl M, Noll C, Babinsky V, Ziegler B, Altenberger C, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res. 2012;18:1619–1629. doi: 10.1158/1078-0432.CCR-11-2450. [DOI] [PubMed] [Google Scholar]
- 40.Baer C, Claus R, Plass C. Genome-Wide Epigenetic Regulation of miRNAs in Cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 41.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mascaux C, Laes JF, Anthoine G, Haller A, Ninane V, Burny A, et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur Respir J. 2009;33:352–359. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- 43.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 45.Chim CS, Wan TS, Wong KY, Fung TK, Drexler HG, Wong KF. Methylation of miR-34a, miR-34b/c, miR-124-1 and miR-203 in Ph-negative myeloproliferative neoplasms. J Transl Med. 2011;9:197. doi: 10.1186/1479-5876-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryant JL, Britson J, Balko JM, Willian M, Timmons R, Frolov A, et al. A microRNA gene expression signature predicts response to erlotinib in epithelial cancer cell lines and targets EMT. Br J Cancer. 2012;106:148–156. doi: 10.1038/bjc.2011.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 48.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.