Abstract
A population of c-kit+ cardiac stem/progenitor cells (CSPC) has been identified in the heart and shown to contribute to myocardial regeneration after infarction. Previously, we have shown the chemokine, stromal cell derived factor 1α (SDF1) is necessary for the myocardial response to infarction where chronic infusion of the CXCR4 antagonist, AMD3100, exacerbated MI. Notably, AMD3100 increased CSPC proliferation. The effect of SDF1 on CSPC proliferation was further investigated in primary cultures of magnetically sorted c-kit+ CSPCs. SDF1 facilitated CSPC quiescence by blocking cell cycle progression at the G0 to G1 transition. SDF1 decreased casein kinase 1α (CK1α) consequently attenuating β-catenin phosphorylation, destabilization, and degradation. Increased levels of β-catenin with SDF1 were effective, increasing TCF/LEF reporter activity. SDF downregulation of CK1α was dependent on proteasomal degradation and decreased mRNA expression. CK1α siRNA knockdown verified SDF1-dependent CSPC quiescence requires CK1α downregulation and stablilization of β-catenin. Conversely, β-catenin knockdown increased CSPC proliferation. SDF1 also increased GSK3β Y216 phosphorylation responsible for increased activity. SDF1 mediated CK1α downregulation and increase in GSK3β activity affected cell cycle through Bmi-1 downregulation, increased cyclin D1 phosphorylation, and decreased cyclin D1 levels. In conclusion, SDF1 exerts a quiescent effect on resident c-kit+ CSPCs by decreasing CK1α levels, increasing GSK3β activity, stabilizing β-catenin, and affecting regulation of the cell cycle through Bmi-1 and cyclin D1. SDF1 dependent quiescence is an important factor in stem and progenitor cell preservation under basal conditions, however, with stress or injury in which SDF1 is elevated, quiescence may limit expansion and contribution to myocardial regeneration.
Keywords: Stromal cell derived factor 1α, CXCR4, Cell cycle, Casein kinase 1α, β-Catenin, Glycogen Synthase Kinase3β
Introduction
A population of resident cardiac stem/progenitor cells (CSPC) has been found to contribute to the regeneration of myocardium after injury, myocardial infarction (MI), or with stress such as pressure overload1-4. This resident CSPC population can be defined by the expression of stem cell markers, c-kit, Sca1, or the multi-drug resistance protein (MDR)1. These stem cells can be modulated by factors such as Notch and its ligand Delta to contribute to the creation of cardiovascular endothelial, smooth muscle, or myocardial cell types5. In particular, cardiac stem cells that express the stem cell factor (SCF) receptor, c-kit, have been characterized extensively and have been shown to be effective in intracoronary transplant improving the myocardial function of patients with heart failure6.
This population of CSPCs has been shown to express the receptor for the chemokine, stromal cell derived factor 1α (SDF1), CXCR47. SDF1-CXCR4 regulation is important in regulating cell survival8, chemotaxis9, angiogenesis10, and proliferation11. We have shown that SDF1 is able to confer myocardial protection in vitro and in vivo through ERK and AKT dependent signaling pathways8. Increased expression of SDF1 has been associated with increased capillary density in the myocardium after MI12, hindlimb ischemia10, and in other models of hypoxia13. In cancer biology, SDF1-CXCR4 has been shown to facilitate tumor metastasis14. SDF1-CXCR4 has also been shown to play a prominent role in the maintenance of the hematopoietic stem cell niche in the bone marrow (in both the perivascular and osteoclastic niche)15-17.
In addition to contributing to the maintenance of the HSC niche, SDF1-CXCR4 interaction has also been shown to play a role in maintenance of HSC quiescence11, 17. In both the conditional CXCR4 and SDF1 knockout mice, deletion of either has resulted in expansion of a SKL (Sca1+/kit+/Lin−) cell population within the bone marrow11, 17. We have also noted, in the myocardium, that chronic blockade of SDF1-CXCR4 with the antagonist AMD3100 results in increased proliferation of c-kit+ CSPCs18 which was also seen in primitive hematopoietic progenitors11, 19. In hematopoietic homeostasis, quiescence is a fundamental stem cell characteristic and its enforcement by SDF1 is an important mechanism of preservation. However, in the myocardium, and other tissues experiencing hypoxia due to injury or stress, increased levels of SDF1 that would necessarily recruit stem and progenitors to facilitate regeneration of damaged tissue, would also confound this effort by limiting progenitor expansion.
CXCR4 is a G-protein coupled receptor that has typically been characterized as coupling through Gαi9. Although CXCR4 regulation of cell cycle has now been reported, the mechanisms associated with this important aspect of cell function are still unknown. Zou and colleagues reported upregulation of p57 by SDF1 may account for this as it was significantly downregulated in Flt3-LSK cells in the CXCR4 KO11. SDF1 has been shown to stabilize β-catenin levels in pancreatic islet cells that was proposed to occur via AKT dependent inhibition of GSK3β activity20. Here β-catenin stabilization had no effect on proliferation. Cardiac Wnt signaling leading to increased β-catenin stabilization was shown to inhibit cardiac side population cell proliferation and impair myocardial response to ischemia reperfusion injury21. β-Catenin levels are regulated by a stabilization complex composed of Axin, adenomatous polyposis coli (APC) and, GSK3β that controls proteasomal degradation. β-catenin levels are increased with attenuated GSK-3β activity that may occur with AKT phosphorylation as GSK3β dependent phosphorylation targets β-catenin for proteosomal degradation. In Wnt signaling, GSK3β activity is a central coordinator of signal input and thus is a key regulator of β-catenin phosphorylation and stability22, 23. Within this signaling scheme casein kinase 1α (CK1α) activity and β-catenin phosphorylation is considered permissive for GSK-3β targeting for ubiquitinization and proteosomal degradation15. CK1α is considered to be constitutively active, widely expressed, without significant regulation24.
Here, we present findings that describe the anti-proliferative, quiescent action of SDF1 on c-kit+ CSPCs and novel mechanisms that regulates β-catenin stability. In this CSPC population, we find that SDF1-CXCR4 action regulates CK1α stability and expression, resulting in decreased levels of this kinase and the consequential decrease in β-catenin phosphorylation. SDF1 also increased GSK3β Y216 phosphorylation to facilitate increased activity. These actions increase β-catenin stability leading to decreased expression and destabilization of Bmi-1 and cyclin D1 that direct CSPC quiescent by blocking cell cycle progression in G0. The implications of SDF1-CXCR4 effects on cell cycle are considered in the context of myocardial function and response to injury.
Methods
CSPC isolation and culture
Procedures involving mice were conducted under the approval of the University of Louisville IACUC in accordance with the NIH Guide for the Care and Use of Laboratory Animals (DHHS publication No. [NIH]85-23, rev. 1996) as previously described8. Mice were anesthetized with sodium pentobarbital (100 mg/kg, ip). The heart was quickly removed, and the aorta cannulated and perfused retrograde by the Langendorff method at a constant flow rate of 3 ml/minute for 5 minutes with a Ca2+-free bicarbonate-based buffer containing (in mM) 120 NaCl, 4.7 KCl, 1.2 MgSO4, 0.6 KH2PO4, 0.6 Na2HPO4, 5.5 glucose, 4.6 NaHCO3, 10 HEPES, 10 2,3-butanedione monoxime, and 30 taurine continuously gassed with 95% O2, 5% CO2 ph 7.4. The enzymatic digestion was initiated with collagenase II perfusion (0.25 mg/mL; Worthington) in buffer containing 12.5 μM Ca2+ for 8 to 10 minutes. The digested ventricular tissue was cut into pieces and gently triturated with a transfer pipette. The cell pellet was finally resuspended in perfusion buffer containing 10% FBS and 1.26 mM Ca2+ for experiments. The non-myocyte population was expanded for 5-6 days and sorted by magnetic beads (CD117 microbeads, Miltenyi) to enrich a c-kit+ population of cardiac stem/progenitor cells which were then used in experiments at passages 2-3 in culture in DMEM/F12 containing 15 mM Hepes, 1% BSA, 10 ng/ml LIF, 1×ITS, 10 ng/ml bFGF, and 20 ng/ml EGF (mSC media). CSPC were characterized for c-kit+ expression by flow cytometry.
Western analysis
Primary CSPCs were grown in culture medium, treated, and lysates prepared with buffer A containing (in mM): 25 TrisHCl, pH 7.5, 0.5 EDTA, 0.5 EGTA, 1% NP-40, 1 DTT to which were freshly added 1 PMSF, 1/100 Halt protease inhibitor cocktail, (Thermo), 1% phosphatase inhibitor cocktail 3 (P0044, Sigma), 1% Phosphatase Inhibitor cocktail 2 (P5736, Sigma). Samples were centrifuged at 14,000 g for 15 minutes and the resulting supernatants were collected, aliquoted and used in Western analysis. The total protein content was measured (Biorad Protein Assay, Biorad), samples denatured for 3 minutes at 95°C, and loaded onto gels for SDS PAGE, followed by transfer to Hybond ECL membrane (GE). After being incubated with primary antibodies at 4°C overnight, proteins were detected with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Cell signaling 1: 5000) and the ECL plus chemiluminescent detection system (GE).
Primary antibodies were used to detect the following: GSK-3 β total, phosphoS9, phosphoY216 (Cell Signaling, 1:2000), phospho β-catenin S45, S33/37/T41 (Cell Signaling, 1:1000), GAPDH (Cell Signaling, 1:5000), CKIα (Cell Signaling, 1:1000), β-catenin (BD Transduction Laboratories, 1:1000), active β-catenin (Millipore,1:500), c-kit (H300, Santa Cruz) ubiquitin (Sigma, 1:500), cyclin D1 (Cell Signaling), and phosphoT286-cyclin D1(Cell Signaling). Densitometry analysis was performed using a Typhoon scanner (GE).
TCF/LEF-Luciferase Assays
CSPCs were transfected with 0.25 μg TOPFlash and FOPFlash luciferase reporters25 containing concatemers of the wild type and mutant TCF/LEF binding sites to determine β-catenin transcriptional activation and 0.025 μg CMV-renilla luciferase using lipofectamine LTX and Plus (Life Technologies). CSPCs were treated 24 hours after transfection with 25 nM SDF1 and vehicle. Luciferase activity was measured after SDF1 treatment for 24 hours using the dual-luciferase reporter assay (Promega).
Immunoprecipitation
CSPC lysates were obtained as stated above for Western analysis. Lysate aliquots of 500 μg total protein were pre-cleared with Protein A-Sepharose beads (30 μl) rotating at 4° C for 30 minutes. Pre-cleared lysates were then incubated with 5 μl of the antibody (anti-β-catenin or anti-CK1α) rotating for 2 hours at 4° C. After incubation, Protein A-Sepharose beads were washed three times with lysis buffer. After the last wash 35 μl of 2× sample loading buffer was added and incubated at 95° C for 3 minutes. Immunoprecipitated samples were resolved by SDS PAGE and ubiquitin, β-catenin, and CK1α detected by Western analysis.
CK1α, GSK3β, and β-catenin siRNA gene silencing
CK1α (24 hours, 20 nmol, siRNA ID s96623, Invitrogen), GSK3β (24 hours, 50 nM, siRNA ID SI01059107, Qiagen), β-catenin (48 hours, 100 nmol, siRNA ID s63418, Invitrogen), and scrambled control (Invitrogen 4404020) siRNA duplexes were transfected into primary CSPCs using Lipofectamine RNAiMAX (Invitrogen) and efficiency of the corresponding gene silencing validated by measuring decreased levels of protein expression by Western analysis and mRNA expression by real time RT-PCR. Cells were then treated with SDF1 (Peprotech) for specified durations.
Flow Cytometry
Cell cycle was analyzed by flow cytometry after CSPCs were treated with SDF1. After treatment, CSPCs were fixed in ethanol at −20° C, washed in PBS, and stained with an anti-Ki67 FITC-conjugated primary antibody (BD, 556026 1:6) for 45 minutes at room temperature (RT). CSPCs stained with the FITC conjugated isotype, IgG1, κ, were used for gating. CSPCs were washed and stained with propidium iodide (PI, 50 μg/ml, Molecular Probes) with RNase A treatment (50 μg/ml) for 15 minutes at RT in the dark. Analysis was performed on the LSR II (BD Biosciences). FACS DIVA and FlowJo software were used to determine the percentage of CSPCs in G0, G1, and S/G2/M.
CSPC death was assessed by Annexin V/PI staining (Invitrogen). CSPCs treated with SDF1, harvested, and washed in cold PBS were stained with Fluorescein conjugated Annexin V and PI for 15 minutes. Stained CSPCs were immediately analyzed by flow cytometry (LSRII, BD).
CSPC c-kit expression was correlated with cell cycle using Ki67. CSPCs were stained for c-kit (BD, APC conjugated) for 30 minutes on ice, fixed with 4% paraformaldehyde for 30 min on ice and permeabilized with 0.1% TitonX-100 for 10 min. Washed CSPCs were stained with a FITC-Ki67 antibody (BD) on ice for 45 minutes. CSPCs were washed and analyzed by flow cytometry (LSRII, BD).
CSPC BrdU labeling proliferation assay
Primary CSPCs were plated at 1×104 cells per well on 96 well plates in triplicate. Prior to treatment CSPCs were cultured for 24 hours in mSC media (no LIF/1% BSA). CSPCs were also treated with siRNAs during this period as specified. CSPCs were then treated as detailed in Results in the presence of BrdU. CSPC proliferation was determined by measuring incorporated BrdU by colorimetric immunoassay according to the manufacturer’s protocol (Calbiochem QIA58).
Real time RT-PCR
Changes in gene expression were determined by quantitative real-time RT-PCR. Total RNA was isolated and purified (Qiagen) and RNA concentration determined by Nano-Drop 2000C spectrophotometer (Thermo Scientific). mRNA was reverse-transcribed and cDNA used in real time PCR reactions prepared with SYBR-Green supermix (Invitrogen).The following qPCR primer sequences were used in reactions: β-catenin, forward, GCAGCGTTATACTCAGAT, reverse, CTCTCAGCAACTCTACAG; Bmi-1, forward, TGACTGTGATGCACTTGAGAAAGTT, reverse, GTAGGCAATGTCCATTAGCGTGTAG; CK1α, forward, CTCACTTCTGCCGCGGGTGG, reverse, GACGCGAAGATGGAGGCGGG; p21Cip1, forward CTGTCTTGCACTCTGGTGTCTGAG, reverse TTTTCTCTTGCAGAAGACCAATCTG; p27Kip1, forward, TTTAATTGGGTCTCAGGCAAACTCT, reverse, CCGTCTGAAACATTTTCTTCTGTTC; p57Kip1, forward, GCAAACGTCTGAGATGAGTTAG, reverse, CATCTCCGGTTCCTGCTACATG; β2 microglobulin, forward, CATACGCCTGCAGAGTTAAGCA, reverse, GATCACATGTCTCGATCCCAGTAG.
Statistical analysis
Data are expressed as mean ± SEM. Statistical significance was assessed by ANOVA followed by Bonferroni/Dunn testing, or unpaired t test. A p-value less than 0.05 was considered statistically significant.
Results
SDF1 promotes CSPC quiescence
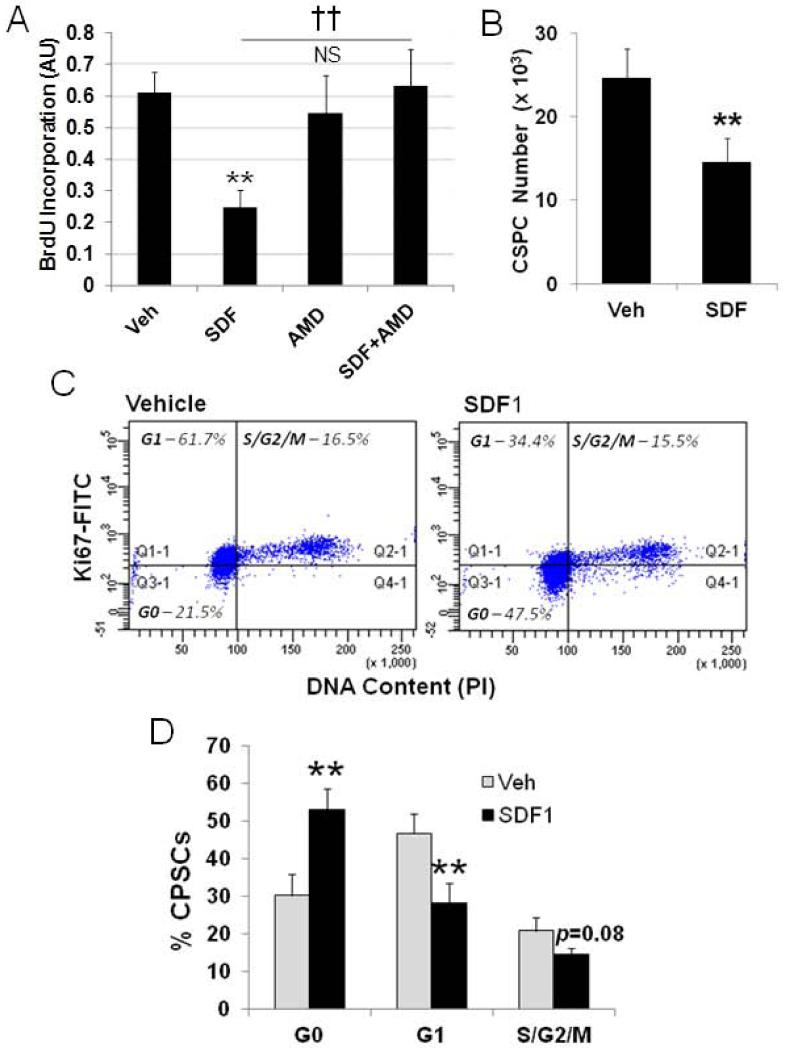
In previous studies we observed increased proliferation of a c-kit+ population of CSPCs in mouse hearts after myocardial infarction (MI) with chronic infusion of the CXCR4 antagonist AMD310018. This finding is congruent with the initial observation that CXCR4 contributes to the quiescence of a primitive hematopoietic stem cell. To examine this further and demonstrate a direct effect of SDF1 we established cultures of c-kit+ CSPCs and examined the effect of SDF1 on their proliferation. CSPCs were isolated from primary cultures of a non-myocyte cardiac cell fraction by magnetic sorting for c-kit+ progenitors26. The c-kit+ CSPCs represented at least 70% of cells in the sorted fraction (Supplementary Figure 1A). CSPCs were used in experiments within passage 2 and up to passage 7 with similar results while retaining c-kit expression (Supplemental Figure 1B). CSPCs were treated with 25 nM SDF1 for 24 hours and at the same time labeled with BrdU. SDF1 treatment resulted in a marked quiescence detected by BrdU incorporation (Figure 1A). SDF1 treatment for 24 hours also decreased the actual number of CSPCs validating decreased proliferation measured by BrdU incorporation (Figure 1B) without change in cell death demonstrated by a low level of Annexin V staining (6%) that was unchanged with SDF1 treatment (Supplemental Figure 2). Decreased BrdU incorporation with SDF1 treatment was antagonized by the CXCR4 antagonist AMD3100 (Figure 1A). AMD3100 alone did not affect CSPC proliferation. Cell cycle analysis demonstrated SDF1 caused CSPCs to exit the cell cycle and accumulate in G0 (Figure 1C and 1D) with a concomitant decline in CSPCs in G1 and S/G2/M (Figure 1C). This arrest at the G0/G1 transition occurred in CSPCs where 65% of c-kit+ CSPCs were Ki67+ (Supplemental Figure 3).
Figure 1. SDF1 attenuates CSPC proliferation through blockade at G1.
Proliferation was measured in cultured CSPCs after treatment with 25 nM SDF1 with and without 40 μM AMD3100. Proliferation was measured by A) BrdU incorporation and directy, B) by cell counts. CSPCs were stained with Ki67 and PI for cell cycle analysis C) Ki67/PI scatter D) Cell cycle plot. Values are the mean ± SE, n=3-5. ** p<0.01 vs Veh, †† p<0.01 vs SDF.
SDF1 increases β-catenin levels
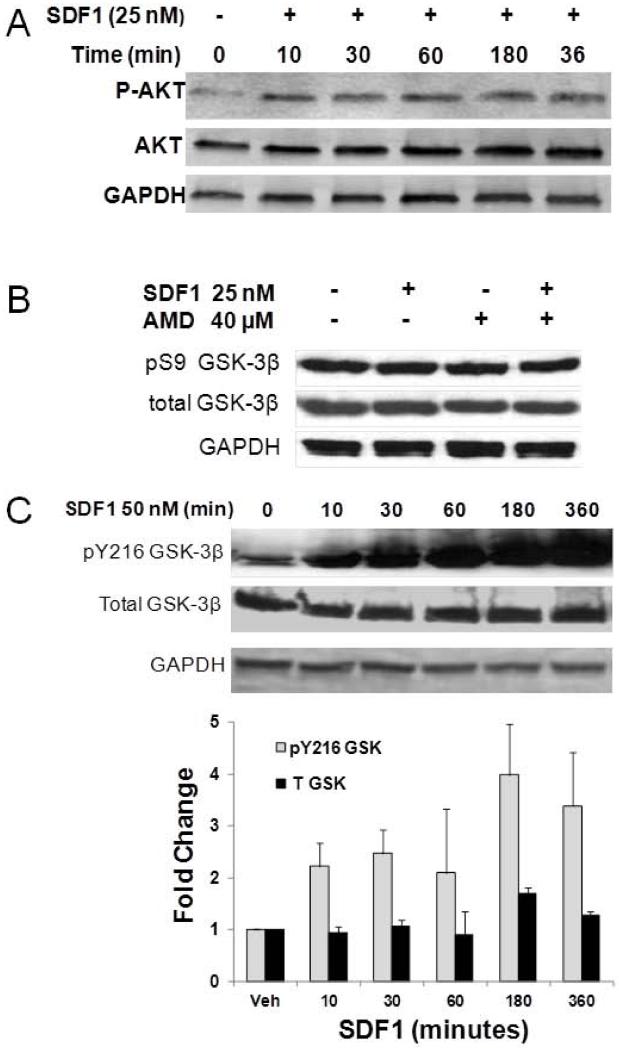
We have previously reported that SDF1 is capable of activating AKT dependent signaling in cardiac myocytes8. As AKT is upstream of GSK3β, we therefore examined whether SDF1-CXCR4 activation of downstream AKT signaling was functional in mouse c-kit+ CSPCs. SDF1 treatment of CSPCs rapidly increased AKT phosphorylation that persisted for up to 6 hours (Figure 2A). SDF1 increased AKT phosphorylation in the low nanomolar range (data not shown).
Figure 2. SDF1 stimulates AKT phosphorylation and increases overall GSK3β activity by Y216 phosphorylation in CSPCs.
A) CSPCs were treated with 25 nM SDF1 for increasing time periods in serum free media. CSPC homogenates were resolved by PAGE and phosphorylated and total AKT detected by Western analysis. Levels of total GSK3β and its S9 (B) and Y216 (C) phosphorylated forms were measured in homogenates after treating CSPCs with 25 nM SDF1 for 24 hours (Panel B) or 50 nM SDF1 for the specified time periods (Panel C) by Western analysis. Values are the mean ± SE, n=2-3.
The downstream AKT target, GSK3β may be phosphorylated to regulate cell cycle. SDF1 activation of AKT would be expected to decrease GSK3β activity through phosphorylation of Ser9. Although SDF1 did increase phosphorylation of AKT, SDF1 did not affect levels of Ser9 GSK3β phosphorylation suggesting SDF1 activation of AKT would not impact signaling via a GSK3β dependent mechanism (Figure 2B). We had reported previously that blockade of SDF1-CXCR4 signaling with the antagonist AMD3100 resulted in decreased Y216 phosphorylation which is considered essential for GSK3β activity27. Conversely, SDF1 treatment increased GSK3β Y216 phosphorylation as would be expected based on the effect of chronic AMD3100 in vivo (Figure 2C). Thus without an effect of upstream AKT on GSK3β S9 phosphorylation, yet an increase in GSK3β Y216 phosphorylation, the overall effect on GSK3β would be to increase its activity. These results also demonstrate AKT activation by SDF1 does not directly regulate GSK3β phosphorylation or activity.
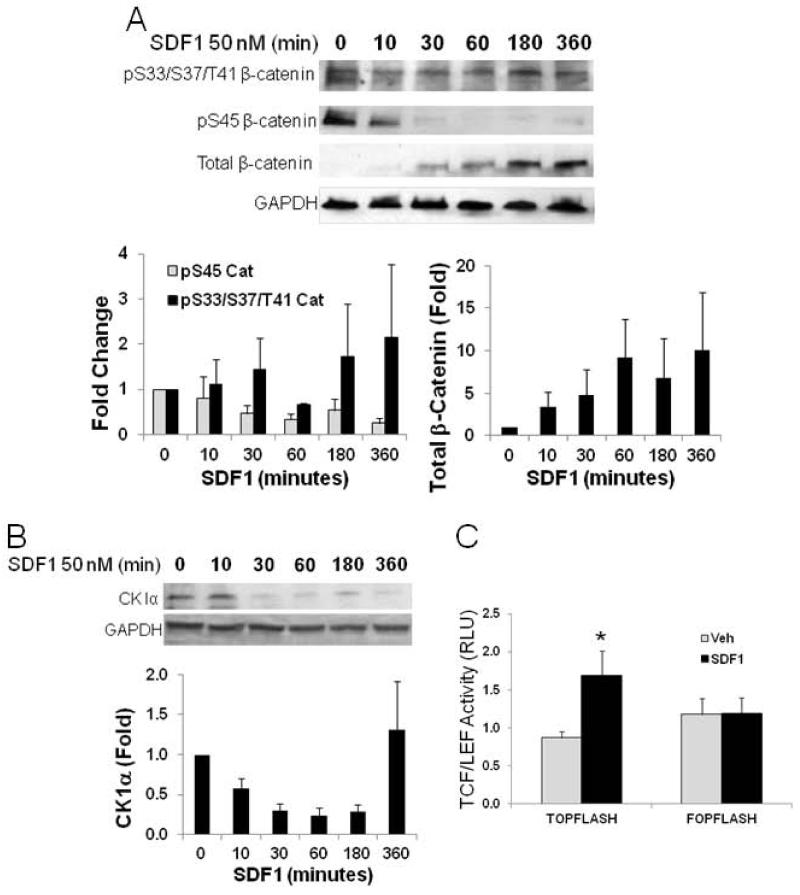
With increased net GSK3β activity, which regulates several important downstream effectors including cell cycle, β-catenin is a direct target being phosphorylated at 3 residues, S33, S37, and T41 which targets it for ubiquitinization and proteasomal degradation28. Surprisingly, CSPCs treated with SDF1 did not show increased phosphorylation at these residues but did show an increase in β catenin (Figure 3A). The lack of correlation between GSK3β and β-catenin regulation may, however, be a consequence of interaction between β-catenin and CK1α. CK1α has been identified as an associate protein of the β-catenin destruction complex and participates in regulation of its stability15. Phosphorylation of β-catenin S45 by CK1α has been found to be a permissive event for further phosphorylation by GSK3β. If CK1α phosphorylation of β-catenin is necessary for GSK3β phosphorylation, this could contribute to the lack of effect of increased GSK3β activity. In CSPCs treated with SDF, levels of CK1α dependent S45 β-catenin phosphorylation declined rapidly within 30 minutes of treatment (Figure 3A). This decline in S45 phosphorylation would effectively blunt the action of GSK3β on β-catenin phosphorylation. CK1α has not previously been shown to be regulated by signaling pathways and thus the means by which levels of β-catenin S45 phosphorylation were affected were investigated initially by examining if levels of expression were changed with SDF1 treatment. As seen in Figure 3B, SDF1 treatment resulted in a marked decline in CK1α levels within 30 min. This decline closely correlates with the loss of CK1α-dependent β-catenin S45 phosphorylation. The overall decline in β-catenin phosphorylation at S45 by CK1α and S33, S37 and T41 by GSK3β would effectively increase its stability as these serve to target β-catenin for ubiquitinization and proteasomal degradation. Indeed, again, with close correlation to CK1α degradation and decreased S45 β-catenin phosphorylation, total levels of β-catenin began to increase at the 30 minute time point after SDF1 treatment (Figure 3A). Increased β-catenin would be expected to localize to the nucleus and increase transcriptional activity. We then investigated the effect of increased β-catenin on transcription with the β-catenin-responsive TCF/LEF reporter25. SDF1 treatment for 24 hours increased TCF/LEF dependent transcriptional activity in CSPCs transfected with the TCF/LEF-Luc reporter (Figure 3C). Increased transcriptional activity with SDF was blocked in the mutant control (Figure 3C). This provides support for increased levels of β-catenin demonstrating function with nuclear translocation and transcriptional activation. The close temporal correlation of these events and decrease in S45 phosphorylation demonstrate the direct regulation of β-catenin by CK1α in response to SDF1 stimulation.
Figure 3. SDF1 regulates levels of CK1α and consequential phosphorylation and expression of β-catenin.
A) SDF1(50 nM) treatment rapidly decreased S45 phosphorylation of β-catenin with little effect on S33, S37, and T41 phosphorylation resulting in increased total levels of β-catenin. B) SDF1 (50 nM) decreased CK1α levels in cultured CSPCs. C) Functional consequences of increased β-catenin levels were confirmed by demonstrating increased β-catenin-dependent activation of the TCF/Lef promoter luciferase reporter, TOPFLASH, and its mutant reporter, FOPFLASH after 25 nM SDF1 treatment for 24 hours. Values are the mean ± SE, n=3. * p <0.05 vs Veh.
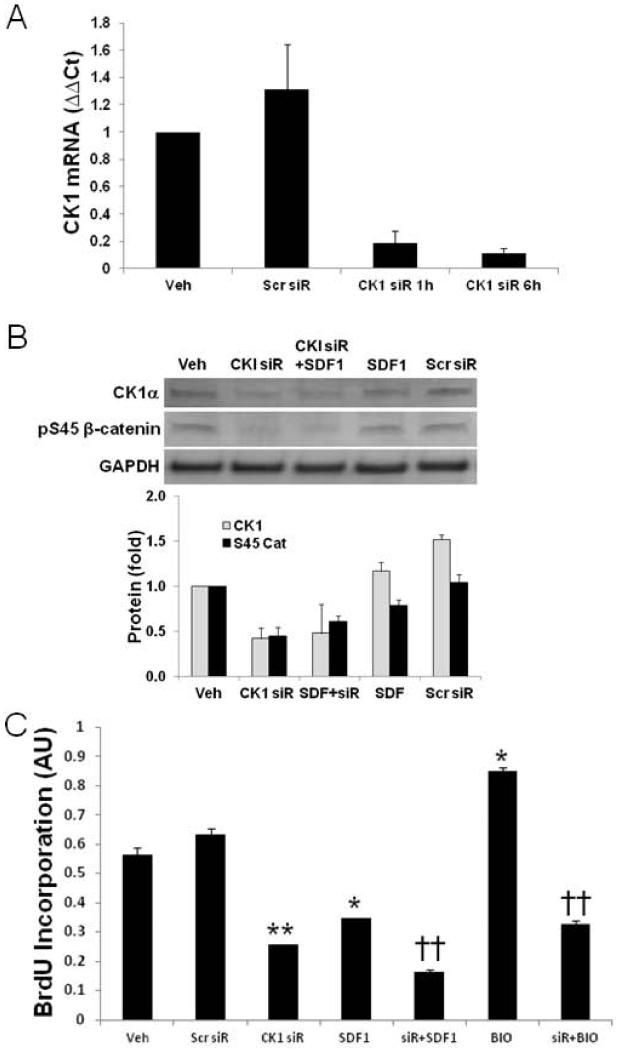
CK1α regulation by external stimuli has not been described. The effect of decreasing CK1α levels on downstream actions mediated by its targets therefore is also unknown. In the context of this study, we examined the effect of CK1α knockdown by siRNA on CSPC cell cycle and levels of β-catenin (Figure 4). CSPCs were treated with siRNA against CK1α for 24 hours and then treated with SDF1 for 24 hours and its effect on proliferation determined by BrdU incorporation. CK1α siRNA effectively decreased CK1α mRNA and protein levels to 9.0±2.8% and 41.6% of scrambled siRNA respectively (Figure 4A and B). Consequently, β-catenin S45 phosphorylation was attenuated to levels below that seen with SDF1 (Figure 4B). Importantly, CK1α knockdown effectively attenuated CSPC proliferation (Figure 4C). CK1α siRNA knockdown was as effective as SDF1 treatment in attenuating CSPC proliferation. Attenuated CSPC proliferation with CK1α siRNA provides supporting evidence that SDF mediated downregulation of CK1α is a primary regulator of cell cycle progression.
Figure 4. CK1α knockdown by siRNA blocks CSPC proliferation.
To assess the role of CK1α in SDF1 proliferation blockade, CK1α was knocked down by siRNA and its effect on CSPC proliferation assessed. A) CK1α siRNA, 20 nM, decreased levels to 9.0±2.8% of scrambled. B) CK1α siRNA knockdown decreased CK1α protein expression and S45 β-catenin phosphorylation. C) CK1α siRNA blocked CSPC proliferation similar to that of 25 nM SDF1 treatment for 24 hours. Values are the mean ± SE, n=2-4. * p<0.05, ** p<0.01 vs Veh or Scr siR, †† p<0.01 vs treated control.
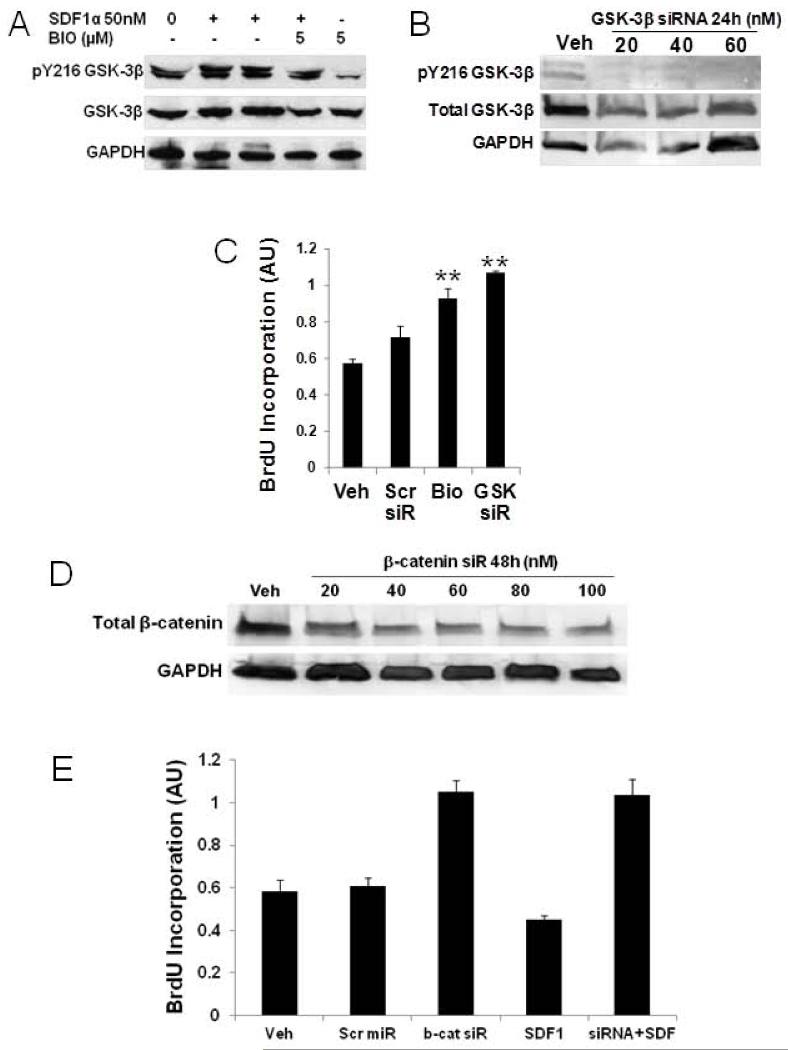
As SDF1-mediated CK1α downregulation is shown above to markedly attenuate β-catenin phosphorylation to facilitate its increased stability and SDF1 leads to increased GSK3β Y216 phosphorylation that increases activity, we were interested in determining whether direct inhibition of GSK3β would have an impact on proliferation. The GSK3β inhibitor, BIO (5 μM), was used to treat CSPCs prior to SDF1 or vehicle. GSK3β is autophosphorylated at Y216 and this phosphorylation is essential for its activity27. BIO blocks GSK3β autophosphorylation rendering it inactive20. Overnight treatment of CSPCs with SDF1 leads to increased levels of Y216 phosphorylation (Figure 5A) and we have shown that chronic infusion of AMD3100 in vivo decreases Y216 phosphorylation18. In CSPCs treated with 5 μM BIO for 12 hours, GSK3β Y216 phosphorylation was almost completely blocked. Thus BIO inhibition of GSK3β Y216 phosphorylation would significantly attenuate its activity and attenuate β-catenin phosphorylation leading to its stabilization. However, as CK1α phosphorylation is a permissive event that is needed for GSK3β to affect β-catenin stability it may not play a significant role through a β-catenin dependent mechanism. BIO inhibition of GSK3β promoted increased CSPC proliferation (Figure 4C and 5C) opposing actions of SDF1 and CK1α siRNA. GSK3β siRNA knockdown effectively decreased GSK3β levels (Figure 5B) and increased CSPC proliferation (Figure 5C), supporting the effect of BIO on CSPC proliferation. Increased CSPC proliferation with BIO inhibition of GSK3β activity and siRNA knockdown suggests that increased GSK3β activity would attenuate cell cycle progression. As SDF1 does not increase GSK3β-dependent β-catenin phosphorylation with increased Y216 phosphorylation, these data demonstrate CK1α is predominant in attenuating cell cycle through β-catenin and that direct effects of SDF1 on GSK3β may support CK1α-independent actions.
Figure 5. GSK3β and β-catenin knockdown increase CSPC proliferation.
A) Bio inhibition of GSK3β blocked Y216 phosphorylation and B) GSK3β siRNA knockdown. C) increased CSPC proliferation. D) β-Catenin siRNA, 100 nM, knockdown decreased levels to 20% of scrambled siRNA. E) β-Catenin knockdown increased levels of CSPC proliferation in cells treated with and without 25 nM SDF1 for 24 hours. Values are the mean ± SE, n=2-4. * p<0.05, ** p<0.01 vs Veh or Scr siR, †† p<0.01 vs treated control.
These findings demonstrate SDF1 stimulation mediates effects on cell cycle that are mechanistically divergent through CK1α and GSK3β. As both of these converge on β-catenin, we sought to determine its effect on proliferation by siRNA knockdown. SDF1 increases β-catenin stability via CK1α, thus, if it were to directly regulate cell cycle, knockdown would result in increased proliferation. β-catenin siRNA knockdown in CPSCs was most effective with 100 nM after 24 hours effectively decreasing β-catenin levels to 20% of control (Figure 5D). CSPCs pretreated with β-catenin siRNA or scrambled siRNA were then treated with 25 nM SDF for 24 hours. First, β-catenin siRNA lead to increased CSPC proliferation which confirms the role of increased β-catenin in SDF1 mediated attenuation of CSPC proliferation (Figure 5E). This action also blocked SDF1 quiescence, directly demonstrating the role of β-catenin in regulating CSPC proliferation.
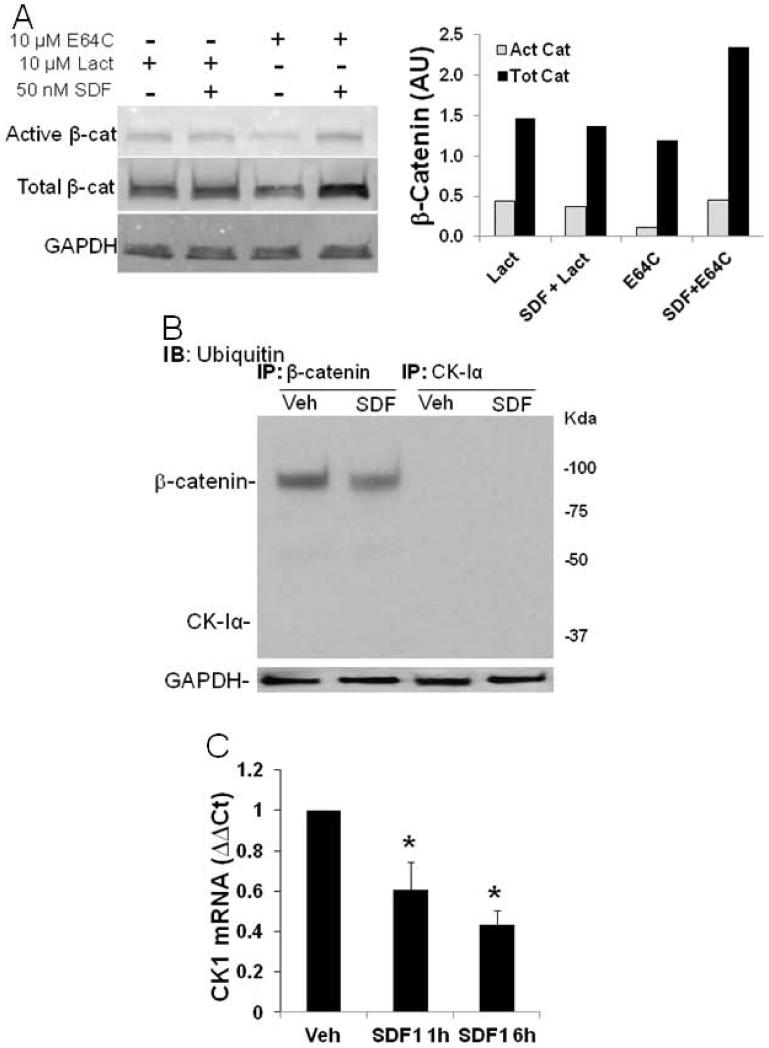
The rapid decrease in CK1α has not previously been reported nor has its regulation by a G-protein coupled receptor. Two degradation mechanisms could account for the rapid SDF1-mediated downregulation of CK1α; proteasomal or lysosomal. To examine the potential involvement of these degradation pathways the proteasomal inhibitor, lactacystin, and the lysosomal inhibitor, E64c, were used to block CK1α degradation to prevent the increase in β-catenin stability. CSPCs were preincubated with 10 μM of lactacystin or E64c for 30 min before SDF1 treatment and levels of active β-catenin (unphosphorylated, stable) and total β-catenin were measured by Western analysis (Figure 6A). Proteosomal inhibition by lactacystin prevented an increase in active or stabilized β-catenin whereas lysosomal inhibition by E64c had no effect on increased β-catenin stability. If a proteosomal mechanism is responsible for CK1α degradation, it would be targeted for degradation by ubiquitin ligases. CK1α and β-catenin were immunoprecipitated (IP) from SDF1 treated CSPC lysates. Ubiquitination of IPed CK1α and β-catenin was then detected by Western analysis. As seen in Figure 6B, β-catenin was ubiquitinated and levels decreased after SDF1 treatment, whereas CK1α was not ubiquitinated. These results suggest CK1α may be degraded indirectly by a proteasomal dependent mechanism where its association with another protein could facilitate its degradation resulting in stabilization of β-catenin.
Figure 6. SDF1 regulates CK1α levels by proteasomal and transcriptional mechanisms.
A) Proteasomal degradation was necessary for decreased CK1α as the proteasomal inhibitor, Lactacystin (10 μmol/L), blocked SDF-dependent β-catenin whereas the lysosomal inhibitor, E64C (10 μmol/L), did not. C) β-Catenin was ubiquitinated, however, CK1α was not. D) CSPC SDF1 treatment, 25 nM for 1 and 6 hours, decreased CK1α mRNA levels. Values are the mean ± SE, n=3. * p<0.05 vs Veh.
As protein degradation via a ubiquitin dependent proteasomal mechanism could not clearly account for decreases in CK1α, changes in mRNA expression were examined. CK1α mRNA expression was significantly and progressively decreased in CSPCs treated with SDF1 for 1 and 6 hours (Figure 6C). These findings provide a mechanism to support SDF1-CXCR4 mediated CK1α downregulation and its effect on CSPC quiescence through decreased transcription or stability of CK1α mRNA.
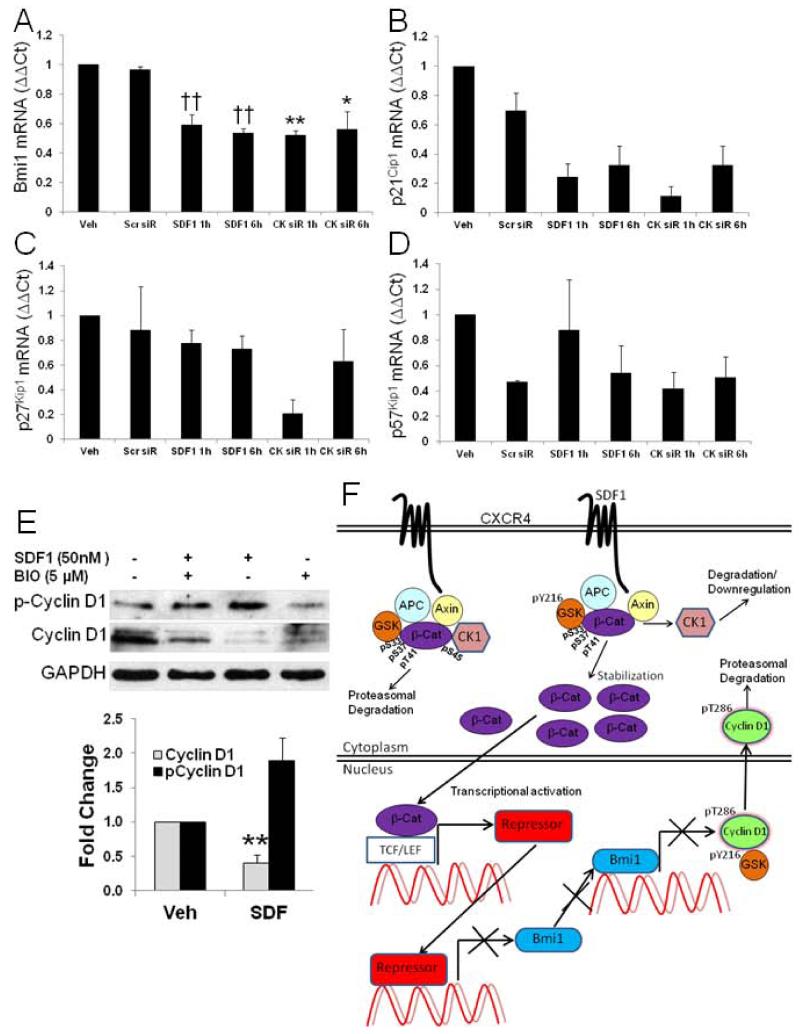
In HSCs where SDF1 has been characterized to exert a quiescent effect, among several cyclin-dependent kinase inhibitors (CKI) and cell cycle regulators examined, SDF1 was shown to exclusively induce the CKI, p57Kip1. To investigate the targets associated with SDF1 mediated quiescence in CSPCs we measured expression of CKIs p21Cip1, p27Kip1, and p57Kip1, and Bmi-1 (Figure 7A-D). The polycomb gene product, Bmi-1, was significantly downregulated by SDF1 progressively at 1 and 6 h time points which supports SDF1 slowdown of cell cycle progression. SDF1 downregulation of Bmi-1 was also replicated with CK1α siRNA knockdown. Surprisingly, p21Cip1 was downregulated with SDF1 at 1 and 6 hours whereas SDF1 had little effect on p27Kip1 or p57Kip1. In addition, levels of p21Cip1 were decreased to a similar extent with CK1α siRNA suggesting these are regulated by SDF1 via a CK1α-dependent mechanism. SDF1 decreased total cyclin D1 levels and increased cyclin D1 phosphorylation that would result in its export from the nucleus and targeting for proteasomal degradation (Figure 7E) effectively limiting G0/G1 transition. These findings suggest SDF1 regulates CSPC quiescence predominantly via a Bmi-1-dependent mechanism through CK1α downregulation and decreased cyclin D1 levels.
Figure 7. SDF1 decreases Bmi1, increases cyclin D1 phosphorylation, and decreases total cyclin D1 to block cell cycle progression.
A-D) Real time RT-PCR measured the effect of 25 nM SDF1 and CK1α siRNA treatment for 1 and 6 hours on cell cycle regulators p21Cip1, p27Kip1, p57Kip1 and Bmi1. E) SDF1 treatment increased cyclin D1 Y286 phosphorylation and decreased total levels. Treatment with 5 μM BIO antagonized the effect of SDF1 on cyclin D1 levels. Values are the mean ± SE, n=2-3, * p<0.05, ** p<0.01 vs Scr siR, †† p<0.01 vs Veh. F) Schematic summary of SDF1 – dependent signaling through CK1α and GSK3β resulting in attenuated Bmi-1 and cyclin D1.
Discussion
Cell cycle regulation is an important mechanism by which stem cells preserve self-renewal and longevity. Limiting stem cell proliferation is a fundamental stem cell characteristic. We present findings that support the role of SDF1 in mediating quiescence in a resident cardiac stem/progenitor cell population of c-kit+ CSPCs by limiting cell cycle progression. We also present novel mechanistic insights that demonstrate SDF1 regulates CK1α expression and GSK3β activity, which then directly affect β-catenin and cyclin D1 stability and CSPC quiescence. Decreased CK1α levels increase β-catenin stability and β-catenin-dependent transcription. SDF1 regulates CK1α through mechanisms dependent on decreased mRNA expression and proteasomal degradation. These events lead to CSPC quiescence by cell cycle arrest in G0 through the downregulation of the polycomb ring finger gene Bmi-1 and the consequential decrease in one of its target genes important in cell cycle progression, cyclin D1. Further, SDF1-dependent GSK3β Y216 phosphorylation increases GSK3β phosphorylation of cyclin D1 signaling its export from the nucleus, ubiquitinization, and degradation. This is the first report to demonstrate GPCR regulation of CK1α expression and GSK3β via Y216 phosphorylation, specifically, SDF1 activation of CXCR4, where both actions converge to facilitate quiescence of resident CSPCs through GSK3β dependent cyclin D1 phosphorylation and Bmi-1 dependent downregulation (see schematic Figure 7F).
The effect of SDF1-CXCR4 signaling to attenuate cell cycle by blocking progression at the G0/G1 transition to facilitate quiescence has been considered an important function to preserve hematopoietic stem cell (HSC) populations in the bone marrow. In this study, we report SDF1 induced quiescence is also demonstrated in a resident myocardial c-kit+ CSPC population. Still unknown in the myocardium is whether SDF1 contributes to the retention of CSPCs as it does for HSCs in the bone marrow through its chemoattractant properties and establishment of a niche environment that facilitates stem cell retention. Unlike the bone marrow environment, little is known regarding the role of SDF1 in maintaining or regulating stem or progenitor cells in the heart. In conjunction with the capacity to maintain stem cells in the bone marrow niche, the property of limiting proliferation of these cells suggests that SDF1-CXCR4 provides further support for its role in stem/progenitor cell maintenance extending to CSPCs resident in the heart.
SDF1 mediated quiescence may serve to preserve stem and progenitor cells in both the bone marrow and myocardium in a healthy state. However, the impact of increased SDF1 production and its effect to counteract expansion of CPSCs after ischemic injury, chronic ischemia, or stress may not be advantageous. For example, the hematologic recovery in response to 5-fluoro-uracil (5FU) chemoablation in SDF1−/− mice was significantly better than that in WT mice17. This was attributed to faster reconstitution of the hematopoietic compartment as bone marrow cellularity and SKL cells were higher in the knockout. Whether 5FU would typically increase SDF1 is not known and thus, if not increased with ablation, basal restrictions on proliferation are sufficient to limit expansion. In the heart, SDF1 levels are elevated transiently after infarction and thus would have an impact on expansion of CSPCs and other resident or transiting stem/lprogenitor cells29. The role of SDF1 quiescence in resident stem/progenitors, however, is complex. Our previous studies with chronic administration of the CXCR4 antagonist, AMD3100, resulted in exacerbated injury18 which was in sharp contrast to the improved response to injury with bolus administration immediately post-MI30, 31. We had shown that expansion of the c-kit+ CSPC population with chronic administration did not benefit the injured myocardium. Thus SDF1 affects additional actions that are blocked with chronic administration but not acute. Attenuated proliferation of side population cells by Wnt3a stimulation in the heart was recently shown to impair the response to injury and illustrates the importance of stem cell expansion after injury to support regeneration21.
Several studies have demonstrated SDF1-CXCR4 regulation of HSC quiescence, however, little is known regarding the responsible mechanism. Here we provide novel insight into the regulation of CSPC proliferation by SDF1-CXCR4 stimulation. In our previous studies SDF1 was shown to activate the MAPKs, ERK, JNK, and p38, and AKT in the myocardium to promote survival signaling against ischemic injury8. The studies presented here extend these by examining the downstream target of AKT, GSK3β and β-catenin. Initial studies showed that GSK3β was marginally affected with SDF1 stimulation via the AKT-dependent phoshorylation of S9; however, levels of β-catenin were significantly increased. Without significant regulation through GSK3β we examined the role of CK1α. CK1α serves a permissive function in β-catenin degradation via its phosphorylation at S45. The marked attenuation of β-catenin S45 phosphorylation suggested CK1α activity was significantly inhibited. Although no experimental evidence has previously been provided to demonstrate CK1α is regulated, we found levels of CK1α were markedly downregulated and this corresponded closely with the decrease in β-catenin S45 phosphorylation and its increased stabilization. Although several CK isoforms exist, CK1α appears to be the predominant isoform regulated by SDF1 that contributes to β-catenin regulation and quiescence. CK1α siRNA knockdown replicated the SDF1 stimulated decrease in β-catenin phosphorylation and quiescence providing direct support for its role in mediating signaling and function downstream of SDF1-CXCR4. Further, β-catenin and GSK3β knockdown increased proliferation as would be predicted. SDF1 down-regulated CK1α via two mechanisms. Proteosomal degradation was necessary for CK1α down regulation, however, this mechanism relies on proteosomal degradation of a CK1α-associated protein as CK1α itself was not ubiquitinated. Levels of CK1α mRNA were also decreased by SDF1. Studies are ongoing to identify the mechanisms by which CK1α levels are regulated by proteosomal degradation and mRNA stability or transcription.
The mechanism by which CK1α downregulation facilitates CSPC quiescence involves downregulation of the polycomb group gene product, Bmi-1, which targets the Ink4a-ARF gene locus encoding transcriptional regulators p16 and p19ARF and cyclin D1. Studies in the conditional CXCR4 KO HSCs had shown significant increases in cyclin D1 that could account for release from quiescence which was accompanied by decreased p57Kip2, a CKI, that would contribute to increased proliferation11. Conversely, SDF1 treatment increased p57Kip2 in these cells. Downregulation of Bmi-1, with both SDF1 treatment and CK1α siRNA, however, is the predominant factor contributing to CSPC quiescence as Bmi-1, noted above, regulates cyclin D1 expression and other CKIs, p21Cip1, p27Kip1, and p57Kip1, were either decreased which would be contrary to their contribution in CSPC quiescence or not affected. Bmi-1 has been shown to play an important role in HSC self-renewal and proliferation32. Conditional deletion of Bmi-1, a transcriptional repressor, resulted in senescence through exit from cell cycle before S phase in mouse embryonic fibroblasts which was associated with increased expression of tumor suppressor genes p16 and p19ARF of the Ink4a locus33.
GSK3β contributes to SDF1 attenuation of cell cycle progression independently of CK1α and β-catenin. SDF1 mediated GSK3β Y216 phosphorylation, in the absence of increased S9 phosphorylation, results in a net increase in activity. These in vitro results extend and support our in vivo findings where chronic AMD3100 antagonism of CXCR4 in a mouse myocardial infarction model decreased GSK3β Y216 phosphorylation18. Although GSK3β autophosphorylates Y216, HSP90 inhibition was able to prevent Y216 phosphorylation27. The mechanism by which SDF1 affects autophosphorylation of GSK3β Y216 or its phosphorylation is unknown. GSK3β targets cyclin D1, cyclin E and Cdc25A for phosphorylation, signaling their ubiquitination, degradation and exit from cell cycle progression34-36. Increased CSPC proliferation with BIO inhibition of GSK3β Y216 phosphorylation and activity or GSK3β knockdown supports SDF1 action increasing activity, cyclin D1 phosphorylation, decreased cyclin D1, and blockade of cell cycle progression. As SDF1 is able to recruit different mechanisms that facilitate quiescence, the relative contribution from each of these is unknown. At least in the case of cyclin D1, both CK1α and GSK3β converge by affecting expression and stability, respectively, resulting in downregulation. These findings demonstrate SDF1 exerts redundant mechanisms to facilitate CSPC quiescence.
Conclusions
These findings provide relevant and novel insight into mechanisms by which SDF1-CXCR4 regulates proliferation of a resident cardiac stem/progenitor cell, the c-kit+ CSPC. This is the first in vitro study to describe SDF1 regulation of CK1α expression and increased GSK3β activity as mechanisms by which SDF1 mediates CSPC quiescence. Further, CK1α levels are shown to have a direct effect on expression of Bmi1. SDF1 actions on CK1α and GSK3β both contribute to downregulation of cyclin D1. These findings demonstrated SDF1 is a physiological regulator of CSPC quiescence where its action blocks cell cycle progression and will have implications for the understanding of myocardial homeostasis and response to ischemic injury and other stimuli that affect SDF1 expression.
Supplementary Material
Supplemental Figure 1. A) Flow cytometry analysis of c-kit+ CPSCs. CSPCs isolated by magnetic sorting at passage P3 were stained for c-kit and c-kit staining analyzed by flow cytometry on a LSRII flow cytometer. B) The percent contribution of c-kit+ CSPCs to the total population remained between 70 and 80% at passages P6 and P8.
Supplemental Figure 2. CSPC Annexin V staining did not change with SDF1 treatment. CSPCs were treated with SDF1 for 24 hours and then staining for the apoptosis marker Annexin V and the nuclear stain PI.
Supplemental Figure 3. c-kit+ CSCPs express the mitosis marker Ki67. CSPCs were stained for c-kit and Ki67 and analyzed by flow cytometry.
Acknowledgements
This work was supported by NIH grants HL091202 and HL074351. We thank Dr. Randall Moon for kindly providing TOPFlash and FOPFlash reporters through Addgene.
Footnotes
Author contribution:
Neviana Dimova: Collection and assembly of data, data analysis and interpretation.
Marcin Wysoczynski: Collection and assembly of data.
Gregg Rokosh: Concept and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclaimers: None
References
- 1.Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh PC, Segers VF, Davis ME, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller P, Kazakov A, Semenov A, et al. Pressure-induced cardiac overload induces upregulation of endothelial and myocardial progenitor cells. Cardiovascular research. 2008;77:151–159. doi: 10.1093/cvr/cvm037. [DOI] [PubMed] [Google Scholar]
- 4.Rupp S, Bauer J, von Gerlach S, et al. Pressure overload leads to an increase of cardiac resident stem cells. Basic Res Cardiol. 2012;107:252. doi: 10.1007/s00395-012-0252-x. [DOI] [PubMed] [Google Scholar]
- 5.Ratajczak MZ, Machalinski B, Wojakowski W, et al. A hypothesis for an embryonic origin of pluripotent Oct-4(+) stem cells in adult bone marrow and other tissues. Leukemia. 2007;21:860–867. doi: 10.1038/sj.leu.2404630. [DOI] [PubMed] [Google Scholar]
- 6.Bolli R, Chugh AR, D’Amario D, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Tillmanns J, Rota M, Hosoda T, et al. Formation of large coronary arteries by cardiac progenitor cells. Proc Natl Acad Sci U S A. 2008;105:1668–1673. doi: 10.1073/pnas.0706315105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X, Dai S, Wu WJ, et al. Stromal cell derived factor-1 alpha confers protection against myocardial ischemia/reperfusion injury: role of the cardiac stromal cell derived factor-1 alpha CXCR4 axis. Circulation. 2007;116:654–663. doi: 10.1161/CIRCULATIONAHA.106.672451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kucia M, Jankowski K, Reca R, et al. CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol Histol. 2004;35:233–245. doi: 10.1023/b:hijo.0000032355.66152.b8. [DOI] [PubMed] [Google Scholar]
- 10.Seeger FH, Rasper T, Koyanagi M, et al. CXCR4 expression determines functional activity of bone marrow-derived mononuclear cells for therapeutic neovascularization in acute ischemia. Arterioscler Thromb Vasc Biol. 2009;29:1802–1809. doi: 10.1161/ATVBAHA.109.194688. [DOI] [PubMed] [Google Scholar]
- 11.Nie Y, Han YC, Zou YR. CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med. 2008;205:777–783. doi: 10.1084/jem.20072513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeger FH, Zeiher AM, Dimmeler S. Cell-enhancement strategies for the treatment of ischemic heart disease. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S110–113. doi: 10.1038/ncpcardio0734. [DOI] [PubMed] [Google Scholar]
- 13.Ceradini DJ, Kulkarni AR, Callaghan MJ, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 14.Kucia M, Reca R, Miekus K, et al. Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells. 2005;23:879–894. doi: 10.1634/stemcells.2004-0342. [DOI] [PubMed] [Google Scholar]
- 15.Itkin T, Lapidot T. SDF-1 keeps HSC quiescent at home. Blood. 2011;117:373–374. doi: 10.1182/blood-2010-09-307843. [DOI] [PubMed] [Google Scholar]
- 16.Levesque JP, Helwani FM, Winkler IG. The endosteal ‘osteoblastic’ niche and its role in hematopoietic stem cell homing and mobilization. Leukemia. 2010;24:1979–1992. doi: 10.1038/leu.2010.214. [DOI] [PubMed] [Google Scholar]
- 17.Tzeng YS, Li H, Kang YL, et al. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117:429–439. doi: 10.1182/blood-2010-01-266833. [DOI] [PubMed] [Google Scholar]
- 18.Dai S, Yuan F, Mu J, et al. Chronic AMD3100 antagonism of SDF-1alpha-CXCR4 exacerbates cardiac dysfunction and remodeling after myocardial infarction. J Mol Cell Cardiol. 2010;49:587–597. doi: 10.1016/j.yjmcc.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cashman J, Clark-Lewis I, Eaves A, et al. Stromal-derived factor 1 inhibits the cycling of very primitive human hematopoietic cells in vitro and in NOD/SCID mice. Blood. 2002;99:792–799. doi: 10.1182/blood.v99.3.792. [DOI] [PubMed] [Google Scholar]
- 20.Meijer L, Skaltsounis AL, Magiatis P, et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chemistry & biology. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Oikonomopoulos A, Sereti KI, Conyers F, et al. Wnt signaling exerts an antiproliferative effect on adult cardiac progenitor cells through IGFBP3. Circ Res. 2011;109:1363–1374. doi: 10.1161/CIRCRESAHA.111.250282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho J, Rameshwar P, Sadoshima J. Distinct roles of GSK-3{alpha} ans GSK-3{beta} in mediating cardiomyocyte differentiation in muring bone marrow derived mesechymal stem cells. J Biol Chem. 2009 doi: 10.1074/jbc.M109.019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda T, Zhai P, Maejima Y, et al. Distinct roles of GSK-3alpha and GSK-3beta phosphorylation in the heart under pressure overload. Proc Natl Acad Sci U S A. 2008;105:20900–20905. doi: 10.1073/pnas.0808315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knippschild U, Gocht A, Wolff S, et al. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cellular signalling. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Veeman MT, Slusarski DC, Kaykas A, et al. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 26.Urbanek K, Cesselli D, Rota M, et al. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci U S A. 2006;103:9226–9231. doi: 10.1073/pnas.0600635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochhead PA, Kinstrie R, Sibbet G, et al. A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Molecular cell. 2006;24:627–633. doi: 10.1016/j.molcel.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 29.Kucia M, Dawn B, Hunt G, et al. Cells expressing early cardiac markers reside in the bone marrow and are mobilized into the peripheral blood after myocardial infarction. Circ Res. 2004;95:1191–1199. doi: 10.1161/01.RES.0000150856.47324.5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jujo K, Hamada H, Iwakura A, et al. CXCR4 blockade augments bone marrow progenitor cell recruitment to the neovasculature and reduces mortality after myocardial infarction. Proc Natl Acad Sci U S A. 2010;107:11008–11013. doi: 10.1073/pnas.0914248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proulx C, El-Helou V, Gosselin H, et al. Antagonism of stromal cell-derived factor-1alpha reduces infarct size and improves ventricular function after myocardial infarction. Pflugers Arch. 2007;455:241–250. doi: 10.1007/s00424-007-0284-5. [DOI] [PubMed] [Google Scholar]
- 32.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs JJ, Kieboom K, Marino S, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 34.Diehl JA, Cheng M, Roussel MF, et al. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang T, Wei Y, Honaker Y, et al. GSK-3 beta targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3 beta inactivation correlates with Cdc25A overproduction in human cancers. Cancer cell. 2008;13:36–47. doi: 10.1016/j.ccr.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welcker M, Singer J, Loeb KR, et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Molecular cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. A) Flow cytometry analysis of c-kit+ CPSCs. CSPCs isolated by magnetic sorting at passage P3 were stained for c-kit and c-kit staining analyzed by flow cytometry on a LSRII flow cytometer. B) The percent contribution of c-kit+ CSPCs to the total population remained between 70 and 80% at passages P6 and P8.
Supplemental Figure 2. CSPC Annexin V staining did not change with SDF1 treatment. CSPCs were treated with SDF1 for 24 hours and then staining for the apoptosis marker Annexin V and the nuclear stain PI.
Supplemental Figure 3. c-kit+ CSCPs express the mitosis marker Ki67. CSPCs were stained for c-kit and Ki67 and analyzed by flow cytometry.