ABSTRACT
Bacterial DNA and live bacteria have been detected in human urine in the absence of clinical infection, challenging the prevailing dogma that urine is normally sterile. Urgency urinary incontinence (UUI) is a poorly understood urinary condition characterized by symptoms that overlap urinary infection, including urinary urgency and increased frequency with urinary incontinence. The recent discovery of the urinary microbiome warrants investigation into whether bacteria contribute to UUI. In this study, we used 16S rRNA gene sequencing to classify bacterial DNA and expanded quantitative urine culture (EQUC) techniques to isolate live bacteria in urine collected by using a transurethral catheter from women with UUI and, in comparison, a cohort without UUI. For these cohorts, we demonstrated that the UUI and non-UUI urinary microbiomes differ by group based on both sequence and culture evidences. Compared to the non-UUI microbiome, sequencing experiments revealed that the UUI microbiome was composed of increased Gardnerella and decreased Lactobacillus. Nine genera (Actinobaculum, Actinomyces, Aerococcus, Arthrobacter, Corynebacterium, Gardnerella, Oligella, Staphylococcus, and Streptococcus) were more frequently cultured from the UUI cohort. Although Lactobacillus was isolated from both cohorts, distinctions existed at the species level, with Lactobacillus gasseri detected more frequently in the UUI cohort and Lactobacillus crispatus most frequently detected in controls. Combined, these data suggest that potentially important differences exist in the urinary microbiomes of women with and without UUI, which have strong implications in prevention, diagnosis, or treatment of UUI.
IMPORTANCE
New evidence indicates that the human urinary tract contains microbial communities; however, the role of these communities in urinary health remains to be elucidated. Urgency urinary incontinence (UUI) is a highly prevalent yet poorly understood urinary condition characterized by urgency, frequency, and urinary incontinence. Given the significant overlap of UUI symptoms with those of urinary tract infections, it is possible that UUI may have a microbial component. We compared the urinary microbiomes of women affected by UUI to those of a comparison group without UUI, using both high-throughput sequencing and extended culture techniques. We identified statistically significant differences in the frequency and abundance of bacteria present. These differences suggest a potential role for the urinary microbiome in female urinary health.
INTRODUCTION
The female urinary tract is a previously overlooked microbial niche. Recent detection of bacterial DNA (1–3) and live bacteria (4, 5) in urine from women with and without lower urinary tract symptoms has provided compelling evidence that the female urinary tract possesses its own unique microbiota. Research into the roles of these bacterial communities in urinary health and disease requires sensitive and specific detection and classification. Although the comparatively low bacterial load in urine challenges such effort, expanded culture conditions have allowed researchers to detect bacteria at lower levels than previously used techniques. For example, we recently reported that bacteria overlooked by standard culture conditions are cultivable by adjusting the growth conditions to include increased urine volume, diverse growth media and atmospheric conditions, and lengthened incubation time (4). Using expanded quantitative urine culture (EQUC), we isolated bacteria from 80% of examined urine samples collected by transurethral catheter, most (92%) of which had been reported as “no growth,” using a standard clinical microbiology urine culture protocol and a 103-CFU/ml threshold (4). Despite this significant advance, many urinary bacteria cannot be cultured under these conditions. Thus, culture-independent methods for bacterial detection, such as high-throughput sequencing of the 16S rRNA gene, have emerged as the predominant research technique, especially as they become increasingly accessible due to declining sequencing cost and improved bioinformatics tools (6).
There is a clinical need for these improved research techniques. For example, urgency urinary incontinence (UUI) is poorly understood chronic urinary condition that is often attributed to abnormal neuromuscular signaling and/or functioning. However, alternative or complementary mechanisms beyond neuromuscular abnormalities must be considered, since UUI pharmacological treatments aimed at the overactive bladder are ineffective in approximately half of the pharmacologically treated UUI population (7, 8).
UUI is common, affecting 1.5 to 22% of the U.S. population, most frequently women and the elderly (8). Affected patients experience a sudden, intense need to urinate with involuntary urine loss that detracts from the patient’s quality of life (9). A proportion of UUI cases resolve over time; however, it is a chronic condition for most women (8). There is a large socioeconomic burden, with the costs of UUI in the United States projected to reach $76.2 billion by 2015 (10).
The clinical diagnosis of UUI requires exclusion of urinary tract infections (UTI); thus, infectious etiology is not considered for UUI. Given the clinical similarly of UUI and UTI symptoms, however, we and others have recently used expanded culture techniques (4, 5) and quantitative PCR (2) to show the presence of bacteria in standard culture-“negative” urine samples collected from UUI patients.
In this analysis, we utilized both 16S rRNA gene sequencing and EQUC to characterize the microbiome in urine obtained by transurethral catheter from women seeking treatment for UUI and a comparison group of women without UUI. Utilizing both techniques, we identified statistically significant differences in the frequency and abundance of bacteria present, suggesting a potential role for the urinary microbiome in female urinary health.
RESULTS
Cohort description.
Table 1 displays the UUI symptoms by cohort. Consistent with the intended composition of the cohorts, the UUI cohort reported more distress on both the urinary and prolapse subscales of the Pelvic Floor Distress Inventory (PFDI) (urinary distress inventory [UDI], 102 [±47] versus 11 [±11] [P < 0.05]; Pelvic Organ Prolapse Distress Inventory [POPDI], 70 [±52] versus 28 [±32] [P < 0.05]). The UUI cohort also reported more-severe overactive bladder (OAB) symptoms (66 [±23] versus 4 [±7]; P < 0.05) and decreased quality of life as measured by health-related quality of life (HRQL) (56 [±25] versus 99 [±3]; P < 0.05) compared to the non-UUI cohort.
TABLE 1 .
Participant demographics and symptoms
| Characteristic | Value for group (na)b or statistic |
||
|---|---|---|---|
| UUI (60) | Non-UUI Control (58) | P valuec | |
| Demographics | |||
| Age (yrs) | 63 (±12) | 49 (±14) | <0.05 |
| Race [no. (%)] | 0.42 | ||
| Caucasian | 50 (83) | 41 (77) | |
| African-American | 10 (17) | 11 (21) | |
| Asian | 0 (0) | 1 (2) | |
| Estrogen status [no. (%)] | <0.05 | ||
| Estrogen positive | 16 (27) | 31 (61) | |
| Estrogen negative | 43 (73) | 20 (39) | |
| Body mass index (kg/m2) | 32 (±8) | 28 (±6) | 0.01 |
| Hypertension [no. (%)] | 21 (35) | 9 (17) | 0.03 |
| Coronary artery disease [no. (%)] | 9 (15) | 1 (2) | 0.02 |
| Symptoms | |||
| Symptom scored | 66 (±23) | 4 (±7) | <0.05 |
| Health-related quality of life (HRQL)d | 56 (±25) | 99 (±3) | <0.05 |
| Urinary Distress Inventory (UDI)e | 102 (±47) | 11 (±11) | <0.05 |
| Pelvic Organ Prolapse Distress Inventory (POPDI)e | 70 (±52) | 28 (±32) | <0.05 |
| Colorectal-Anal Distress Inventory (CRADI)e | 71 (±61) | 43 (±62) | 0.02 |
n, no. of subjects.
Mean ± SD or no. (%).
Pearson’s chi-square and Fisher’s exact tests were used with categorical variables. Student’s t test was used with continuous variables.
Based on OAB questionnaire.
Based on Pelvic Floor Disease Inventory.
Table 1 also displays the participant demographics by cohort. Marital status, diabetes, smoking, and prior pelvic surgeries did not differ by group. However, the UUI cohort was heavier (body mass index [BMI], 32 [±8] versus 28 [±6]; P = 0.01), less likely to be using estrogen (27% versus 61%; P < 0.05), and older (63 [±12] versus 49 [±14]; P < 0.05).
Sequence-based characterization of female urinary microbiome.
Bacterial DNA was detected in similar proportions of urine samples (UUI, 63.9% [23/36]; non-UUI, 65.8% [25/38]). The lack of bacterial detection in the remaining samples may be due to low bacterial load, insufficient bacterial lysis, and/or primer bias rather than being conclusive evidence of no bacteria in these urine samples.
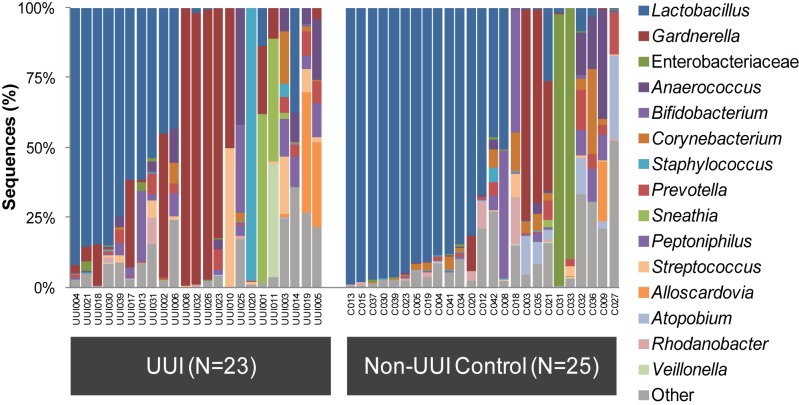
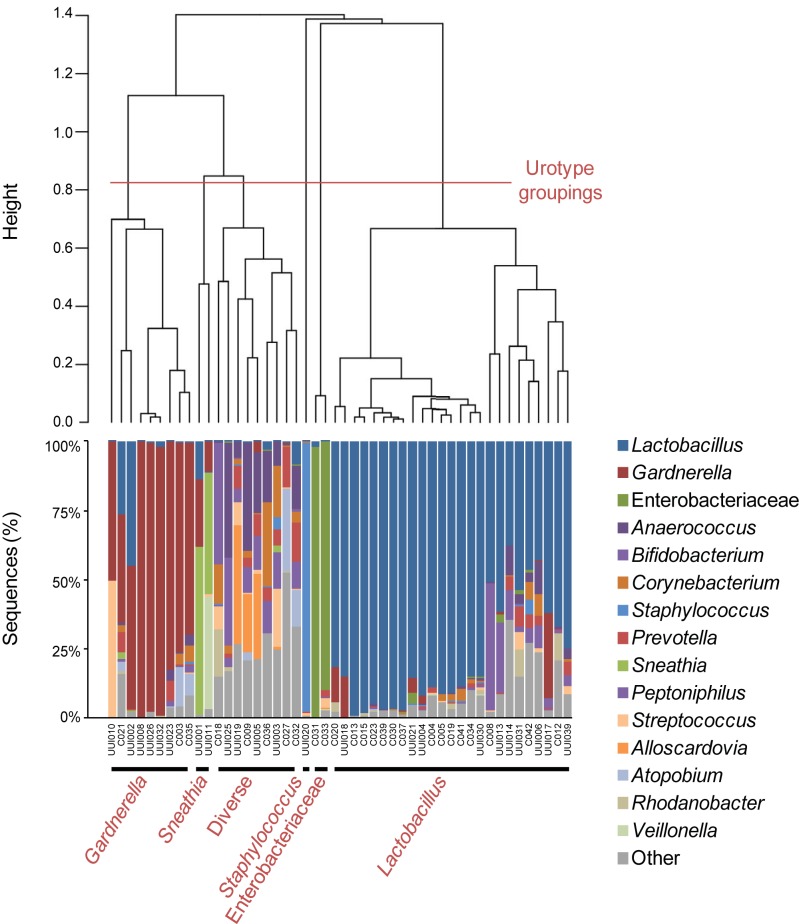
Further analyses were performed on the samples with detectable bacterial DNA (for UUI, n = 23; for non-UUI, n = 25). The sequences from these samples were classified into 22 phyla, 34 classes, 69 orders, 150 families, and 386 genera. The most abundant phyla detected were Firmicutes, with a median abundance of 60%, followed by Actinobacteria (16%), Proteobacteria (1.5%), and Bacteroidetes (0.6%). From each sample, the majority of sequences could be classified to the genus level. The two exceptions (C031 and C033) were predominantly Enterobacteriaceae (Fig. 1). In the remainder of the samples, the percentage of sequences per sample that could not be classified to the genus level ranged from 0.02 to 24.1%. At the genus level, the majority of urine samples were dominated by one or two bacterial families or genera, most frequently Lactobacillus and Gardnerella (Fig. 1). Urine samples with the same dominant taxa clustered together in a dendrogram generated via hierarchical clustering of the Euclidean distance between urine samples (Fig. 2). We observed six groups, now termed urotypes, which were named based on either the dominant family or genus (Gardnerella, Sneathia, Staphylococcus, Enterobacteriacae, and Lactobacillus) or the lack of a dominant family or genus (“diverse”). The most frequent urotype in both cohorts was Lactobacillus (43% UUI and 60% non-UUI controls), followed by Gardnerella (26% UUI and 12% non-UUI controls) (Table 2). Three urotypes were present in both cohorts (Gardnerella, Lactobacillus, and diverse). The Staphylococcus and Sneathia urotypes were present only in the UUI cohort, whereas the Enterobacteriaceae urotype was present only in the non-UUI cohort; however, these differences were not statistically significant at our current sample size (Fig. 1 and Table 2).
FIG 1 .
Urinary microbiome profile by cohort based on 16S rRNA gene V4 sequencing. Stacked bar plots depict the sequence abundances of the 15 most abundant genus- or family-level taxa in the UUI and non-UUI cohorts. Taxa were ranked according to mean abundance across all samples. The y axis represents the percentage of sequences for a particular bacterial taxa; the x axis represents the study participants separated by cohort. The family Enterobacteriaceae could not be classified to the genus level. The remainder of sequences were combined in the category labeled Other.
FIG 2 .
Clustering of the urinary microbiome into urotypes. The dendrogram was based on hierarchical clustering of the Euclidean distance between samples in the combined UUI and non-UUI cohorts. The dashed line depicts where the clades were divided into 6 urotypes: Gardnerella, Sneathia, Diverse, Staphylococcus, Enterobacteriaceae, and Lactobacillus. The stacked bar plot below the dendrogram depicts the sequence abundances of the overall most abundant taxa.
TABLE 2 .
Urotype frequency by cohort
| Urotype | No. (%) of participants for group (na) |
P valuec | |
|---|---|---|---|
| UUI (23)b | Non-UUI control (25)b | ||
| Gardnerella | 6 (26) | 3 (12) | 0.23 |
| Sneathia | 2 (9) | 0 (0) | 0.22 |
| Diverse | 4 (17) | 5 (20) | 0.99 |
| Staphylococcus | 1 (4) | 0 (0) | 0.48 |
| Enterobacteriaceae | 0 (0) | 2 (8) | 0.49 |
| Lactobacillus | 10 (43) | 15 (60) | 0.39 |
n, no. of subjects.
Analysis performed on the subset of samples with detectable bacterial DNA.
Fisher’s exact test.
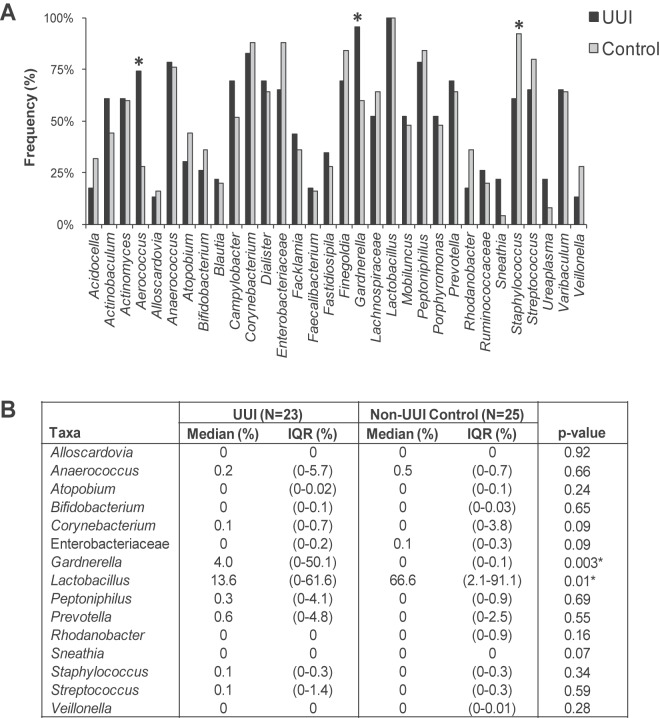
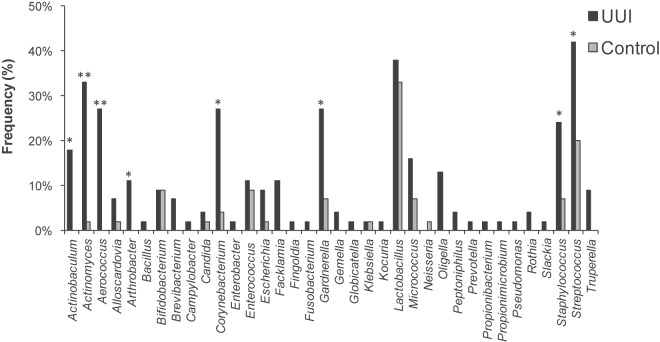
We calculated the frequency that each genus was observed in each cohort (Fig. 3A). While Lactobacillus was detected in every sample in both cohorts, some genera were observed more frequently in one cohort than in the other. For example, Gardnerella (99% UUI, 60% non-UUI; P = 0.003) and Aerococcus (74% UUI, 28% non-UUI; P = 0.002) were detected more frequently in the UUI cohort than in the non-UUI one. Conversely, Staphylococcus (61% UUI, 92% non-UUI; P = 0.01) was observed less frequently in the UUI cohort than among non-UUI women. Actinobaculum (61% UUI, 44% non-UUI; P = 0.24) and Sneathia (22% UUI, 4% non-UUI; P = 0.09) exhibited a trend toward increased detection in the UUI cohort; however, this difference was not statistically significant at our current sample size.
FIG 3 .
Comparison of sequence-based urinary microbiome by cohort. The frequency (A) and median sequence abundance (B) of the overall most abundant taxa detected by sequencing were calculated. The families Enterobacteriaceae, Lachnospiraceae, and Ruminococcaceae could not be classified to the genus level. In panel A, a combination of Pearson chi-square and Fisher’s exact tests was used to compare the frequency of genera detected by sequencing between the cohorts. “*” represents a P value of <0.05. In panel B, a Wilcoxon rank sum test was used to compare the median sequence abundances between the cohorts. IQR, interquartile range. “*” represents P values of <0.05.
By calculating frequency, all samples are weighted similarly regardless of sequence abundance. For example, urine samples collected from control participants C013 and C036 both contained Lactobacillus sequences; however, the sequence abundances differed. For C013, Lactobacillus represented more than 99% of the sequences; for C036, Lactobacillus represented less than 5% of the total sequences (Fig. 1). To further investigate these differences, the median sequence abundance was calculated and compared between the cohorts (Fig. 3B). The UUI cohort exhibited decreased Lactobacillus (14% UUI, 67% non-UUI; P = 0.01) and increased Gardnerella (4% UUI, 0% non-UUI; P = 0.003) sequence abundances compared to those for the control cohort. Figure S2 in the supplemental material displays the distribution of the 15 most abundant taxa detected by sequencing.
To measure the richness of the UUI and non-UUI urinary microbiomes, the number of observed operational taxonomic units (OTUs) and the Chao1 estimator were calculated. To measure diversity, the Shannon index and inverse Simpson index were calculated. Overall, the urinary microbiome exhibited an average of 97 OTUs per urine, a Chao1 index of 7,210, an inverse Simpson’s index of 1.72, and a Shannon index of 3.93 (Table 3). There were no statistically significant differences between the UUI and non-UUI control cohorts based on these estimators of richness and diversity.
TABLE 3 .
Richness and diversity measures of urinary microbiome
| Metric | Total [mean (SD)] | UUI (n = 23)a |
Non-UUI control (n = 25)a |
P valueb | ||
|---|---|---|---|---|---|---|
| Mean (SD) | Min–Max | Mean (SD) | Min–Max | |||
| No. of observed OTUS (richness) | 97.02 (40.6) | 86.35 (36.89) | 25–155 | 106.84 (42.17) | 39–201 | 0.08 |
| Chao1 estimator (richness) | 7,210 (5,331) | 5,977 (4,647) | 2,688–20,021 | 8,344 (5,750) | 1,697–22,997 | 0.12 |
| Shannon index (diversity) | 1.72 (0.96) | 1.70 (0.91) | 0.17–3.68 | 1.74 (1.02) | 0.30–3.83 | 0.88 |
| Inverse Simpson index (diversity) | 3.93 (4.28) | 3.69 (3.39) | 1.03–16.08 | 4.15 (5.07) | 1.07–20.40 | 0.71 |
Analysis performed on the subset of samples with detectable bacterial DNA. Min, minimum; Max, maximum.
Student’s t test.
Culture-based characterization of the female urinary microbiota.
Of the urine specimens assayed via EQUC, 71/90 (78.9%) grew bacterial species, while 64 of the 71 (90.1%) that grew bacterial species were deemed culture negative (no growth) by standard culture. For the UUI and control cohorts, standard culture had false-negative rates of 90.3% and 90.0%, respectively. This highlights the limitations of the standard clinical microbiology protocol.
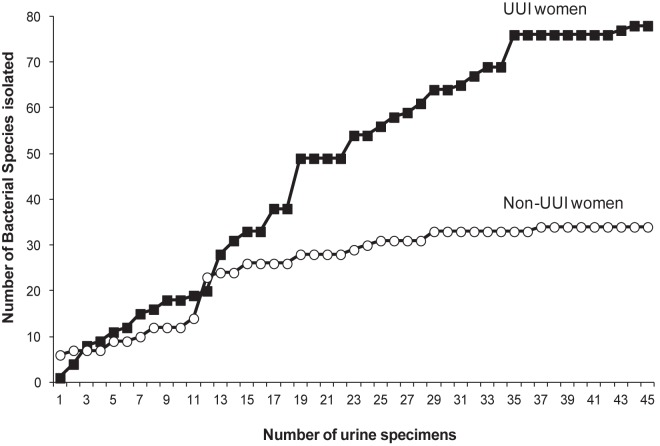
Rarefaction curves comparing the number of bacteria species to the number of urine samples assayed reveal that the UUI cohort curve began to plateau at a higher number of isolates than the non-UUI cohort curve (Fig. 4), indicating that these two cohorts possess different urinary microbiota. In support of this supposition, there was a statistically significant difference between the median numbers of bacterial isolates cultured from the UUI women and those for the non-UUI women (4 [interquantile ratio {IQR} = 1 to 7] versus 1 [IQR = 1 to 7]; P < 0.001).
FIG 4 .
Rarefaction curves of the cultured bacterial species by cohort. The plot depicts the number of species cultured via EQUC by the number of urine samples assayed.
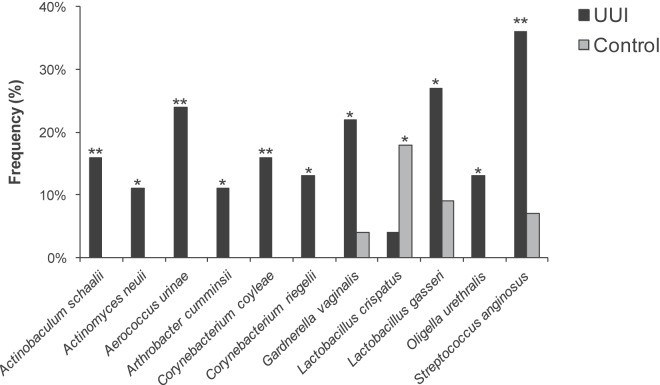
The frequencies at which we cultured each genus were compared between cohorts. Nine genera (Actinobaculum, Actinomyces, Aerococcus, Arthrobacter, Corynebacterium, Gardnerella, Oligella, Staphylococcus, and Streptococcus) were more frequently isolated from the UUI cohort than from controls (Fig. 5). Four of these genera were isolated solely from the UUI cohort (Actinobaculum, Aerococcus, Arthrobacter, and Oligella); the concentrations ranged from 10 CFU/ml to 100,000 CFU/ml. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), the majority of the cultured isolates were classified to the species level, one taxonomic level further than was achievable with V4 sequencing. This finer level of resolution revealed additional differences between the cohorts. For example, the genus Lactobacillus was frequently detected in both cohorts; however, when examination at the species level was done, distinct differences were detected between cohorts. Lactobacillus gasseri was more frequently isolated from the UUI cohort (27% UUI, 9% non-UUI; P = 0.02), whereas Lactobacillus crispatus was more frequently isolated from the control cohort (4% UUI, 18% non-UUI; P = 0.037) (Fig. 6). Also, Actinobaculum schaalii, Actinomyces neuii, Aerococcus urinae, Arthrobacter cumminsii, Corynebacterium coyleae, Gardnerella vaginalis, Oligella urethralis, and Streptococcus anginosus were more frequently isolated from UUI women (see Table S2 in the supplemental material).
FIG 5 .
Genus-level comparison of cultured urinary microbiota by cohort. The Pearson chi-square and Fisher’s exact tests were used to compare the frequencies of the genera isolated from urine via EQUC. *, P <0.05; **, P < 0.001.
FIG 6 .
Species-level comparison of cultured urinary microbiota by cohort. The Pearson chi-square and Fisher’s exact tests were used to compare the frequencies of the species isolated from urine via EQUC. *, P < 0.05; **, P < 0.01.
Comparison of culture versus sequencing for defining the urinary microbiome.
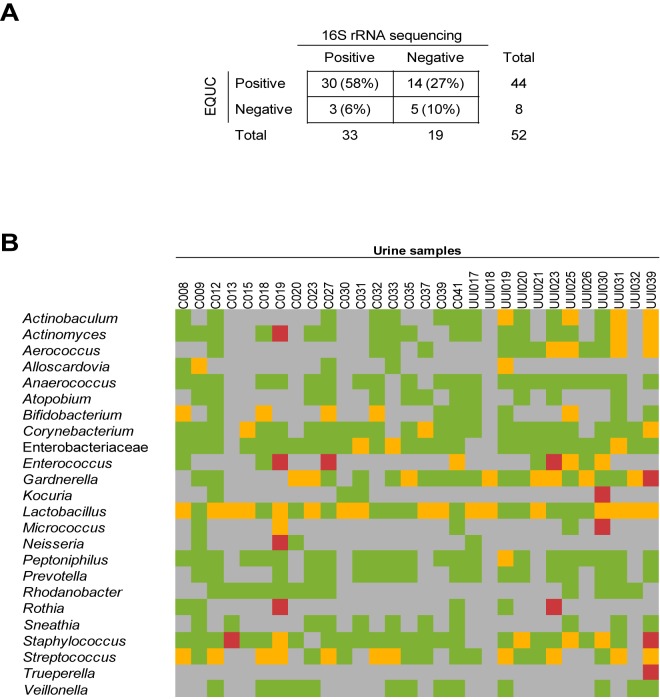
More than half (58%; 30/52) of specimens examined by both EQUC and 16S rRNA gene sequencing tested positive for bacteria by both techniques. Bacteria were cultured from 14 (27%) sequence-negative urine samples, while bacterial DNA was sequenced from 3 (6%) EQUC-negative urine samples. Altogether, bacteria were detected in 90% (47/52) of urine samples by EQUC or sequencing or both. Only 10% (5/52) of the urine samples tested negative for bacteria by both approaches (Fig. 7A).
FIG 7 .
Comparison of taxa detected by 16S rRNA gene sequencing and EQUC. (A) Comparison of sequence status and EQUC status for the 52 urine samples that were assayed by both methods. (B) Comparison of the taxa detected by sequencing and culture of the sequence-positive, EQUC-positive urine samples (n = 30). Each square was color coded based on whether the taxa were detected by sequence only (green), EQUC only (red), sequence and EQUC (yellow), or neither sequence nor EQUC (gray).
Of the 30 urine samples that were positive by both sequencing and EQUC, there was considerable overlap in terms of the bacterial taxa detected (Fig. 7B). A total of 18 different genera were cultured, the majority of which were detected by sequencing, thus providing additional evidence of their presence in the urinary microbiome and indicating that many of the sequenced genera represent live bacteria. In some urine samples, EQUC detected genera that were not detected by sequencing; however, these nine genera were detected by sequencing in other urine samples, demonstrating that these genera could be amplified by the universal primers. Trueperella was the only cultured genus that was not detected at the sequencing level in any of the urine samples. In contrast, some genera, such as Atopobium, were detected via sequencing but not by culture, suggesting that even the expanded culture technique was limited.
DISCUSSION
This analysis directly compared the urinary microbiome of women with UUI to that of women without UUI symptoms. We used two independent yet complementary techniques, 16S rRNA gene sequencing and extended culture, to characterize the female urinary microbiome. Both techniques identified statistically significant differences in the frequency and abundance of bacteria. These differences suggest a potential role for the urinary microbiome in female urinary health and warrant further study.
Each technique detected similar but not identical microbiome profiles. High-throughput sequencing provided the broader view of the bacteria present in the bladder, regardless of ability to cultivate the organisms. On the other hand, the cultured isolates could be identified to the species level, thus providing a finer level of resolution for the female urinary microbiome. Furthermore, these cultured isolates can be utilized in future studies to further investigate their potential for symbiosis or pathogenesis. Combined, these complementary techniques yielded overlapping results and provided the most complete description of the female urinary microbiome to date.
In both cohorts, one or two genera dominated the majority of the sequence profiles. The two most frequently detected genera, by both sequencing and culture, were Lactobacillus and Gardnerella. The former are lactic acid-producing, facultative anaerobic bacteria known to play protective roles in the vaginal tract by decreasing pH and producing various bacteriostatic/cidal compounds (11, 12). The latter are also facultative anaerobic bacteria frequently isolated in the vaginal tract, often in association with bacterial vaginosis. In this study, Lactobacillus was detected at similar frequencies in both cohorts but displayed lower median sequence abundance for women with UUI than for those without UUI. Furthermore, at the species level, the Lactobacillus species differed between cohorts. Whereas L. gasseri was more frequently cultured in samples from the UUI cohort, L. crispatus was more frequently cultured in samples from controls. Why the distribution of these two Lactobacillus species differs between women with and without UUI remains unknown, but it is possible that these two members of the normal vagina flora perform distinct functions in the bladder. A similar scenario seems to be developing for Gardnerella, which was detected more frequently and in increased sequence abundance for the UUI cohort than for the non-UUI cohort. Since Gardnerella was detected in the urine of women without UUI, it is unlikely that the simple presence of Gardnerella indicates a dysbiotic environment. G. vaginalis can be cultured from the vaginal tracts of women with and without bacterial vaginosis (13, 14). G. vaginalis strains isolated from these two groups of women vary in their ability to adhere and induce cytotoxicity, suggesting the possibility of pathogenic and symbiotic strains of G. vaginalis (14). Whether the isolates identified by MALDI-TOF MS as Gardnerella species fall into the latter category remains to be determined.
Several bacterial genera were more frequently sequenced and cultured from the urine of women with UUI, including Actinobaculum, Actinomyces, Aerococcus, Arthrobacter, and Oligella. Interestingly, many of these genera contain emerging uropathogens, including Actinobaculum schaalii, Aerococcus urinae, Oligella ureolytica, and Arthrobacter cumminsii (15–19). Whether these bacteria contribute to UUI is unknown at this time.
In addition to Lactobacillus and Gardnerella, many of the genera detected in the urinary microbiome are often found in the vaginal tract, including Bifidobacterium, Enterococcus, Actinomyces, Prevotella, and Atopobium (13, 20). Since the urine samples in this study were collected via transurethral catheter, as opposed to a voided approach, these genera are likely true inhabitants of the urinary tract and not vaginal contamination. This contention is supported by our previous demonstration that the microbiome sequence profile of catheterized urine is distinct from that of voided urine and instead closely resembles that of urine collected via suprapubic aspiration, which bypasses the vagina and urethra (1). Genera found in both the urinary and vaginal tracts could suggest a shared urogenital microbiome. Alternatively, there could be differences at the species or strain levels, such that the vaginal and urinary community members differ. It is easy to imagine that the specific conditions of these quite different environmental niches could select for different traits.
Two genera, Corynebacterium and Streptococcus, were cultured more frequently from UUI women yet were detected at similar frequencies by sequencing. A third genus, Staphylococcus, was more frequently cultured from UUI women but more frequently detected by sequencing in the control cohort. The simplest explanation for these discrepancies would be that certain species of these genera do not grow under the current EQUC conditions. Efforts to further extend this new protocol are planned.
Although the urinary microbiome shares a number of genera in common with other microbial sites within the human body, two key differences set the urinary microbiome apart. Compared to the colon, which contains 1011 to 1012 CFU/g (21), on average, the urinary tract microbiome consists of <104 CFU/ml total bacteria. In our study, the median amount of bacteria per urine was 85 CFU/ml. The urinary microbiome, with a median inverse Simpson index of 2.3 and Shannon index of 1.5, is also less diverse than other microbial sites in the human body, such as the skin, mouth, and gastrointestinal tract (22, 23). Taken together, these findings suggest that the bladder is a relatively unique microbial site within the human body and may be more akin to other low-abundance sites, such as the eye (24).
Changes in microbial diversity within a niche have been linked to disease. Examples include decreased fecal microbiome diversity associated with Clostridium difficile infection (25) and increased vaginal microbiome diversity associated with bacterial vaginosis (26). In this study, we did not detect large differences in sequence-based diversity between the two cohorts. It is possible that we were unable to detect a difference due to the lack of power with our small sample size. Alternatively, the amount of diversity within the urinary microbiome of UUI women may not differ from that for non-UUI women. Instead, the key distinction might be the bacterial types that are present. Although we did not detect differences in richness or diversity by sequence, we detected differences in the species richness between the cohorts by culture. A greater number of bacterial species were cultured from the UUI cohort than from the non-UUI control cohort. This finding further demonstrates the power of utilizing multiple approaches to define a microbial community.
The urine of UUI women was more likely to contain Actinomyces, Aerococcus, and Gardnerella and less likely to contain Lactobacillus than urine collected from women without UUI. The mechanism behind this observation is unknown. One possibility is that the UUI bladder selects for some bacteria over others, and as such, the presence of these organisms in the bladder could serve as a marker for dysbiosis. Another possibility is that these bacteria contribute to UUI symptoms, a supposition supported by the observation that each of the genera associated with the UUI cohort contains at least one reported pathogenic species. Taken together, this study, along with others, will eventually allow us to define a core or common urinary microbiome that can be used to detect alterations to that community.
A limitation of this analysis is that our UUI and non-UUI cohorts differed in several characteristics that may have clinical relevance for the female urinary microbiome. Thus, we are unable to say whether age, BMI, or hormonal status affects our findings, and it is possible that our findings are related to these differences rather than to urinary symptomatology. Studies that describe the female urinary microbiome in large, well-characterized populations have not been published. Future studies, with larger sample sizes, will be required to evaluate these potentially important differences. Studies describing the longitudinal stability of the female urinary microbiome (including populations undergoing treatment for urinary disorders) are also lacking. Such studies should be prioritized given the emerging evidence that the female urinary microbiome may yield important clinical information.
MATERIALS AND METHODS
Study design and population.
Following Loyola institutional review board (IRB) approval, participants gave verbal and written consent for chart abstraction and urine collection with analysis for research purposes. Participants were recruited from the clinical practice of the Female Pelvic Medicine and Reconstructive Surgery Center of Loyola University Medical Center between August 2012 and February 2014. These included women undergoing UUI treatment (UUI cohort) and a comparison group of women not bothered by urinary symptoms (non-UUI control cohort). All women were screened for potential study participation using the validated symptom questionnaire, the Pelvic Floor Distress Inventory (PFDI) (27, 28). Exclusion criteria for both cohorts included current UTI (based on urine dipstick) or history of recurrent UTI, antibiotic exposure in the past 4 weeks for any reason, immunologic deficiency, neurological disease known to affect the lower urinary tract, pelvic malignancy or radiation, untreated symptomatic pelvic organ prolapse (POP) greater than POP-Q stage II (vaginal protrusion more than 1 cm outside of the vaginal hymen), or pregnancy. Clinical and demographic information were abstracted from the electronic medical record. Enrolled participants completed the long form of the PFDI and the Overactive Bladder Questionnaire (OAB-q) (29). Premenopausal woman and postmenopausal women taking any form of estrogen replacement were considered estrogen positive.
Sample collection.
Urine was collected aseptically via transurethral catheter. A portion of the urine sample was placed in a BD Vacutainer Plus C&S preservative tube for culturing. A separate portion for sequencing was placed at 4° C for <4 h following collection; 10% AssayAssure (Thermo Scientific; Waltham, MA) was added before storage at −80° C.
The first urine samples collected (14 UUI samples and 8 non-UUI control samples) were screened exclusively by 16S rRNA gene sequencing; this was before we implemented EQUC. Subsequent urine samples (22 UUI and 30 non-UUI control samples) were screened by both 16S rRNA gene sequencing and EQUC. The final urine samples (23 UUI and 15 non-UUI control samples) were screened only by EQUC without high-throughput sequencing. Thus, we used EQUC for 45 UUI and 45 non-UUI samples and 16S rRNA gene sequencing for 36 UUI and 38 non-UUI samples.
Urine culture.
For standard urine culture, we struck 0.001 ml of urine onto 5% sheep blood (BAP) and MacConkey agars (BD BBL prepared plated media), which were incubated aerobically at 35°C for 24 h. Each separate morphological colony type was counted and identified in any amount. The detection level was 1,000 CFU/ml, represented by 1 colony of growth on either plate. If no growth was observed, the culture was reported as “no growth,” indicating no growth of bacteria at the lowest dilution, i.e., 1:1,000.
For EQUC, we struck 0.1 ml of urine onto BAP, chocolate and colistin, naladixic acid (CNA) agars (BD BBL prepared plated media), which were incubated in 5% CO2 at 35°C for 48 h (see Fig. S1 in the supplemental material). A second set of BAPs was inoculated with 0.1 ml of urine and incubated in room atmosphere at 35°C and 30°C for 48 h. We also inoculated 0.1 ml of urine onto each of two CDC anaerobe 5% sheep blood agar plates (BD BBL prepared plated media) and incubated either in a Campy gas mixture (5% O2, 10% CO2, and 85% N) or under anaerobic conditions at 35°C for 48 h. The detection level was 10 CFU/ml, represented by 1 colony of growth on any of the plates. Finally, to detect any bacterial species that may be present at quantities lower than 10 CFU/ml, 1.0 ml of urine was placed in thioglycolate medium (BD BBL prepared tubed media) and incubated aerobically at 35°C for 5 days. If growth was visually detected in the thioglycolate medium, the medium was mixed, and a few drops were plated on BAP and CDC Anaerobe 5% sheep blood agars for isolation and incubated aerobically and anaerobically at 35°C for 48 h. Each morphologically distinct colony type was isolated on a different plate of the same medium to prepare a pure culture that was used for identification. Matrix-assisted laser desorption ionization–time of flight mass spectrophotometry (MALDI-TOF MS) with the MALDI Biotyper 3.0 software program (Bruker Daltonics, Billerica, MA) was used to identify the bacterial isolates, as described elsewhere (4). To determine the false-negative rate, the following equation was used: the number of false negatives (EQUC positive, standard negative) divided by the sum of the number of true positives (EQUC positive) and false negatives (EQUC positive, routine negative).
DNA isolation from urine.
We used a previously validated DNA extraction protocol developed for the Human Microbiome Project. The protocol includes the addition of the peptidoglycan-degrading enzymes mutanolysin and lysozyme, which ensure robust lysis of Gram-positive and Gram-negative species, to isolate genomic DNA from urine samples (30). Briefly, 1 ml of urine was centrifuged at 13,500 rpm for 10 min, and the resulting pellet was resuspended in 200 µl of filter-sterilized buffer consisting of 20 mM Tris-Cl (pH 8), 2 mM EDTA, 1.2% Triton X-100, and 20 µg/ml lysozyme and supplemented with 30 µl of filter-sterilized mutanolysin (5,000 U/ml; Sigma-Aldrich, St. Louis, MO). The mixture was incubated for 1 h at 37°C, and the lysates were processed through the DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The DNA was eluted into 50 µl of buffer AE, pH 8.0, and stored at −20° C.
16S rRNA gene library generation and MiSeq sequencing.
Sequencing was performed using a MiSeq desktop sequencer (Illumina, San Diego, CA). First, a 16S rRNA gene amplicon library was generated via two consecutive PCR amplifications. In the first reaction, the variable 4 region (V4) of the 16S rRNA gene was amplified using the universal primers 515F and 806R, which were modified to encode the Illumina MiSeq sequencing primer sequence at the 5′ end (see Table S1 in the supplemental material). Reaction mixtures were incubated at 94°C for 2 min to denature the DNA template and amplified for 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s. To ensure complete amplification, samples were incubated at 72°C for an additional 10 min. Ten-microliter aliquots of each reaction mixture were run on a 1% agarose gel. Samples containing a band of approximately 360 bp were considered PCR positive and subjected to further library preparation. Samples with no visible amplified product were considered PCR negative and not processed further. The PCR-positive reaction mixtures were diluted 1:50 and amplified for an additional 10 cycles, utilizing primers encoding the required adapter sequences for Illumina MiSeq sequencing and an 8-nucleotide (nt) sample index (see Table S1), using the PCR conditions described above. Unincorporated nucleotides and remaining primers were removed via use of the QIAquick PCR purification kit (Qiagen, Valencia, CA), and the DNA concentration of the eluted product was determined by Nanodrop spectroscopy (Thermo Scientific; Waltham, MA). One hundred nanograms of each sample amplicon was pooled and run through a 1% agarose gel. The final product, which includes the V4 region and the adapter sequences (390 to 450 bp in length depending upon the length of the V4 region), was gel extracted via the QIAquick gel extraction kit and further purified via Agencourt AMPure XP-PCR magnetic beads (Beckman Coulter, Pasadena, CA). The final concentration of the pooled DNA was determined via Nanodrop spectroscopy and diluted in EBT (Elution Buffer with Tris; Illumina, San Diego, CA) to 2 nM. An equal volume of 0.2 N NaOH was added, incubated at room temperature for 5 min, and quenched with hybridization buffer (Illumina, San Diego, CA) to a final concentration of 8 pM. The Human Microbiome Project mock community HM-782D (BEI Resources, ATCC, Manassas, VA), a standard used to optimize our approach, was mixed 1:1 with randomly generated PhiX libraries, which are added to help focus the cameras on the sequencing clusters. This mixture was added to the sample library at equal volumes and placed in the 2 × 250 bp sequencing reagent cartridge according to the manufacturer’s instructions (Illumina, San Diego, CA).
Care was taken to avoid bacterial DNA contamination by utilizing DNA-free reagents when applicable, filter sterilizing all solutions through a 0.2 µM filter, and working in a PCR-clean hood. To control for the introduction of contaminating DNA, negative controls for extraction (no urine) and PCR (no template) were included in each experiment. The extraction negative control for each experiment was sequenced to identify spurious genera likely introduced from contaminated reagents and materials.
Sequence analysis.
The Illumina MiSeq postsequencing software preprocessed sequences by removing primers and sequence adaptors. Using the open-source software program mothur (v 1.31.2), the paired-end reads were assembled and contigs of incorrect length (<285 bp or >300 bp) and/or contigs containing ambiguous bases were removed (31, 32). These modified sequences were aligned to the SILVA reference database, and any potential chimeric sequences were detected and removed using the program UCHIME (33). The remaining sequences were taxonomically classified, using a naive Bayesian classifier (34) and the mothur-formatted RDP training set v9, and clustered into operational taxonomic units (OTUs), an operational definition of a species-level cluster based on sequence similarity, using a 97% cutoff. The software program METAGENassist was used to link OTU nomenclature to taxonomic assignments (35).
All samples were processed in duplicate, and the percent reads of the replicates were averaged for downstream analysis. Stacked bar plots based on sequence abundance were produced for each sample. Euclidean distance was calculated between samples, and the complete method was used for hierarchical clustering via R software, version 2.15.1 (36). Richness and diversity metrics, including the number of observed OTUs, Chao1 estimator, Shannon index, and inverse Simpson’s index, were calculated using mothur and were based on subsampling to the number of sequences in the sample with the least coverage. Urine is a low-biomass environment, and thus the sequencing results are more likely to be influenced by extraneous DNA arising from reagents and resources; therefore, only reads representing >0.01% of the sample total were included in the stacked bar plots, dendrogram, and frequency and abundance analyses.
Statistical analysis.
Statistical analyses, comparing participant demographics and symptoms, were performed using the SPSS software program, version 19. For continuous variables, Student’s t tests were applied. For categorical variables, Pearson chi-square and Fisher’s exact tests were performed. Results were considered significant when the P value was less than 0.05.
Statistical analyses of the microbiome data were performed using the SAS software program, version 9.3. The Wilcoxon rank sum tests were used to compare the median abundances for the 15 most abundant sequenced taxa and all cultured genera between groups. The frequencies of detected genera were compared between groups, using either the Pearson chi-square or Fisher’s exact test, depending on assumption validity. No adjustments for multiple comparisons were made, since these analyses were considered descriptive.
SUPPLEMENTAL MATERIAL
Outline of EQUC procedure. Download
Distribution of taxa detected by sequencing. The distribution of the top 15 most abundant taxa detected by sequencing is displayed as box plots. Download
16S rRNA gene V4 amplicon library primers
Species that are statistically significantly different in the two cohorts
ACKNOWLEDGMENTS
We thank Mary Tulke and Bozena Zemaitaitis for their assistance in recruitment of participants and sample collection and Kathleen McKinley for her clinical microbiology expertise. We acknowledge and thank the Loyola University Chicago Health Sciences Division’s Office of Informatics and Systems Development for their expertise and for the computational resources utilized in support of this research. The following reagent was obtained through BEI Resources, NIAID, NIH, as part of the Human Microbiome Project: Genomic DNA from Microbial Mock Community A (Even, Low Concentration), HM-782D.
Loyola University Chicago Stritch School of Medicine’s research computing facility was developed through grant funds awarded by the Department of Health and Human Services, award number 1G20RR030939-01. This study was supported by a grant from the Falk Foundation (LU 202567), by NIH grant R21DK097435-01A1, and by Astellas Medical and Scientific Affairs and is registered at http://www.clinicaltrials.gov as NCT01642277.
Footnotes
Citation Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, Kliethermes S, Schreckenberger PC, Brubaker L, Gai X, Wolfe AJ. 2014. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio 5(4):e01283-14. doi:10.1128/mBio.01283-14.
REFERENCES
- 1. Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE, Brubaker L. 2012. Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol. 50:1376–1383. 10.1128/JCM.05852-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brubaker L, Nager CW, Richter HE, Visco A, Nygaard I, Barber MD, Schaffer J, Meikle S, Wallace D, Shibata N, Wolfe AJ. Urinary bacteria in adult women with urgency urinary incontinence. Int. Urogynecol. J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, Huang ST, Ljungberg I, Sprague BM, Lucas SK, Torralba M, Nelson KE, Groah SL. 2012. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J. Transl. Med. 10:174. 10.1186/1479-5876-10-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC. 2014. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 52:871–876. 10.1128/JCM.02876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL, Malone-Lee J. 2013. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J. Clin. Microbiol. 51:2054–2062. 10.1128/JCM.03314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Riesenfeld CS, Schloss PD, Handelsman J. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38:525–552. 10.1146/annurev.genet.38.072902.091216 [DOI] [PubMed] [Google Scholar]
- 7. Nitti VW, Kopp Z, Lin AT, Moore KH, Oefelein M, Mills IW. 2010. Can we predict which patient will fail drug treatment for overactive bladder? A think tank discussion. Neurourol. Urodyn. 29:652–657. 10.1002/nau.20910 [DOI] [PubMed] [Google Scholar]
- 8. Hartmann KE, McPheeters ML, Biller DH, Ward RM, McKoy JN, Jerome RN, Micucci SR, Meints L, Fisher JA, Scott TA, Slaughter JC, Blume JD. August 2009. Treatment of overactive bladder in women. Evidence report/technological assessment no. 187. (Prepared by the Vanderbilt Evidence-based Practice Center under Contract No. 290-2007-10065-I.) AHRQ publication no. 09-E017 Agency for Healthcare Research and Quality, Rockville, MD: [PMC free article] [PubMed] [Google Scholar]
- 9. Haylen BT, de Ridder D, Freeman RM, Swift SE, Berghmans B, Lee J, Monga A, Petri E, Rizk DE, Sand PK, Schaer GN, International Urogynecological Association. International Continence Society 2010. An International Urogynecological Association (IUGA)/international Continence Society (ICS) joint report on the terminology for female pelvic floor dysfunction. Neurourol. Urodyn. 29:4–20. 10.1007/s00192-009-0976-9 [DOI] [PubMed] [Google Scholar]
- 10. Coyne KS, Wein A, Nicholson S, Kvasz M, Chen CI, Milsom I. 2014. Economic burden of urgency urinary incontinence in the United States: a systematic review. J. Manag. Care Pharm. 20:130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Redondo-Lopez V, Cook RL, Sobel JD. 1990. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev. Infect. Dis. 12:856–872. 10.1093/clinids/12.5.856 [DOI] [PubMed] [Google Scholar]
- 12. Kaewsrichan J, Peeyananjarassri K, Kongprasertkit J. 2006. Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol. Med. Microbiol. 48:75–83. 10.1111/j.1574-695X.2006.00124.x [DOI] [PubMed] [Google Scholar]
- 13. Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102:7952–7957. 10.1073/pnas.0503236102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harwich MD, Jr, Alves JM, Buck GA, Strauss JF, III, Patterson JL, Oki AT, Girerd PH, Jefferson KK. 2010. Drawing the line between commensal and pathogenic Gardnerella vaginalis through genome analysis and virulence studies. BMC Genomics 11:375. 10.1186/1471-2164-11-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmermann P, Berlinger L, Liniger B, Grunt S, Agyeman P, Ritz N. 2012. Actinobaculum schaalii an emerging pediatric pathogen? BMC Infect. Dis. 12:201. 10.1186/1471-2334-12-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bank S, Jensen A, Hansen TM, Søby KM, Prag J. 2010. Actinobaculum schaalii, a common uropathogen in elderly patients, Denmark. Emerg. Infect. Dis. 16:76–80. 10.3201/eid1601.090761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rasmussen M. 2013. Aerococci and aerococcal infections. J. Infect. 66:467–474. 10.1016/j.jinf.2012.12.006 [DOI] [PubMed] [Google Scholar]
- 18. Dabkowski J, Dodds P, Hughes K, Bush M. 2013. A persistent, symptomatic urinary tract infection with multiple “negative” urine cultures. Conn. Med. 77:27–29 [PubMed] [Google Scholar]
- 19. Funke G, Pagano-Niederer M, Sjödén B, Falsen E. 1998. Characteristics of Arthrobacter cumminsii, the most frequently encountered Arthrobacter species in human clinical specimens. J. Clin. Microbiol. 36:1539–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U. S. A. 108:4680–4687. 10.1073/pnas.1002611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O’Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep. 7:688–693. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Microbiome Human Project 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong Q, Brulc JM, Iovieno A, Bates B, Garoutte A, Miller D, Revanna KV, Gao X, Antonopoulos DA, Slepak VZ, Shestopalov VI. 2011. Diversity of bacteria at healthy human conjunctiva. Invest. Ophthalmol. Vis. Sci. 52:5408–5413. 10.1167/iovs.10-6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197:435–438. 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- 26. Liu MB, Xu SR, He Y, Deng GH, Sheng HF, Huang XM, Ouyang CY, Zhou HW. 2013. Diverse vaginal microbiomes in reproductive-age women with vulvovaginal candidiasis. PLoS One 8:e79812. 10.1371/journal.pone.0079812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barber MD, Kuchibhatla MN, Pieper CF, Bump RC. 2001. Psychometric evaluation of 2 comprehensive condition-specific quality of life instruments for women with pelvic floor disorders. Am. J. Obstet. Gynecol. 185:1388–1395. 10.1067/mob.2001.118659 [DOI] [PubMed] [Google Scholar]
- 28. Uebersax JS, Wyman JF, Shumaker SA, McClish DK, Fantl JA. 1995. Short forms to assess life quality and symptom distress for urinary incontinence in women: the Incontinence Impact Questionnaire and the urogenital Distress Inventory. Continence Programs Women Research Group. Neurourol. Urodyn. 14:131–139 [DOI] [PubMed] [Google Scholar]
- 29. Coyne K, Revicki D, Hunt T, Corey R, Stewart W, Bentkover J, Kurth H, Abrams P. 2002. Psychometric validation of an overactive bladder symptom and health-related quality of life questionnaire: the OAB-q. Qual. Life Res. 11:563–574. 10.1023/A:1016370925601 [DOI] [PubMed] [Google Scholar]
- 30. Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. 2012. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 7:e33865. 10.1371/journal.pone.0033865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, Sinelnikov I, Budwill K, Nesbø CL, Wishart DS. 2012. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 40:W88–W95. 10.1093/nar/gkr734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. R Development Core Team 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Outline of EQUC procedure. Download
Distribution of taxa detected by sequencing. The distribution of the top 15 most abundant taxa detected by sequencing is displayed as box plots. Download
16S rRNA gene V4 amplicon library primers
Species that are statistically significantly different in the two cohorts