Abstract
Imine-based reactions are useful for a wide range of bioconjugation applications. Although aniline is known to catalyze the oxime ligation reaction under physiological conditions, it suffers from slow reaction kinetics, specifically when a ketone is being used or when hydrazone-oxime exchange is performed. Here, we report on the discovery of a new catalyst that is up to 15 times more efficient than aniline. That catalyst, m-phenylenediamine (mPDA), was initially used to analyze the kinetics of oxime ligation on aldehyde- and ketone-containing small molecules. While mPDA is only modestly more effective than aniline when used in equal concentrations (~ 2-fold), its much greater aqueous solubility relative to aniline allows it to be used at higher concentrations, resulting in significantly more efficient catalysis. In the context of protein labeling, it was first used to site-specifically label an aldehyde-functionalized protein through oxime ligation, and its kinetics were compared to reaction with aniline. Next, a protein was labeled with an aldehyde-containing substrate in crude cell lysate, captured with hydrazide-functionalized beads and then the kinetics of immobilized protein release via hydrazone-oxime exchange were analyzed. Our results show that mPDA can release and label 15 times more protein than aniline can in 3 h. Then, using the new catalyst, ciliary neurotrophic factor, a protein with therapeutic potential, was successfully labeled with a fluorophore in only 5 min. Finally, a protein containing the unnatural amino acid, p-acetyl phenylalanine, a ketone-containing residue, was prepared and PEGylated efficiently via oxime ligation using mPDA. This new catalyst should have a significant impact on the field of bioconjugation, where oxime ligation and hydrazone-oxime exchange are commonly employed.
Introduction
Imine-based reactions are widely used to link complex biomolecules due to their high chemoselectivity and reversibility.1–8 When there is an oxygen or nitrogen atom adjacent to a nitrogen as in the cases of hydrazines and alkoxyamines, the reaction favors imine formation;9, 10 in the absence of such a substituent, the equilibrium favors formation of free amine and free aldehyde or ketone. Among all imine-based reactions, oximes are the most stable imines10 and as a result have found widespread use in applications such as protein labeling,11–15 analyzing protein-protein interactions and in vivo cell imaging.11, 12 Although aniline is known to catalyze the oxime formation reaction, it suffers from relatively slow reaction kinetics, especially when a ketone is being used11,18 or when transoximization is conducted.13 In the case of ketones under physiological conditions, even in presence of aniline, it takes several hours for the reaction to be complete although faster rates can be achieved if higher reactant concentrations are employed. Moreover, in the case of hydrazone-oxime exchange, the reaction rates are orders of magnitudes slower. Therefore, discovery of an improved catalyst would significantly improve the utility and broaden the scope of this valuable bioorthogonal reaction. Here, we introduce a new catalyst, m-phenylenediamine (mPDA) that can accelerate oxime ligation several times faster compared with aniline-catalyzed oxime ligation. That feature, in concert with the substantially greater aqueous solubility of m-phenylenediamine allows rate accelerations of approximately 15-fold to be obtained.
Experimental Section
Dansyl fluorescence assay
Reaction mixtures contained phosphate buffer (PB) (100 mM, pH 7.0), varying concentrations (10 µM to 300 µM) of aminooxy-dansyl (1), 0.08 % (w/v) n-dodecyl-β-D-maltoside, 50 µM aldehyde (citral or dodecanal) and varying concentrations of catalysts, in a final volume of 250 µL. The reaction mixtures were equilibrated at rt for 1 min, initiated by the addition of the aldehyde, and monitored for an increase in fluorescence (λex=340 nm, λem=505 nm) for approximately 50 min.
Catalyst screening
m-Phenylediamine, o-phenylenediamine, p-phenylediamine, o-aminophenol, m-aminophenol, p-aminophenol, o-aminobenzoic acid and aniline were analyzed for catalytic activity in the oxime ligation reaction. Reaction mixtures contained PB (100 mM, pH 7.0), 100 µM aminooxy-dansyl (1), 0.08 % (w/v) n-dodecyl-β-D-maltoside, 50 µM aldehyde (citral or dodecanal) and 25 µM catalyst, in a final volume of 200 µL. The reaction mixtures were equilibrated at rt for 1 min, initiated by the addition of the aminooxy reagent, and monitored for an increase in fluorescence (λex=340 nm, λem=505 nm) for 50 min.
Effect of the catalyst concentration on the kobs
Reaction mixtures contained PB (100 mM, pH 7.0), 100 µM aminooxy-dansyl (1), 0.08 % (w/v) n-dodecyl-β-D-maltoside, 30 µM aldehyde (citral) and varying concentrations of catalysts (25 to 50 mM) in a final volume of 200 µL. The reaction mixtures were equilibrated at rt for 1 min, initiated by the addition of the aldehyde, and monitored for an increase in fluorescence (λex=340 nm, λem=505 nm) for approximately 25 min.
Kinetic analysis of oxime ligation between 2-pentanone and aminooxy-dansyl 1
Reaction mixtures contained Tris·HCl (50 mM, pH 7.5), aminooxy-dansyl 1 (150 µM), 0.4 % (w/v) n-dodecyl-β-D-maltoside, 5 mM ketone (2-pentanone) and varying concentrations of catalysts, in a final volume of 200 µL. The reaction mixtures were equilibrated at rt for 1 min, initiated by the addition of the aminooxy, and monitored for an increase in fluorescence (λex=340 nm, λem=505 nm) for approximately 4 h.
Enzymatic incorporation of 2 into GFP-CVIA (3)
Enzymatic reaction mixtures (50 mL) contained Tris·HCl (50 mM, pH 7.5), MgCl2 (10 mM), KCl (30 mM), ZnCl2 (10 µM), DTT (5.0 mM), GFP-CVIA (3, 2.4 µM), 2 (50 µM), and PFTase (200 nM). After incubation at rt overnight, the reaction mixture was concentrated using an Amicon Centriprep centrifugation device (10,000 MW cut-off). Next, excess 2 was removed with a NAP-5 (Amersham) column using Tris·HCl (50 mM, pH 7.5) as the eluent. The subsequent protein concentration was calculated by UV absorbance at 488 nm (ε=55,000 M−1·cm−1).
Kinetic analysis of protein labeling via oxime ligation
Reaction mixtures contained PB (100 mM, pH 7.0), 10 µM GFP-aldehyde 4a, 50 µM aminooxy-dansyl 1 and varying concentrations of m-phenylenediamine or 100 mM aniline, in a final volume of 250 µL. The reaction mixtures were equilibrated at rt for 1 min, initiated by the addition of the catalyst, and monitored for an increase in fluorescence (λex=340 nm, λem=505 nm) for approximately 100 min.
Crude prenylation, immobilization and subsequent labeling and release of GFP-CVIA
A pellet of cells expressing GFP-CVIA were suspended in buffer (20 mM Tris·HCl pH 7.5, 1 mM EDTA), sonicated and clarified by centrifugation. The GFP concentration present in the crude soluble protein mixture was calculated by UV absorbance at 488 nm. Next, prenylation was performed by adding PFTase (200 nM), 2 (50 µM), Tris·HCl (50 mM, pH 7.5), MgCl2 (10 mM), KCl (30 mM), ZnCl2 (10 µM) and DTT (5.0 mM) to a solution of 3, to achieve a final concentration of 2.0 µM in the crude mixture. After overnight incubation at rt, the reaction mixture was filtered and concentrated using an Amicon Centriprep centrifugation device (10,000 MW cut-off). Next, excess 2 was removed through a NAP-5 (Amersham) column using Tris·HCl (50 mM, pH 7.5) as the eluting solvent. The subsequent GFP concentration in the crude mixture was calculated by UV absorbance at 488 nm. Hydrazide agarose beads (Thermo Scientific, hydrazide loading: 16 µmol/mL) (300 µL) were washed with PB (0.1 M, 3×500 µL, pH 7.0). PB (30 µL, 1 M, pH 7.0) was added to the beads followed by addition of 4a in the crude mixture (200 µL, 70 µM). Immobilization was initiated by adding aniline (100 mM) or m-phenylenediamine (40 mM). After 1 h with constant agitation, the beads were washed thoroughly with PB (0.3 M, pH 7.3) and KCl (3×300 µL, 1 M) to remove non-specifically bound proteins followed by incubation with aminooxy fluorophore 5 (0.7 mM) and either m-phenylenediamine (700 mM) or aniline (100 mM) with constant agitation. The supernatant was then analyzed via SDS-PAGE and in-gel fluorescence analysis to compare the amount of protein released from the beads with either catalyst.
LC-MS analysis of proteins for determination of prenylation and labeling efficiency
Purified prenylated GFP (4a) and pure GFP-CVIA (3) were analyzed by LC-MS to ensure complete prenylation. Proteins were stored in Tris·HCl (50 mM, pH 7.5) prior to injection into the LC-MS instrument. Crude reaction mixtures of GFP-aldehyde 4a and aminooxy 1 catalyzed by either aniline or m-phenylenediamine were analyzed by LC-MS to ensure complete ligation in both cases of the catalysts. The LC-MS method used was a gradient of 0–100% solvent A (H2O, 0.1% HCO2H) to B (CH3CN, 0.1% HCO2H) in 25 min.
Kinetic analysis of hydrazone-oxime exchange
The hydrazide beads containing immobilized GFP were incubated in PB (0.3 M, pH 7.0) with aminooxy-alexafluor-488 5 (1 mM) and either aniline (100 mM) or m-phenylenediamine (750 mM) with constant agitation of the resulting mixtures at rt. The solutions were centrifuged at different time points (every 1 h for mPDA and every 2 h for aniline for a total of 8 h) and the amounts of released protein in the solutions were analyzed via SDS-PAGE. Gels were visualized by staining with Coomassie blue after being scanned via in-gel fluorescence imaging of alexafluor-488.
PEGylation from immobilized GFP-beads
Immobilization was performed as described above. Beads were washed thoroughly with PB (0.3 M, pH 7.3) and KCl (3×300 µL, 1 M) to remove non-specifically bound proteins. Next, the beads were incubated with aminooxy PEG 10 (5 mM) and m-phenylenediamine (200 mM) for 2 h with constant agitation of the solution at rt. MALDI-MS analysis of the supernatant indicated the successful PEGylation and release of the aldehyde-GFP from the hydrazide-beads.
Prenylation of CNTF-CVIA with aldehyde analog 2
Enzymatic reaction mixtures (20 mL) contained Tris·HCl (50 mM, pH 7.5), MgCl2 (10 mM), KCl (30 mM), ZnCl2 (10 µM), DTT (5.0 mM), CNTF-CVIA (2.4 µM), 2 (50 µM), and PFTase (200 nM). After incubation at rt for 90 min, the reaction mixture was concentrated using an Amicon Centriprep centrifugation device (10,000 MW cut-off). Next, excess 2 was removed with a NAP-5 (Amersham) column using Tris·HCl (50 mM, pH 7.5) as the eluent. Purified prenylated CNTF (9) and pure CNTF-CVIA were analyzed by LC-MS to ensure complete prenylation. Proteins were stored in Tris·HCl (50 mM, pH 7.5) prior to injection into the LC-MS instrument.
Coupling reaction between aldehyde-labeled CNTF-CVIA (9) with alexafluor-488 (5)
Alexafluor-488 (5) (4.2 µL of 3.2 mM solution in DMSO) was added to 42 µL of 9 (stock solution of 60 µM in Tris·HCl (50 mM, pH 7.5)). PB (2 M, pH 7, 2.5 µL) was added and the reaction was initiated by adding 50 mM m-phenylenediamine (stock solution of 1.5 M in 0.3 M PB, pH 7.0) and was allowed to proceed for 30 min at rt. LS-MS analysis of the sample showed only oxime ligated protein and no free aldehyde was detected indicating a complete reaction in both prenylation and oxime ligation reactions.
Rate analysis of the coupling reaction between aldehyde-CNTF 9 and aminooxy 5
Alexafluor-488 (5) (8 µL of 3.1 mM solution in DMSO) was added to 6 µL of PB (1 M, pH 7) and 2 µL of m-phenylenediamine (stock solution of 1.5 M in 0.3 M PB, pH 7.0) or 2 µL water as the control reaction. Reactions were performed at rt and were initiated by adding 20 µL of 9 (stock solution of 60 µM in Tris·HCl (50 mM, pH 7.5)). To monitor the reactions, 15 µL aliquots were withdrawn at 5 min intervals, added to 15 µL of loading buffer, flash frozen by liquid nitrogen and subjected to SDS-PAGE analysis. Samples were heated at 98 °C for 4 min prior to gel analysis.
PEGylation of DHFR2 M174pAcF 11 protein with aminooxy-PEG 12 using mPDA
DHFR2 fusion protein with the unnatural amino acid p-acetyl phenylalanine (DHFR2 M174pAcF) (7 µM) was reacted with aminooxy-PEG 12 (3 kDa) (5 mM) in PB (0.1 M, pH 7) in the presence of either 100 mM aniline, 500 mM mPDA or no catalyst at rt. The amounts of PEGylated protein in the solutions were analyzed at different time points via SDS-PAGE. Gels were visualized by staining with Coomassie blue. Densitometry analyses on the stained gels were performed using the program ImageJ v1.46.
General procedure for MALDI analysis of protein samples
The sample was adsorbed onto a zip-tip (C4 column) via repeated cycles of aspiration and ejection (5–10 cycles of 10 µL each) using a pipettor. Next, in order to remove excess buffer and reagents, the zip-tip was washed 5×10 µL with solvent A (H2O containing 0.1% TFA; v/v) and the proteins eluted with 2 µL of a mixture of A and B (25:75) (solvent B: CH3CN containing 0.1% TFA; v/v). Next 0.7 µL of the eluted material was added to a MALDI plate and 0.7 µL of matrix was added on top of the sample plate and both were mixed thoroughly to form crystals. A saturated solution of sinapinic acid (3,5-dimeth-oxy-4-hydroxy-cinnamic acid) was used as the matrix.
Results and Discussion
Development of an assay to analyse kinetic of the oxime ligation reaction
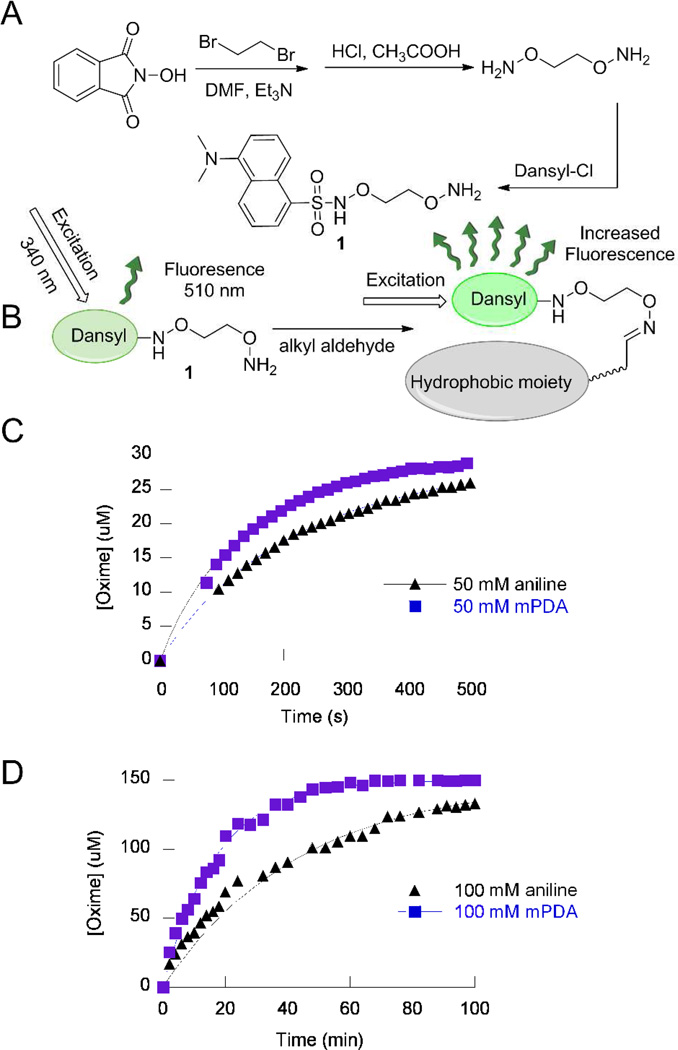
In order to study catalysis of oxime ligation, a continuous fluorescence assay was developed that enabled easy monitoring of the reaction. The dansyl chromophore manifests environmentally sensitive fluorescence that increases in the presence of hydrophobic groups.14 Thus, it was postulated that reaction of an aminooxy dansyl compound with a hydrophobic aldehyde or ketone would result in an increase in dansyl fluorescence that could easily be used to monitor the kinetics of oxime ligation. Therefore, an aminooxy-derivatized dansyl-containing compound, 1, was designed and synthesized in three steps (Figure 1A). Incubation of 1 in the presence of citral, dodecanal or 2-pentanone resulted in a time dependent increase in fluorescence indicative of oxime formation. The formation of these products was confirmed by HPLC and MS analysis.
Figure 1.
A) Synthesis of dansyl-aminooxy 1. B) Schematic representation of dansyl-fluorescence assay. When the dansyl moiety is in close proximity to a hydrophobic group, its fluorescence increases. “Hydrophobic moiety” represents the alkyl chain of the aldehyde- or ketone-containing reactants. C) Analysis of the reaction of 1 (100 µM) with citral (30 µM), a conjugated hydrophobic aldehyde, in presence of either 50 mM mPDA (squares) or 50 mM aniline (triangles) with kobs of 78.2 s−1M−1and 48.9 s−1M−1 respectively. D) Analysis of the reaction of 1 (150 µM) with 2-pentanone (5 mM), a non-conjugated ketone, in presence of either 100 mM mPDA (squares) or 100 mM aniline (triangles) with kobs of 0.20 s−1M−1and 0.082 s−1M−1 respectively.
Screening for a new catalyst suitable for oxime ligation reaction
After validating the continuous fluorescence assay, a number of different compounds were examined to identify new putative oxime ligation reaction catalysts. Two key features were considered in screening for such catalysts. First, the basicity of the intermediate Schiff base imine formed15 should not be significantly lower than the original amine (catalyst) basicity. Second, the catalyst should have high water solubility. Aromatic amines that are conjugated, such as aniline, meet only the first requirement in that their Schiff bases have basicities close to their free amines.16 Based on the idea that the presence of electron donating groups on an aromatic ring would render the corresponding Schiff base more basic, it was reasoned that such compounds would be more efficient reaction catalysts. Thus, o-, m- and p-aminophenols, o-aminobenzoic acid and o-, m- and p-phenylenediamines were examined. The latter three compounds have the advantage of having two amino groups that theoretically increases the probability of formation of an intermediate Schiff base that could result in higher catalytic efficiency. After using the fluorescence assay described above to screen all seven proposed catalysts, and fitting the data to the second order kinetic model previously employed by Dawson and coworkers,15 a range of results were obtained and are summarized in Table 1 and Figure S1. Given the variations in rate observed with the different catalysts, it is likely that a complex interplay of factors including inductive, steric and resonance effects together with hydrogen bonding (in the case of ortho substituents) participate in controlling their reactivity. o-Aminophenol was found to have a solubility too low to be useful. o-Phenylenediamine, m-aminophenol, and o-aminobenzoate were found to be equal to or less efficient than aniline while p-aminophenol, m-phenylenediamine and p-phenylenediamine were found to be more efficient compared with aniline. Of those latter three, m-phenylenediamine (mPDA) was the most efficient catalyst (1.7-fold compared to aniline) and had high water solubility. Hence mPDA was selected for further study.
Table 1.
Kinetic analysis of oxime ligation reactions between the aldehyde, citral, and 1 with different catalysts.
| Catalysta | kobsb (s−1 M−1) |
kobs /kobs(aniline) |
t1/2 (min) |
Approximate Solubilityc |
|---|---|---|---|---|
| Aniline | 24.4±0.5 | - | 5.54 | 100 mM |
| o-Aminophenol | - | - | - | Insoluble |
| m-Aminophenol | 23.9±0.5 | 0.98 | 5.65 | 100 mM |
| p-Aminophenol | 33.5±0.9 | 1.37 | 4.04 | 50 mM |
| o-Aminobenzoic acid | 24.8±0.8 | 1.01 | 5.45 | very soluble (>2 M) |
| o-Phenylene-diamine | 18.1±0.3 | 0.74 | 7.47 | very soluble (>1 M) |
| m-Phenylene-diamine | 41.5±1.2 | 1.70 | 3.26 | very soluble (>2 M) |
| p-Phenylene-diamine | 29.2±0.7 | 1.20 | 4.63 | very soluble (>1 M) |
Reactions were performed using citral (50 µM), aminooxy-dansyl 1 (100 µM) and catalyst (25 mM).
The kobs values were obtained by fitting the experimental data to Equation S3 using Kaleidagraph v4.1.3. The values are provided ± the standard error obtained from the curve fit.
Solubility in phosphate buffer (0.1 M), pH 7.0, at rt.
Kinetic analysis of oxime ligation reactions using aniline and m-phenylenediamine (mPDA)
Next, the reactions between several aldehyde- and ketone-containing compounds and alkoxyamine 1 were studied using aniline and mPDA as catalysts. Two hydrophobic aldehydes compatible with the fluorescence-based assay, citral and dodecanal (conjugated and nonconjugated aldehydes, respectively) were selected for initial experimentation. Analyses were performed using 50 mM catalyst (aniline or mPDA), 100 µM 1 and 30 µM aldehyde in phosphate buffer at pH 7.3. Interestingly, analysis of those reactions showed that both aldehydes were labeled with dansyl-aminooxy 1 within a few minutes. An equal concentration of mPDA was almost twice as efficient (Figure 1C, blue squares) as aniline (black triangles) consistent with what was observed in the initial screening; when no catalyst was employed, oxime formation required more than 80 min to achieve comparable levels of conversion under the same conditions (Figure S2-A, green circles). Significantly, citral, a conjugated aldehyde, reacted almost two times slower than dodecanal did (Figure S2-B). Given the importance of ketone-containing compounds as targets for bioorthogonal reactions, 2-pentanone was also studied as an example and was found to react at a rate at least two orders of magnitude slower than either of the two aldehydes (kobs for oxime ligation of citral and 2-pentanone with aminooxy-dansyl 1 were 48.6 s−1M−1 and 0.082 s−1M−1 using 50 mM and 100 mM aniline as a catalyst, respectively; Table 1 and Table 3). This highlights the role of carbonyl group reactivity in oxime formation kinetics. Aldehydes react more rapidly than ketones and non-conjugated aldehydes are more reactive than conjugated ones. Dawson and coworkers originally established that the kinetics of oxime ligation reactions catalyzed by aniline fit a second order model that is first order in both aldehyde and alkoxyamine.15 They also showed that the apparent second order rate constant for this process varied with aniline concentration. Wen-jun et al. extended those observations by demonstrating that the apparent second order rate constant varied linearly with aniline.17 Accordingly, we analyzed the rate data for the citral model reaction described here and confirmed that both the aniline- and mPDA-catalyzed reactions vary linearly with catalyst concentration (Figure S4 and S5; Table 2). A linear dependence on reaction rate with catalyst concentration was also observed in the model reaction involving the ketone, 2-pentanone (Table 3 and Figure S3). Overall, the results of these model studies indicate that the increase in reaction rate using mPDA is approximately 2-fold higher than that obtained with aniline when the catalysts are employed at equal concentrations. However, the fact that the rate of oxime formation is first order in catalyst, coupled with the much greater solubility of mPDA suggested that it should be possible to obtain much greater rate acceleration with mPDA by employing it at concentrations substantially above the solubility limit of aniline (~ 100 mM). That feature is apparent from the data shown in Table 3 for 2-pentanone and was investigated in more detail using aldehyde- and ketone-containing proteins as described below.
Table 3.
Kinetic analysis of oxime ligation reaction between the ketone, 2-pentanone, and 1 at different catalyst concentrations.
| [Catalyst]a (mM) |
kobsb (s−1 M−1) |
kobs /kobs(aniline) |
t1/2 (min) |
|---|---|---|---|
| 100 aniline | 0.082±0.0007 | - | 28.5 |
| 100 mPDA | 0.20±0.0016 | 2.41 | 11.2 |
| 300 mPDA | 0.36±0.0042 | 4.45 | 6.4 |
| 500 mPDA | 0.73±0.0071 | 8.89 | 3.2 |
| 700 mPDA | 1.17±0.013 | 14.3 | 2.0 |
| 900 mPDA | 1.69±0.022 | 20.7 | 1.4 |
Reactions were performed using 2-pentanone (5.0 mM), aminooxy-dansyl 1 (150 µM) and the catalysts at the concentrations indicated.
The kobs values were obtained by fitting the experimental data to Equation S3 using Kaleidagraph v4.1.3. The values are provided ± the standard error obtained from the curve fit.
Table 2.
Kinetic analysis of oxime ligation reaction, between the aldehyde, citral, and 1, with varying concentrations of catalysts.
| Anilinea | kobsb (s−1 M−1) |
kobs/[cat.] (ms−1 M−2) |
t1/2 (min) |
mPDAa | kobsb (s−1 M−1) |
kobs/[cat.] (ms−1 M−2) |
t1/2 (min) |
kobs(mPDA) /kobs(aniline) |
|---|---|---|---|---|---|---|---|---|
| 25 mM | 24.3±0.2 | 0.97 | 5.20 | 25 mM | 38.7±0.1 | 1.55 | 3.26 | 1.59 |
| 35 mM | 33.5±0.2 | 0.96 | 3.77 | 35 mM | 58.3±0.3 | 1.66 | 2.17 | 1.74 |
| 42 mM | 36.9±0.3 | 0.88 | 3.42 | 42 mM | 65.6±0.6 | 1.56 | 1.92 | 1.78 |
| 50 mM | 48.6±0.2 | 0.97 | 2.60 | 50 mM | 78.2±0.7 | 1.56 | 1.62 | 1.61 |
Reactions were performed using citral (30 µM), aminooxy-dansyl 1 (100 µM) and varying concentrations of catalysts (25 to 50 mM).
The kobs values were obtained by fitting the experimental data to Equation S3 using Kaleidagraph v4.1.3. The values are provided ± the standard error obtained from the curve fit.
Kinetic analysis of oxime ligation on proteins
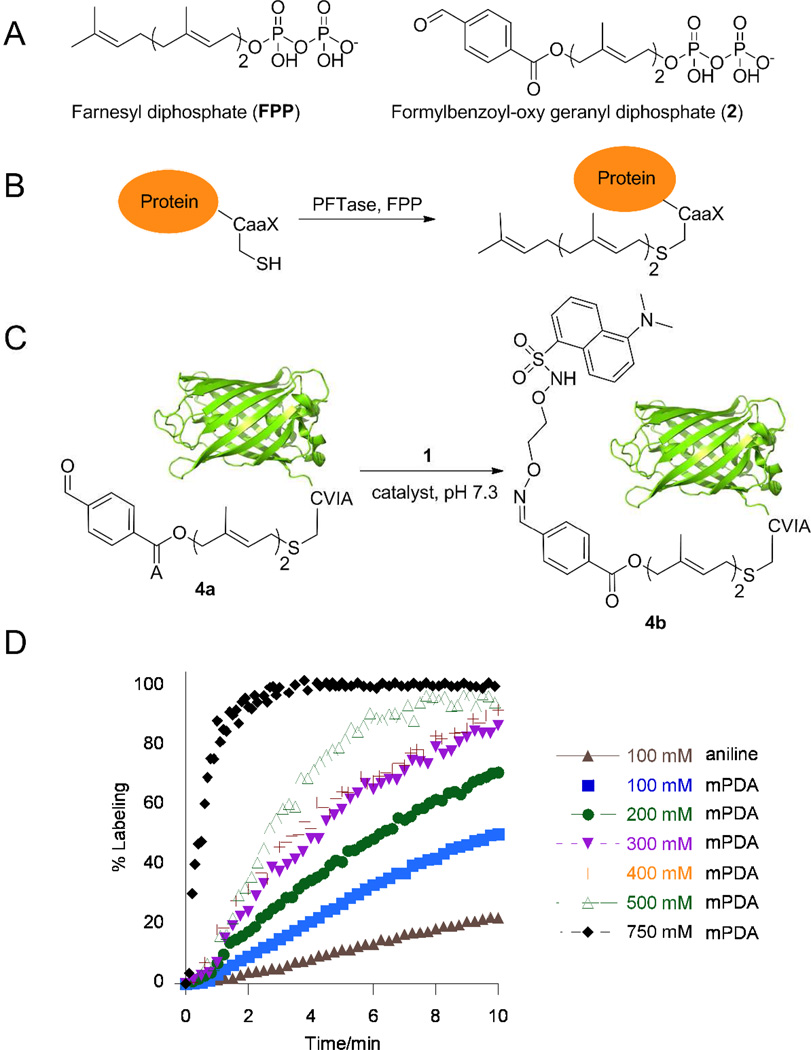
To examine the kinetics of oxime ligation on proteins, the enzyme protein farnesyltransferase (PFTase) was employed to introduce aldehyde functionality into proteins. In previous work, a number of groups have exploited the high specificity of PFTase to site-specifically modify proteins. PFTase catalyzes the transfer of an isoprenoid group from farnesyl pyrophosphate (FPP, Figure 2A) to a cysteineyl sulfur atom present in a tetrapeptide sequence (denoted as a CaaX-box) positioned at the C-terminus of a protein. Importantly, CaaX-box sequences such as CVIA can be appended to the C-termini of many proteins rendering them efficient substrates for PFTase. Since PFTase can tolerate many simple modifications of the isoprenoid substrate, it can be used to introduce a variety of different functional groups into proteins; chemoselective reaction with the resulting functionalized protein can then be used for a wide range of applications. We previously showed that aldehyde-containing FPP analogues could be successfully incorporated into the proteins using this strategy and used such aldehyde-modified proteins here.
Figure 2.
Kinetic analysis of oxime ligation with an aldehyde-functionalized protein. A) Structures of FPP and compound 2. B) Schematic representation of a prenylation reaction with a protein containing a CaaX box at its C-terminus. C) Schematic representation of oxime ligation with an aldehyde-functionalized protein 4a using either aniline or mPDA as the catalyst. D) Kinetic analysis of oxime ligation between 4a (10 µM) and 1 (50 µM) using either aniline or different concentrations of mPDA. Note: at high mPDA/aminooxy ratios, Schiff base formation is competitive with oxime ligation. Hence, the % labeling reported here for oxime formation is relative to the value at equilibrium and not complete conversion (vide infra). The solubility limit for aniline is approximately 100 mM under these conditions.
GFP-CVIA (3) was employed as a model protein to perform oxime kinetic analysis (Figure 2C). The protein was tagged with analogue 218 using PFTase to yield aldehyde-labeled protein 4a that was used in subsequent ligation reactions. Dansyl-aminooxy 1 was added to a solution of 10 µM aldehyde-functionalized protein 4a and the reactions were initiated by the addition of either aniline or mPDA catalyst. As was observed with the simple aldehyde model compounds described above, oxime formation between 1 and 4a resulted in a significant increase in dansyl group fluorescence that was then used to measure the reaction rate; no fluorescence change was observed when unprenylated GFP-CVIA (3) was treated with 1 (Figure S7). Additionally, analysis of the initial protein 3, aldehyde-functionalized protein 4a, and oxime-labeled protein 4b via LC-MS gave species at 27,335 Da, 27,619 Da, 27,926 Da, confirming that prenylation and the following oxime labeling were proceeding as expected (Figure S6). Interestingly, examiniation of the reaction rates using 4a via the fluorescence assay showed that both catalysts were somewhat less effective with the protein aldehyde than they were with the simple model aldehydes (Figure S7). That observation suggests that the reactivity of the aldehyde can be modulated by the protein to which it is attached and is consistent with previous observations reported by Dawson and Bertozzi where varying extents of acceleration by aniline were reported.5, 11 Since mPDA, in contrast to aniline, has a high solubility in water at pH 7 (approximately 100 mM for aniline compared to >2 M for mPDA), we decided to examine the effect of employing higher concentrations of the new catalyst on the oxime ligation reaction using a protein-aldehyde substrate. Consistent with the first-order kinetics observed in the model reactions discussed above, a plot of rate versus mPDA concentration for oxime-protein ligation was also linear (Figure S8). Thus, the ability to employ higher concentrations of mPDA compared with aniline due to its greater solubility resulted in significant rate acceleration; for example inclusion of mPDA at 750 mM resulted in essentially complete labeling of the aldehyde-functionalized protein 4a in about 90 s (Figure 2D), which is an impressive result compared with the minimal reaction (<7%) obtained with 100 mM aniline during that same time period. Kinetic analysis revealed that when aniline and mPDA were used at the same concentrations (100 mM), mPDA catalyzed the reaction ~2.5 times more efficiently than aniline (compare kobs of 10.3 M−1·s−1 for aniline versus 27.0 M−1·s−1 for mPDA, Table S1). More importantly the rate was greater than 10 times higher when mPDA was 500 mM and approximately 15 times higher when mPDA was 750 mM (Figure S8 and Table S1).
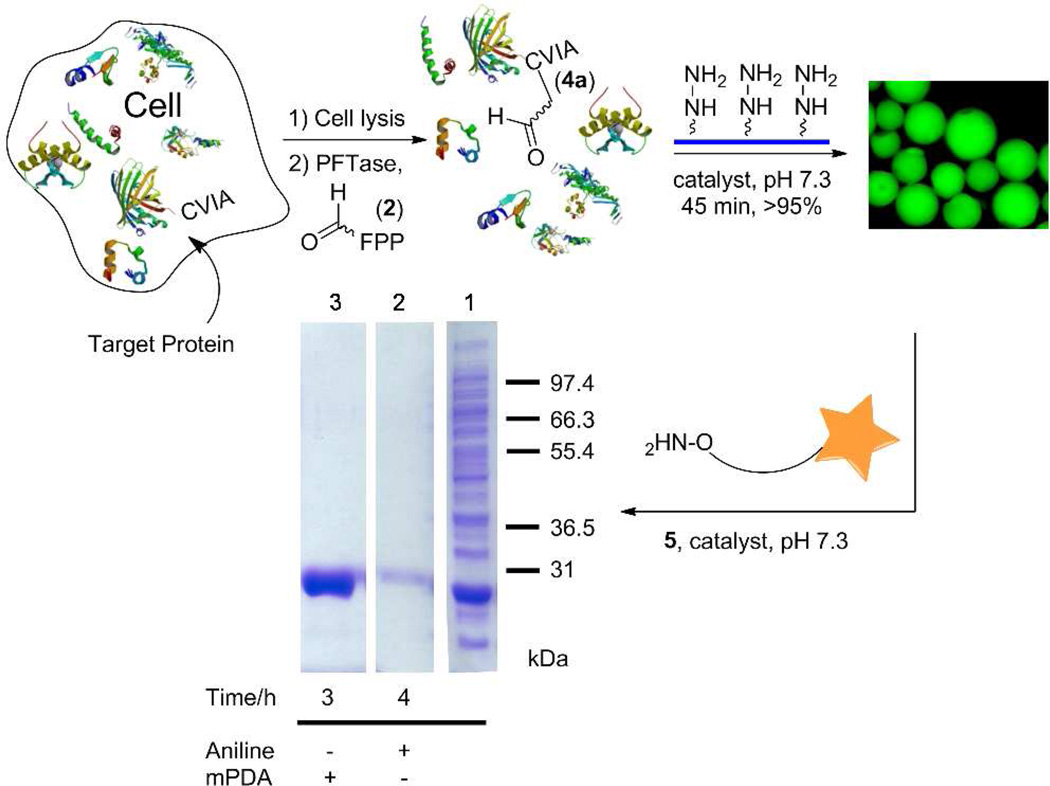
Capture and release strategy to purify and label proteins from crude cell extract using mPDA
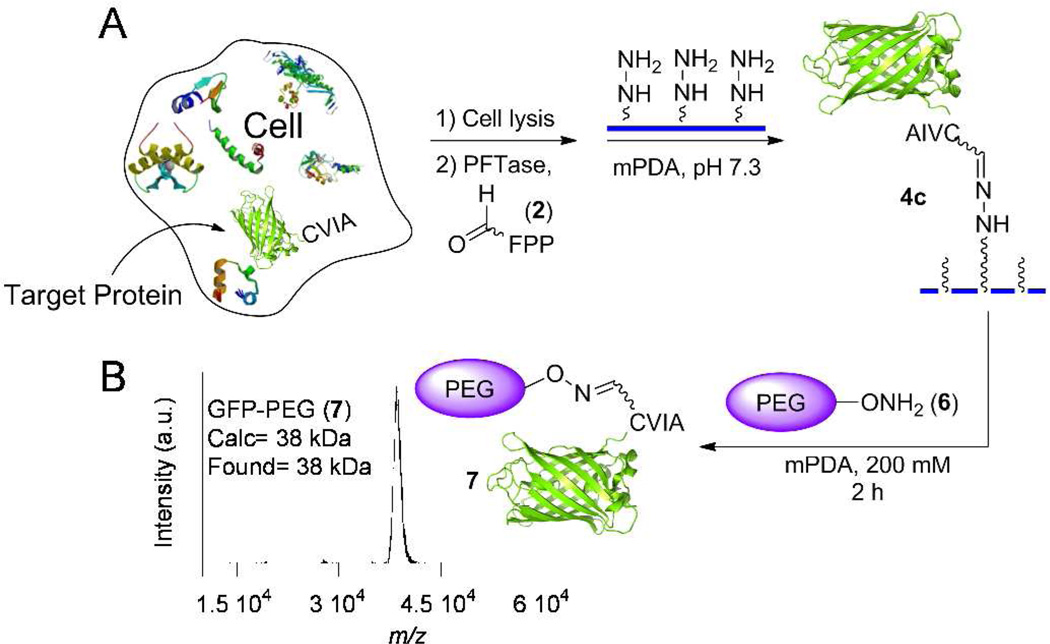
For many protein labeling applications, the target polypeptide is not always pure or present in high abundance; therefore, specific modification strategies that function in a crude mixture are necessary. In previous work, we reported on a method for site-specifically functionalizing a protein in a crude cell mixture, and labeling and purifying the modified protein using aniline without any additional purification.18 Although that process was successful, it required long reaction times to achieve significant levels of conversion. Accordingly, we next re-evaluated our aforementioned method for protein labeling with the new, more effective, mPDA catalyst discovered here. Thus, E. coli cells expressing GFP-CVIA 3 were lysed and enzymatically prenylated using PFTase and substrate 2. LC-ESI/MS analysis of the reaction sample confirmed incorporation of the aldehyde functionality into the GFP-CVIA 3 in the crude cell lysate. The reaction mixture was then concentrated and excess 2 was removed via size exclusion column chromatography. Aldehyde-GFP 4a was then selectively immobilized from the crude cell lysate onto hydrazide-functionalized beads using either 100 mM aniline or 40 mM mPDA as the catalyst. Immobilization was complete in less than 45 min in both cases at which point the beads became highly fluorescent. Next, the beads were thoroughly washed to remove any non-specifically bound proteins and then treated with aminooxy-alexafluor-488 5 in the presence of 100 mM aniline or 700 mM mPDA and the amount of released and fluorescently labeled protein was measured. SDS-PAGE analysis of the supernatant solution showed a single band attributable to the released, labeled GFP 4d (Figure 3). Qualitatively, from that data, it is clear that the efficiency of protein release using mPDA is substantially greater than with aniline. A more thorough kinetic analysis revealed that mPDA, under these reaction conditions, can release immobilized protein from the beads with an initial velocity approximately 15 times faster than aniline (compare kobs of 0.0159 h−1 for 100 mM aniline with 0.237 h−1 for 750 mM mPDA, Table S2, Figure 4). The in-gel fluorescence analysis further confirmed that the released protein was actually labeled with fluorophore 5 with either catalyst (Figure 4). Overall, this important result indicates that a protein can be enzymatically modified in a crude mixture, selectively immobilized onto hydrazide beads in the presence of many other proteins and then successfully labeled and released back into the solution in high yield using an aminooxy-fluorophore in less than 8 h. It is worth noting that when the mPDA/aminooxy reagent ratio is very high (>250), Schiff base formation (as an end product) is competitive with oxime ligation. Thus, we recommend that mPDA/aminooxy reagent ratios of less than 250 be used so that competitive Schiff base formation becomes negligible.
Figure 3.
Purification and selective labeling of a protein using mPDA. Target protein was site-specifically tagged by aldehyde-FPP analog via PFTase followed by capture of the aldehyde-functionalized protein in the crude cell lysate by hydrazide functionalized surfaces. Prenylation in the crude was confirmed by LC-MS analysis on the crude extract after prenylation. The immobilized protein was then released into the solution and fluorescently labeled by addition of aminooxy-fluorophore 5 (0.7 mM), using either mPDA (700 mM) or aniline (100 mM, its approximate solubility limit) as the catalyst. SDS-PAGE analysis (visualized by staining with Coomassie blue): Right lane (1): crude cell lysate, middle lane (2): released protein (after immobilization) obtained using aniline for 4 h; Left lane (3): released protein after immobilization obtained using mPDA for 3 h.
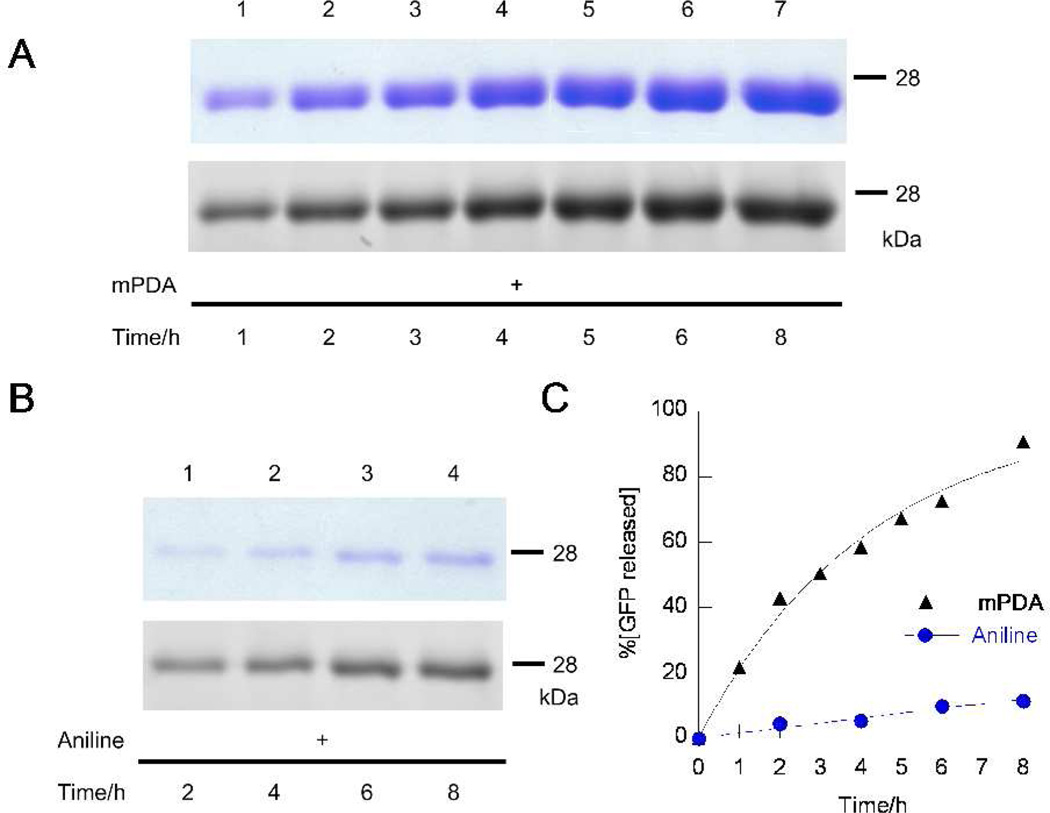
Figure 4.
Kinetic analysis of the release of the hydrazone immobilized protein into fluorescently labeled oxime protein via hydrazone-oxime exchange reaction. Immobilized protein was incubated with aminooxy fluorophore 5 (1 mM) and catalyst, followed by analysis of the amount of released protein in the solution via SDS-PAGE. In (A) and (B), the upper bands were visualized by staining with Coomassie blue while the lower bands were detected via in-gel fluorescence imaging of Alexafluor-488. A) Time course of protein release in 1–8 h using mPDA (750 mM) as the catalyst. B) Time course of protein release using aniline (100 mM, its approximate solubility limit). C) Comparison of protein release rates using aniline or mPDA obtained by densitometric analysis of the SDS-PAGE results shown in (A) and (B) using the Coomassie blue stained gels.
Capture and release strategy to purify and PEGylate proteins from crude cell extract using the catalyst
The attachment of polyethylene glycol (PEG) chains to proteins is the most widely employed method for increasing the half-life of protein-based therapeutic agents in blood.19, 20 Site-specific methods for protein PEGylation can minimize deleterious effects associated with nonselective PEGylation.21 In previous work, we described a method for site-specific protein PEGylation based on the capture and release strategy outlined above using aniline as a catalyst.18 In that work, PEGylation was achieved by releasing the captured protein (obtained by enzymatic aldehyde incorporation in crude extract followed by immobilization via hydrazide formation) using an aminooxy-functionalized PEG polymer. Given the significant rate enhancement observed with mPDA, we elected to evaluate the utility of this new catalyst for rapid protein PEGylation. Thus, 3 was prenylated with 2 using PFTase in crude E. coli extract followed by capture using hydrazide-functionalized agarose. After washing the material to remove nonspecifically bound proteins, the desired PEGylated protein (7) was eluted via treatment with 5 mM aminooxy-PEG 6 in the presence of 200 mM mPDA (Figure 5). Analysis of that material by MALDI-MS (Figure 5) showed a product with a mass of 38 kDa (and nothing else) consistent with the formation of highly pure PEGylated protein 7 from crude lysate in less than 2 h; the broad peak observed is consistent with the attachment of a polydisperse polymer to a monodisperse protein. It is also important to note that no species resulting from the Schiff base formation of mPDA or addition of multiple PEG chains were observed, consistent with the selective nature of the chemistry employed here.
Figure 5.
A) Use of PFTase-catalyzed protein modification for site-specific PEGylation from crude cell lysate using the mPDA. The target protein was prenylated, captured using hydrazide beads and then simultaneously released into the solution and PEGylated by the addition of aminooxy-PEG (6, 5 mM using 200 mM mPDA as a catalyst. B) MALDI analysis of the released material in less than 2 h confirmed the formation and release of the highly pure PEGylated GFP (7) into the solution.
Rapid labeling of CNTF, a protein of biomedical importance, using mPDA
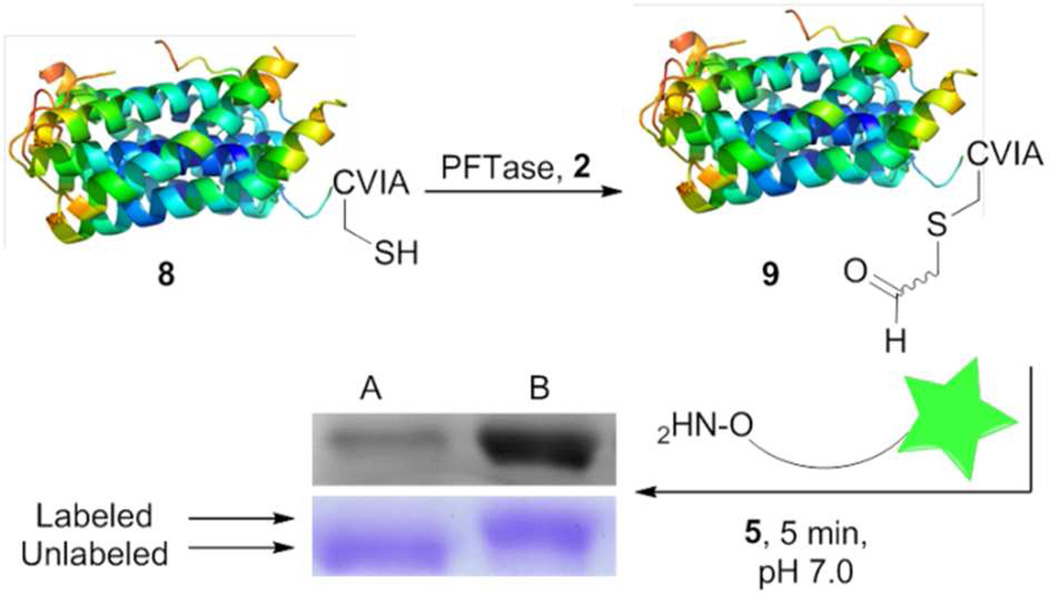
Having established the utility of this method for rapid C-terminal site-specific modification with a model protein, GFP, we decided to illustrate its use by modifying a protein of biomedical importance. Ciliary neurotrophic factor (CNTF) is a member of a class of proteins that stimulate neurite outgrowth and promote neuron survival during inflammatory events.22 Recently, CNTF has been studied extensively as a possible therapeutic agent for slowing retinal degeneration.23, 24 Thus, we elected to investigate the preparation of a fluorescent form of CNTF using the new catalyst reported here. To accomplish this, purified CNTF−CVIA (8), a form of CNTF engineered to contain a C-terminal CAAX box, was prenylated with analog 2 under conditions established above for GFP followed by labeling using aminooxy fluorophore 5 and mPDA as the catalyst. LC-MS analysis (Figure S9) confirmed successful prenylation and subsequent labeling of CNTF; as expected, both the enzymatic prenylation and the subsequent oxime ligation proceed with essentially complete conversion. Next, we analyzed how fast the protein is indeed labeled when the new catalyst is used. The aldehyde-labeled protein 9 (50 µM) was incubated with alkoxyamine 5 (600 µM), with or without mPDA as the catalyst (80 mM) and the reaction was monitored via SDS-PAGE as a function of time. In-gel fluorescence analysis of the resulting gel showed a substantially higher labeling efficiency when mPDA was used as the catalyst relative to when no catalyst was used (Figure 6). Inspection of the Coomassie blue stained gel revealed that the labeled protein manifests a decrease in electrophoretic mobility compared to the prenylated protein due to their mass difference allowing the two forms to be distinguished. Accordingly, Coomassie blue visualization of the gel showed that while almost all of the protein was labeled when mPDA was used, the amount of labeling that was obtained in the absence of mPDA was very small when the reaction was performed within the same time period (Figure 6).
Figure 6.
Prenylation of CNTF-CVIA (8) with aldehyde analog 2 followed by fluorophore labeling of the prenylated protein (50 µM) via oxime ligation with 5 (250 µM) for 5 min. A and B are SDS-PAGE analysis of the labeled protein. A) No catalyst was used for labeling. B) mPDA was used as the catalyst (80 mM) for labeling. Upper bands are detected via in-gel fluorescence analysis and lower bands are visualized by staining with Coomassie blue. The fluorescently labeled protein has a higher mass and hence shows a decrease in its electrophoretic mobility compared to the precursor prenylated protein.
PEGylation of a protein containing a ketone using mPDA
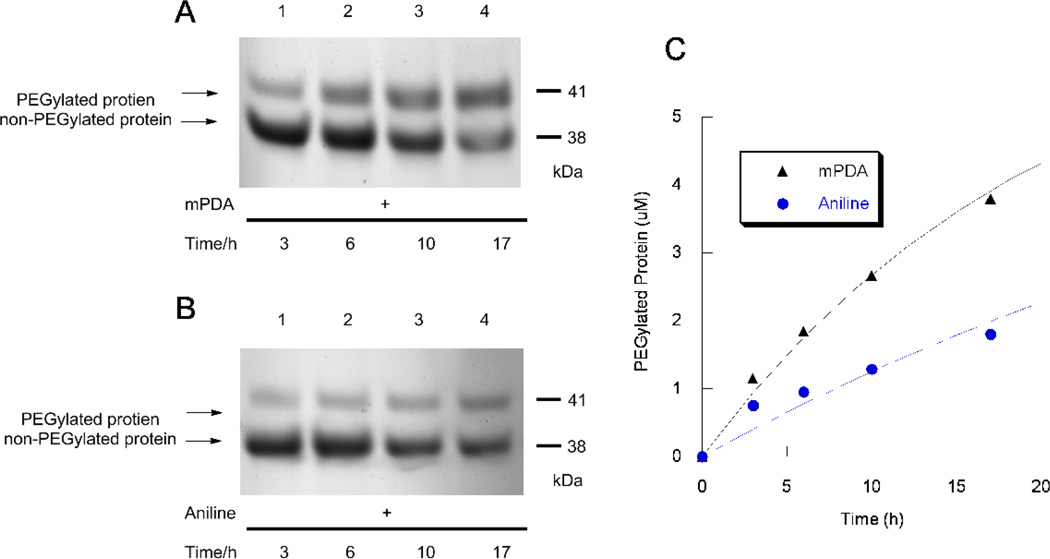
In addition to aldehydes, ketone-containing proteins have also been used for a number of bioconjugation applications including protein PEGylation and spin-labeling.25 The incorporation of the non-natural amino acid p-acetyl phenylalanine into polypeptides via suppressor t-RNA technology is a versatile approach for site-selective introduction of ketones into proteins.26, 27 However, given the attenuated reactivity of ketones, oxime formation is typically carried out under conditions more extreme than those used for comparable reactions with aldehydes. For example, a recent report from Schultz and coworkers employed 100 µM of a ketone-containing antibody and 5 mM alkoxyamine at pH 4.5, 37 °C to achieve near completion in 24 h.28 To probe the utility of mPDA for catalyzing oxime formation with a ketone-containing protein, a p-acetyl phenylalanine-containing protein, (DHFR2 M174pAcF, 11) was produced in E. coli and purified. Aminooxy-PEG 12 (3 kDa, 5 mM) was reacted with DHFR2 M174pAcF (11, 7 µM) in PB (0.1 M, pH 7) in the presence of either 100 mM aniline, 500 mM mPDA or no catalyst. The extent of PEGylation was analyzed at different times via SDS-PAGE and the protein bands were visualized by staining with Coomassie blue. Densitometric analysis of the gels revealed that while aniline did not have a significant effect on the reaction rate (Figure S10), approximately 2.5 fold increase in the rate was observed when mPDA was used as the catalyst (Figure 7). It should be noted that while the protein concentration in this experiment (7.0 µM) was comparable to that used in the work described above with aldehyde-containing proteins (10 µM), it was necessary to increase the concentration of the aminooxy-PEG reagent to compensate for the decreased reactivity of the ketone (compare 0.1–1.0 mM for the aldehyde with 5 mM for the ketone). The presence of elevated concentrations of the aminooxy compound increases the rate for uncatalyzed oxime formation thereby reducing the net effect of the catalyst. Nevertheless, using mPDA, it was still possible to obtain substantial oxime formation under conditions significantly milder (7 µM protein-ketone, 5 mM aminooxy-PEG, pH 7.0, rt) than those reported by other groups.25, 28, 29
Figure 7.
Kinetic analysis of PEGylation of the protein DHFR2 M174pAcF which contains the unnatural amino acid p-acetyl phenylalanine with aminooxy-PEG (3 kDa). Protein (7 µM) was incubated with aminooxy-PEG (5 mM) with either 100 mM aniline or 500 mM catalyst. Next the samples were analyzed via SDS-PAGE and the protein was visualized by staining with Coomassie blue. A) Time course of protein PEGylation in 1–17 h using mPDA (500 mM) as the catalyst. B) Time course of protein PEGylation using aniline (100 mM, its approximate solubility limit) as the catalyst. C) Comparison of protein PEGylation rates using aniline or mPDA obtained by densitometric analysis of the SDS-PAGE results shown in (A) and (B) using the Coomassie blue stained gels.
Effect of mPDA on protein structure and function
Since the mPDA catalyst described here is typically employed at elevated concentrations (relative to aniline), we decided to study its effect on protein structure and enzymatic function using two different assays. In the first assay, GFP was treated with different concentrations of mPDA followed by gel filtration chromatography to remove the catalyst. Circular dichroism spectroscopy of the resulting samples showed no significant differences suggesting that exposure to mPDA does not cause substantial denaturation or irreversible protein unfolding (Figure S12). In the second assay, the effect of mPDA on enzymatic activity was examined. PFTase was treated with various concentrations of mPDA followed by dilution (to reduce the mPDA concentration by a factor of 20) and activity measurement using the aforementioned spectrofluorometric assay. Analysis of that data showed no significant loss of enzymatic activity following treatment with mPDA (Figure S8). Overall, these results suggest that mPDA can be used to catalyze oxime-ligation reactions with proteins without deleterious effects on protein structure or enzymatic activity.
Conclusion
In conclusion, an analysis of the oxime ligation reaction for bioconjugation purposes was performed and resulted in the discovery of several new catalysts for oxime formation and hydrazone-oxime exchange with, m-phenylenediamine (mPDA), being the most efficient. mPDA is a highly efficient catalyst that accelerates oxime formation by orders of magnitude under physiological conditions; compared with aniline, it can catalyze the reaction approximately two-fold faster when employed at equal concentrations. Importantly, mPDA has the advantage of being significantly more soluble than aniline. That feature, in concert with greater catalytic efficiency, allows oxime ligation and hydrazone-oxime exchange to be performed up to 15-fold faster with mPDA compared with aniline; this result is reminiscent of observations made in mechanistic studies of native chemical ligation where replacement of thiophenol with 4-carboxymethyl thiophenol led to significant rate enhancement in transthioesterification due to the much greater solubility of the latter compound.30 The acceleration obtained with mPDA is particularly useful for bioconjugation where high concentrations of proteins cannot typically be obtained to increase the reaction rate. To showcase the utility of this new catalyst, we demonstrated that a protein can be enzymatically labeled with an aldehyde moiety in crude cellular extract, selectively immobilized onto hydrazide beads, and released back into the solution as a fluorescently labeled or PEGylated protein via mPDA catalyzed hydrazone-oxime exchange in a few hours. The amount of protein released in 3 h from the beads was up to 15 times higher in the case of mPDA compared to aniline as the catalyst. While the decreased reaction times achievable with mPDA could be useful in many contexts, this catalyst may be particularly useful for the preparation of materials containing short half-life radionuclides employed for various biomedical imaging applications.31–33 Overall, this new catalyst should be useful for a wide range of bioconjugation applications that require rapid reactions under mild, biocompatible reaction conditions.
Supplementary Material
Acknowledgement
We thank Dr. Joseph Dalluge for helpful discussions regarding mass spectrometry. This work was supported by the National Institutes of Health (GM058842 and GM084152), University of Minnesota Endowment Funding and the Minnesota Supercomputer Institute. We also thank Dr. Peter Schultz’s group at Scripps Research Institute for providing the plasmid pEVOL_pAcF.
Abbreviations
- mPDA
m-phenylenediamine
- PB
phosphate buffer
- DHFR
dihydrofolate reductase
- rt
room temperature
- PFTase
protein farnesyl transferase
- FPP
farnesyl pyrophosphate
- PEG
poly ethylene glycol
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- pAcF
p-acetylphenylalanine
- CNTF
ciliary neurotrophic factor
- LC-MS
liquid chromatography-mass spectrometry
- MALDI
matrix-assisted laser desorption ionization.
Footnotes
Supporting Information Available: Synthetic procedures and additional data and figures. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Maynard HD, Broyer RM, Kolodziej CM. Protein and Peptide Conjugation to Polymers and Surfaces Using Oxime Chemistry. In: Lahann J, editor. Click Chemistry for Biotechnology and Materials Science. Chichester, UK: John Wiley & Sons, Ltd; 2009. pp. 53–68. [Google Scholar]
- 2.Singh Y, Renaudet O, Defrancq E, Dumy P. Preparation of a Multitopic Glycopeptide–Oligonucleotide Conjugate. Org. Lett. 2005;7:1359–1362. doi: 10.1021/ol050134n. [DOI] [PubMed] [Google Scholar]
- 3.Sukhorukov AY, Ioffe SL. Chemistry of Six-Membered Cyclic Oxime Ethers. Application in the Synthesis of Bioactive Compounds. Chem. Rev. 2011;111:5004–5041. doi: 10.1021/cr100292w. [DOI] [PubMed] [Google Scholar]
- 4.Kurpiers T, Mootz HD. Bioorthogonal Ligation in the Spotlight. Angew. Chem. Int. Ed. 2009;48:1729–1731. doi: 10.1002/anie.200805454. [DOI] [PubMed] [Google Scholar]
- 5.Dirksen A, Dawson PE. Rapid Oxime and Hydrazone Ligations with Aromatic Aldehydes for Biomolecular Labeling. Bioconjugate Chem. 2008;19:2543–2548. doi: 10.1021/bc800310p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S, Yousaf MN. An Interfacial Oxime Reaction To Immobilize Ligands and Cells in Patterns and Gradients to Photoactive Surfaces. Langmuir. 2008;24:6201–6207. doi: 10.1021/la8005663. [DOI] [PubMed] [Google Scholar]
- 7.Von Delius M, Geertsema EM, Leigh DA. A synthetic small molecule that can walk down a track. Nat Chem. 2010;2:96–101. doi: 10.1038/nchem.481. [DOI] [PubMed] [Google Scholar]
- 8.Lempens EHM, Helms BA, Merkx M, Meijer EW. Efficient and Chemoselective Surface Immobilization of Proteins by Using Aniline-Catalyzed Oxime Chemistry. ChemBioChem. 2009;10:658–662. doi: 10.1002/cbic.200900028. [DOI] [PubMed] [Google Scholar]
- 9.Sander EG, Jencks WP. Equilibria for additions to the carbonyl group. J. Am. Chem. Soc. 1968;90:6154–6162. [Google Scholar]
- 10.Kalia J, Raines RT. Hydrolytic Stability of Hydrazones and Oximes. Angew. Chem. Int. Ed. 2008;47:7523–7526. doi: 10.1002/anie.200802651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc. Natl. Acad. Sci. 2010;107:10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Meth. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 13.Dirksen A, Yegneswaran S, Dawson PE. Bisaryl Hydrazones as Exchangeable Biocompatible Linkers. Angew. Chem. Int. Ed. 2010:2023–2027. doi: 10.1002/anie.200906756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pompliano DL, Gomez RP, Anthony NJ. Intramolecular fluorescence enhancement: a continuous assay of Ras farnesyl:protein transferase. J. Am. Chem. Soc. 1992;114:7945–7946. [Google Scholar]
- 15.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic Catalysis of Oxime Ligation. Angew. Chem. Int. Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 16.Cordes EH, Jencks WP. J. Am. Chem. Soc. 1962;84:832–837. [Google Scholar]
- 17.Wen-jun W, Chen-ming C, Chen J, Xin W, George-peng W. Kinetic Studies on Aniline-catalyzed Carbohydrate Oxime Formation via Real-time NMR. Chem. Res. Chinese Universities. 2011;27:886–890. [Google Scholar]
- 18.Rashidian M, Song JM, Pricer RE, Distefano MD. Chemoenzymatic Reversible Immobilization and Labeling of Proteins without Prior Purification. J. Am. Chem. Soc. 2012;134:8455–8467. doi: 10.1021/ja211308s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jevsevar S, Kunstelj M, Porekar VG. PEGylation of therapeutic proteins. Biotechnol J. 2010;5:113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- 20.Veronese FM. Peptide and protein PEGylation. Biomaterials. 2001;22:405–417. doi: 10.1016/s0142-9612(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 21.Thom J, Anderson D, McGregor J, Cotton G. Recombinant protein hydrazides: application to site-specific protein PEGylation. Bioconjug. Chem. 2011;22:1017–1020. doi: 10.1021/bc2001374. [DOI] [PubMed] [Google Scholar]
- 22.Ip NY, Yancopoulos GD. The neurotrophins and CNTF: two families of collaborative neurotrophic factors. Annu. Rev. Neurosci. 1996;19:491–515. doi: 10.1146/annurev.ne.19.030196.002423. [DOI] [PubMed] [Google Scholar]
- 23.Rhee KD, Yang X-J. Function and mechanism of CNTF/LIF signaling in retinogenesis. Adv. Exp. Med. Biol. 2010;664:647–654. doi: 10.1007/978-1-4419-1399-9_74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen R, Tao W, Li Y, Sieving PA. CNTF and retina. Prog Retin Eye Res. 2012;31:136–151. doi: 10.1016/j.preteyeres.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleissner MR, Brustad EM, Kalai T, Altenbach C, Cascio D, Peters FB, Hideg K, Peuker S, Schultz PG, Hubbell WL. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proceedings of the National Academy of Sciences. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Schultz PG. Expanding the Genetic Code. Angew. Chem. Int. Ed. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 27.Guo J, Melancon CE, Lee HS, Groff D, Schultz PG. Evolution of Amber Suppressor tRNAs for Efficient Bacterial Production of Proteins Containing Nonnatural Amino Acids. Angew. Chem. Int. Ed. 2009;48:9148–9151. doi: 10.1002/anie.200904035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CH, Axup JY, Dubrovska A, Kazane SA, Hutchins BA, Wold ED, Smider VV, Schultz PG. Synthesis of Bispecific Antibodies using Genetically Encoded Unnatural Amino Acids. J. Am. Chem. Soc. 2012 doi: 10.1021/ja303904e. 120606110334009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi L, Sun H, Wu Y-W, Triola G, Waldmann H, Goody RS. A Highly Efficient Strategy for Modification of Proteins at the C Terminus. Angew. Chem. Int. Ed. 2010;49:9417–9421. doi: 10.1002/anie.201003834. [DOI] [PubMed] [Google Scholar]
- 30.Johnson ECB, Kent SBH. Insights into the Mechanism and Catalysis of the Native Chemical Ligation Reaction. J. Am. Chem. Soc. 2006;128:6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- 31.Weissleder R. Molecular imaging in cancer. Science. 2006;312:1168–1171. doi: 10.1126/science.1125949. [DOI] [PubMed] [Google Scholar]
- 32.Schlemmer H-PW, Pichler BJ, Schmand M, Burbar Z, Michel C, Ladebeck R, Jattke K, Townsend D, Nahmias C, Jacob PK, Heiss W-D, Claussen CD. Simultaneous MR/PET Imaging of the Human Brain: Feasibility Study. Radiology. 2008;248:1028–1035. doi: 10.1148/radiol.2483071927. [DOI] [PubMed] [Google Scholar]
- 33.Nahrendorf M, Zhang H, Hembrador S, Panizzi P, Sosnovik DE, Aikawa E, Libby P, Swirski FK, Weissleder R. Nanoparticle PET-CT Imaging of Macrophages in Inflammatory Atherosclerosis. Circulation. 2008;117:379–387. doi: 10.1161/CIRCULATIONAHA.107.741181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.