Abstract
The MDM2 and MDMX (also known as HDMX and MDM4) proteins are deregulated in many human cancers and exert their oncogenic activity predominantly by inhibiting the p53 tumour suppressor. However, the MDM proteins modulate and respond to many other signalling networks in which they are embedded. Recent mechanistic studies and animal models have demonstrated how functional interactions in these networks are crucial for maintaining normal tissue homeostasis, and for determining responses to oncogenic and therapeutic challenges. This Review highlights the progress made and pitfalls encountered as the field continues to search for MDM-targeted antitumour agents.
Thirty years of research have shown that the tumour suppressor p53 has a crucial role in many physiological processes, and that it is mutated or functionally inactivated in most human cancers. In a substantial proportion of cancers TP53 (which encodes p53) is wild type but the protein is inactivated; this offers an attractive strategy for cancer therapy based on p53 reactivation. Although clinically approved, p53 activators are still a dream; recent studies in cancer patients have provided proof-of-concept for this approach. Such activators are the product of basic research conducted over the past 20 years that has led to the appreciation of MDM2 and MDMX (also known as HDMX and MDM4) as the two major negative regulators of p53, which now seem to be ‘druggable’ using a variety of strategies.
In this Review, we highlight the major advances in our understanding of the biological function of MDM2 and MDMX, and evaluate the evidence that they are oncogenic. We discuss the physiological roles of MDM2 and MDMX and their associated key signalling pathways, as studies in this area have provided important insights into potential clinical benefits and toxicities that are likely to arise from using MDM2 and MDMX antagonists. Finally, we review the current status of small-molecule and peptidic MDM2 and MDMX inhibitors and emphasize how systems biology approaches have provided rationales for developing novel combination strategies. The emerging picture is one of context: MDM2 and MDMX should be considered as two of many crucial factors that contribute to tumour development. Thus, their misregulation sets the stage for additional genomic and epigenetic alterations that lead to cancer. Such a perspective should stimulate approaches to identify and to treat patients whose tumours are particularly susceptible to the targeting of defective MDM2–MDMX–p53 circuitry.
The core pathway
Most p53 mutants in human tumours are transactivation-deficient, suggesting that blocking p53-dependent transcription is a crucial event in tumorigenesis1. Consistent with this, inhibition of p53 transcriptional activation was the first functional role ascribed to MDM2 (Ref. 2). Amplification of MDM genes or altered expression of MDM proteins is a feature of many tumours3–10 (Table 1). In many cases, the frequency of MDM protein deregulation is higher in tumours that retain wild-type p53. Taken together, these observations indicate that a major oncogenic role of MDM proteins is to block p53 transcriptional activity.
Table 1. Frequency of MDM gene or protein alterations in selected human cancers.
| Tumour type | MDM2* | MDMX* | Detection‡ | Refs | ||||
|---|---|---|---|---|---|---|---|---|
| Overall | p53 wild-type | p53 mutant | Overall* | p53 wild-type | p53 mutant | |||
| Glioblastoma§ | 14% | 20% | 0% | 7% | 8% | 0% | Genome | 5 |
| Well-differentiated liposarcoma§ | 70% | 70% | 0% | 0% | 0% | 0% | Genome | 8 |
| Cutaneous melanoma║ | 37% | 46% | 0% | 68% | 85% | 100% | Protein | 10 |
| Breast | 31% | 80% | 14% | 38% | 80% | 24% | Protein | 6 |
| Oesophagus | 18% | 21% | 14% | NA | NA | NA | Genome | 3 |
| Osteosarcoma | 16% | 14% | 14% | 35% | NA | NA | Genome | 7,8 |
| Colorectal | 9% | 65% | 35% | 49% | NA | NA | Genome (MDM2); protein (MDMX) | 4,9 |
NA, not available.
The overall frequency at which alterations were present for both MDM2 and MDMX are listed. All numbers are converted to percentages for clarity. Where possible, the fraction of tumour samples with wild-type or mutant p53 that displayed MDM2 or MDMX amplification is listed.
Detection method was either immunofluorescence or western blot (protein) or gene amplification (genome).
Glioblastoma and well-differentiated liposarcoma show clear mutual exclusivity for MDM deregulation and p53 mutation, whereas this is less clear in other tumour types (for example, colorectal cancer).
Note that the sample size with validated mutant p53 in the cutaneous melanoma study was small (n = 3) and so more studies are required.
Data correlating amplification status with p53 status were not available in these studies.
Although both MDM2 and MDMX can inhibit p53 transactivation function by engaging its amino-terminal transactivation domain via related N-terminal hydrophobic pockets2,11,12, key differences between MDM2 and MDMX affect their ability to regulate p53, as well as their biochemical functions. For example, although p53-responsive elements have been found in both the MDM2 and MDMX promoters, MDM2 is more broadly responsive to p53 activation. By contrast, HDMXL, which is an MDMX protein with an 18-amino acid N-terminal extension, is induced by p53 under more selective conditions13,14. MDM2 homo-oligomers have E3 ubiquitin ligase activity, which depends on an intact carboxy-terminal RING domain15. On binding, MDM2 ubiquitylates p53 and leads to its proteasomal degradation; this keeps p53 levels and activity low in unstressed cells. By contrast, MDMX does not homo-oligomerize and has no intrinsic ubiquitin ligase function, although it can increase or decrease MDM2 ubiquitin ligase activity depending on MDMX abundance16. Hetero-oligomerization of MDM2 and MDMX via their RING domains is crucial for the suppression of p53 activity during embryonic development17,18. Furthermore, aromatic residues that are present in the RING-proximal C-terminal domains of both MDM2 and MDMX are required for the recruitment of E2 ubiquitin-conjugating enzymes19–21. Thus, hetero-oligomerization of MDM2 and MDMX may create a more effective p53 E3 ubiquitin ligase complex, or a more effective inhibitor of p53-dependent transactivation; determining whether these two functions are separable will require additional in vivo models.
Given these findings, we focus on the p53–MDM2–MDMX network, as perturbing this pathway has clear implications for tumorigenesis and presents exciting opportunities for cancer therapy. However, it is important to emphasize that both MDM proteins are reported to have p53-independent roles (Box 1). Such functions may explain the apparent selection for deregulation of MDM2 or MDMX in some tumours that express mutant p53.
Box 1. p53-independent roles of MDM proteins.
Roles for the MDM proteins beyond p53 regulation are elusive, partly owing to the lack of a strong overt phenotype in Trp53–MDM double-knockout mice and cells in culture. However, MDM2 is reported to ubiquitylate numerous targets in addition to p53, and both MDM2 and MDMX are overexpressed in some p53-mutant tumours. This suggests that both proteins may have p53-independent roles in tumorigenesis, as discussed in recent reviews61,134,135. Intriguingly, MDM2 can regulate gene expression and DNA repair by interacting with chromatin or chromatin-associated factors. For example, MDM2 associates with and ubiquitylates oestrogen receptor and androgen receptor136–140. This leads to changes in the expression of hormone-responsive genes and enhances cell proliferation in some contexts137,141. MDM2 may also have a role in the modification of the cellular epigenetic status. For example, MDM2-dependent degradation of RB can increase the levels and activity of the DNA methyltransferase DNMT3A142. This is associated with the silencing of tumour suppressor genes, and suggests that targeting this p53-independent function of MDM2 is a potential therapeutic strategy. MDM2 also interacts with the histone methyltransferase SUV39H1. Depending on the context, this leads to SUV39H1 degradation or to the repression of p53 target genes143,144. Given that loss of SUV39H1 is associated with epigenetic reprogramming, genomic instability and tumorigenesis145,146, further studies into the links between MDM2 and chromatin modifiers are warranted. MDM2 may also engender genomic instability, a hallmark of many cancers, by interacting with and inhibiting proteins that are involved in the DNA damage response134. In contrast to observations with SUV39H1, however, this does not require MDM2 ubiquitin ligase function147. Despite its homology with MDM2, no clear role for MDMX in the regulation of epigenetic modulators has been demonstrated. However, MDMX is reported to interact with and inhibit transcription factors of the E2F and SMAD families148,149. Therefore, MDMX may have a broader influence on gene expression beyond the modulation of p53-dependent transcription. Additionally, MDMX may participate in a ‘failsafe’ mechanism to preserve genome integrity150, although the molecular mechanism (or mechanisms) by which this occurs are unclear.
MDM transcriptional regulation
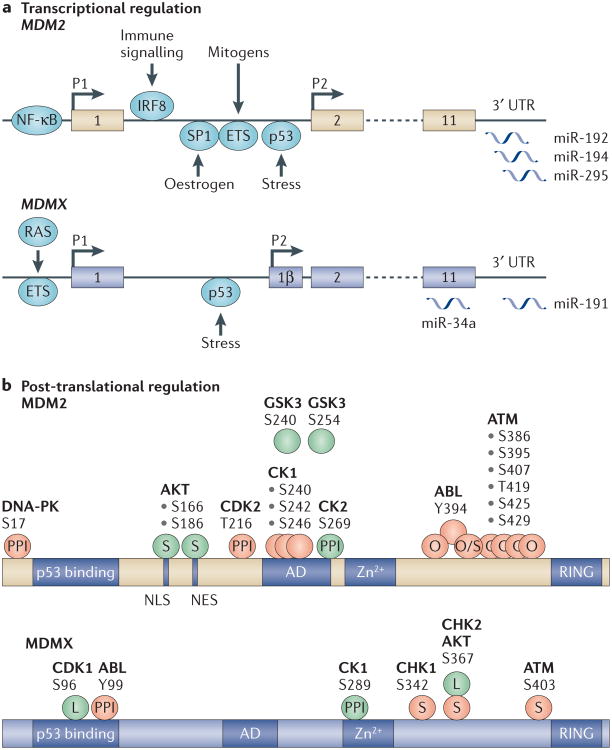
Gene amplification can lead to increased MDM2 or MDMX protein expression. However, many tumours exhibit high MDM2 and MDMX protein levels without increased copy number. These include melanoma10, Ewing's sarcoma22, colon carcinoma9 and retinoblastoma12,23. Thus, the gene amplification criterion may underestimate the number of tumours in which MDM2 or MDMX overexpression contributes to cancer initiation, maintenance or progression. Studies of tissue-specific regulation of MDM transcription and translation might provide clues to how such high MDM protein levels can be attained. For example, the post-translational stabilization of MDM2 and MDMX in some tumours may occur via the activation of cancer-specific signalling pathways, or the hijacking of normal signalling modules that regulate MDM2 and MDMX levels. As identifying the components of such pathways should lead to the development of novel strategies for reducing MDM2 and MDMX abundance, we review below the mechanisms by which MDM proteins are induced or stabilized in normal tissue homeostasis and cancer (Fig. 1).
Figure 1. Transcriptional and post-translational regulation of MDM2 an d MDMX.
a | Transcriptional regulation of MDM2 and MDMX is shown. The first coding exon for both MDM2 and MDMX is exon 2. An ATG in an additional exon (1β) in MDMX can give rise to a longer MDMX-L protein. p53-induced transcription is mediated by the P2 promoter for both MDM2 and MDMX, whereas basal transcription initiates from the P1 promoter. Additional transcription factors (shown in blue) can positively modulate the expression of MDM2 and MDMX from both the P1 and the P2 promoters. Several microRNAs (miRNAs) have been proposed to block translation of either MDM2 or MDMX mRNA. In contrast to the situation with MDM2, one of the miRNAs that targets MDMX mRNA (miR-34a) binds to the coding sequence, rather than to the 3′ untranslated region (UTR). b | In addition to transcriptional regulation, both MDM2 and MDMX are subject to diverse post-translational modifications that affect their ability to bind to p53, to bind to each other and to interact with other cellular proteins that can dramatically affect their stability and that of p53. For simplicity, we focus on phosphorylation to show the diversity of the sites that are modified and the consequences of their modification. Shown in green are phosphorylation sites that are reported to increase MDM2- or MDMX-dependent inhibition of p53. Phosphorylation sites shown in orange are reported to inhibit MDM2- and MDMX-dependent p53 inhibition. In many cases, the precise mechanisms by which phosphorylation modulates the relationship between p53 and MDM2 or MDMX is unclear. Phosphorylation can disrupt the interaction with p53 (by inhibiting protein–protein interaction (PPI)), can modulate the stability of MDM2 or MDMX (S), change oligomerization status (O), modulate stability and oligomerization (O/S) or can alter subcellular localization (L). Residue numbering is for the human proteins, and it should be noted that not all kinase consensus sites are shared between humans and mice. AD, acidic domain; ATM, ataxia-telangiectasia mutated; CDK, cyclin-dependent kinase; CK, casein kinase; DNA-PK, DNA-dependent protein kinase; GSK3, glycogen synthase kinase 3; IRF8, interferon regulatory factor 8; NES, nuclear export signal; NF-κB, nuclear factor-κB; NLS, nuclear localization signal; Zn2+, zinc finger domain.
Proliferating cells are more sensitive than resting cells to MDM2 and MDMX depletion24. It is not surprising, therefore, that links between mitogenic signalling and MDM proteins are emerging. The RAS signalling pathway converges on the activation of ETS transcription factors, and overexpression of RAS leads to ETS-dependent upregulation of both MDM2 and MDMX expression9,25. The subsequent inhibition of p53 may increase the likelihood of RAS-expressing cells escaping p53-dependent arrest. As colon carcinomas and melanomas frequently harbour activated RAS and high levels of MDM2 or MDMX, combinations of RAS pathway inhibitors and MDM2 and/or MDMX antagonists might be particularly effective in such tumours (discussed below).
The activity of MYC, another potent mitogen and an oncogene, is also sensed by p53. In Mdm2+/−- and Mdmx+/−-heterozygous mice, the threshold for p53 activation in response to MYC is lowered, resulting in lower rates of MYC-induced lymphoma26,27. Conversely, one might expect MYC and MDM2 or MDMX to collaborate during tumorigenesis. Intriguingly, MDMX levels seem to be increased in B cells that overexpress MYC before tumorigenesis28. However, whether this is a cause or a consequence of MYC-induced transformation of B cells is unclear. Although potential MYC-binding sites have been found in MDMX (M.W., unpublished observations), there is little evidence supporting MYC-dependent increases in MDMX mRNA. However, there are binding sites for the B cell-specific transcription factor interferon-regulatory factor 8 (IRF8) in both MDM2 and MDMX29 (M.W., unpublished observations). Furthermore, there is genetic evidence that MDM2 is upregulated by IRF8 during germinal centre B cell proliferation29. The induction of B cell proliferation during homeostasis (or following oncogene activation) and the ensuing IRF8-dependent increase in MDM proteins may limit p53 activity as cells undergo genetic recombination events.
Much more remains to be discovered regarding the regulation of MDM2 and MDMX transcription. Although activated p53 was thought to exclusively upregulate MDM2, new data have revealed p53-dependent increases in MDMX expression under certain conditions14. These mechanisms may contribute to the inhibition of p53 in response to both physiological stresses and p53-targeted therapeutics. Mutation of the p53 response elements in the Mdm2 and Mdmx promoters in vivo will be required to establish their role in the p53 response. Additionally, the factors that control tissue-specific transcriptional regulation of MDM2 and MDMX have not been systematically investigated. For example, it will be interesting to determine the tissue-specific and stress-specific factors that control the expression of MDM2 from the P2 promoter. Originally identified as p53-responsive, it is now clear that other transcription factors can modulate expression from this promoter30,31.
MDM post-transcriptional regulation
Additional layers of post-transcriptional control of MDM proteins are continually being revealed. Endogenous mechanisms include regulation by microRNAs (miRNAs) and by post-translational modifications (PTMs). Intriguingly, viruses may subvert these mechanisms as part of their life cycle. We discuss below each of these regulatory circuits as they relate to cancer.
PTMs
Multiple kinases are known to modify both MDM proteins and (perhaps unsurprisingly) the downstream effects cluster into those that inhibit and those that activate p53. The AKT kinases regulate cell growth and survival, and they are activated in human cancers; among their targets are MDM2 and MDMX. Phosphorylation of MDM2 at Ser166 and Ser186 (Ref. 32), and of MDMX at Ser367 (Ref. 33), leads to their stabilization, and is associated with p53 inhibition. Casein kinase 1α (CK1α)-mediated phosphorylation of MDMX increases its affinity for p53 and may therefore reduce p53 activity34. Following genotoxic stress, DNA-dependent protein kinase (DNA-PK) phosphorylates Ser17 of MDM2 (Ref. 35), and ABL (also known as ABL1) phosphorylates Tyr99 of MDMX36. In each case, this leads to the dissociation of p53 from its negative regulators.
Following DNA damage, the ataxia-telangiectasia mutated (ATM) and CHK kinases also phosphorylate multiple serine residues in or close to the RING domains of MDM2 and MDMX37. This can lead either to the dissociation of MDM oligomers or to the destabilization of MDM2 or MDMX38–41. The downstream consequences of the ensuing p53 activation are determined in a tissue-specific manner42. Conversely, dephosphorylation of these residues may inhibit p53. Indeed, protein phosphatase 1D (PPM1D) can dephosphorylate both MDM2 and MDMX, thereby contributing to its oncogenic function43,44. The CK1δ kinase phosphorylates MDM2 in the acidic domain, and this also leads to p53 activation45. Mutation of some of these phosphorylation sites is associated with tumorigenesis in knock-in mouse models46,47 (discussed below).
miRNA links to p53 pathway control by MDM2 and MDMX
Recent studies have added numerous miRNAs to the gene constellation that p53 regulates48,49, which in turn contribute to its activation. Importantly, several studies have indicated that some p53-induced miRNAs contribute to the downregulation of MDM2 and MDMX. Although perhaps expected in normal cells, it was surprising to find that in multiple myelomas in which p53 is wild type, p53 can induce the expression of mir-192, mir-194 and mir-215, which subsequently downregulate MDM2 expression50. This would be expected to activate p53. However, hypermethylation of the promoter region of all three miRNAs impairs MDM2 downregulation on p53 activation, which should blunt p53 function in these tumours50.
In addition to genotoxin-induced degradation of MDMX, MDMX mRNA levels is also reduced on DNA damage. This results, in part, from the action of mir-34a51. The mir-34a target site colocalizes with a single nucleotide polymorphism (SNP) in exon 11 of the MDMX mRNA52. Sequence alignment indicates that the C allele of the SNP would disrupt mir-34a binding52. In addition, a SNP (SNP34091) in the MDMX mRNA 3′ untranslated region (UTR) was recently shown to be an illegitimate target site of miR-191 that was nevertheless targeted53. However, the effects of these miRNAs in modulating MDMX levels, and therefore p53 regulation in vivo, require additional study.
Although the data suggest that p53, microRNAs, MDM2 and MDMX form regulatory circuits to facilitate the rapid activation of p53 on stress, a recent mir-34-knockout mouse model suggests caution in such an interpretation. These studies have revealed that the mir-34 family does not have a substantial effect on p53 tumour suppressor functions in vivo54. Thus, the biological importance of the p53-miRNA regulatory network for tumour suppression and stem cell biology remains to be clarified, and additional in vivo models should illuminate this48.
The study of virus-induced cellular miRNAs is also beginning to reveal some surprises. For example, p53-independent induction of mir-34a by Epstein-Barr virus (EBV) seems to be necessary for the transformation of B lymphocytes55. This again underscores the context-dependent effects of mir-34a induction. EBV infection also leads to the expression of miRNAs that have seed complementarity to both MDM2 and MDMX55. However, the downregulation of MDM2 and MDMX would be expected to induce growth inhibition; thus it is likely that additional cellular transcripts with target sites that are similar to the MDM mRNAs are the primary targets. Whether other viruses might use miRNAs to modulate MDM2 or MDMX mRNA levels to inhibit the p53 feedback loop is unclear.
Regardless, virus-induced upregulation of MDM2 by both transcriptional and post-transcriptional mechanisms has been reported. EBV is associated with several tumour types, and it expresses latency antigens that stabilize MDM2 (Ref. 56). MDMX levels are also fairly high in EBV-infected B cells, although the underlying mechanism is unclear. Kaposi's sarcoma-associated herpes virus (KSHV) expresses vIRF4, a factor that binds MDM2 and that seems to switch its ubiquitin ligase activity to p53 degradation57. Both EBV and lymphoid cells exhibit B cell tropism, and it is clear that small changes in the levels of MDM proteins in these cells render them susceptible to transformation58. Tumour-associated SNPs in the MDM2 promoter are also correlated with the increased risk of hepatitis C-induced cancer59. Together, these data suggest that targeting MDM proteins in virus-associated malignancies may have therapeutic benefit. In support of this, the MDM2 antagonist nutlin 3a induces cell death in KSHV-positive tumour cells60.
MDM2 and MDMX as oncogenes: the evidence
Cell-based studies
MDM2 and MDMX have oncogenic properties in vitro, but this is only manifest in the context of other oncogenic lesions. For example, attempts to use primary cells to demonstrate that MDM2 alone has transforming activity have been unsuccessful. This is probably because MDM2 overexpression at supraphysiological levels is toxic to many normal cells, suggesting that it may have other cellular targets61. MDM2 is also inherently unstable, making it difficult to express the protein at levels that are sufficient to inactivate p53 and to induce tumorigenesis. By contrast, the overexpression of MDM2 in immortalized mouse NIH3T3 cells renders them tumorigenic62. Thus, the effects of MDM2 are manifest in a background of pre-existing genetic changes. Conversely, MDMX has a long half-life and can generally be expressed at extremely high levels. MDMX can immortalize primary mouse embryonic fibroblasts and can accelerate the growth of human fibroblasts63,64. However, despite its ability to inhibit p53, MDMX overexpression alone is insufficient for the robust transformation of either human or mouse cells.
In vivo studies
Tables 2,3 list the tissue-specific effects of Mdm2- or Mdmx-knockout mice, as well as several in vivo models that have shed light on their oncogenic roles. Early transgenic mouse models showed that MDM2 overexpression could induce carcinoma65 and lymphomas or sarcomas66. MDM2 overexpression does not seem to accelerate the onset of tumorigenesis in Trp53 (which encodes p53 in mice)-null animals. Together, these data provide evidence that a major oncogenic effect of MDM2 is mediated through p53 inhibition. It is possible that positional effects and a lack of wild-type gene architecture contribute to the phenotypes of transgenic animals. However, a more recent mouse model used a knock-in of a human tumour-associated MDM2 SNP at the endogenous Mdm2 locus. Data from this model confirm that a subtle increase in MDM2 levels engenders tumorigenesis at a rate that is similar to that observed in Trp53-hemizygous animals67. Conversely, haploinsufficiency of MDM2 delays the onset of MYC-induced lymphoma in vivo27.
Table 2. Phenotypes of mice with modifications at the Mdm2 and Mdmx loci.
| Tissue | Genotype | p53 regulation strategy* | Phenotype‡ | Refs |
|---|---|---|---|---|
| CNS | Mdm2-/- | Nestin-Cre;Trp53LSL/- | Embryonic lethal; apoptosis | 24 |
| Mdmx-/- | Nestin-Cre;Trp53LSL/- | Apoptosis and arrest | ||
| Mdm2FM/FM | Nestin-Cre | Embryonic lethal; apoptosis | 109 | |
| MdmxFX/FX | Nestin-Cre | Embryonic lethal; apoptosis and arrest | ||
| Mdm2-/- | Trp53ER/- | No effect | 81 | |
| Mdmx-/- | Trp53ER/- | Apoptosis only in SVZ | ||
| Intestine | Mdm2FM/FM | Villin-Cre | Apoptosis | 164 |
| MdmxFX/FX | Villin-Cre | Apoptosis in proliferating cells | 165 | |
| Mdm2-/- | Trp53ER/- | Apoptosis | 81 | |
| Mdmx-/- | Trp53ER/- | Apoptosis | 85 | |
| Smooth muscle | Mdm2FM/FM | Sm22-CreERT2 | Cell death | 166 |
| MdmxFX/FX | Sm22-CreERT2 | No effect | ||
| Erythrocyte | Mdm2lox/lox | EporGFP-Cre/+ | Embryonic lethal; apoptosis | 167 |
| Mdmxlox/lox | EporGFP-Cre/+ | Arrest only in fetal erythropoiesis | ||
| Heart | Mdm2FM/- | Myhc-Cre | Embryonic lethal; apoptosis | 168 |
| MdmxFX/- | Myhc-Cre | Apoptosis | 169 | |
| Mdm2-/- | Trp53ER/- | No effect | 81 | |
| Mdmx-/- | Trp53ER/- | No effect | 85 | |
| Thymus and spleen | Mdm2-/- | Trp53ER/- | Apoptosis | 81 |
| Mdmx-/- | Trp53ER/- | Apoptosis | 85 | |
| Testis | Mdm2-/- | Trp53ER/- | Arrest | 81 |
| Mdmx-/- | Trp53ER/- | Not reported | 85 | |
| Lung, kidney and liver | Mdm2-/- | Trp53ER/- | No effect | 81 |
| Mdmx-/- | Trp53ER/- | No effect | 85 |
CNS, central nervous system; Epor, erythropoietin receptor; FM, floxed Mdm2 allele; FX, floxed Mdmx allele; GFP, green fluorescent protein; Myhc, myosin heavy chain; Sm22, smooth muscle protein 22 (also known as transgelin); SVZ, subventricular zone.
Knockout of MDM2 or MDMX is embryonic lethal, and therefore either MDM knockout or p53 expression must be rendered conditional. This is usually achieved using Cre-mediated recombination. Note that the Trp53LSL and Trp53ER systems are to some extent ‘leaky’. Thus, adult animals can only be obtained by reducing Trp53 to hemizygosity.
Phenotype is for the adult tissue, except where the knockout was embryonic lethal.
Table 3. Phenotypes of mice with alterations of MDM2 and MDMX.
| Mouse model | Effect | Phenotype | Refs |
|---|---|---|---|
| Knock-in mice | |||
| MDM2-S394A | Removes ATM phosphorylation site | Radioresistant and accelerated spontaneous tumours | 47 |
| MDM2-C305F | Disrupts interaction with ribosomal proteins | Accelerated MYC-induced lymphoma | 170 |
| MDM2-C462A | Disrupts RING domain and MDMX binding; ubiquitin ligase activity lost | Embryonic lethal | 171 |
| MDM2 (SNP309G/G) | Increases MDM2 expression | Accelerated spontaneous tumorigenesis | 67 |
| MDMX RING deletion | Removes all RING-associated functions | Homozygote is embryonic lethal | 18 |
| MDMX-C462A | Disrupts RING domain and MDM2 binding | Homozygote is embryonic lethal | 17 |
| MDMX-3SA | Removes AKT, ATM and CHK2 phosphorylation sites | No accelerated spontaneous tumorigenesis; accelerated MYC-induced lymphoma | 46 |
| Overexpression models | |||
| MDM2 transgenic | BLG promoter-driven mammary gland expression of wild-type MDM2 cDNA | Inhibits mammary gland development and increased mammary gland tumours | 65 |
| MDM2 transgenic | Entire wild-type MDM2 gene and promoter construct | Increased spontaneous tumours | 66 |
| MDM2 S166D/S186D | MMTV-driven MDM2 cDNA mimicking constitutive AKT phosphorylation | No increase in spontaneous tumours; accelerated ERBB2-induced tumours | 172 |
| MDM4 transgenic | Non-targeted transgenic; Cre-activated wild-type cDNA | Accelerated spontaneous tumorigenesis | 68 |
| HA-tagged MDM4 | Knock-in of Cre-activated wild-type cDNA at ROSA26 locus | Homozygous overexpression is embryonic lethal; no increase in spontaneous or MYC-induced tumours | 28 |
ATM, ataxia-telangiectasia mutated; BLG, β-lactoglobulin; MMTV, mouse mammary tumour virus.
Given the many functional similarities between MDM2 and MDMX, and the frequent overexpression of MDMX in some human cancers, it was logical to predict that MDMX overexpression in vivo would be tumorigenic. However, mouse models of MDMX overexpression have yielded conflicting results. In one study, transgenic mice that ubiquitously expressed MDMX developed tumours with a latency that was comparable to that of Trp53-hemizygous mice68. However, a subsequent study found no evidence for enhanced tumorigenicity following high-level MDMX transgene expression in many tissues28. Differences in transgene introduction strategies and mouse genetic backgrounds may contribute to these disparate results, which underscores the idea that the context in which MDMX is overexpressed is probably a major determinant of its oncogenic activity.
On the basis of the finding that MDM proteins are the targets of multiple kinases, two mouse models have been generated to test the effect of MDMX and MDM2 PTMs on p53 regulation and the subsequent biological sequelae in different tissues. In the first mouse model, Wang et al.46 mutated three serines in the C terminus of MDMX to alanine (MDMX3SA). Phosphorylation of these residues by ATM and CHK2 triggers MDMX degradation. Therefore, MDMX3SA is resistant to DNA damage-induced degradation, which ultimately attenuates p53 function. Consistent with this, MDMX3SA mice are extremely radioresistant. Surprisingly, although there was no increase in spontaneous tumour formation in MDMX3SA animals, these mice were more prone to Myc-induced tumorigenesis69 (discussed below). These data reveal that MDMX modification is important for preventing oncogene-induced tumorigenesis, but does not affect the tumorigenicity that is associated with background mutagenic events. By contrast, the mutation of Ser394 to alanine (Ser394Ala) in mouse MDM2 in vivo removes an ATM phosphorylation site, engenders radioresistance and increases the rate of spontaneous tumorigenesis to that observed in Trp53-hemizygous animals47. Furthermore, the predominance of T cell lymphomas in the MDM2-Ser394Ala mice recapitulates the range of tumour types found in Trp53-deficient mice. Together, these data indicate that the phosphorylation of both MDM2 and MDMX C termini is required for the p53 response to high levels of ionizing radiation. However, p53 sensing of endogenous (and potentially oncogenic) DNA damage in vivo seems to specifically require modification of the MDM2 C terminus.
Cooperating lesions
Both the cell-based and the in vivo data discussed above indicate that the full oncogenic potential of MDM2 and MDMX is only revealed in the presence of additional genetic lesions. In vitro culture and mouse models have provided some insight into the nature of these lesions. The transduction of normal human and mouse fibroblasts with oncogenic RAS variants leads to senescence, which is generally p53-dependent. However, co-expression of MDMX leads to cellular transformation in vitro63,64,70. Importantly, this requires the p53-binding domain of MDMX, suggesting that p53-inhibitory activity of MDMX is crucial for transformation63. A recent in vivo model of melanoma has also provided evidence for the cooperation between MDMX and RAS during tumorigenesis. Following melanocyte-specific expression of MDMX and RAS, the time to onset of melanomas was reduced and the tumours that developed were more aggressive10. Furthermore, the finding that melanomagenesis in Trp53-hemizygous mice is accelerated by MDMX overexpression confirms that p53 is the main target of MDMX in this cancer. The recapitulation of some of the genetic and clinical features of melanoma in this study should inspire additional in vivo models to dissect the tissue-specific role of MDMX in other tumour types.
Similar to RAS, the oncogenic activity of MYC can be limited by p53 activation. This was clearly established in mouse models in which the deletion of Trp53 decreased the time to onset of MYC-driven B cell lymphoma71. Although p53 mutations are commonly found in human and mouse lymphomas, a substantial proportion retains the wild-type Trp53 allele. This suggests that there are additional ways to subvert p53 activation in B cell neoplasms. An obvious candidate is MDMX, and two mouse models have addressed its potential role in MYC-induced lymphomagenesis. Marine and colleagues28 found that MYC-induced lymphomas were not accelerated in mice with ubiquitous overexpression of wild-type Mdmx. By contrast, Wahl and colleagues46 reported a dramatic acceleration of MYC-induced lymphomas in the presence of the MDMX3SA mutant. As MDMX levels were lower in this study, the amount of MDMX present cannot explain the variation in results. As MDMX3SA mice have an attenuated response to DNA damage, and because MYC is known to induce genotoxic stress, disruption of the MYC–DNA damage–MDMX circuit might contribute to increased tumorigenesis in MDMX3SA mice. Intriguingly, one of the ATM target sites (MDMX-Ser367) was mutated in a squamous cell carcinoma sample, and this occurred in the context of TP53 heterozygosity72. However, further clinical and experimental studies are required to establish the importance of MDMX phosphorylation in human cancer. Although there are correlations between high levels of NMYC and MDM2 in neuroblastoma cell lines73 there is no direct in vivo evidence that MYC and elevated MDM2 levels cooperate to accelerate tumorigenesis.
In addition to their cooperation with oncogenes, the expression of MDM proteins can accelerate tumorigenesis that is caused by the loss of tumour suppressors. For example, retinoblastoma (which is caused by the loss of proteins of the RB tumour suppressor family) is accelerated by MDMX overexpression12. This is important from a clinical perspective because retinoblastomas retain wild-type TP53 and may therefore benefit from treatment with MDM2 and/or MDMX antagonists. Loss of the PTEN tumour suppressor leads to the activation of the AKT kinases and increases the level of MDM2 (Ref. 74). This is associated with an attenuated p53 response. Although AKT has multiple downstream targets in addition to MDM2, tumour cells with activated AKT pathways are sensitive to MDM2 antagonists75,76.
Together, these data indicate that the oncogenic stress that is generated by the loss of tumour suppressors may lead to the selection of tumour cells with elevated MDM2 or MDMX levels. In this regard, understanding the homeostatic roles of MDM proteins will provide insights into the mechanisms by which they are deregulated during cancer (Box 2). Furthermore, the failure of MDM2 and MDMX overexpression in isolation to robustly induce tumours may indicate that ‘too much is not enough’. Rather, their full oncogenic potential may only be unleashed in the context of other neoplastic changes.
Box 2. Homeostatic roles of MDM proteins.
Ancestral roles of the p53 family were probably geared towards reproductive fitness. Together with other family members, p53 ensures that cells with DNA damage are eliminated during development and enhances the rate of embryonic implantation. Loss of MDM proteins activates p53 in most tissues studied, although the onset of p53-dependent apoptosis seems to be confined to radiosensitive tissues such as the thymus and spleen24,81. Thus, the MDM proteins probably evolved to block spurious p53 activation. Given that lymphoid cells generate DNA strand breaks during somatic recombination, MDM proteins in these cells may buffer against ‘damage’-induced p53 activation. Interestingly, T cell activation is accompanied by the increased transcription of both MDM2 and MDMX, suggesting that p53–MDM2–MDMX autoregulatory feedback loops are important in this tissue151.
Intestinal cells are sensitive to p53, probably because they are highly proliferative. Moreover, recent data have suggested that intestinal DNA damage leads to p53 activation via modulation of MDM2 (Ref. 47). Consistent with this, intestinal loss of MDM2 induces apoptosis; together with haematopoietic cell depletion, this is thought to cause the death of Mdm2-deficient animals81. Therefore, as in the haematopoietic system, the role of MDM proteins is likely to restrict p53 activation unless genomic integrity is threatened.
The subventricular zone (SVZ) of the adult brain is the putative site of proliferation for adult neurogenesis and the location of specific neuronal stem cells152. Levels of MDM proteins are relatively high in the brain, and the deletion of either MDM gene in the SVZ leads to p53-dependent apoptosis24,81,85. This underscores the importance of MDM proteins in the protection of proliferative cells during homeostasis. Strikingly, MDM2 and MDMX are amplified or overexpressed in glioblastomas at high frequency153. As glioblastomas are derived from putative stem cells in the SVZ152, p53 inhibition by MDM proteins may contribute to the onset or the maintenance of these tumours.
p53-induced mRNAs and microRNAs are associated with liver pathology154,155, and excessive p53 activation in adipocytes can contribute to insulin resistance154. Thus, MDM proteins may serve a protective role in these tissues by blocking p53 activation. MDM2 may also modulate adipocyte differentiation in a p53-independent manner156. The additional roles of MDM2 in adipocytes may come at a price, however, as amplification of MDM2 in these cells is associated with liposarcoma157.
p53 reactivation in cancer therapy
Around 22 million cancer patients have defects in p53 signalling77. Although ∼50% of these patients harbour mutant p53, which has lost its tumour suppressor function, the other 50% retain a wild-type TP53 allele. The function of p53 is attenuated by MDM2, MDMX and other signalling modules. Mouse models clearly demonstrate that the inactivation of p53 promotes tumorigenesis, whereas restoration of p53 causes the regression of established tumours78–80. Although deletion of Mdm2 can cause p53 activation in both tumour tissue and normal tissue81, and p53 upregulation in the haematopoietic system engenders myeloablation82,83, the MDM2 antagonist nutlin 3a (discussed below) is well-tolerated in mouse preclinical models84. Mouse tissues also seem to tolerate the p53 activation that accompanies Mdmx deletion85. These data indicate that wild-type p53 is a valid therapeutic target, and that reactivation of p53 via selective targeting of either MDM2 or MDMX is a viable strategy for cancer therapy.
Several pharmacological strategies have been proposed for the activation of wild-type p53 (Table 4). However, interfering with MDM2 and MDMX is the most direct approach. First, reducing MDM2 and MDMX abundance in cancer cells should enhance p53 activity. Second, inhibitors of MDM2 E3 ubiquitin ligase function should increase p53 level and activity. Third, protein–protein interaction (PPI) antagonists that selectively disrupt p53–MDM2 or p53–MDMX N-terminal interaction should activate p53. In addition, attenuation of MDM2 ubiquitin ligase activity should increase p53 levels and activity. This may be achieved using PPI antagonists of the heterodimerization of MDM2 and MDMX RING domains, or of the interaction between MDM2 and E2 ubiquitin-conjugation enzymes (designated MDM2/E2). There are also more indirect approaches, which is discussed in Box 3. Additionally, as the pharmacological reactivation of mutant p53 is an attractive, though challenging, strategy, we refer readers to reviews that discuss this option in detail77,86.
Table 4. Approaches to target MDM2 and MDMX.
| Targeting approach | Compound | Class | Target | Proposed working mechanism | Remarks |
|---|---|---|---|---|---|
| Modulating protein expression | NSC207895 (Ref. 88) | Small molecule | MDMX | Inhibits MDMX transcription | Highly related to DNA damage agents89 |
| 17-AAG92 | Small molecule | HSP90 | HSP90 inhibitor | Destabilizes MDMX through an undefined mechanism92 | |
| Targeting protein–protein interaction | Nutlin 3a96 | Small molecule | MDM2 N-terminal p53-binding pocket | Disrupts p53–MDM2 interaction | The derivative, RG7112, is in Phase I clinical trial |
| MI-219 (Ref. 97) | Small molecule | MDM2 N-terminal p53-binding pocket | Disrupts p53–MDM2 interaction | Preclinical development | |
| SJ-172550 (Ref. 105) | Small molecule | MDMX N-terminal p53-binding pocket | Disrupts p53–MDMX interaction | Forms adducts with cysteine in MDM2 and MDMX106 | |
| RO-5963 (Ref. 103) | Small molecule | Both MDM2 and MDMX N-terminal p53-binding pocket | Disrupts p53–MDM2 and p53–MDMX interactions | Causes MDM2 and MDMX homodimers and heterodimers103 | |
| WK 298 (Ref. 173) | Small molecule | MDMX | Disrupts p53–MDMX interaction | No cellular activity173 | |
| AM-8553 (Ref. 174) | Small molecule | MDM2 N-terminal p53-binding pocket | Disrupts p53–MDM2 interaction | ||
| SAH-p53-8 (Ref. 110) | Peptidic compound | Both MDM2 and MDMX N-terminal p53-binding pocket | Disrupts p53–MDM2 and p53–MDMX interactions | Cellular uptake requires pinocytosis113 | |
| PMI peptide112 | Peptidic compound | Both MDM2 and MDMX N-terminal p53-binding pocket | Disrupts p53–MDM2 and p53–MDMX interactions | No cellular activity175 | |
| pDI peptide111 | Peptide | Both MDM2 and MDMX N-terminal p53-binding pocket | Disrupts p53–MDM2 and p53–MDMX interactions | Cellular activity only in context of adenoviral delivery111 | |
| RITA158 | Small molecule | p53 N-terminal domain | Disrupts p53–MDM2 interaction | Showed p53-independent activity161 | |
| Targeting E3 ubiquitin ligase activity | HLI98 (Ref. 115) | Small molecule | MDM2 | Inhibits MDM2 ubiquitin ligase activity | Showed p53-independent activity115 |
| MPD116 | Small molecule | MDM2 RING domain | Inhibits MDM2 ubiquitin ligase activity | Based on HLI compound. Potent derivative showed p53-independent activity116 | |
| MEL23 and MEL24 (Ref. 117) | Small molecule | MDM2 | Inhibits MDM2 ubiquitin ligase activity | ||
| Activating p53 via other mechanisms | JNJ-26854165 | Small molecule | MDM2 | Inhibits p53–MDM2–proteasome interaction | Showed p53-independent activity162 |
HSP90, heat shock protein 90; RITA, reactivation of p53 and induction of tumour cell apoptosis; SAH-p53-8, stabilized α-helix of p53 variant 8.
Box 3. Alternative MDM2- and MDMX-dependent p53-activating approaches.
The small-molecule compound reactivation of p53 and induction of tumour cell apoptosis (RITA; also known as NSC 652287) was found to directly bind p53 and to block the p53–MDM2 interaction158. RITA was shown to cause p53-dependent accumulation and activation via a high-throughput cell proliferation assay using TP53+/+ and TP53-/- HCT116 cell lines. Its proposed mode of action was via binding to the p53 amino-terminal domain, leading to a p53 conformational change and the dissociation of MDM2 (Ref. 158). However, in addition to its ability to bind to p53, RITA causes protein–DNA crosslinks159 and is also metabolized to a reactive species160. Given these additional undesired properties, it is perhaps not surprising that RITA is cytotoxic in cells with both mutant and truncated p53 (Ref. 161).
JNJ-26854165, a tryptamine derivative, was reported to activate p53 by preventing the MDM2–p53 complex from binding to the proteasome, thereby blocking p53 degradation. However, preclinical studies have revealed activity in p53 wild-type and p53-mutant cancer cells and a general genotoxic effect, indicating that this compound is not p53-specific162. Although JNJ-26854165 entered Phase I clinical trials (see the ClinicalTrials.gov website; see Further information), the programme was halted owing to cardiotoxicity and an MDM2-independent mechanism of action (W. Hait, personal communication).
The MDMX3SA and MDM2S394A mouse models46,47 show that p53 is not activated by DNA damage signals if MDM2 and MDM4 are not phosphorylated correctly. Small-molecule therapeutics that mimic the effect of MDM2S394A or MDMX3SA may be useful both in basic research and in the clinic. For example, in patients undergoing radiotherapy, acute p53-dependent apoptosis in normal radiosensitive tissues is a major source of side effects. Transient inhibition of p53 reduces cell death in normal tissues without compromising p53 tumour suppressor function163. Therefore small molecules that block DNA damage-induced MDM protein phosphorylation should transiently inhibit p53 during radiotherapy, thereby reducing unwanted side effects.
Modulating protein expression
A straightforward strategy to reduce MDM2 and MDMX protein levels in cancer cells is to specifically target them using small interfering RNA (siRNA), short hairpin RNA (shRNA) or miRNA approaches. Unfortunately, siRNA therapy is still hampered by delivery and cellular uptake issues87, making this approach currently unrealistic. A benzofuroxan derivative (NSC207895) that selectively inhibits MDMX expression has been identified88. This molecule downregulates MDMX levels and causes p53-dependent transactivation of proapoptotic genes in several cancer cell lines. NSC207895 seems to repress the MDMX promoter, although the underlying molecular mechanism of promoter-specific targeting has not yet been revealed88. However, NSC207895 also clustered with known DNA-damaging agents, such as methyl methanesulphonate (MMS) and camptothecin, in a cross-species chemogenomic profiling screen89. As DNA-damaging agents induce MDMX degradation90 it is possible that the effects of NSC207895 on MDMX protein levels and p53 activation involve more than the repression of MDMX transcription.
The molecular chaperone heat shock protein 90 (HSP90) is highly expressed in many cancers91. Treatment with the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin (17-AAG; also known as tanespimycin) destabilizes MDMX in several cancer cell lines. Co-treatment of such cancer cells with 17-AAG to downregulate MDMX, and nutlin 3a to antagonize the MDM2–p53 interaction, yielded synergistic cytotoxic effects, probably owing to enhanced p53 activation92. Currently, no compounds are reported to selectively reduce MDM2 transcription and protein abundance.
Targeting protein–protein interactions
Genome-wide protein–protein network studies have provided a detailed map of disease-associated PPIs93,94. However, flat, featureless and extended PPI surfaces are considered undruggable with small-molecule compounds95. Small-molecule PPI antagonists that disrupt the p53–MDM2 interaction have been identified, including nutlin 3a96 and MI-219 (Ref. 97). These compounds reactivate wild-type p53 by competing with it for binding to the hydrophobic cleft in the MDM2 N terminus. RG7112 (also known as RO5045337), a derivative of nutlin 3a with better potency and pharmacological properties, is in Phase I clinical trials (see the ClinicalTrials.gov website; see Further information), and the results of the first proof-of-mechanism study for RG7112 in patients have just been published98. A substantial proportion of liposarcomas express wild-type p53 and have amplified MDM2; thus, patients with liposarcomas of this class provide a suitable population in which to assess the ability of RG7112 to activate p53 and elicit downstream responses, as well as to determine the side effects from the use of this compound. The results of the analysis must be considered preliminary as only a small number of patients (20) were enrolled. However, the data indicate that RG7112 reaches its target in a solid tumour, and increases the level of macrophage inhibitory cytokine 1 (MIC1; also known as GDF15), which is a secreted protein product of a p53 target gene. Interestingly, although RG7112 treatment generally correlated with increased p53 and p21 levels, and decreased proliferation, the data did not reach significance. As the tumours were not microdissected, the modest results might reflect the use of heterogeneous tumour tissue in which only a small proportion of cells responded to RG7112 at the time of analysis. Notably, p53 pathway reactivation resulted in at least one adverse event in each patient, the most frequent being related to haematological toxicities. The authors suggested that RG7112 may be considered in neoadjuvant therapy when combined with existing clinically approved cytotoxic agents. However, the radiosensitivity of the haematopoietic system of mice to p53 activation83 suggests that such an approach would probably exacerbate the toxicity that is associated with RG7112. Rather, we suggest that combining non-genotoxic molecularly targeted therapeutic agents with p53 agonists might be a safer alternative.
Although nutlin 3a and MI-219 robustly activate p53 in cancer cells with overexpressed MDM2, they do not consistently elicit p53-dependent effects in cancer cells that overexpress MDMX99–102. This was surprising because the sequence of the MDMX N-terminal p53-binding domain is very similar to that of MDM2. However, subtle structural differences in the MDMX N-terminal p53-binding pocket dramatically reduce the binding affinity of both nutlin 3a and MI-219 for MDMX; this is supported by in vitro binding studies showing that nutlin 3a is 500-fold less potent against MDMX compared with MDM2 (Ref. 103). Despite this, a customized ocular formulation of nutlin 3a enhanced the efficacy of topotecan in a p53-dependent manner when it was subconjunctivally administered to MDMX-overexpressing retino-blastomas in situ104. However, it is not clear to what extent this synergy depends on the disruption of MDMX–p53 complexes versus MDM2–p53 complexes at the high dose of nutlin 3a used, and whether downregulation of MDMX by topotecan may also contribute. The small molecule SJ-172550 was identified as an MDMX–p53-selective antagonist105 that exhibited additive cytotoxicity with nutlin 3a in MDMX-amplified retinoblastoma cells. However, subsequent analyses showed that SJ-172550 forms covalent adducts with cysteine residues in the p53-binding domain of both MDM2 and MDMX. The thiol reactivity precludes this chemical scaffold from further development and optimization as a selective MDMX inhibitor106. Although a selective MDMX small-molecule inhibitor does not currently exist, it is conceivable that MDMX-selective inhibitory polypeptides could be computationally designed. The extended length of such molecules may overcome some of the limitations that are associated with the use of small peptides. The expression of MDMX-targeted polypeptides would be a valuable addition to the study of MDMX function in normal and cancer cells.
Mouse genetic models strongly suggest that MDM2 and MDMX are the two major p53 antagonists in vivo107,108 and that they exert non-overlapping inhibitory activities towards p53 (Refs 24,109). Therefore, dual antagonists for p53–MDM2 and p53–MDMX may effectively reactivate p53 in cancer. RO-5963 was identified as a dual inhibitor of p53–MDM2 and p53–MDMX103. Structural analyses suggest that the compound induces p53 activity via an unusual mechanism: it triggers the formation of MDM2–MDMX homodimers and heterodimers via interactions with their N-termini103. Intriguingly, although RO-5963 showed better in vitro binding affinity for MDM2 than nutlin 3a (IC50 values of 17.3 nM and 18.7 nM, respectively) it has poorer pharmacological properties. For example, at equimolar doses, nutlin 3a is a more potent p53 activator in cultured cancer cells103. Furthermore, in contrast to nutlin 3a, RO-5963 kills cells with both high MDM2 levels and high MDMX levels. This suggests that the disruption of p53–MDMX, rather than p53–MDM2, complexes may contribute to its cellular efficacy103. Further in vivo studies are required to understand whether this dual inhibitor will be useful in the clinic given its apparent weaker antagonism of p53–MDM2.
Peptide antagonists of p53–MDM2 and p53–MDMX interactions have been developed using structure-based rational design and phage display methods110–112. Although peptides and mini-proteins can antagonize PPIs more efficiently owing to their larger interaction surfaces, most of them are unstable in vivo and are poorly internalized, which compromises their ability to antagonize intracellular PPIs. To overcome these drawbacks, stapled peptides with improved cellular uptake and stability have been developed113. The stabilized α-helix of p53 variant 8 (SAH-p53-8) peptide has nanomolar binding affinity to the N-terminal p53-binding pocket of both MDM2 and MDMX in vitro. The IC50 of SAH-p53-8 for MDM2 is tenfold better than that of nutlin 3a, and the peptide also disrupts p53–MDMX interactions110. Additionally, in vitro assays indicate that SAH-p53-8 has a higher affinity for MDMX compared with MDM2 (Ref. 110). However, SAH-p53-8 is far less efficient at disrupting the p53–MDM2 interaction in cells and it requires nutlin 3a for optimal activity in cells that overexpress both MDM2 and MDMX. One interpretation is that SAH-p53-8 is a poorer antagonist of the MDM2–p53 interaction in cells with high MDM2 levels10,110, possibly because its effective intracellular concentration is much lower than that of nutlin 3a. Indeed, cellular uptake of stapled peptides such as SAH-p53-8 requires pinocytosis113, which is less effective than the uptake of small molecules through passive diffusion. Furthermore, SAH-p53-8 uptake is attenuated in the presence of serum (Y.-C.L. and G.M.W., unpublished observations). Inefficient uptake probably accounted for the need to use SAH-p53-8 at high concentrations (15–30 μM) to induce cytoxicity in a melanoma model10. Also, pharmacological properties of SAH-p53-8 might affect its affinity for MDM2 in vitro versus in the cellular microenvironment, as occurs with ABT-737, a peptidomimetic that antagonizes the interaction of BCL-XL with pro-apoptic BH3 family members114. Despite these issues, it is noteworthy that SAH-p53-8 is effective at high concentrations in a mouse model of melanoma in which MDMX is overexpressed10. In this model, SAH-p53-8 treatment increased cytotoxicity and sensitized melanomas to the chemotherapeutic agent cisplatin and the BRAF-V600E inhibitor vemurafenib (also known as PLX4032). This suggests a potential clinical utility of combined stapled peptides combining p53 agonists and BRAF inhibitors.
Targeting ubiquitin ligase activity
Small molecules that inhibit MDM2 ubiquitin ligase activity have been identified in high-throughput screens. These include HLI98 (Ref. 115), the derivative MPD compounds116, MEL23 and MEL24 (Ref. 117). Treating cells with these compounds stabilized p53 and MDM2 and subsequently led to p53 activation. MPD37 (the most potent among such compounds) was shown to bind the MDM2 RING domain, although whether this prevents MDM2–MDMX heterodimer formation is unclear116. These MDM2 E3 ubiquitin ligase inhibitors exhibit some p53-independent cytotoxicity, especially at higher concentrations, which may be due to the inhibition of other cellular RING domain E3 ubiquitin ligases.
An important gap in our knowledge of MDM2–p53 regulation is the identity of the E2 ubiquitin-conjugating enzyme or enzymes involved in the ubiquitylation of p53 in vivo. Although several E2s can transfer ubiquitin to p53 in vitro 118,119, the intracellular E2s that are required for p53 ubiquitylation have not been identified. This is probably due to the low affinity and the transient nature of E2–E3 interactions120,121. Conceivably, MDM2 might use different E2s to regulate itself, MDMX and/or p53. Alternatively, different developmental stages or different tissues might require discrete MDM2–E2 pairs to modulate p53 activities. Also, discrete MDM2–E2 pairs might be used in normal or malignant tissues. Thus, the identification of the bona fide intracellular E2s will undoubtedly bring us a more complete picture of p53–MDM2 regulation. Once specific E2s are identified, the relatively weak nature of MDM2–E2 interactions may render them particularly susceptible to selective PPI antagonists122. This in turn would prevent p53 ubiquitylation and trigger p53 activation.
Combination approaches
Conventional single-agent cancer therapy increases the likelihood of resistance. To minimize the emergence of resistant clones and to achieve maximal therapeutic response, combinations of different classes of therapeutic agents123 are commonly used. Indeed, the MDM2 antagonist nutlin 3a has been combined with non-targeted genotoxic agents to augment efficacy (reviewed in Ref. 124). We discuss below recent rational combinations of targeted agents and MDM2–MDMX antagonists.
The PI3K–AKT pathway usually has an anti-apoptotic role to promote cancer cell survival. This may be partly due to AKT-dependent stabilization of MDM2 (Refs 125,126). Thus, targeting both the AKT and p53 axes simultaneously might be particularly effective in cancer cells. Consistent with this, inhibition of the PI3K–AKT pathway sensitizes acute lymphoblastic leukaemia (ALL) cell lines to nutlin 3a-induced p53 activation127. More than 80% of acute myeloid leukaemias (AMLs) express constitutively activated MAPK, and 50% overexpress MDM2 (Ref. 128). Nutlin 3a synergizes with AZD6244 (also known as selumetinib), an inhibitor of downstream MAPK signalling, to induce apoptosis in AML128, providing proof-of-concept for this rational combination treatment.
Early combination regimens were mainly determined empirically. However, in the post-genomic era, several research institutes have approached combinatorial targeted therapy in a systematic manner129,130. In one such study130, as expected, p53 mutation status was the strongest indicator of nutlin 3a resistance; interestingly, BRAF mutation was associated with nutlin 3a sensitivity in the same study130. This suggests that targeting BRAF, as well as p53 pathways, should generate additive or synergistic effects. Indeed, co-treatment of melanomas with the p53-activating stapled peptide SAH-p53-8 and the BRAF inhibitor PLX4032 significantly enhanced cytotoxicity when compared with single-agent treatment10.
Insensitivity to MDM2 antagonists can arise from a variety of mechanisms, including p53 mutation, the presence of dominant survival pathways or the inhibition of death-inducing pathways. In an effort to identify genes that protect cells from p53-induced apoptosis, a genome-wide siRNA screen revealed that knocking down ATM and MET (also known as HGFR) kinases exhibited synthetic lethality with nutlin 3a131. At first this was surprising, because ATM kinase activates p53 in response to DNA damage. However, context is likely to be the key, and the protective role of ATM may be separate from its function as a DNA damage-activated kinase. Together, these results indicate that ATM and MET affect multiple downstream regulators in order to modulate the cellular response to MDM2 antagonists. Although a detailed molecular mechanism was not elucidated, it seems that the MET and MDM2 signalling pathways are used during development for cell survival132. This underscores the idea that signalling networks in normal cells are frequently usurped during tumorigenesis to promote cancer cell growth.
Concluding remarks
The intricacies of the MDM2–MDMX–p53 regulatory circuit have attracted the attention of academics, translational scientists and drug companies. Collaborations between these groups have inspired the implementation of numerous drug discovery projects and the design of clinical trials133. Ultimately, this will benefit cancer patients, although currently only one MDM2 antagonist, RG7112 (see the ClinicalTrials.gov website; see Further information), is in clinical trials and many challenges still lie ahead. For example, with the obvious exception of p53, we still do not understand how MDM2 and MDMX expression is regulated during development or in adult tissues during homeostasis. Insight here may reveal signalling pathways that can be targeted for therapeutic benefit. Are MDM2 and MDMX involved in the modulation of pathologies other than cancer? Given the role of p53 in the control of core metabolic pathways, this is entirely possible. Finally, how can we effectively select patients for clinical trials of MDM2- and MDMX- targeted therapies? A reliance simply on MDM gene amplification is inadequate. Combined screening for additional markers such as the loss of miRNA clusters that regulate MDM2 or MDMX, the presence of deubiquitylases that stabilize these oncogenes or druggable kinases that cooperate with MDM proteins should all be considered.
At a glance.
MDM2 and MDMX are RING domain proteins that exert their oncogenic effects primarily by inhibiting the p53 tumour suppressor protein.
Each protein is overexpressed in diverse tumour types by mechanisms including gene amplification and post-translational stabilization; this is generally more frequent in tumours with a wild-type TP53 allele.
Despite their similar structures, only MDM2 has intrinsic E3 ubiquitin ligase activity. Although MDM2 alone can inhibit p53, its RING-dependent heterodimerization with MDMX has an important role in p53 inhibition.
Both MDM2 and MDMX interact with multiple other partners. Aberrant interactions with these partners may also affect gene expression and genome stability.
Structure-based drug design has yielded several MDM antagonists that block MDM–p53 interactions, leading to p53 activation. At least one agent has progressed to clinical trials.
Systems biology studies are providing the rationale for using MDM protein antagonists in combination with both approved and experimental pathway-targeted anticancer drugs.
Acknowledgments
Studies relevant to the topics discussed here were supported by grants from the US National Institutes of Health (R01-CA61449 and R03-MH089489-01), Cancer Center Support Grant CA014195 and a sanofi-aventis sponsored research grant awarded to G.M.W.
Glossary
- Antagonists
Chemical substances that interfere with or inhibit the physiological activity of other biological entities such as proteins or enzymes
- miRNAs
Derived from an RNA polymerase II transcribed precursor, miRNAs are a class of non-protein coding mRNA that reduces the expression of cellular proteins through various mechanisms
- Hemizygous
A genetic status in which one allelic copy of a gene is deleted or otherwise inactivated
- Haploinsufficiency
A genetic status in which a single wild-type copy of an allelic pair is present, but the level of expression of the product is insufficient to give wild-type function
- Myeloablation
The depletion of bone marrow cells
- Neoadjuvant therapy
Administration of therapeutic agents to reduce tumour volume before giving a primary treatment such as surgery
- IC50
The half-maximal inhibitory concentration, which is the concentration of a compound causing 50% inhibition of biological or biochemical function
Footnotes
Competing interests statement: The authors declare no competing financial interests.
References
- 1.Bieging KT, Attardi LD. Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 2012;22:97–106. doi: 10.1016/j.tcb.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. The first demonstration that MDM2 could inhibit p53 activity. [DOI] [PubMed] [Google Scholar]
- 3.Shibagaki I, et al. p53 mutation, murine double minute 2 amplification, and human papillomavirus infection are frequently involved but not associated with each other in esophageal squamous cell carcinoma. Clin Cancer Res. 1995;1:769–773. [PubMed] [Google Scholar]
- 4.Forslund A, et al. MDM2 gene amplification is correlated to tumor progression but not to the presence of SNP309 or TP53 mutational status in primary colorectal cancers. Mol Cancer Res. 2008;6:205–211. doi: 10.1158/1541-7786.MCR-07-0239. [DOI] [PubMed] [Google Scholar]
- 5.Ito M, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam S, et al. Role of Mdm4 in drug sensitivity of breast cancer cells. Oncogene. 2010;29:2415–2426. doi: 10.1038/onc.2009.522. [DOI] [PubMed] [Google Scholar]
- 7.Mejia-Guerrero S, et al. Characterization of the 12q15 MDM2 and 12q13-14 CDK4 amplicons and clinical correlations in osteosarcoma. Genes Chromosomes Cancer. 2010;49:518–525. doi: 10.1002/gcc.20761. [DOI] [PubMed] [Google Scholar]
- 8.Ito M, et al. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res. 2011;17:416–426. doi: 10.1158/1078-0432.CCR-10-2050. [DOI] [PubMed] [Google Scholar]
- 9.Gilkes DM, et al. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol. 2008;28:1999–2010. doi: 10.1128/MCB.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gembarska A, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nature Med. 2012;18:1239–1247. doi: 10.1038/nm.2863. The first report showing that MDMX is associated with aggressive melanoma in vivo and can be targeted with a dual MDM2 and MDMX inhibitor peptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. The structure that described the hydrophobic p53-binding pocket of MDM2, on which many drug discovery efforts are now based. [DOI] [PubMed] [Google Scholar]
- 12.Laurie NA, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 13.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 14.Phillips A, et al. HDMX-L is expressed from a functional p53-responsive promoter in the first intron of the HDMX gene and participates in an autoregulatory feedback loop to control p53 activity. J Biol Chem. 2010;285:29111–29127. doi: 10.1074/jbc.M110.129726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. This paper revealed that MDM2 controls the stability of both itself and p53, and that the RING domain was required for these functions. [DOI] [PubMed] [Google Scholar]
- 16.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci USA. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pishas KI, et al. Nutlin-3a is a potential therapeutic for ewing sarcoma. Clin Cancer Res. 2011;17:494–504. doi: 10.1158/1078-0432.CCR-10-1587. [DOI] [PubMed] [Google Scholar]
- 23.McEvoy J, et al. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS ONE. 2012;7:e42739. doi: 10.1371/journal.pone.0042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francoz S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ries S, et al. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell. 2000;103:321–330. doi: 10.1016/s0092-8674(00)00123-9. [DOI] [PubMed] [Google Scholar]
- 26.Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haploinsufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 2003;22:1442–1450. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Clercq S, et al. Widespread overexpression of epitope-tagged Mdm4 does not accelerate tumor formation in vivo. Mol Cell Biol. 2010;30:5394–5405. doi: 10.1128/MCB.00330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou JX, et al. IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol. 2009;183:3188–3194. doi: 10.4049/jimmunol.0803693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarocchi M, et al. Carcinogen-induced hepatic tumors in KLF6+/− mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology. 2011;54:522–531. doi: 10.1002/hep.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, et al. Transcription factor NFAT1 activates the mdm2 oncogene independent of p53. J Biol Chem. 2012;287:30468–30476. doi: 10.1074/jbc.M112.373738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Pajares V, Kim MM, Yuan ZM. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem. 2008;283:13707–13713. doi: 10.1074/jbc.M710030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by casein kinase 1 α. Mol Cell Biol. 2005;25:6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 36.Zuckerman V, et al. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem. 2009;284:4031–4039. doi: 10.1074/jbc.M809211200. [DOI] [PubMed] [Google Scholar]
- 37.Meek DW, Hupp TR. The regulation of MDM2 by multisite phosphorylation-opportunities for molecular-based intervention to target tumours? Semin Cancer Biol. 2010;20:19–28. doi: 10.1016/j.semcancer.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Maya R, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411–3422. doi: 10.1038/sj.emboj.7600812. This paper revealed the correlation between phosphorylation and degradation of MDMX and activation of p53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereg Y, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol. 2006;26:6819–6831. doi: 10.1128/MCB.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28:3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson JG, Post SM, Lozano G. Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. J Pathol. 2011;223:127–136. doi: 10.1002/path.2783. [DOI] [PubMed] [Google Scholar]
- 43.Lu X, Nguyen TA, Zhang X, Donehower LA. The Wip1 phosphatase and Mdm2: cracking the “Wip” on p53 stability. Cell Cycle. 2008;7:164–168. doi: 10.4161/cc.7.2.5299. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, et al. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69:7960–7968. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter M, et al. Protein kinase CK1δ phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry. 2004;43:16356–16364. doi: 10.1021/bi0489255. [DOI] [PubMed] [Google Scholar]
- 46.Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage- activated kinases. Cancer Cell. 2009;16:33–43. doi: 10.1016/j.ccr.2009.05.008. The first report that MDMX phosphorylation is crucial for the response to ionizing radiation in vivo and that MDMX cooperates with another oncogene, MYC, to accelerate cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell. 2012;21:668–679. doi: 10.1016/j.ccr.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CP, Choi YJ, Hicks GG, He L. The emerging functions of the p53-miRNA network in stem cell biology. Cell Cycle. 2012;11:2063–2072. doi: 10.4161/cc.20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pichiorri F, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18:367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Markey M, Berberich SJ. Full-length hdmX transcripts decrease following genotoxic stress. Oncogene. 2008;27:6657–6666. doi: 10.1038/onc.2008.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandke P, et al. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS ONE. 2012;7:e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wynendaele J, et al. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9649. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 54.Concepcion CP, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. This article forces a re-evaluation of the contexts in which miR-34 contributes to the p53 response in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forte E, et al. The Epstein-Barr virus (EBV)-induced tumor suppressor microRNA MiR-34a is growthpromoting in EBV-infected B cells. J Virol. 2012;86:6889–6898. doi: 10.1128/JVI.07056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forte E, Luftig MA. MDM2-dependent inhibition of p53 is required for Epstein-Barr virus B-cell growth transformation and infected-cell survival. J Virol. 2009;83:2491–2499. doi: 10.1128/JVI.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee HR, et al. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J Virol. 2009;83:6739–6747. doi: 10.1128/JVI.02353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P, et al. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27:1590–1598. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- 59.Dharel N, et al. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867–4871. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 60.Sarek G, et al. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest. 2007;117:1019–1028. doi: 10.1172/JCI30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 62.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danovi D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lenos K, et al. Oncogenic functions of hMDMX in in vitro transformation of primary human fibroblasts and embryonic retinoblasts. Mol Cancer. 2011;10:111. doi: 10.1186/1476-4598-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lundgren K, et al. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- 66.Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci USA. 1998;95:15608–15612. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Post SM, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–230. doi: 10.1016/j.ccr.2010.07.010. In vivo evidence that an MDM2 SNP is associated with increased cancer incidence in women is causally associated with tumorigenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong S, et al. Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer Res. 2010;70:7148–7154. doi: 10.1158/0008-5472.CAN-10-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SS, et al. Insertion of Myc into Igh accelerates peritoneal plasmacytomas in mice. Cancer Res. 2005;65:7644–7652. doi: 10.1158/0008-5472.CAN-05-1222. [DOI] [PubMed] [Google Scholar]
- 70.Miller KR, Kelley K, Tuttle R, Berberich SJ. HdmX overexpression inhibits oncogene induced cellular senescence. Cell Cycle. 2010;9:3376–3382. doi: 10.4161/cc.9.16.12779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu B, et al. Evidence that c-myc mediated apoptosis does not require wild-type p53 during lymphomagenesis. Oncogene. 1995;11:175–179. [PubMed] [Google Scholar]
- 72.Catalogue of Somatic Mutations in Cancer. Wellcome Trust Sanger Institute. [online] http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- 73.Slack A, Lozano G, Shohet JM. MDM2 as MYCN transcriptional target: implications for neuroblastoma pathogenesis. Cancer Lett. 2005;228:21–27. doi: 10.1016/j.canlet.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 74.Mayo LD, Dixon JE, Durden DL, Tonks NK, Donner DB. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J Biol Chem. 2002;277:5484–5489. doi: 10.1074/jbc.M108302200. [DOI] [PubMed] [Google Scholar]
- 75.Cipriano R, Patton JT, Mayo LD, Jackson MW. Inactivation of p53 signaling by p73 or PTEN ablation results in a transformed phenotype that remains susceptible to Nutlin-3 mediated apoptosis. Cell Cycle. 2010;9:1373–1379. doi: 10.4161/cc.9.7.11193. [DOI] [PubMed] [Google Scholar]
- 76.Tabe Y, et al. MDM2 antagonist nutlin-3 displays antiproliferative and proapoptotic activity in mantle cell lymphoma. Clin Cancer Res. 2009;15:933–942. doi: 10.1158/1078-0432.CCR-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nature Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 78.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 79.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 80.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 82.Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6:309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang YV, et al. Fine-tuning p53 activity through C-terminal modification significantly contributes to HSC homeostasis and mouse radiosensitivity. Genes Dev. 2011;25:1426–1438. doi: 10.1101/gad.2024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tovar C, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia D, et al. Validation of MdmX as a therapeutic target for reactivating p53 in tumors. Genes Dev. 2011;25:1746–1757. doi: 10.1101/gad.16722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harb Perspect Biol. 2010;2:a001222. doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nature Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, Ma X, Ren S, Buolamwini JK, Yan C. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther. 2011;10:69–79. doi: 10.1158/1535-7163.MCT-10-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kapitzky L, et al. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol. 2010;6:451. doi: 10.1038/msb.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pereg Y, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 92.Vaseva AV, Yallowitz AR, Marchenko ND, Xu S, Moll UM. Blockade of Hsp90 by 17AAG antagonizes MDMX and synergizes with Nutlin to induce p53-mediated apoptosis in solid tumors. Cell Death Dis. 2011;2:e156. doi: 10.1038/cddis.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, et al. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nature Biotechnol. 2012;30:159–164. doi: 10.1038/nbt.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nature Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 95.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 96.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. This paper describes the first small-molecule MDM2 antagonist that inhibits the p53–MDM2 interaction and activates p53 in a non-genotoxic manner. [DOI] [PubMed] [Google Scholar]
- 97.Ding K, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006;49:3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 98.Ray-Coquard I, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13:1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 99.Patton JT, et al. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- 100.Wade M, Rodewald LW, Espinosa JM, Wahl GM. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–1982. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 102.Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 103.Graves B, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brennan RC, et al. Targeting the p53 pathway in retinoblastoma with subconjunctival Nutlin-3a. Cancer Res. 2011;71:4205–4213. doi: 10.1158/0008-5472.CAN-11-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reed D, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786–10796. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bista M, et al. On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53. PLoS ONE. 2012;7:e37518. doi: 10.1371/journal.pone.0037518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 108.Parant J, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature Genet. 2001;29:92–95. doi: 10.1038/ng714. References 107 and 108 demonstrate that deletions of Mdm2 and Mdmx cause embryonic lethality that can be rescued by Trp53 deletion, thus providing genetic evidence that MDM2 and MDM4 are crucial p53 repressors with non-overlapping functions. [DOI] [PubMed] [Google Scholar]
- 109.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 forp53 inhibition in central nervous system development. Proc Natl Acad Sci USA. 2006;103:3226–3231. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernal F, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 112.Pazgier M, et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci USA. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 114.Aranovich A, et al. Differences in the mechanisms of proapoptotic BH3 proteins binding to Bcl-XL and Bcl-2 quantified in live MCF-7 cells. Mol Cell. 2012;45:754–763. doi: 10.1016/j.molcel.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 115.Yang Y, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 116.Roxburgh P, et al. Small molecules that bind the Mdm2 RING stabilize and activate p53. Carcinogenesis. 2012;33:791–798. doi: 10.1093/carcin/bgs092. [DOI] [PubMed] [Google Scholar]
- 117.Herman AG, et al. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 2011;1:312–325. doi: 10.1158/2159-8290.CD-11-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shloush J, et al. Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2. J Biol Chem. 2011;286:4796–4808. doi: 10.1074/jbc.M110.157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saville MK, et al. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 120.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 122.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 123.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nature Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 124.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Feng J, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt- dependent phosphorylation. J Biol Chem. 2004;279:35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- 126.Jackson MW, et al. Hdm2 nuclear export, regulated by insulin-like growth factor-I/MAPK/p90Rsk signaling, mediates the transformation of human cells. J Biol Chem. 2006;281:16814–16820. doi: 10.1074/jbc.M511617200. [DOI] [PubMed] [Google Scholar]
- 127.Zhu N, Gu L, Li F, Zhou M. Inhibition of the Akt/survivin pathway synergizes the antileukemia effect of nutlin-3 in acute lymphoblastic leukemia cells. Mol Cancer Ther. 2008;7:1101–1109. doi: 10.1158/1535-7163.MCT-08-0179. [DOI] [PubMed] [Google Scholar]
- 128.Zhang W, et al. Blockade of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase and murine double minute synergistically induces Apoptosis in acute myeloid leukemia via BH3-only proteins Puma and Bim. Cancer Res. 2010;70:2424–2434. doi: 10.1158/0008-5472.CAN-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sullivan KD, et al. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nature Chem Biol. 2012;8:646–654. doi: 10.1038/nchembio.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moumen A, Patane S, Porras A, Dono R, Maina F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134:1443–1451. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- 133.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nature Rev Clin Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 134.Bouska A, Eischen CM. Murine double minute 2: p53-independent roads lead to genome instability or death. Trends Biochem Sci. 2009;34:279–286. doi: 10.1016/j.tibs.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 135.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 2011;30:468–479. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim K, et al. MDM2 regulates estrogen receptor a and estrogen responsiveness in breast cancer cells. J Mol Endocrinol. 2011;46:67–79. doi: 10.1677/JME-10-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin HK, Wang L, Hu Y, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21:4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Linn DE, et al. Differential regulation of androgen receptor by PIM-1 kinases via phosphorylation-dependent recruitment of distinct ubiquitin E3 ligases. J Biol Chem. 2012;287:22959–22968. doi: 10.1074/jbc.M111.338350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sanchez M, Picard N, Sauve K, Tremblay A. Coordinate regulation of estrogen receptor β degradation by Mdm2 and CREB-binding protein in response to growth signals. Oncogene. 2012 Feb 20; doi: 10.1038/onc.2012.19. [DOI] [PubMed] [Google Scholar]
- 141.Saji S, et al. MDM2 enhances the function of estrogen receptor a in human breast cancer cells. Biochem Biophys Res Commun. 2001;281:259–265. doi: 10.1006/bbrc.2001.4339. [DOI] [PubMed] [Google Scholar]
- 142.Tang YA, et al. MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin Cancer Res. 2012;18:4325–4333. doi: 10.1158/1078-0432.CCR-11-2617. [DOI] [PubMed] [Google Scholar]
- 143.Chen L, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. EMBO J. 2010;29:2538–2552. doi: 10.1038/emboj.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mungamuri SK, et al. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nature Struct Mol Biol. 2012;19:478–484. doi: 10.1038/nsmb.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Onder TT, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 147.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28:4862–4874. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kadakia M, Brown TL, McGorry MM, Berberich SJ. MdmX inhibits Smad transactivation. Oncogene. 2002;21:8776–8785. doi: 10.1038/sj.onc.1205993. [DOI] [PubMed] [Google Scholar]
- 149.Wunderlich M, Ghosh M, Weghorst K, Berberich SJ. MdmX represses E2F1 transactivation. Cell Cycle. 2004;3:472–478. [PubMed] [Google Scholar]
- 150.Matijasevic Z, Krzywicka-Racka A, Sluder G, Jones SN. MdmX regulates transformation and chromosomal stability in p53-deficient cells. Cell Cycle. 2008;7:2967–2973. doi: 10.4161/cc.7.19.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Busuttil V, et al. NF-κB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci USA. 2010;107:18061–18066. doi: 10.1073/pnas.1006163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2010;96:169–179. doi: 10.1007/s11060-009-9959-4. [DOI] [PubMed] [Google Scholar]
- 154.Minamino T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nature Med. 2009;15:1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 155.Tomita K, et al. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J Hepatol. 2012;57:837–843. doi: 10.1016/j.jhep.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 156.Hallenborg P, et al. Mdm2 controls CREB-dependent transactivation and initiation of adipocyte differentiation. Cell Death Differ. 2012;19:1381–1389. doi: 10.1038/cdd.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Crago AM, Singer S. Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol. 2011;23:373–378. doi: 10.1097/CCO.0b013e32834796e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Issaeva N, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nature Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 159.Nieves-Neira W, et al. DNA protein cross-links produced by NSC 652287, a novel thiophene derivative active against human renal cancer cells. Mol Pharmacol. 1999;56:478–484. doi: 10.1124/mol.56.3.478. [DOI] [PubMed] [Google Scholar]
- 160.Rivera MI, et al. Selective toxicity of the tricyclic thiophene NSC 652287 in renal carcinoma cell lines: differential accumulation and metabolism. Biochem Pharmacol. 1999;57:1283–1295. doi: 10.1016/s0006-2952(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 161.Di Conza G, et al. IGF-1R/MDM2 relationship confers enhanced sensitivity to RITA in Ewing sarcoma cells. Mol Cancer Ther. 2012;11:1247–1256. doi: 10.1158/1535-7163.MCT-11-0913. [DOI] [PubMed] [Google Scholar]
- 162.Kojima K, Burks JK, Arts J, Andreeff M. The novel tryptamine derivative JNJ-26854165 induces wild-type p53- and E2F1-mediated apoptosis in acute myeloid and lymphoid leukemias. Mol Cancer Ther. 2010;9:2545–2557. doi: 10.1158/1535-7163.MCT-10-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 164.Valentin-Vega YA, Okano H, Lozano G. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 2008;15:1772–1781. doi: 10.1038/cdd.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Valentin-Vega YA, Box N, Terzian T, Lozano G. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation. 2009;77:442–449. doi: 10.1016/j.diff.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boesten LS, et al. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ. 2006;13:2089–2098. doi: 10.1038/sj.cdd.4401973. [DOI] [PubMed] [Google Scholar]
- 167.Maetens M, et al. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- 168.Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Fernandez-Garcia B, Lozano G. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. Circulation. 2007;115:2925–2930. doi: 10.1161/CIRCULATIONAHA.107.689901. [DOI] [PubMed] [Google Scholar]
- 170.Macias E, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 172.Cheng X, et al. Activation of murine double minute 2 by Akt in mammary epithelium delays mammary involution and accelerates mammary tumorigenesis. Cancer Res. 2010;70:7684–7689. doi: 10.1158/0008-5472.CAN-09-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Popowicz GM, et al. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle. 2010;9:1104–1111. doi: 10.4161/cc.9.6.10956. [DOI] [PubMed] [Google Scholar]
- 174.Rew Y, et al. Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem. 2012;55:4936–4954. doi: 10.1021/jm300354j. [DOI] [PubMed] [Google Scholar]
- 175.Liu M, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc Natl Acad Sci USA. 2010;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]