Abstract
The mammary gland undergoes dynamic changes throughout life. In the mouse, these begin with initial morphogenesis of the gland in the mid-gestation embryo followed by hormonally regulated changes during puberty and later in adulthood. The adult mammary gland contains a hierarchy of cell types with varying potentials for self-maintenance and differentiation. These include cells able to produce complete, functional mammary glands in vivo and that contain daughter cells with the same remarkable regenerative potential, as well as cells with more limited clonogenic activity in vitro. Here we review how applying in vitro and in vivo methods for quantifying these cells in adult mammary tissue to fetal mammary cells has enabled the first cells fulfilling the functional criteria of transplantable, isolated mammary stem cells to be identified a few days before birth. Thereafter, the number of these cells increases rapidly. Populations containing these fetal stem cells display growth and gene expression programs that differ from their adult counterparts but share signatures characteristic of certain types of breast cancer. Such observations reinforce growing evidence of important differences between tissue-specific fetal and adult cells with stem cell properties and emphasize the merits of investigating their molecular basis.
Keywords: Stem cells, Progenitors, Mammary gland, Fetal, Development, Transcriptomes, Breast cancer
Introduction
The mouse mammary gland undergoes extensive cellular remodeling and changes in morphology throughout life starting from the first emergence of placodes in the embryonic surface ectoderm. Reciprocal interactions between the developing mammary epithelial cells and the underlying mesenchyme are thought to regulate the production of diverse cell types and their organization into a primordial gland via multiple signaling factors including fibroblast growth factors (FGF), Wnts and others [1]. After birth the gland grows isometrically until the onset of puberty when ovarian and pituitary hormones initiate dynamic changes that continue until a complete gland structure is attained in early adulthood [2]. Subsequent fluctuations in the size and composition of the gland are driven hormonally by the estrous cycle, pregnancy, and the onset and cessation of lactation [3, 4], as well as by additional, poorly understood aging-associated mechanisms [5].
Histological studies of the normal adult mammary gland have led to its description as a bi-layered structure consisting of an inner layer of “luminal” cells, and an outer layer of “basal” cells encased by a laminin- and collagen-rich basement membrane [3]. The luminal compartment consists of different subsets of estrogen receptor-positive and -negative progenitors and cells that secrete milk in response to hormonal stimuli [6, 7]. The functionally distinct basal layer contains myoepithelial cells with contractile properties, and most of the cells with demonstrable stem cell activity. These are termed mammary repopulating units (MRUs) based on their ability as single cells to regenerate complete bi-layered gland structures containing both myoepithelial and luminal elements [8, 9]. Basal cells are also thought to play important roles in maintaining tissue integrity and in controlling ductal growth and differentiation [10].
Luminal and basal cells of the adult mammary epithelium are distinguished from one another not only by their different locations in the gland, but also by their differential and variable expression of several proteins. In both mouse and humans cytokeratins (CKs) 8, 18 and 19, MUC1 and prominin-1 (CD133) are markers of luminal cells [11], although CK8/18 expression decreases during lactation [12]. Similarly, CK14, a prominent basal cell marker, is expressed in some luminal cells from birth until puberty [12, 13]. Other basal cell markers include CK5, CD10, THY-1 (CD90) and p63 [11].
Surface markers that typify adult basal and luminal cells appear less specific in the developing mammary gland. For example, many fetal mammary cells express basal (CK14) and luminal (CK8) CKs simultaneously [13-15]. This suggests that fetal cells exist in a transcriptionally distinct state. The extensive proliferative and regenerative potential of fetal mammary cells has been appreciated for decades based on studies involving the transplantation of intact fetal rudiments into adult mice [16]. More recently, the growth and differentiation properties of isolated fetal mammary cells and features of their molecular regulation have been reported [14]. Here, we review the methods that have allowed primitive cells in the adult mammary gland to be identified, and the insights gained from their application to fetal mammary cells.
Stem Cell Concepts
The concept of a molecularly defined “stem cell state” in adult tissues has been largely fuelled by studies of the blood-forming system where, more than 50 years ago, it was demonstrated that single cells could generate multiple mature myeloid blood cell types as well as daughter cells with the same latent multi-lineage differentiation capability [17, 18]. Subsequent experiments showed that single cells with an even broader range of potentialities could regenerate and sustain the entire blood-forming system through a multi-step, hierarchical process [19]. Embryonic stem cell differentiation involves an analogously ordered process of lineage restriction wherein cells successively lose the ability to activate particular programs [20]. An important aspect of the mechanism that determines such ordered changes appears to be mediated by chromatin modifying factors that place epigenetic marks at specific sites in the genome. These serve in some cases to prime, but not activate, specific genes for transcription and to make other differentiation-associated genes inaccessible [21]. As a result, an undifferentiated, but poised state can be created and transmitted through successive divisions, providing a possible explanation for how stem cell self-renewal divisions might be achieved [22].
Until recently, such sequential patterns of lineage restriction that stem cells undergo have been assumed to reflect the operation of essentially irreversible events that regulate co-ordinated changes in patterns of gene expression. However, while this concept may prove to be the physiologic norm, notable exceptions have been described. Examples include the de-differentiation achieved through nuclear cloning [23], the generation of induced pluripotent stem cells from relatively mature cells [24], and the direct reprogramming of cells across tissue types [25]. These findings raise the possibility that, even in vivo, cells might display more plastic behavior than implicit in hierarchical models of differentiation, particularly in the face of unusual in vivo stimuli or as a result of mutations.
Evidence of Mammary Cells with Multi-Lineage and Self-Renewal Potential
Following the initial demonstration of the gland-generating activity of cells in mouse mammary tissue fragments transplanted into “cleared” pre-pubertal mammary gland fat pads [26], it was shown that this result could also be obtained with transplants of tissue fragments taken from many parts of the adult gland, from various developmental stages, and in some cases could support 9 serial transplant generations [27-29]. Even intact mammary buds from day 13 embryos could produce glands when transplanted into the cleared fat pad of adults [16]. The first evidence that this activity could be attributed to a single cell came from experiments using mouse mammary tumor virus insertion sites as endogenous clonal markers [30]. In humans, the existence of analogous cells was inferred from the demonstration of the same X-chromosome inactivation pattern in adjacent ducts and lobules in normal adult tissue, implying their clonal derivation and maintenance [31].
Another technique that allows the origin of cells generated in vivo to be tracked without transplantation is lineage-tracing. This technique relies on the use of an inserted reporter gene to “mark” cells irreversibly at some specified time in development. This then allows their progeny to be subsequently traced without requiring that they be removed or physically manipulated. Typically, permanently activated expression of a fluorescent protein is elicited using a presumed cell-type specific promoter, or a specific signaling pathway response gene, to direct the expression of Cre-recombinase (CRE). Modifications of this technology allow the timing of the CRE expression to be further controlled, thus enabling the onset and duration of the marking to be more finely regulated; e.g., using a tetracycline/doxycyline regulated (r)tTA-tetO system [32]. However, the physiologically relevant lineage-tracing power afforded by this methodology is necessarily constrained by several caveats. One is the potentially promiscuous or unanticipated expression of the gene promoter used to activate CRE due to the fact that even a low level of promoter activity may activate sufficient CRE to enable expression of the marker protein. A second is the inability of lineage-tracing per se to identify clonal outputs of the marked cells.
Experiments that have used lineage-tracing to understand the embryonic origin of the different types of cells present in the adult mammary gland illustrate these points. For example, CK14 activation of CRE in the late-stage, embryonic day 17.5 (E17.5) mammary rudiment has confirmed that the targeted cells generate both basal and luminal cells present in the adult. However, initiating the same trace soon after birth marks cells that subsequently give rise primarily to basal cells [15]. These results imply that programs responsible for multi-potency are present in the embryonic mammary gland, but that after birth, they are either not used or are rapidly lost. However, our understanding of the mammary lineage specificity or consistency of CK14 expression during development may also be incomplete. The use of another gene, Axin2, to drive expression of CRE has produced an even more complex picture. Axin2-expressing mammary rudiment cells present at either an earlier time (E12.5), or at E17.5, were found to contribute primarily to the adult luminal compartment, whereas cells marked at a pre-pubescent stage (postnatal days 14 to 16) generated primarily basal cells [33]. These results exemplify the difficulties in interpreting data based exclusively on promoter-driven expression of a marker in cells whose numbers or biological and molecular properties are not yet clearly established.
Quantitative In Vivo Assays for Mammary Stem Cells in the Adult Mammary Gland
Alternative approaches that enable the robust identification, isolation and quantitation of mammary epithelial cells with stem cell properties rely on methods that detect the extensive growth and differentiation properties of individual starting cells either in vivo or in vitro. These methods have certain advantages but, like lineage-tracing, also rely on certain assumptions that impact data interpretation. Examples include the assumption that the generation of a single cell suspension does not alter the growth, survival or differentiation properties of the cells subsequently evaluated, or that the assay conditions are neither grossly suboptimal nor physiologically irrelevant.
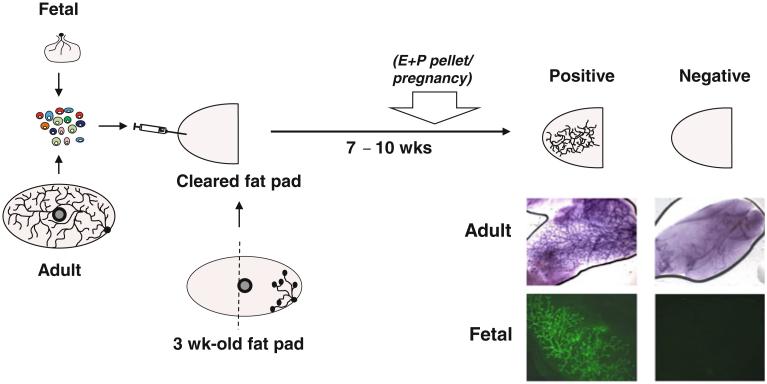
Advances in the development of reagents that allow mammary glands to be dissociated into viable single-cell suspensions was an important step in making it possible to perform cleared fat pad transplants with isolated mammary cells. This, in turn, enabled the application of Poisson statistics to quantify MRUs based on their ability to form a complete tree-like structure within 7–10 weeks in recipients of limiting numbers of cells (Fig. 1) [8, 9]. Importantly, these studies also demonstrated the ability of the original MRUs to generate daughter MRUs with the same individual regenerative potential detectable in secondary transplants. Thus, MRUs exhibit properties expected of mammary stem cells.
Fig. 1.
Schematic representation of the protocol for detecting fetal and adult MRUs. Cells from mammary glands are dissociated into a single-cell suspension and then transplanted into the cleared fat pad of a pubertal female mouse. Seven to ten weeks later, glands are removed and scored for the presence or absence of a large positive tree-structure. Photomicrographs show carmine-stained examples of positive and negative glands injected with adult cells (top) and examples of positive and negative glands injected with green fluorescent protein+ fetal cells (bottom). MRU detection can sometimes be increased by inducing pregnancy or implanting an estrogen and progesterone pellet (E + P pellet) 3 weeks prior to sacrifice
This methodology has now been widely used, although the MRU frequencies reported are varied (Table 1). This is likely due in part to mouse strain differences as well as the variable inclusion of non-epithelial cells in different test cell suspensions, which could be overcome by use of EpCAM as a marker of input epithelial cell content [7, 34], or the measurement of absolute numbers of MRUs per gland. Other factors, such as the inclusion of Matrigel in the transplant inoculum, the presence of macrophages in the cleared fat pad [35], or the extent to which the host is hormonally activated (Fig. 1) can also have large effects on MRU detection frequencies (Table 1). Coupling cell fractionation strategies with a subsequent MRU assay has shown that most adult mouse MRU activity resides in basal cells expressing high levels of CD49f and CD29 with reduced or absent expression of luminal cell markers (MUC1, EpCAM, CD24, CD14 and, in certain mouse strains, CD117/c-kit). These properties have allowed the isolation of adult MRUs at a purity of ~1–2 % [7-9, 36-39].
Table 1.
Examples of different MRU frequencies measured in suspensions of unfractionated fetal and adult mammary tissue and from different strains of mice and the effect of including Matrigel in the inoculum
| Stage | Mouse strain | Frequency of MRU (95 % CI) no Matrigel |
Frequency of MRU (95 % CI) +Matrigel |
Composition of inoculum solution |
References |
|---|---|---|---|---|---|
| Adult | FVB | 1/1,400 (1/600–1/3,000) | HF | [8] | |
| Adult | FVB | 1/106 (1/70–1/160) | PBS: Matrigel (1:1) | [85] | |
| Adult | BALB/c | 1/7,800 (1/1,900–1/31,000) | 5 μg/ml Matrigel | [37] | |
| Adult | BL/6J | 1/2,000 (1/900–1/4,500) | HF | [8] | |
| Adult | BL/6J × CD-1 | 1/30,000 (1/11,000–1/80,000) | 1/300 (1/150–1/500) | HF or HF: Matrigel (1:1) | [14] |
| Fetala | BL/6J × CD-1 | 1/800 (1/400–1/1,600) | 1/60 (1/40–1/80) | HF or HF: Matrigel (1:1) | [14] |
HF: 2 % FBS in Hanks’ Balanced salt solution
Similar frequencies obtained by Makarem et al. unpublished
A similar approach to detect and quantify human “MRUs” in highly immunodeficient recipients as hosts has also been devised. This involves either suspending the test cells in fibroblast-containing collagen gels that are then placed under the kidney capsule, or injecting the test cells with fibroblasts into pre-cleared mammary fat pads [40, 41]. Human MRU activity is then identified in the subrenal capsule assay by harvesting the cells 4 weeks later and determining whether mammary cells with colony-forming cell (CFC) activity in a secondary in vitro assay can be detected, based on the assumption that their presence would reflect their derivation from a more primitive mammary cell. In the cleared fat pad assay, human MRU activity is indicated by the appearance after 7 weeks of a branched gland-like structure. In either case, the original MRUs thus defined, like their mouse counterparts, share markers of basal cells (low levels of EpCAM and high levels of CD49f) and are found in this subset at a frequency of ~0.01–0.1 %.
Collectively, these observations indicate that MRU assays identify mammary cells with features of basal cells that display extensive regenerative potential in a transplanted host. However, their detection is also dependent on numerous, and still poorly characterized, microenvironmental/niche parameters as well as the genetics and epigenetics of the cells being assayed.
Quantitative In Vitro Assays for Primitive Mammary Cells in the Adult Mammary Gland
In vivo mammary repopulation assays are relatively costly, time consuming and may under- (or over-) estimate the frequency of cells that exist in a “stem cell state” in vivo due to technical difficulties or other biological requirements/limitations of the assays used to detect them. Such considerations have driven the development of in vitro 2D or 3D assays for quantifying and investigating many aspects of primitive mammary cell behavior, particularly for those of human origin. In vitro methods to detect the clonogenic potential of mammary epithelial cells were first described for rat and human cells [42-45] and later found to be also applicable to mouse cells. Originally, these methods relied on plating cells at low density under conditions that support the growth of mammary cells adhering to plastic (2D cultures) using media containing epidermal growth factor (EGF) and added irradiated fibroblasts to achieve maximal cloning efficiencies [8, 9, 46]. In the adult mouse, robust colony growth in these 2D assays is restricted to cells with a luminal phenotype unless the O2 concentration is reduced from 20 % to ~5 % and a Rho-kinase inhibitor is added [46, 47]. The addition of a Rho-kinase inhibitor also enhances the detection of human basal and luminal CFCs in 2D assays, but lower O2 conditions can reduce colony yields (N. Kannan and C. Eaves, unpublished observations).
In the adult mouse, the luminal CFCs detected in these 2D assays make up ~5–10 % of the epithelial (EpCAM+) cells and are CD24/EpCAM++CD49f+. With the recent addition of antibodies to CD61, CD49b, Sca-1, and c-kit, luminal CFC enrichments to purities of 40–50 % are achieved [6, 7, 38]. However, the progeny they generate in the CFC assay express CKs associated with both luminal and basal cells in vivo [6]. Thus, 2D CFC assay cultures of mouse cells appear to elicit an abnormal gene expression profile; thus serving to emphasize the principle that expression of the genes considered as “lineage markers” in the mammary gland may not be as rigidly controlled as predicted from in vivo analyses. The basal CFCs account for an additional 5–10 % of the total adult mouse mammary epithelial compartment with CFC activity. It is not yet known how strictly the phenotype of the basal CFC progeny resembles the cells they generate in vivo. Thus, overall, cells detectable as CFCs in 2D assays are at least 100-fold more numerous than cells detected as MRUs (Fig. 2) [47], with the possibility of some overlap, particularly in the basal cell population since this is the subset that contains most of the MRUs.
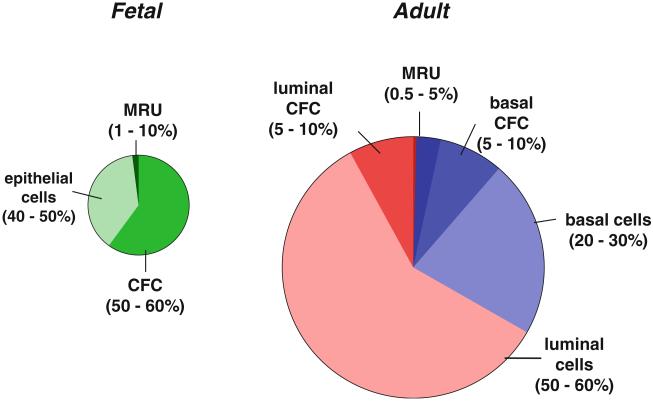
Fig. 2.
Schematic representation of the relative distribution of epithelial (EpCAM+) cell types within the smaller E18.5 fetal and virgin female adult mammary gland (i.e., excluding stromal cells, endothelial cells and blood cells). The smaller fetal gland is artificially enlarged here by ~5-fold. In both the fetal and adult mammary gland, the MRU content is smaller than the CFC content, shown here as separate entities, recognizing that they may overlap to some extent. However, the proportion of all epithelial cells that are detectable as CFCs (in 2D assays) or MRUs (in transplantation assays) is higher in the fetal gland than in the adult gland
Isolated mouse and human mammary cells can also be stimulated to proliferate under non-adherent conditions in liquid cultures. However, because of the very strong tendency of mammary epithelial cells to adhere to one another, the resultant clusters obtained (usually referred to as mammospheres [48]) represent a variable mixture of aggregation and proliferation. Thus, unless such cultures are initiated with a single isolated cell or the cells are immobilized in suspension using a permissive semi-solid medium such as Matrigel [8, 49], the number of structures obtained cannot be used to quantify clonogenic cells.
Numerical and Phenotypic Characterization of Primitive Cells in the Fetal Mammary Gland
Diverse experimental strategies indicate the presence of cells with innate stem cell properties in the fetal mammary gland. The precise timing and molecular regulation of their emergence, however, remain poorly understood, as do the mechanisms and extent of the phenotypic changes of these cells later during post-natal mammary gland development. In contrast to the ability of intact embryonic mammary rudiments obtained as early as E12.5 to produce gland-like structures when transplanted into the cleared fat pad [14, 16, 50], dissociated cells with this activity (MRUs) are barely measurable at E15.5, after which their numbers increase dramatically to E18.5 (Fig. 3a) [14, 47]. These findings suggest that critical properties required for the growth of primitive fetal mammary cells following their dissociation and transplantation into a cleared fat pad are not acquired until a few days before birth. Expression of this activity by fetal mammary cells is also promoted by the addition of Matrigel to the transplant inoculum allowing MRU frequencies of 1 % of unseparated cells and up to ~10 % of purified populations to be achieved (Table 1) [14]. 2D and 3D in vitro assays also allow the detection of CFCs in suspensions of the E18.5 fetal mammary rudiment. Interestingly, these analyses show that the ratio of CFCs to MRUs is somewhat higher in the fetal rudiment than in the adult gland (Fig. 2).
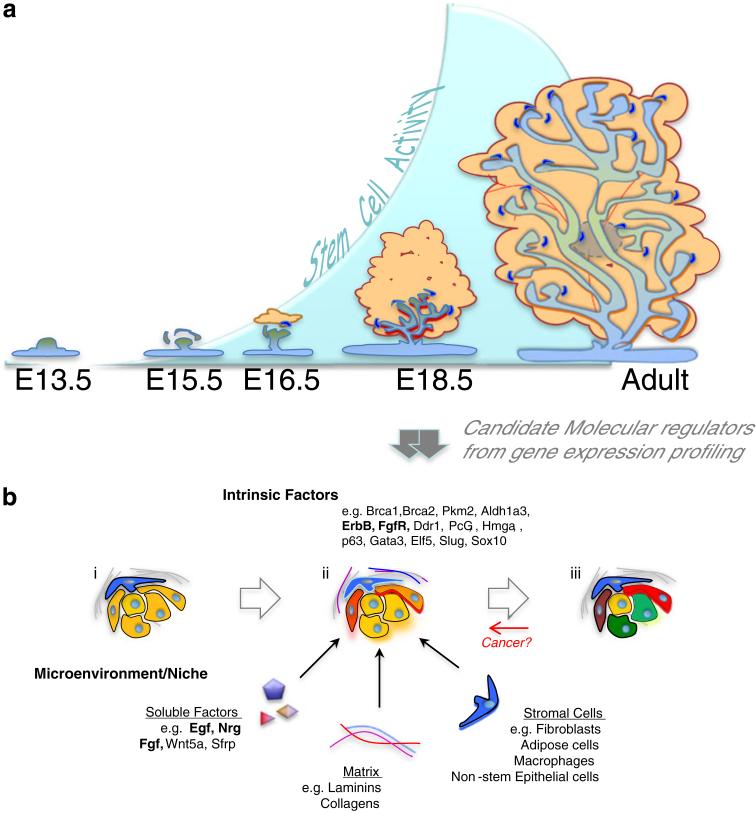
Fig. 3.
The mammary stem cell state is dynamic and context dependent. a Recent work has revealed that detectable MRU activity increases dramatically as the context of the fetal mammary epithelium changes in late gestation. b Candidate molecular regulators of the mammary stem cell state have been identified from molecular profiling of the late fetal mammary epithelium and associated stroma, but very few of these (bold) have been functionally and quantitatively tested for a role in mammary stem cell regulation. An emerging view is that when cells with intrinsic mammary stem cell potential (yellow cells) encounter appropriate microenvironmental stimuli (glowing cells) in the late stage embryo, they acquire properties that confer on them a stem cell state that can be expressed in isolation (ii). This does not occur at an earlier developmental stage either because critical response components or extrinsic stimulators are lacking (i), nor later under homeostatic conditions when more progeny become committed to specific lineages (green and red) (iii). The acquisition of an active stem state may also involve stimulation of non-stem cells in the niche (blue cells). Importantly, such molecular regulators may provide therapeutic targets if cancers achieve their robust stem like proliferative potential by resurrecting aspects of the fetal context
Fetal MRUs and CFCs detected in vitro express the same CD49f/CD29 basal markers as adult MRUs and basal CFCs, but differ in their higher expression of CD24 [14] and EpCAM [47]. Thus, new surface markers will be needed to separate basal CFCs from MRUs in the fetal as well as the adult mammary gland. Also colonies produced in vitro from fetal populations enriched in MRU activity frequently co-express luminal and basal markers and only occasionally express markers of a single lineage [14]. In addition, the fetal mammary gland lacks a distinct CD24++/EpCAM++CD49f+/CD29+ population that, in the adult, contains cells with luminal progenitor activity. Thus, the same multi-step differentiation process apparent in the adult mammary gland does not fit the cell types detected in the developing mammary rudiment.
Characterization of the Transcriptome of Primitive Cells in the Fetal Mammary Gland
The development of protocols to obtain mammary subpopulations that are highly enriched in MRUs and/or CFCs has provided an important opportunity to identify genes whose expression may be relevant to the functional state and developmentally-determined unique biological properties of these cells. Similar analysis of the separately purified stromal elements has enabled candidate “niche” regulators to be identified and tested functionally (Fig. 3b) [14].
Unsupervised analysis of array data generated from the MRU/CFC-enriched fraction of E18.5 fetal mammary cells has detected the expression of genes associated with diverse aspects of normal cell biology and cancer [14]. For instance, Brca1 and Brca2, which are implicated in the etiology of breast cancers [51] and have multiple roles, including DNA repair [52], are both expressed in the fetal MRU/CFC-enriched fraction. Transcripts encoding important metabolic regulators such as Pkm2 and Aldh1a3 are also present. Pkm2 contributes to balancing cellular glucose usage under hypoxic conditions [53] and Aldh1a3 is most highly expressed in the luminal progenitor subset of the adult mammary gland of both mice and humans [7, 54].
Candidate autocrine/paracrine mammary stem cell regulators include ligands that activate ErbB or FGF receptors. Expression profiling showed all 4 FGF receptors (most prominently FGFR2 and FGFR3) and all 4 ErbB family receptors to be expressed in the fetal mammary rudiment with specifically elevated expression of ErbB2-4 in the stem cell-enriched population [14], The finding that recombinant basic FGF and the ErbB ligands, EGF and Neuregulin 1, stimulate proliferation of E18.5 fetal mammary cells, while ErbB inhibition blocks their growth in vitro [14], extends previous studies implicating both FGF and ErbB receptor families in regulating the development of the fetal mammary rudiment [55-58]. Array analysis of the gene expression profile of the stromal cells that are closely associated with the developing E18.5 fetal rudiment also showed that they contain Fgf7 and Fgf10 transcripts. The fetal mammary rudiment thus shows features of the developing hair follicle, where FGF7/10 released from the mesenchymal dermal papilla has been found to act on stem cells in the bulge [59]. We also found Fgf7 and Fgf10 to be more highly expressed in the E18.5 fetal mammary stroma than in the stroma co-isolated with adult mammary tissue or earlier stages of the developing fetal mammary gland, raising questions as to their possibly distinct versus overlapping roles in stimulating primitive mammary epithelial cells at E18.5.
Surprisingly, our transcription analyses did not reveal evidence of Wnt signaling as driving fetal mammary stem cell activity, contrary to what might be anticipated from a recent report that Wnt signaling can promote the extensive proliferation of adult mammary MRUs in vitro [49]. In fact, the most prominent Wnt-related genes expressed in the fetal mammary gland at E18.5, when MRU frequencies reach a peak, are the negative regulators Sfrp1 and Sfrp5 in the MRU-enriched subset, and the non-canonical Wnt5a in the accompanying stromal cells. Additional studies have failed to detect significant effects of canonical Wnt ligands or activators on the growth of primitive fetal mammary cells in vitro [47] (B. Spike and G. Wahl, unpublished observations).
Discoidin domain receptor tyrosine kinase 1 (Ddr1) is another gene whose expression is up-regulated in the fetal mammary MRU-enriched fraction [14, 60], in agreement with previous experiments [61]. These showed Ddr1 to be a critical regulator of mammary development—its absence causing delayed and abnormal ductal branches with abnormal differentiation. Ddr1 is a unique tyrosine kinase receptor that recognizes collagen as a ligand, and expression of collagens, (Col1a1 and Col9a1) and laminins (α1, α2, and α4) are also all elevated in E18.5 fetal mammary tissue relative to the stem cell-poor E15.5 rudiment or the adult mammary gland (where expression of laminin α3 is greater) [14]. Taken together, these findings suggest that interactions of primitive fetal mammary cells with the basement membrane or other sites of extracellular matrix protein deposition may be of particular importance to their development and/or expansion.
Functional Properties of Primitive Mammary Cells During Development
Our more recent observations reveal that fetal mammary cells may possess greater self-renewal capacity and more robust production of progenitors than adult cells. This conclusion is based on in vitro experiments indicating a ~3-fold greater expansion of MRUs by fetal as compared to adult mammary epithelial cells over a 7-day period [47]. These observations are reminiscent of the changing self-renewal activity displayed by stem cells in other tissues [62-65], and related evidence for their regulation by common pathways; e.g., involving Bmi-1 and Hmga2 [64, 66–68]. Interestingly, expression of Hmga2 is higher in fetal mammary cells relative to the adult gland, with maximal levels at E15.5. However, the decreasing expression of Hmga2 that occurs between E15.5 and E18.5 is accompanied by an increased expression of Hmga1 [14], another epigenetic regulator with reprogramming activity and able to promote embryonic stem cell self-renewal [69]. Expression of Bmi-1 declines slightly during the latter stage of embryogenesis, but expression of Ezh2 and Eed, additional polycomb complex genes important for the maintenance of fetal and adult mammary cells, remains elevated in a few cells [14] (C. Dravis, B. Spike and G. Wahl, unpublished observations). Both the Let7-HMGA2 pathway [70] and Ezh2 [71] have also been associated with the poorly differentiated basal-like breast cancer subtype. Thus, it is inviting to speculate that these regulators may contribute to the augmented growth and potency of primitive fetal as compared to adult mammary cells. Hence when activated in the adult, they may contribute to the perturbed growth characteristic of mammary gland neoplasia.
P63 is an important transcription factor implicated in mammary development as p63 null mice lack mammary glands [72] and a role of p63 in maintaining many types of stem cells, is well established [73, 74]. However, p63 expression is reduced in the fetal relative to the adult mammary epithelium, in both of which it marks a large fraction of the basal compartment, including the MRU-enriched subset [14, 75]. In contrast, the luminal transcription factor, Gata-3, which plays a major role in controlling the size of the adult luminal compartment [76, 77] and is highly expressed by the fetal stem cell enriched population at E18.5 [14], is required for the formation of the embryonic mammary primordia [76] and has also been implicated in adult-derived MRU regulation [76].
Elf5 and Slug (Snail2) are 2 functionally antagonistic transcription factors also implicated in controlling adult mammary stem cell properties; with Elf5 being highly specific to glandular epithelia, and Slug a transcriptional repressor of the epithelial state. Loss of Elf5 in the mammary gland results in blocked alveologenesis and an expansion of cells with stem cell properties, which may be a consequence of its failure to repress Slug, a reported regulator of the stem cell state in the adult gland [78, 79].
Variable Control and Expression of the Mammary Stem Cell State
Given the extent of control that allows organisms to develop in a reproducible fashion, it may be useful to conceptualize a distinction between mechanisms that establish and mechanisms that stabilize stem cell states. Accumulating evidence indicates that the acquisition of programs that endow cells with capacities for self-renewal and/or altered differentiation potentialities may be activated under specific conditions but usually involve an external intervention that causes a major alteration of transcriptome control [24, 25]. In addition, there are increasing examples where perturbations of homeostatic physiology can unmask additional cellular potentialities that are not observed in the unperturbed state. In mice, such examples include the ability of bulge stem cells to regenerate the epidermis when the skin is wounded [80, 81], or the stem cell activity elicited from intestinal progenitors of the secretory lineage in sublethally irradiated mice [82], or upon specific ablation of the usually active Lgr5+ population at the base of the crypt [83].
Recent studies have provided evidence of a similarly increased spectrum of activity of mammary cells under various conditions of perturbation. Some adult mouse mammary progenitors assumed to be luminally-restricted, can be stimulated to differentiate into multiple lineages after being “passaged” in vivo under the kidney capsule in collagen or in collagen/Matrigel gels [7]. It has also been found that adult basal and luminal cells exhibit increased potential to repopulate a mammary gland following constitutive Met activation [84], or after transient co-expression of Slug and Sox9 [79], or expansion in vitro in the presence of Matrigel and fibroblast feeders [47]. Interestingly, the frequency of fetal and adult cells that can produce MRUs in this latter type of Matrigel culture is >20-fold and >100-fold greater, respectively, than the frequency of MRUs detectable in the cells used to initiate the cultures. This suggests that Matrigel and fibroblast feeders may rapidly convert cells with latent MRU potential to an active state. Continuing studies of the specific cell types in the mammary gland throughout its development, including comparison of their molecular profiles, single-cell analyses and functional tests of stem cell properties, should facilitate the identification of relevant signaling networks that underlie the genesis of the stem cell state during embryogenesis.
Conclusions and Future Directions
It is becoming clear that the mammary stem cell state is dynamic and subject to both cell intrinsic expression programs and to a myriad of cell non-autonomous signals unique to different stages of mammary gland development. The application of assays developed for adult mammary stem cells (MRUs) to single-cell suspensions prepared from fetal mammary tissue has elucidated the time at which cells with overt MRU activity first become detectable. The isolation and initial molecular characterization of these cells is beginning to illuminate factors that contribute to their regulation and regeneration. Thus, the stage is set for exciting investigations that should elucidate the genes and pathways involved in establishing and modulating the expression of stem cell properties by normal mammary cells. These will benefit from a combination of lineage-tracing, transplantation in vivo, and assessment of responses in vitro, following manipulations of candidate genes and their products now identifiable by high through-put screening methods.
A key challenge for the future will be the deployment of molecular methods to investigate the biological heterogeneity of mammary stem cells and provide a more concrete under-standing of the mammary stem cell state. Such heterogeneity is already evident at the single-cell level and appears further compounded by developmentally regulated changes that affect their properties. Significant differences between mechanisms regulating primitive human and mouse mammary cells will also warrant continuing attention. However, we anticipate that the mouse will remain a critical model where unique insights will be gained from further molecular analyses of the first mammary stem cells to appear in the embryo. Further elucidation of the role of interactions of fetal mammary cells and the surrounding stroma is also likely to impact our understanding of normal and transformed adult mammary stem cells and provide a precedent for analyzing other types of epithelial stem cells.
Acknowledgments
CE received support from the Canadian Breast Cancer Research Alliance with funds from the Canadian Cancer Society and from the British Columbia and Yukon Division of the Canadian Breast Cancer Foundation (BC-Y CBCF). MM held a Canadian Institutes of Health Research Studentship and NK a BC-Y CBCF Fellowship. CD was supported by T32 post-doctoral training grant 2T32CA009370; BTS was partially supported by T32 grant CA009523 and GW received support from the Breast Cancer Research Foundation, Susan G. Komen for the Cure, the Department of Defense BCRP, and many studies were enabled by Cores supported by Cancer Center Support Grant NCI 5P30CA014195. We also wish to acknowledge the many other important contributors to this field whose papers we could not reference due to space constraints.
Abbreviations
- CD
Cluster designation
- CFC
Colony-forming cell
- CK
Cytokeratin
- CRE
Cre-recombinase
- D
Dimensional
- E
Embryonic day
- EGF
Epidermal growth factor
- EpCAM
Epithelial cell adhesion molecule
- FGF
Fibroblast growth factor
- MRU
Mammary repopulating unit
- MUC1
Mucin 1
Contributor Information
Maisam Makarem, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Avenue, Vancouver, BC V5Z 1L3, Canada.
Benjamin T. Spike, Gene Expression Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, USA
Christopher Dravis, Gene Expression Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, USA.
Nagarajan Kannan, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Avenue, Vancouver, BC V5Z 1L3, Canada.
Geoffrey M. Wahl, Gene Expression Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, USA
References
- 1.Howard BA. In the beginning: the establishment of the mammary lineage during embryogenesis. Semin Cell Dev Biol. 2012;23(5):574–82. doi: 10.1016/j.semcdb.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Brisken C, O’Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010;2(12):a003178. doi: 10.1101/cshperspect.a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. 2000;5(2):227–41. doi: 10.1023/a:1026499523505. [DOI] [PubMed] [Google Scholar]
- 4.Fata JE, Chaudhary V, Khokha R. Cellular turnover in the mammary gland is correlated with systemic levels of progesterone and not 17beta-estradiol during the estrous cycle. Biol Reprod. 2001;65(3):680–8. doi: 10.1095/biolreprod65.3.680. [DOI] [PubMed] [Google Scholar]
- 5.Kannan N, Huda N, Tu L, Droumeva R, Aubert G, Chavez E, et al. The luminal progenitor compartment of the human mammary gland constitutes a unique site of telomere dysfunction. Stem Cell Reports. 2013 doi: 10.1016/j.stemcr.2013.04.003. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176(1):19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shehata M, Teschendorff A, Sharp G, Novcic N, Russell A, Avril S, et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14(5):R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 9.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 10.Deugnier MA, Teuliere J, Faraldo MM, Thiery JP, Glukhova MA. The importance of being a myoepithelial cell. Breast Cancer Res. 2002;4(6):224–30. doi: 10.1186/bcr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23(22):2563–77. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikaelian I, Hovick M, Silva KA, Burzenski LM, Shultz LD, Ackert-Bicknell CL, et al. Expression of terminal differentiation proteins defines stages of mouse mammary gland development. Vet Pathol. 2006;43(1):36–49. doi: 10.1354/vp.43-1-36. [DOI] [PubMed] [Google Scholar]
- 13.Sun P, Yuan Y, Li A, Li B, Dai X. Cytokeratin expression during mouse embryonic and early postnatal mammary gland development. Histochem Cell Biol. 2010;133(2):213–21. doi: 10.1007/s00418-009-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10(2):183–97. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–93. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 16.Sakakura T, Nishizuka Y, Dawe CJ. Capacity of mammary fat pads of adult C3H/HeMs mice to interact morphogenetically with fetal mammary epithelium. J Natl Cancer Inst. 1979;63(3):733–6. doi: 10.1093/jnci/63.3.733. [DOI] [PubMed] [Google Scholar]
- 17.Till JE, Mc CE. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–22. [PubMed] [Google Scholar]
- 18.Siminovitch L, McCulloch EA, Till JE. The distribution of colony-forming cells among spleen colonies. J Cell Physiol. 1963;62:327–36. doi: 10.1002/jcp.1030620313. [DOI] [PubMed] [Google Scholar]
- 19.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120–36. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Ang YS, Gaspar-Maia A, Lemischka IR, Bernstein E. Stem cells and reprogramming: breaking the epigenetic barrier? Trends Pharmacol Sci. 2011;32(7):394–401. doi: 10.1016/j.tips.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 23.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9(6):504–16. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Deome KB, Faulkin LJ, Jr, Bern HA, Blair PB. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19(5):515–20. [PubMed] [Google Scholar]
- 27.Hoshino K, Gardner WU. Transplantability and life span of mammary gland during serial transplantation in mice. Nature. 1967;213(5072):193–4. doi: 10.1038/213193a0. [DOI] [PubMed] [Google Scholar]
- 28.Young LJ, Medina D, DeOme KB, Daniel CW. The influence of host and tissue age on life span and growth rate of serially transplanted mouse mammary gland. Exp Gerontol. 1971;6(1):49–56. doi: 10.1016/0531-5565(71)90048-9. [DOI] [PubMed] [Google Scholar]
- 29.Daniel CW, De Ome KB, Young JT, Blair PB, Faulkin LJ., Jr The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc Natl Acad Sci U S A. 1968;61(1):53–60. doi: 10.1073/pnas.61.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125(10):1921–30. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 31.Tsai YC, Lu Y, Nichols PW, Zlotnikov G, Jones PA, Smith HS. Contiguous patches of normal human mammary epithelium derived from a single stem cell: implications for breast carcinogenesis. Cancer Res. 1996;56(2):402–4. [PubMed] [Google Scholar]
- 32.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89(12):5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11(3):387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 34.Smalley MJ, Kendrick H, Sheridan JM, Regan JL, Prater MD, Lindeman GJ, et al. Isolation of mouse mammary epithelial sub-populations: a comparison of leading methods. J Mammary Gland Biol Neoplasia. 2012 doi: 10.1007/s10911-012-9257-1. [DOI] [PubMed] [Google Scholar]
- 35.Gyorki DE, Asselin-Labat ML, van Rooijen N, Lindeman GJ, Visvader JE. Resident macrophages influence stem cell activity in the mammary gland. Breast Cancer Res. 2009;11(4):R62. doi: 10.1186/bcr2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaillant F, Lindeman GJ, Visvader JE. Jekyll or Hyde: does Matrigel provide a more or less physiological environment in mammary repopulating assays? Breast Cancer Res. 2011;13(3):108. doi: 10.1186/bcr2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, et al. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One. 2009;4(8):e6594. doi: 10.1371/journal.pone.0006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regan JL, Kendrick H, Magnay FA, Vafaizadeh V, Groner B, Smalley MJ. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene. 2012;31(7):869–83. doi: 10.1038/onc.2011.289. [DOI] [PubMed] [Google Scholar]
- 39.Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465(7299):803–7. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 40.Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14(12):1384–9. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 41.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–13. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 42.Emerman JT, Stingl J, Petersen A, Shpall EJ, Eaves CJ. Selective growth of freshly isolated human breast epithelial cells cultured at low concentrations in the presence or absence of bone marrow cells. Breast Cancer Res Treat. 1996;41(2):147–59. doi: 10.1007/BF01807160. [DOI] [PubMed] [Google Scholar]
- 43.Dundas SR, Ormerod MG, Gusterson BA, O’Hare MJ. Characterization of luminal and basal cells flow-sorted from the adult rat mammary parenchyma. J Cell Sci. 1991;100:459–71. doi: 10.1242/jcs.100.3.459. Pt 3. [DOI] [PubMed] [Google Scholar]
- 44.Stingl J, Eaves CJ, Zandieh I, Emerman JT. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat. 2001;67(2):93–109. doi: 10.1023/a:1010615124301. [DOI] [PubMed] [Google Scholar]
- 45.Stingl J, Eaves CJ, Emerman JT. Characterization of normal human breast epithelial cell subpopulations isolated by flourescence-activated cell sorting and their clonogenic growth in vitro. In: Ip MM, Asch BB, editors. Methods in mammary gland biology & breast cancer research. Kluwer Academic/Plenum Publishers; New York: pp. 177–193. [Google Scholar]
- 46.Smalley MJ, Titley J, O’Hare MJ. Clonal characterization of mouse mammary luminal epithelial and myoepithelial cells separated by fluorescence-activated cell sorting. In vitro cellular & developmental biology. Animal. 1998;34(9):711–21. doi: 10.1007/s11626-998-0067-0. [DOI] [PubMed] [Google Scholar]
- 47.Makarem M, Kannan N, Nguyen LV, Knapp DJHF, Balani S, Prater MD, et al. Developmental changes in the in vitro activated regenerative activity of primitive mammary epithelial cells. 2013 doi: 10.1371/journal.pbio.1001630. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6(6):568–77. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakakura T, Nishizuka Y, Dawe CJ. Mesenchyme-dependent morphogenesis and epithelium-specific cytodifferentiation in mouse mammary gland. Science. 1976;194(4272):1439–41. doi: 10.1126/science.827022. [DOI] [PubMed] [Google Scholar]
- 51.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4(9):665–76. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 52.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12(1):68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2(7):551–6. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eirew P, Kannan N, Knapp DJ, Vaillant F, Emerman JT, Lindeman GJ, et al. Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem Cells. 2012;30(2):344–8. doi: 10.1002/stem.1001. [DOI] [PubMed] [Google Scholar]
- 55.Wansbury O, Panchal H, James M, Parry S, Ashworth A, Howard B. Dynamic expression of Erbb pathway members during early mammary gland morphogenesis. J Investig Dermatol. 2008;128(4):1009–21. doi: 10.1038/sj.jid.5701118. [DOI] [PubMed] [Google Scholar]
- 56.Howard B, Panchal H, McCarthy A, Ashworth A. Identification of the scaramanga gene implicates Neuregulin3 in mammary gland specification. Genes Dev. 2005;19(17):2078–90. doi: 10.1101/gad.338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Panchal H, Wansbury O, Parry S, Ashworth A, Howard B. Neuregulin3 alters cell fate in the epidermis and mammary gland. BMC Dev Biol. 2007;7:105. doi: 10.1186/1471-213X-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, et al. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129(1):53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- 59.Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4(2):155–69. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wansbury O, Mackay A, Kogata N, Mitsopoulos C, Kendrick H, Davidson K, et al. Transcriptome analysis of embryonic mammary cells reveals insights into mammary lineage establishment. Breast Cancer Res. 2011;(4):13, R79. doi: 10.1186/bcr2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel WF, Aszodi A, Alves F, Pawson T. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol Cell Biol. 2001;21(8):2906–17. doi: 10.1128/MCB.21.8.2906-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kruger GM, Mosher JT, Bixby S, Joseph N, Iwashita T, Morrison SJ. Neural crest stem cells persist in the adult gut but undergo changes in self-renewal, neuronal subtype potential, and factor responsiveness. Neuron. 2002;35(4):657–69. doi: 10.1016/s0896-6273(02)00827-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130(3):470–83. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 2008;135(2):227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118(25):6553–61. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 66.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19(12):1438–43. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425(6961):962–7. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, et al. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18(14):1094–9. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- 69.Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J, et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One. 2012;7(11):e48533. doi: 10.1371/journal.pone.0048533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pietersen AM, Horlings HM, Hauptmann M, Langerod A, Ajouaou A, Cornelissen-Steijger P, et al. EZH2 and BMI1 inversely correlate with prognosis and TP53 mutation in breast cancer. Breast Cancer Res. 2008;(6):10, R109. doi: 10.1186/bcr2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 73.Blanpain C, Fuchs E. p63: revving up epithelial stem-cell potential. Nat Cell Biol. 2007;9(7):731–3. doi: 10.1038/ncb0707-731. [DOI] [PubMed] [Google Scholar]
- 74.Nekulova M, Holcakova J, Coates P, Vojtesek B. The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett. 2011;16(2):296–327. doi: 10.2478/s11658-011-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asselin-Labat ML, Shackleton M, Stingl J, Vaillant F, Forrest NC, Eaves CJ, et al. Steroid hormone receptor status of mouse mammary stem cells. J Natl Cancer Inst. 2006;98(14):1011–4. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]
- 76.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9(2):201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 77.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127(5):1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, Sinha S. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 2012;30(7):1496–508. doi: 10.1002/stem.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447(7142):316–20. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 81.Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 2007;21(7):1358–66. doi: 10.1096/fj.06-6926com. [DOI] [PubMed] [Google Scholar]
- 82.van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14(10):1099–104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–9. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gastaldi S, Sassi F, Accornero P, Torti D, Galimi F, Migliardi G, et al. Met signaling regulates growth, repopulating potential and basal cell-fate commitment of mammary luminal progenitors: implications for basal-like breast cancer. Oncogene. 2012 doi: 10.1038/onc.2012.154. [DOI] [PubMed] [Google Scholar]
- 85.Moraes RC, Zhang X, Harrington N, Fung JY, Wu MF, Hilsenbeck SG, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134(6):1231–42. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]