Abstract
To investigate the genetic mechanism of mercury accumulation in maize (Zea mays L.), a population of 194 recombinant inbred lines derived from an elite hybrid Yuyu 22, was used to identify quantitative trait loci (QTLs) for mercury accumulation at two locations. The results showed that the average Hg concentration in the different tissues of maize followed the order: leaves > bracts > stems > axis > kernels. Twenty-three QTLs for mercury accumulation in five tissues were detected on chromosomes 1, 4, 7, 8, 9 and 10, which explained 6.44% to 26.60% of the phenotype variance. The QTLs included five QTLs for Hg concentration in kernels, three QTLs for Hg concentration in the axis, six QTLs for Hg concentration in stems, four QTLs for Hg concentration in bracts and five QTLs for Hg concentration in leaves. Interestingly, three QTLs, qKHC9a, qKHC9b, and qBHC9 were in linkage with two QTLs for drought tolerance. In addition, qLHC1 was in linkage with two QTLs for arsenic accumulation. The study demonstrated the concentration of Hg in Hg-contaminated paddy soil could be reduced, and maize production maintained simultaneously by selecting and breeding maize Hg pollution-safe cultivars (PSCs).
Introduction
Mercury (Hg) is a non-essential element in higher plants, and is one of the most hazardous heavy metals; it can accumulate in living organisms and cause serious damage [1]. With the development of industry and modern agriculture, Hg pollution has become a worldwide environmental problem [2], [3]. Numerous cases of mercury pollution in soils have been reported throughout the world [4]. Generally, Hg concentration in unpolluted arable soil is 20–150 µg kg−1 [5]. However, the Hg concentration in the soils of paddy fields is much higher, especially in the Philippines and Japan; the average Hg concentration is 24 mg kg−1 and 146 mg kg−1, respectively, for land irrigated with water contaminated with Hg [6]. The concentration of Hg in paddy soil is as high as 45.9 mg kg−1 in some area of China [7].
At high concentrations in soil, Hg can poison plant cells and cause physiological disorders [8], producing detrimental effects on plant growth and metabolism [9], [10]. Hg accumulation in roots prevents the uptake and transport of other mineral nutrients [11], and excess Hg in solution culture can inhibit biomass production [5]. Hg toxicity is not only associated with water uptake and transpiration [12], but also with the decrease in chlorophyll content and photosynthetic efficiency [13]. In addition, Hg stress is believed to trigger the production of reactive oxygen species (ROS), causing oxidative stress and membrane lipid peroxidation in plants [14], [15]. Hg in soil can accumulate in the edible parts of vegetables and crops, and is then transferred to humans via the food chain [16]. Hg is also toxic to humans, causing impaired health in adults. Hg has toxicological effects on the developing central nervous system, on the general physiological systems of children, and adverse effects on the cardiovascular system [17] and fetal brain development [18].
Hg is more easily absorbed and transported in straw and grain than other heavy metals [19]. Previous studies have shown that different genotypes in rice differ widely in their tolerance to Hg toxicity [14]. Wang et al. identified three quantitative trait loci (QTLs) for Hg tolerance in rice seedlings using a recombinant inbred line (RIL) population [20]. Using doubled haploid (DH) lines, three QTLs for Hg accumulation in rice were determined by Yu et al. [14]. Rugh et al. expressed the merApe9 gene in Arabidopsis thaliana, and found that the transgenic seedlings showed low levels of toxicity during the growth and flowering stages because merApe9 converted toxic Hg2+ to the less toxic Hg0 [21]. Shen et al. isolated three novel HO genes from rapeseed, and HO-1 has been tested for its ability to regulate plant tolerance to Hg-induced oxidative stress [22]. Wei et al. found that the overexpression of HO-1 confers algal tolerance to excess Hg, whereas silencing of HO-1 had adverse effects on algal growth [23]. Recently, Chen et al. reported that overexpression of the MTH1745 gene could enhance mercury tolerance in transgenic rice [24]. In maize, there have been some studies on the physiological and biochemical responses to Hg tolerance. The net photosynthesis rate and carboxylation efficiency of maize leaves exposed to 50 ng m−3 Hg (in the air) were significantly reduced [25]. In vivo, HgCl2 inhibited 5-aminolevulinic acid dehydratase activity in excised greening leaf segments of maize, and the inhibition could be alleviated by the addition of KNO3 [26]. Additionally, Hg had a strong toxic effect in the roots of maize seedlings, as inferred from the observed higher proportion of oxidized glutathione (GSSG), enhanced carbonyl content and the negative effects on growth [27].
Maize is one of the most important staple crops in the world, and is used as feedstuff and raw-food material in many countries. Maize is used as a staple food for more than 1.2 billion people in sub-Saharan Africa and Latin America [28]. Maize planted in Hg-contaminated paddy soil can accumulate Hg in the grains, which is transferred to consumers by the food chain (soil-plant-human or soil-plant-animal-human), posing a human health risk. However the Hg concentration and distribution pattern in the different tissues of maize and the genetic basis for Hg accumulation remain unclear. Therefore, the objectives of the study were to: (i) examine the accumulation and distribution of Hg in different tissues of maize, (ii) detect QTLs for Hg accumulation in the tissues of maize under Hg-contaminated paddy soil treatment, and to dissect the genetic bases of Hg accumulation in maize.
Materials and Methods
The experimental population
A RIL population including 194 inbred lines was used in the study. This population was derived from a cross between two inbred lines, Zong3 and 87-1; the result was an elite hybrid, Yuyu22, which was grown on about 2.7 million hectares per year from 2001 to 2004 in China [29]. The parent Zong3 was selected from a synthetic population with Chinese domestic germplasm; while the inbred line 87-1 was selected from an exotic germplasm. In 2012, the RIL population, both parents and the hybrid were evaluated in Xixian and Changge, Henan Province, China. The experimental materials followed a randomized complete block design with three replications. Each plot included fifteen plants with one 4-m-long and 0.67-m-wide row. The density was 45,000 plants per hectare.
The mercury content in soil
The experimental materials were evaluated in Xixian country of Xinyang city in Henan province of China (E114°95′–114°72′, N32°35′–32°44′) and Changge country of Xuchang city in Henan province of China (E113°34′–114°08′, N34°09′–34°20′), with an annual average temperature of 15.2°C and 14.3°C; and annual average rainfall of 946 mm and 711.1 mm. The field studies did not involve endangered or protected species, and no specific permissions were required for these locations/activities. The average value of agricultural soil Hg exposure in the Xixian experimental area is 457.57±31.30 µg kg−1Hg (pH 6.5), as a result of irrigation with mercury-rich surface water. The average value of agricultural soil Hg exposure in the Changge experimental area is 345.40±22.24 µg kg−1Hg (pH 6.5), which was used as a control.
Quantification of mercury content in the different tissues of maize
Five consecutive plants per row were harvested at physiological maturity stage and at natural withering for mercury content measurements in the different tissues of maize. The whole plant was separated into five tissues: kernels, axis, stems, bracts and leaves, which were ground into powders. A 0.5-g sample of each tissue was digested in 5 ml of HNO3/HClO4 (80/20 v/v) on a heating block (Digestion Systems of AIM500, A. I. Scientific, Australia). An atomic fluorescence spectrometry (AFS-3000, Beijing Haiguang Analytical Instrument Co, Beijing, China) was used to determine the Hg concentration. Data analyses using the PROC MIXED procedure were performed using SAS 8.0 statistical software.
Molecular Linkage Construction and QTL Mapping
A total of 263 SSR markers that covered the whole genome of maize were used to construct the genetic linkage map for the RIL population, which spanned 2.361 cM with an average interval of 9 cM between markers [29]. The composite interval mapping method in software QTL Cartographer 2.5 [30] was employed for QTL mapping of measured traits, using the average data of three replications. Model 6 of the Zmapqtl module was used, with scanning intervals of 2 cM between markers and putative QTLs and a 10-cM window. The number of marker cofactors for background control was set by forward–backward stepwise regression with five controlling markers. The logarithm of odds (LOD) threshold was set for each trait by randomly permuting 1,000 times at a significance level of P = 0.05.
Results
Hg concentrations in different maize tissues at two locations
The Hg concentration in the five tissues varied widely in the RIL population (Table 1, Fig 1). In Xixian, the Hg concentration in the kernels (KHC) for the parent Zong3 was higher (3.53 µg kg−1) than that in the parent 87-1 (2.14 µg kg−1); the same trend for the two parents was observed in Changge. The KHC was higher in the hybrid Yuyu22 (4.62 µg kg−1, 2.74 µg kg−1) than in the two parents in both locations. The average KHC of the RIL population was 2.99±1.25 µg kg−1 (range 0.73–7.47 µg kg−1) and 2.37±1.52 µg kg−1 (range 0.26–7.18 µg kg−1) in Xixian and Changge, respectively.
Table 1. Hg concentration in the five measured tissues in the RIL population.
| Location | Population | Trait | KHC (µg kg−1) | AHC (µg kg−1) | SHC (µg kg−1) | BHC (µg kg−1) | LHC (µg kg−1) |
| Xixian | P1 | Mean | 3.53 | 2.89 | 3.71 | 8.80 | 30.20 |
| P2 | Mean | 2.14 | 1.51 | 6.02 | 6.21 | 43.20 | |
| F1 | Mean | 4.62 | 3.40 | 5.15 | 5.95 | 43.83 | |
| RIL | Mean | 2.99±1.25 | 3.84±1.49 | 5.36±1.30 | 7.59±1.81 | 41.06±6.65 | |
| Range | 0.73–7.47 | 1.08–8.37 | 1.82–9.02 | 4.07–15.56 | 27.30–61.45 | ||
| Skewness | 1.05 | 0.83 | −0.14 | 1.54 | 0.28 | ||
| Kurtosis | 1.51 | 0.66 | 0.34 | 4.99 | −0.06 | ||
| Changge | P1 | Mean | 1.96 | 1.86 | 4.70 | 9.18 | 43.14 |
| P2 | Mean | 0.91 | 3.71 | 2.83 | 8.23 | 37.31 | |
| F1 | Mean | 2.74 | 3.32 | 3.67 | 7.14 | 45.23 | |
| RIL | Mean | 2.37±1.52 | 2.71±1.76 | 4.26±2.32 | 5.83±1.84 | 39.60±11.79 | |
| Range | 0.26–7.18 | 0.18–8.18 | 0.47–11.83 | 2.06–14.03 | 18.64–74.50 | ||
| Skewness | 1.11 | 0.94 | 0.76 | 1.11 | 0.92 | ||
| Kurtosis | 0.90 | 0.25 | 0.56 | 2.51 | 0.46 |
Note: KHC, Hg concentration in kernels; AHC, Hg concentration in the axis; SHC, Hg concentration in stems; BHC, Hg concentration in bracts; LHC, Hg concentration in leaves.
Figure 1. Histogram of Hg concentration in the five tissues of the RIL population.
The Hg concentration in the axis (AHC) for the parent Zong3 was higher (2.89 µg kg−1) than that of the parent 87-1(1.51 µg kg−1) in Xixian. However, in Changge, the value of AHC for the parent Zong3 was lower (1.86 µg kg−1) than that of the parent 87-1(3.71 µg kg−1). The hybrid had a higher AHC (3.40 µg kg−1) in Xixian and a mid-parent value (3.32 µg kg−1) in Changge, compared with its parents. The average AHC for the RIL population was 3.84±1.49 µg kg−1 (range 1.08–8.37 µg kg−1) in Xixian and 2.71±1.76 µg kg−1 (range 0.18–8.18 µg kg−1) in Changge.
For the Hg concentration in the stem (SHC), the value of the parent Zong3 was lower (3.71 µg kg−1) than that of the parent 87-1(6.02 µg kg−1) in Xixian, but the opposite trend was found in Changge. The hybrid showed a mid-parent value (5.15 µg kg−1, 3.67 µg kg−1) at both locations. For the RIL population, the average SHC was 5.36±1.30 µg kg−1 (range 1.82–9.02 µg kg−1) in Xixian and 4.26±2.32 µg kg−1 (range 0.47–11.83 µg kg−1) in Changge.
For the Hg concentration in the bract (BHC), the parent Zong3 had higher values (8.80 µg kg−1, 9.18 µg kg−1) than the parent 87-1 (6.21 µg kg−1, 8.23 µg kg−1) at both locations. In addition, the BHC (5.95 µg kg−1, 7.14 µg kg−1) for the hybrid expressed the same low-parent performance at both locations. The average BHC for the RIL population was 7.59±1.81 µg kg−1 (range 4.07–15.56 µg kg−1) in Xixian and 5.83±1.84 µg kg−1 (range 2.06–14.03 µg kg−1) in Changge.
The parent Zong3 had a lower Hg concentration (30.20 µg kg−1) than the parent 87-1 (43.20 µg kg−1) in the leaves in Xixian, but the value of the parent Zong3 (43.14 µg kg−1) was higher than the parent 87-1 (37.31 µg kg−1) in Changge. The Hg concentration in the leaves (LHC) for the hybrid was 43.83 µg kg−1 in Xixian and 45.23 µg kg−1 in Changge. For the LHC of the RIL populations, the average value was 41.06±6.65 µg kg−1 (range 27.30–61.45 µg kg−1) in Xixian and 39.60±11.79 µg kg−1 (range 18.64–74.50 µg kg−1) in Changge.
Among the different tissues of maize, the kernels had the lowest Hg concentration, followed by the axis, stems, bracts and leaves, and both locations showed the same trend. According to variance analysis, the Hg concentration for the five measured tissues (kernels, axis, stems, bracts, and leaves) in the RIL population exhibited significant variations between genotypes and environments, as well as in the interaction of genotypes and environments (Table 2, P<0.01). For the Hg concentration in five measured tissues in Xixian, the KHC and AHC displayed significant positive relationships, yet the BHC positively correlated with LHC. However, there were no significant relationships among the Hg concentrations in the five measured tissues in the RIL population in Changge by the phenotypic relationship analysis (Table 3, P<0.01).
Table 2. Variance analysis of the five measured tissues in the RIL population.
| Kernel | Axis | Stem | Bract | Leave | |
| L | 135.49** | 161.05** | 59.1** | 205.2** | 5.43* |
| G | 7.03** | 2.68** | 7.17** | 3.07** | 6.71** |
| L×G | 7.37** | 2.83** | 6.46** | 3.88** | 8.48** |
Note: *significant at P<0.05, **significant at P<0.01.
Table 3. Correlation coefficients among five measured tissues in the RIL population.
| Kernel | Axis | Stem | Bract | Leave | |
| Kernel | 0.25** | 0.16 | −0.07 | −0.03 | |
| Axis | 0.12 | 0.04 | 0.09 | 0.01 | |
| Stem | −0.07 | −0.01 | 0.05 | 0.05 | |
| Bract | −0.05 | −0.11 | −0.03 | 0.27** | |
| Leave | −0.05 | −0.04 | 0.09 | 0.15 |
Note: **significant at P<0.01.
Correlation coefficients in Xixian are above the diagonal, while those in Changge are below the diagonal.
QTL analysis for Hg concentration in the five tissues of maize
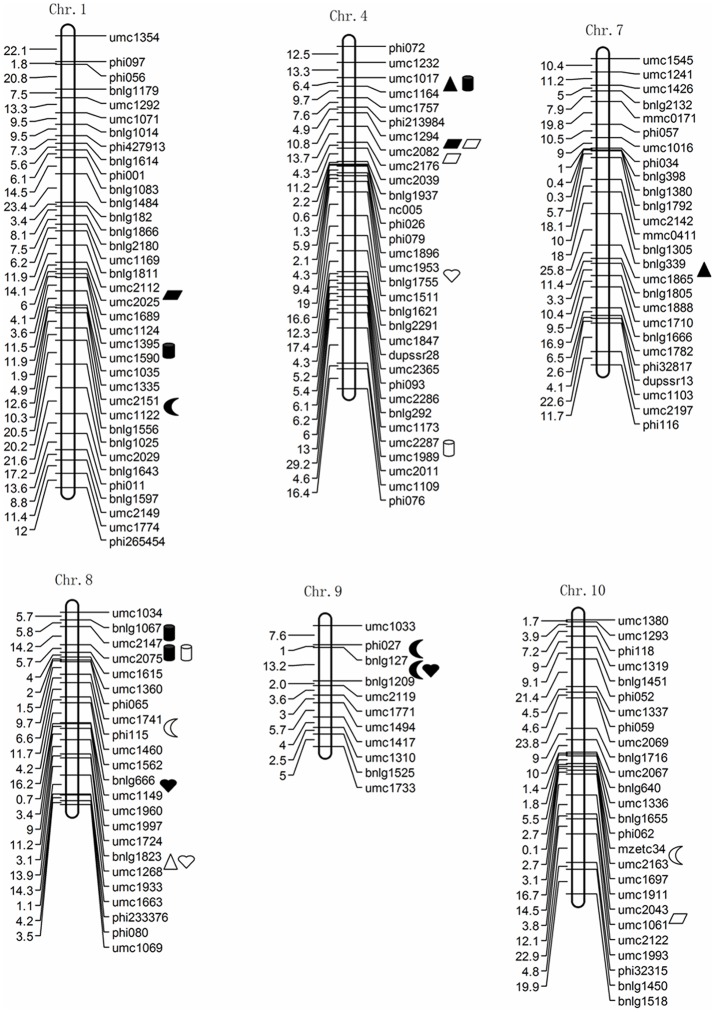
Twenty-three QTLs were detected for Hg concentration in the five tissues of maize in the RIL population at the two locations (Table 4, Fig 2). These QTLs were distributed on chromosomes 1, 4, 7, 8, 9 and 10. Five QTLs for Hg concentration in the kernels were identified in the two environments. In Xixian, there were three QTLs; two of them, qKHC9a and qKHC9b was adjacent and explained 10.75% and 10.85% of the phenotypic variance, respectively. These two were derived from the parent 87-1. In Changge, two QTL qKHC8 and qKHC10 were identified, with 13.00% and 8.42% contribution rate to total phenotypic variance, which came from the parent Zong3 and 87-1, respectively.
Table 4. QTLs detected for Hg concentration in five maize tissues.
| Location | Tissue | QTLa | Location | Flanking-markers | LODb | Ac | R2d |
| Xixian | kernel | qKHC1 | 238.51 | umc2151-umc1122 | 3.66 | 0.51 | 15.11 |
| qKHC9a | 9.61 | phi027-bnlg127 | 3.06 | −0.42 | 10.75 | ||
| qKHC9b | 18.91 | bnlg127-bnlg1209 | 2.50 | −0.42 | 10.85 | ||
| axis | qAHC4 | 25.81 | umc1017-umc1164 | 2.86 | 0.44 | 8.18 | |
| qAHC7 | 145.31 | bnlg339-umc1865 | 3.08 | 0.70 | 22.10 | ||
| stem | qSHC1 | 211.31 | umc1395-umc1590 | 2.50 | 0.47 | 11.76 | |
| qSHC4a | 30.81 | umc1017-umc1164 | 3.62 | 0.43 | 10.94 | ||
| qSHC8a | 6.71 | bnlg1067-umc2147 | 5.88 | 0.56 | 18.24 | ||
| qSHC8b | 16.51 | umc2147-umc2075 | 4.20 | 0.59 | 20.63 | ||
| bract | qBHC8a | 71.11 | bnlg666-umc1149 | 2.65 | 0.52 | 7.80 | |
| qBHC9 | 14.91 | bnlg127-bnlg1209 | 2.85 | 0.53 | 8.30 | ||
| leave | qLHC1 | 178.51 | umc2112-umc2025 | 2.64 | −2.07 | 9.30 | |
| qLHC4a | 57.41 | umc1294-umc2082 | 3.19 | −3.08 | 16.46 | ||
| Changge | kernel | qKHC8 | 46.91 | umc1741-phi115 | 3.18 | 0.69 | 13.00 |
| qKHC10 | 115.61 | mzetc34-umc2163 | 3.16 | −0.46 | 8.42 | ||
| axis | qAHC8 | 111.61 | bnlg1823-umc1268 | 2.55 | −0.52 | 6.69 | |
| stem | qSHC4b | 221.71 | umc2287-umc1989 | 3.76 | −0.88 | 13.94 | |
| qSHC8b | 25.51 | umc2147-umc2075 | 2.71 | 0.61 | 6.70 | ||
| bract | qBHC4 | 109.51 | umc1953-bnlg1755 | 2.81 | 0.53 | 7.34 | |
| qBHC8b | 114.61 | bnlg1823-umc1268 | 2.66 | 0.48 | 6.44 | ||
| leave | qLHC4a | 59.41 | umc1294-umc2082 | 3.55 | 4.63 | 14.38 | |
| qLHC4b | 69.21 | umc2082-umc2176 | 2.51 | 3.94 | 10.39 | ||
| qLHC10 | 138.21 | umc2043-umc1061 | 3.38 | 3.73 | 9.22 |
Notes: aQTL detected for Hg concentration in the five maize tissues; bLOD for each QTL; cAdditive effect; positive values indicate that Zong3 alleles increase rates; dR2, contribution ratio.
Figure 2. Chromosomal locations of QTLs for Hg concentration for five maize tissues.
Note: Moon QTL detected for Hg content in kernels, Triangle QTL detected for Hg content in the axis, Cylinder QTL detected for Hg content in stems, Heart QTL detected for Hg content in bracts, Quadrangle QTL for Hg content in leaves. Black indicates a QTL detected in Xixian, and lucidity indicates a QTL detected in Changge.
For the Hg concentration in the axis, two QTLs, qAHC4 and qAHC7 were detected in Xixian and qAHC8 was detected in Changge, which contributed 8.18%, 22.10% and 6.69% of the phenotypic variance in the Hg concentration in the axis, respectively. The alleles of qAHC4 and qAHC7 were derived from the parent Zong3, and the qAHC8 allele came from the parent 87-1.
Six QTLs associated with SHC were detected at the two locations, which were located on chromosomes 1, 4 and 8. The QTLs contributed 6.70%–20.63% of the total phenotypic variance. Among the QTLs, qSHC8b was found at both environments simultaneously, accounting for 20.63% and 6.70% of the total variance, and the effects resulted from the parent Zong3.
Four QTLs were associated with BHC in this study. In Xixian, two QTLs, qBHC8a and qBHC9, contributed 7.80% and 8.30% of phenotypic variance, respectively. In Changge, the QTLs qBHC4 and qBHC8b explained 7.34% and 6.44% of the phenotypic variance of BHC, respectively. All these QTLs were derived from the high performance parent, Zong3.
For the Hg concentration in the leaves, there were five QTLs at both locations, which explained 9.22%–16.46% of the phenotypic variance. QTL qLHC4a was found at both locations simultaneously, and was responsible for 16.46% and 14.38% of the total variance, respectively.
Discussion
Hg pollution has aroused global concern because of its toxicity to organisms, persistence in the environment and long-range transport [25]. In some contaminated areas, food chain transfer is very common [31]–[34]. In rice, the Hg concentration is much higher in roots than in shoots [14]. Similar results were obtained from other plant species [35], [36]. Meng et al. reported that Hg levels in rice tissues followed the trend: root > stalk> leaf > husk > seed [37], which is consistent with previous studies [38]–[40]. Liu et al. found that the Hg content in the different tissues of maize had features as follows: root > leaf > stalk > grain [41]. In this study, we observed a similar distribution in Hg concentration in different maize tissues (leaves > bracts > stems >axis > kernels). These results demonstrated that the Hg concentration in kernels was lower than that in the other tissues, and the mechanism of Hg accumulation and distribution in different tissues of maize was possibly related to the plant detoxification mechanism. In this study, the Hg concentration in soil in Xixian was higher (457.57±31.30 µg kg−1) than that in Changge (345.40±22.24 µg kg−1). Compared with the average Hg concentration for the five measured tissues in the two locations, the average value in the RIL population was higher in Xixian than in Changge for all tissues. These phenomena indicated that the Hg concentrations in the different tissues of maize were mainly reflected in the Hg concentration in the soil.
Heavy metals are increasingly becoming environmental concerns because of their release into ecosystems, which pose a threat to human health [23]. Low-cost and ecologically sustainable strategies are needed to restore heavy metal-contaminated soils [28]. However, physical or chemical methods to address heavy metal contamination are environmentally invasive, expensive and inefficient, especially for large scale clean up [42]. Phytoremediation is considered a cost-effective and environmentally friendly approach to remove heavy metals from contaminated soils [43]; however, its application is limited because it is time consuming, there is a lack of suitable plant species and production during remediation is unprofitable [44]. Yu et al. raised the concept of pollution-safe cultivars (PSCs), in which a specific pollutant was accumulated by the edible part at a low level for safe consumption, even when planted in contaminated soil [45]. As an alternative choice, there has been increasing interest in selecting and breeding PSCs [46]. In this study, we found that Hg accumulated in high concentrations in the axis, bracts, stems and leaves, which represent the main biomass products in maize, while the kernels contained the lowest concentration among the five tested tissues. For KHC in the RIL population, two parents and the hybrid, the highest value was 4.62 µg kg−1 in Xixian and 2.74 µg kg−1 in Changge; however, the values were much lower than 20 µg kg−1, which is the maximum recommended by the Chinese National Standard Agency [47]. Maize is the most commonly planted crop worldwide, with broad adaptability. Thus, there is the potential to decrease the concentration of Hg in Hg-contaminated paddy soil, and maintain maize production simultaneously by selecting and breeding maize Hg PSCs.
There are many forms of Hg in soil, including the elemental (Hg0), ionic (Hg2+), hydroxide (Hg(OH)2), methyl (MeHg), and sulfide (HgS) [48], [49]. The environment and the genotype control the uptake of Hg species in rice [50], and rice varieties differ widely in their absorption and tolerance of Hg toxicity [14]. In peas, six Hg2+ responsive genes were identified using suppression subtractive hybridization [51]. Their gene products were involved in the salicylic acid (SA) biological defense system, the biosynthetic pathway of isoflavonoids, antioxidant reactions, sulfur metabolism, and cell wall rigidity [51]. Heidenreich et al. profiled the transcriptome of A. thaliana exposed to Hg2+, and identified Hg-induced genes that encoded proteins involved in chlorophyll synthesis, cell wall metabolism, and P450-mediated biosynthesis of secondary metabolites [52]. In addition, Yamaguchi et al. found that Hg and other heavy metals could induce certain genes at the same time [53]. Recently, several other genes related to Hg accumulation have been identified [54], [55]. In rice, 20 proteins that were differentially expressed in roots under Hg treatment were identified using 2-D electrophoresis. They were involved in cellular functions, including redox and hormone homeostasis, chaperone activity, metabolism, and transcription regulation [56]. Wang et al. identified three QTLs for relative root length in rice under Hg stress, which coincided with QTLs for drought, Zn2+, Fe2+, Cd2+, Cu2+ and Al3+ toxicity tolerance in rice [19]. In this study, out of the 23 QTLs for Hg concentration in different maize tissues, three (qKHC9a, qKHC9b, qBHC9) were adjacent to two QTLs for drought tolerance using the F2:3 populations derived from the same parents Zong3 and 87-1 [57]; one QTL, qLHC1, was in linkage with two QTLs for arsenic accumulation using RIL populations derived from a cross between two maize inbred lines, Huang-C and Xu178 [58]. These results suggested that the QTLs related to Hg accumulation had pleiotropic effects.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by the State Key Basic Research and Development Plan of China (2014CB138203) and Scientific Personnel Innovation Fund of Henan Province in China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang L, Wong MH (2007) Environmental mercury contamination in China: Sources and impacts. Environment International 33: 108–121. [DOI] [PubMed] [Google Scholar]
- 2. Chen J, Shiyab S, Han FX, Monts DL, Waggoner CA, et al. (2009) Bioaccumulation and physiological effects of mercury in Pteris vittata and Nephrolepis exaltata. Ecotoxicology 18: 110–121. [DOI] [PubMed] [Google Scholar]
- 3. Elbaz A, Wei YY, Meng Q, Zheng Q, Yang ZM (2010) Mercury induced oxidative stress and impact on antioxidant enzymes in Chlamydomonas reinhardtii. Ecotoxicology 9: 1285–1293. [DOI] [PubMed] [Google Scholar]
- 4. Luis R, Jesusa R, Isaac A, Laura RC (2007) Capability of selected crop plants for shoot mercury accumulation from polluted soils: Phytoremediation Perspectives. International Journal of Phytoremediation 9: 1–13. [DOI] [PubMed] [Google Scholar]
- 5. Du X, Zhu YG, Liu WJ, Zhao XS (2005) Uptake of mercury (Hg) by seedlings of rice (Oryza sativa L.) grown in solution culture and interactions with arsenate uptake. Environmental and Experimental Botany 54: 1–7. [Google Scholar]
- 6. Li P, Feng XB, Qiu GL, Shang LH, Li ZG (2009) Mercury pollution in Asia: A review of the contaminated sites. Journal of Hazardous Materials 168: 591–601. [DOI] [PubMed] [Google Scholar]
- 7.China National Environmental Monitoring Station (1990) Background elemental concentrations of Chinese soils. China Environmental Science Press, Beijing, pp. 8–10.
- 8. Zhou ZS, Huang SQ, Guo K, Mehta SK, Zhang PC, et al. (2007) Metabolic adaptations to mercury-induced oxidative stress in roots of Medicago sativa L. Journal of Inorganic Biochemistry 101: 1–9. [DOI] [PubMed] [Google Scholar]
- 9. Patra M, Sharma A (2000) Mercury toxicity in plants. Botanical Review 66: 379–422. [Google Scholar]
- 10. Tamas L, Mistrik I, Huttova J, Haluskova L, Valentovicova K, et al. (2010) Role of reactive oxygen species-generating enzymes and hydrogen peroxide during cadmium, mercury and osmotic stresses in barley root tip. Planta 231: 221–231. [DOI] [PubMed] [Google Scholar]
- 11. Boening DW (2000) Ecological effects, transport, and fate of mercury: A general review. Chemosphere 40: 1335–1351. [DOI] [PubMed] [Google Scholar]
- 12. Zhang WH, Tyerman SD (1999) Inhibition of water channels by HgCl2 in intact wheat root cells. Plant Physiology 120: 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xylander M, Hagen C, Braune W (1996) Mercury increases light susceptibility in the green alga Haematococcus lacustris. Bot. Acta 109: 222–228. [Google Scholar]
- 14. Yu JY, Hu HT, Wang CC, Y L (2011) QTL analysis of mercury tolerance and accumulation at the seedling stage in rice (Oryza sativa L.). Journal of Food Agriculture and Environment 9(2): 748–752. [Google Scholar]
- 15. Han Y, Xuan W, Yu T, Fang WB, Lou TL, et al. (2007) Exogenous hematin alleviates mercury-induced oxidative damage in the roots of Medicago sativa. Journal of Integrative Plant Biology 49: 1703–1713. [Google Scholar]
- 16. Zheng N, Wang QC, Zheng DM (2007) Mercury contamination and health risk to crops around the zinc smelting plant in Huludao City, northeastern China. Environmental Geochemistry and Health 29: 385–393. [DOI] [PubMed] [Google Scholar]
- 17. Abbas ES, Esmail A, Sharif JS, Seyed MG (2012) Hair mercury levels in six Iranian sub-populations for estimation of methylmercury exposure: A mini-review. Iranian Journal of Toxicology 15: 541–547. [Google Scholar]
- 18. Drum DA (2009) Are toxic biometals destroying your children's future? Biometals 22: 697–700. [DOI] [PubMed] [Google Scholar]
- 19. Liu WX, Shen LF, Liu JW, Wang YW, Li SR (2007) Uptake of toxic heavy metals by rice (Oryza sativa L.) cultivated in the agricultural soil near Zhengzhou city, People's Republic of China. Bulletin of Environmental Contamination and Toxicology 79: 209–213. [DOI] [PubMed] [Google Scholar]
- 20. Wang CC, Wang T, Mu P, Li ZC, Yang L (2013) Quantitative trait loci for mercury tolerance in rice seedlings. Rice Science 20(3): 238–242. [Google Scholar]
- 21. Rugh CL, Wilde HD, Stack NM, Thompson DM, Meagher R B (1996) Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. PNAS 93: 3182–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen Q, Jiang M, Li H, Che LL, Yang ZM (2011) Expression of a Brassica napus heme oxygenase confers plant tolerance to mercury toxicity. Plant Cell and Environment 34: 752–763. [DOI] [PubMed] [Google Scholar]
- 23. Wei YY, Zheng Q, Liu ZP, Yang ZM (2011) Regulation of tolerance of Chlamydomonas reinhardtii to heavy metal toxicity by heme oxygenase-1 and carbon monoxide. Plant and Cell Physiology 52(9): 1665–1675. [DOI] [PubMed] [Google Scholar]
- 24. Chen Z, Pan YH, Wang SS, Ding YF, Yang Wj, et al. (2012) Overexpression of a protein disulfide isomerase-like protein from Methanothermobacter thermoautotrophicum enhances mercury tolerance in transgenic rice. Plant Science 197: 10–20. [DOI] [PubMed] [Google Scholar]
- 25. Niu ZC, Zhang XS, Wang ZW, Ci ZJ (2011) Field controlled experiments on the physiological responses of maize (Zea mays L.) leaves to low-level air and soil mercury exposures. Enviroment Pollution 159: 2684–2689. [DOI] [PubMed] [Google Scholar]
- 26. Priyanka G, Meeta J, Juliana S, Rekha G (2013) Inhibition of 5-aminolevulinic acid dehydratase by mercury in excised greening maize leaf segments. Plant Physiology and Biochemistry 62: 63–69. [DOI] [PubMed] [Google Scholar]
- 27. Rubén RÁ, Cristina OV, Ana ÁF, Francisca FC, Luis EH (2006) A stress responses of Zea mays to cadmium and mercury. Plant and Soil 279: 41–50. [Google Scholar]
- 28. Wuana RA, Okieimen FE (2010) Phytoremediation Potential of Maize (Zea mays L.) A Review. African Journal of General Agriculture 6 (4): 275–287. [Google Scholar]
- 29. Tang JH, Teng WT, Yan JB, Ma XQ, Meng YJ, et al. (2007) Genetic dissection of plant height by molecular markers using a population of recombinant inbred lines in maize. Euphytica 155: 117–124. [Google Scholar]
- 30. Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qiu GL, Feng XB, Li P, Wang SF, Li GH, et al. (2008) Methylmercury accumulation in rice (Oryza sativa L.) grown at abandoned mercury mines in Guizhou, China. Journal of Agricultural and Food Chemistry 56: 2465–2468. [DOI] [PubMed] [Google Scholar]
- 32. Horvat M, Nolde N, Fajon V, Jereb V, Logar M, et al. (2003) Total mercury, methylmercury and selenium in mercury polluted areas in the province Guizhou, China. Science of the Total Enviroment 304: 231–256. [DOI] [PubMed] [Google Scholar]
- 33. Meng B, Feng XB, Qiu GL, Cai Y, Wang DY, et al. (2010) Distribution patterns of inorganic mercury and methylmercury in tissues of rice (Oryza sativa L.) plants and possible bioaccumulation pathways. Journal of Agricultural and Food Chemistry 58: 4951–4958. [DOI] [PubMed] [Google Scholar]
- 34. Zhang H, Feng XB, Larssen T, Shang LH, Li P (2010) Bioaccumulation of methylmercury versus inorganic mercury in rice (Oryza sativa L.) grain. Environmental Science & Technology 44: 4499–4504. [DOI] [PubMed] [Google Scholar]
- 35. Israr M, Sahi S, Datta R, Sarkar D (2006) Bioaccumulation and physiological effects of mercury in Sesbania drummondii. Chemosphere 65: 591–598. [DOI] [PubMed] [Google Scholar]
- 36. Wang Y, Greger M (2004) Clonal differences in mercury tolerance, accumulation and distribution in willow. Journal of Environmental Quality 33: 1779–1785. [DOI] [PubMed] [Google Scholar]
- 37. Meng M, LI B, Shao JJ, Wang TH, He Bin, et al. (2014) Accumulation of total mercury and methylmercury in rice plants collected from different mining areas in China. Environmental Pollution 184: 179–186. [DOI] [PubMed] [Google Scholar]
- 38. Sierra MJ, Millán R, Esteban E (2008) Potential use of Solanum melongena in agricultural areas with high mercury background concentrations. Food and Chemical Toxicology 46: 2143–2149. [DOI] [PubMed] [Google Scholar]
- 39. Sierra MJ, Millán R, Cardona AI, Schmid T (2011) Potential cultivation of Hordeum Vulgare L. in soils with high mercury background concentrations. International Journal of Phytoremediation 13: 765–778. [DOI] [PubMed] [Google Scholar]
- 40. Zornoza P, Millán R, Sierra MJ, Seco A, Esteban E (2009) Efficiency of the white lupin in the removal of mercury from contaminated soils: soil and hydroponic experiments. Journal of Environmental Sciences 22(3): 421–427. [DOI] [PubMed] [Google Scholar]
- 41. Liu R, Wang Q, Lv X, Li Z, Wang Y (2004) Distribution and stock of mercury in typical wetland plant in the Sanjiang Plain. Chinese Journal of Applied Ecology 15: 287–290. [PubMed] [Google Scholar]
- 42. Kärenlampi S, Schat H, Vangronsveld J, Verkleij JAC, Lelie D, et al. (2000) Genetic engineering in the improvement of plants for phytoremediation of metal polluted soil. Environmental Pollution 107: 225–231. [DOI] [PubMed] [Google Scholar]
- 43. Kramer U (2005) Phytoremediation: novel approaches to cleaning up polluted soils. Current Opinion in Biotechnology 16: 133–141. [DOI] [PubMed] [Google Scholar]
- 44. Pilon-Smits EAH, Freeman JL (2006) Environmental cleanup using plants: biotechnological advances and ecological considerations. Frontiers in Ecology and the Environment 4 (4): 203–210. [Google Scholar]
- 45. Yu H, Wang JL, Fang W, Yuan J, Yang ZY (2006) Cadmium accumulation in different rice cultivars and screening for pollution-safe cultivars of rice. Science of the Total Enviroment 370(2–3): 302–309. [DOI] [PubMed] [Google Scholar]
- 46. Grant CA, Clarke JM, Duguid S, Chaney RL (2008) Selection and breeding of plant cultivars to minimize cadmium accumulation. Science of the Total Environment 390(2–3): 301–310. [DOI] [PubMed] [Google Scholar]
- 47.Chinese National Standard Agency (1994) Tolerance limit of mercury in foods. GB 2762–94. Beijing, China.
- 48. Heaton ACP, Rugh CL, Wang NJ, Meagher RB (2005) Physiological responses of transgenic merA-tobacco (Nicotiana tabacum) to foliar and root mercury exposure. Water Air and Soil Pollution 161: 137–155. [Google Scholar]
- 49. Han FX, Su Y, Monts DL, Waggoner CA, Plodinec MJ (2006) Binding, distribution, and plant uptake of mercury in a soil from Oak Ridge, Tennessee, USA. Science of the Total Enviroment 368: 753–768. [DOI] [PubMed] [Google Scholar]
- 50. Rothenberg SE, Feng XB, Zhou WJ, Tu M, Jin BW, et al. (2012) Environment and genotype controls on mercury accumulation in rice (Oryza sativa L.) cultivated along a contamination gradient in Guizhou, China. Science of the Total Enviroment 426: 272–280. [DOI] [PubMed] [Google Scholar]
- 51. Sävenstrand H, Strid Å (2004) Six genes strongly regulated by mercury in Pisum sativum roots. Plant Physiology and Biochemistry 42: 135–142. [DOI] [PubMed] [Google Scholar]
- 52. Heidenreich B, Mayer K, Sandermann JR, Ernst D (2001) Mercury-induced genes in Arabidopsis thaliana: identification of induced genes upon long-term mercuric ion exposure. Plant Cell and Environment 24: 1227–1234. [Google Scholar]
- 53. Yamaguchi H, Fukuoka H, Arao T, Ohyama A, Nunome T, et al. (2010) Gene expression analysis in cadmium-stressed roots of a low cadmium-accumulating solanaceous plant, Solanum torvum. Journal of Experimental Botany 61: 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hsieh JL, Chen CY, Chiu MH, Chein MF, Chang JS, et al. (2009) Expressing a bacterial mercuric ion binding protein in plant for phytoremediation of heavy metals. Journal of Hazardous Materials 161: 920–925. [DOI] [PubMed] [Google Scholar]
- 55. Ruiz ON, Alvarez D, Torres C, Roman L, Daniell H (2011) Metallothionein expression in chloroplasts enhances mercury accumulation and phytoremediation capability. Plant Biotechnology Journal 9: 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen YA, Chi WC, Huang LT, Lin CY, Quynh Nguyeh TT, et al. (2012) Mercury-induced biochemical and proteomic changes in rice roots. Plant Physiology and Biochemistry 55: 23–32. [DOI] [PubMed] [Google Scholar]
- 57. Liu Y, Subhash C, Yan JB, Song CP, Zhao JR, et al. (2011) Maize leaf temperature responses to drought: Thermal imaging and quantitative trait loci (QTL) mapping. Enviroment and Experimental Botany 71: 158–165. [Google Scholar]
- 58. Ding D, Li WH, Song GL, Qi HY, Liu JB, et al. (2011) Identification of QTLs for arsenic accumulation in maize (Zea mays L.) using a RIL population. PLOS ONE 6: e25646–25652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.