Abstract
The class of [NiFe]–hydrogenases is characterized by a bimetallic cofactor comprising low–spin nickel and iron ions, the latter of which is modified with a single carbon monoxide (CO) and two cyanide (CN−) molecules. Generation of these ligands in vivo requires a complex maturation apparatus in which the HypC–HypD complex acts as a ‘construction site’ for the Fe–(CN)2CO portion of the cofactor. The order of addition of the CO and CN– ligands determines the ultimate structure and catalytic efficiency of the cofactor; however much debate surrounds the succession of events. Here, we present an FT–IR spectroscopic analysis of HypC–HypD isolated from a hydrogenase–competent wild–type strain of Escherichia coli. In contrast to previously reported samples, HypC–HypD showed spectral contributions indicative of an electron–rich Fe–CO cofactor, at the same time lacking any Fe–CN– signatures. This immature iron site binds external CO and undergoes oxidative damage when in contact with O2. Binding of CO protects the site against loss of spectral features associated with O2 damage. Our findings strongly suggest that CO ligation precedes cyanation in vivo. Furthermore, the results provide a rationale for the deleterious effects of O2 on in vivo cofactor biosynthesis.
Introduction
Hydrogenases are ancient and widespread iron–sulfur enzymes [1]. All hydrogenases exploit low–spin transition metal ions to catalyze proton reduction and heterolytic dihydrogen oxidation following H2 ↔ H+ + H− ↔ 2 H+ + 2 e−. Hydrogenases have inspired a great wealth of models [2] that exploit abundant chemical elements over noble metal catalysts like platinum. [NiFe]–, [FeFe]–, and [Fe]–hydrogenases can be distinguished based on their active site metal composition [3]. A common motif in the organometallic redox chemistry of [NiFe]– and [FeFe]–hydrogenases is a modification of the active site cofactor with carbon monoxide (CO) and cyanide (CN−). These are found coordinated exclusively with the iron ions and allow for efficient hydrogen turnover at minimal overpotential [4]. Generation of CO and CN− in vivo is associated with reactive intermediates, thus evolution has established complex maturation pathways to synthesize and coordinate these diatomic ligands [5]. The catalytic cofactor of [NiFe]–hydrogenases consists of a nickel ion and an iron ion. Four cysteinyl residues coordinate the nickel site, which typically adopts the Ni+3 oxidation state in the catalytically inactive, oxidized ‘Ni–A’ and ‘Ni–B’ species [6], [7]. Carbon monoxide and O2 inhibit [NiFe]–hydrogenase activity although there are prominent exceptions [8]–[10]. Two of the aforementioned cysteine residues coordinate not only nickel but the iron ion as well. All [NiFe]–hydrogenase have a precise 2∶1 ratio of CN– to CO [11] yet it is completely unclear how this is maintained. The sixth coordination site in the iron ion is shared with nickel and was found to be occupied by hydrogen and oxygen species [3], [6], [7]. We refer to the Ni–Fe cofactor as ‘[NiFe]–(CN)2CO’ in the following.
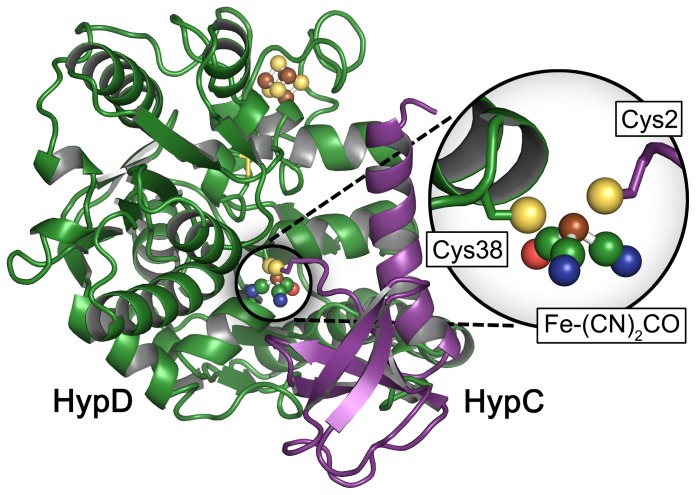
In vivo generation of the [NiFe]–(CN)2CO cofactor (‘maturation’) has been the subject of intensive research [3], [5], [12], [13]. Minimally six accessory proteins are required specifically for cofactor biosynthesis [11], including HypA through HypF. The iron ion is modified with the three diatomic ligands prior to translocation onto the hydrogenase large subunit apo–protein. The natural source of iron has not yet been clearly identified; however, metallo–chaperone activity of HypC suggests that the small OB–fold protein is involved in iron acquisition. Furthermore, IR signatures of bound CO2 were detected and shown to be lost upon specific release of iron from the protein [14]. HypC forms a tight complex with HypD [15], [16], a redox active iron–sulfur protein that is the central construction site of the Fe–(CN)2CO moiety of the cofactor. Although no crystal structure of HypD with an intact iron site has been resolved, FT–IR analysis clearly proves the existence of the modified iron ion [17], [18]. Figure 1 shows a schematic representation of the HypC–HypD (HypCD) complex carrying the mature metal site as calculated by Albareda and co–workers [19]. The model consists of a R. leguminosarum HypC homology structure superimposed onto the crystal structure of HypCD from T. kodakarensis [20]. Fe–(CN)2CO was added to the N–terminus of HypC (Cys2) with the iron ion in close proximity to Cys38 of HypD and optimized in an aqueous environment [19].
Figure 1. Mixed crystal structure/ homology model of the HypCD complex.
The model consists of a R. leguminosarum HypC homology structure superimposed onto the crystal structure of HypCD from T. kodakarensis [20]. Fe–(CN)2CO was added to the N–terminus of HypC (Cys2) with the iron ion in close proximity to Cys38 of HypD and optimized in an aqueous environment. The Fe–(CN)2CO cofactor has been added to the model in silico [19].
While CO2 is a promising candidate as precursor of the CO ligand on HypD [14], the role of HypE and HypF in generation of the CN− ligands has been unequivocally demonstrated [21], [22]. HypF catalyzes an ATP–dependent activation of carbamoylphosphate to carbamoyladenylate. This carbamoyl moiety is transferred to the C–terminal cysteinyl residue of HypE, yielding a thiocarbamate, which is dehydrated to thiocyanate at the expense of another ATP hydrolysis step. The crystal structure of a HypCDE complex suggests that thiocyanate from HypE is transferred to the iron site on HypD via substrate channeling [15], [23]. The order of addition of the diatomic ligands still needs to be resolved.
In this study, we isolated a mixed–state sample of HypCD from the E. coli K–12 wild–type strain MC4100. The protein preparation exhibits more complex infrared signatures than previously analyzed HypCD samples from hydrogenase–inactive strains [14], [17], [18], [24]. Gas treatments rendered specific changes as probed by attenuated total reflection (ATR) Fourier–transform infrared (FT–IR) absorption spectroscopy. The simultaneously extant populations reacted differently to O2 and CO, identifying a sample fraction devoid of iron–cyanide contributions. The IR analysis presented allows us to propose an order of events for the in vivo modification of the iron site on HypD. Furthermore, it explains how maturation is inhibited by O2.
Materials and Methods
The strains used in this study were E. coli MC4100 [25] and BL21(DE3) [26]. Plasmid pT–hypDEFCStrep [16] was used as the source of HypCStrep–HypD (HypCD in the following). For overproduction of HypCD cells were grown in 3 l of modified TB medium [18] with 100 g l–1 ampicillin in a 5 l conical flasks on rotary shakers at 37°C with slow rotation until an OD of 0.4 at 600 nm was reached. This ensured that the cultures were strongly O2–limited. At that point gene expression was induced by addition of 0.2 mg l−1 anhydrotetracycline and the culture was incubated at 30°C for a further 3 to 5 h up to OD600 = 1.0. Cells were harvested by centrifugation at 4°C at 15 000 g for 20 min and resuspended anaerobically at a ratio of 1 g per 3 ml buffer W (100 mM Tris–HCl, 150 mM NaCl, pH 8.0) including 2 mM sodium dithionite, 5 g ml−1 DNase, and 0.2 mM PMSF. Cells were disrupted by sonication (30 W for 5 min with 0.5 s pulses). Unbroken cells and debris were removed by centrifugation for 30 min at 50 000 g at 4°C. The supernatant derived from 10 g wet weight of cells was used for anaerobic purification of the HypCD complex. Varying sub–stoichiometric amounts of HypE were associated with the 1∶1 HypCD complex and in the interest of clarity we will henceforth refer to the complex as HypCD. Proteins were isolated using a 5 ml gravity–flow Streptactin sepharose column (IBA, Göttingen, Germany). Unbound proteins were removed by washing with five volumes of buffer W. Recombinant HypCD was eluted with buffer W including 5 mM desthiobiotin. Afterward the elutant was removed by passage through a series of Hi–Prep PD10 desalting columns (GE Healthcare). Proteins were concentrated by centrifugation at 7 500 g using centrifugal filters (Amicon Ultra 50 K, Millipore, Eschborn, Germany).
Fourier–transform IR spectroscopy was conducted on a rapid scan Tensor27 (Bruker Optik, Ettlingen, Germany) equipped with a three–reflection silicon crystal ATR cell (Smith Detection, Warrington, USA) as described earlier [24]. The spectrometer is situated in an anaerobic gas chamber (Coy Laboratories, Grass Lake, USA) with a N2/ H2 gas composition of 99∶1 and O2 levels below 0.2 ppm. Nitrogen is provided with 99.995% purity from a N2 generator (Inmatec, Herrsching, Germany). Carbon monoxide (12CO) was purchased from Linde Gas (Unterschleißheim, Germany). Isotopically labelled 13CO (99% 13CO with <5% 18O) was provided by Sigma Aldrich.
Gas treatments of HypCD were performed on humid films. First, a 1 µl drop of concentrated sample (∼30 g/l HypCD) was placed on top of the ATR silicon crystal and dried under water–free N2 to judge the amount of protein and integrity of the active site cofactor. Absolute spectra consist of 2.000 scans and are subtracted for liquid water contribution by spline functions. The crystal was covered by a gas–tight, three–way acrylic glass cell that allows for gas treatment without contamination of the anaerobic atmosphere. Second, dry N2 was passed through distilled water. The aerosol was fed to the ATR gas cell in order to temper the protein film and make it available for further gas treatments. Nitrogen was replaced by air or CO to induce changes in the HypCD spectrum. Eventually, difference spectra (minimally 10.000 scans) were recorded on dry films. Nitrogen was re–routed from water to a 1% (v/v) solution of H2O2 to trigger the oxidation process as reported earlier [24].
Results
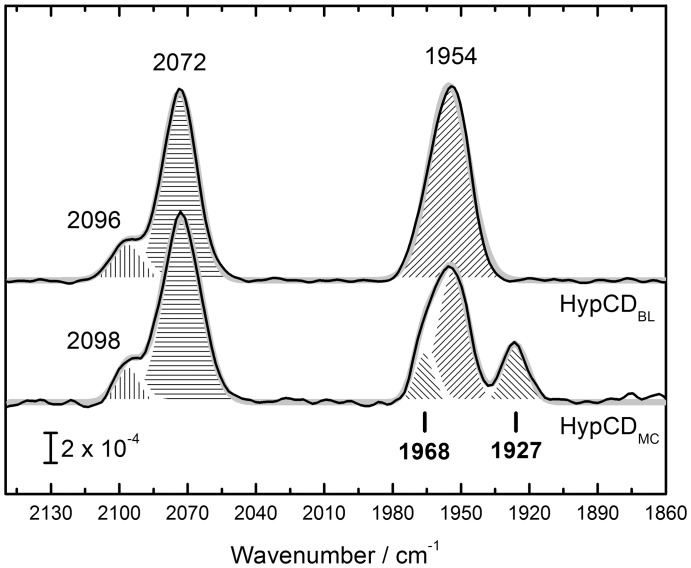
The HypCD protein complex was over–produced and purified anaerobically via Streptactin sepharose affinity chromatography from extracts of E. coli MC4100 (referred to as HypCDMC) by making use of the C–terminal strep–tag on HypC. Strain MC4100 typically synthesizes three [NiFe]–hydrogenases after anaerobic growth and has the full complement of accessory proteins (HypABCDEF) required for maturation [25], [27]. Figure 2 shows FT–IR spectra comparing HypCDMC and HypCDBL isolated from E. coli strain BL21(DE3). This strain is defective in synthesis of active hydrogenases [28] and was transformed with plasmid phypCDEF to overproduce the accessory proteins. HypCDBL exhibits contributions from two bands at 2072 and 2096 cm−1 and a single contribution at 1954 cm−1. The latter signal has been attributed to Fe(II)–CO, while both former bands stem from Fe(II)–CN1 and Fe(II)–CN2 [17], [18]. In the spectrum of HypCDMC, three bands fit with 2072, 2098, and 1954 cm−1, corresponding to Fe(II)–CN1, Fe(II)–CN2, and Fe(II)–CO, respectively. A new peak appeared at lower wavenumbers with a maximum at 1927 cm−1. The main Fe(II)–CO peak shows a clear shoulder at higher wavenumbers, fitting best with 1968 cm−1.
Figure 2. Comparison of the IR spectra of HypCD from E. coli strains BL21(DE3) and MC 4100.
HypCDBL shows the characteristic signature [17], [18] of one CO (1954 cm−1, diagonal hatch) and two CN− ligands (2072, 2096 cm−1, straight hatch). HypCDMC has a novel contribution in the CO region (1927 cm−1) and shows a shoulder blue–shifted to the CO peak at 1954 cm−1. The latter shoulder fits best to a Gaussian peak with a center wavenumber of 1968 cm−1. The bold gray line in the background represents an overall fit of the spectrum.
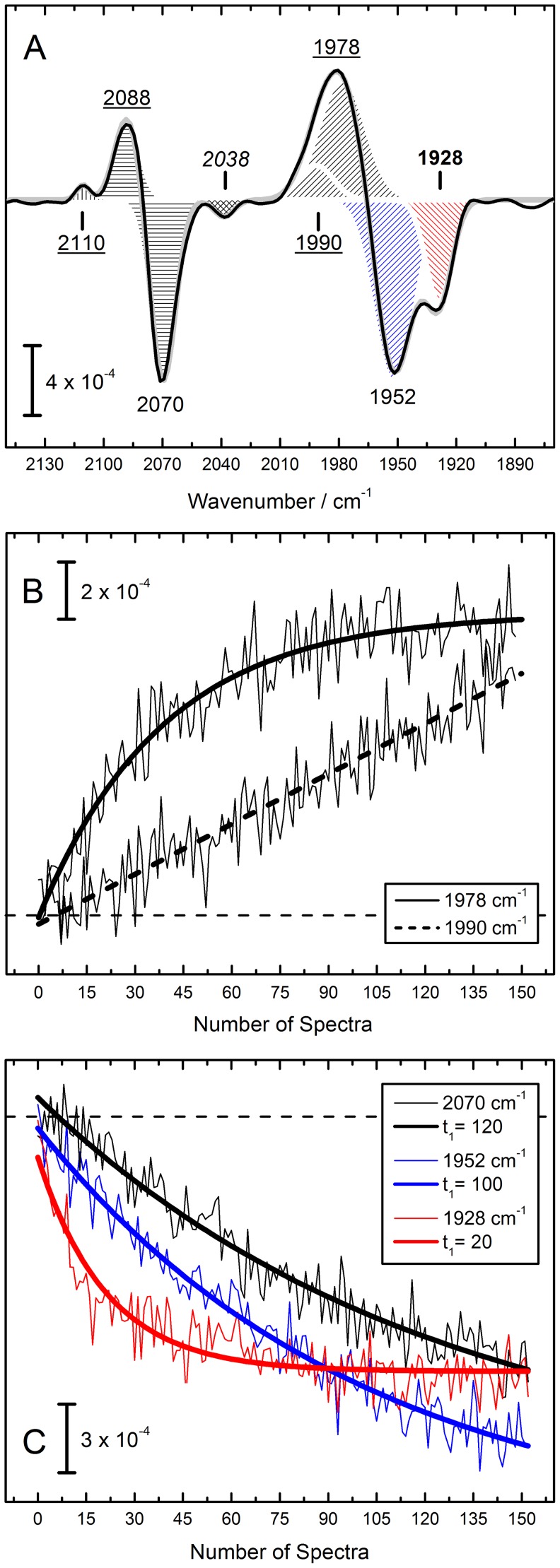
HypCDBL has been shown previously to react with H2O2 [24], with rapid oxidation of the iron cofactor due to Fenton chemistry [29], [30]. We performed a kinetic analysis of the effect of H2O2 on HypCDMC to identify coupled vibrations. Figure 3A shows the final difference spectrum of HypCDMC subjected to H2O2 oxidation. Negative peaks represent the state before the treatment. Strong bands appear at 1928, 1952, and 2070 cm−1. One negative band fits to 2038 cm−1. Peaks at 1968 and 2096 cm−1 are not visible because they are masked by large positive contributions at 1978/1990 cm−1 and 2088/2110 cm−1. While the peaks at 2088/2110 cm−1 can readily be assigned to Fe(III)–CN1 and CN2 [24], the 1978/1990 cm−1 peaks were identified based on their deviating kinetic behavior in the oxidation process.
Figure 3. H2O2 IR difference spectrum of HypCDMC.
(Fig. 3A) Negative features represent the as–isolated state (1928 and 1952 cm−1 for CO in red and blue hatch, respectively, and 2070 cm−1 for CN−), positive features are attributed to the oxidized, instable Fe(III)–(CN)2CO state with 1978/1992 cm−1 attributable to CO and 2088 and 2110 cm−1 attributable to the CN− ligands [24]. A new band arises at 2038 cm−1 (cross hatch). (Fig. 3B) While the contribution at 1978 cm−1 exponentially increases after contact of the sample with H2O2, the band at 1990 cm−1 obeys a rather linear kinetic behavior. (Fig. 3C) The previously identified contributions at 1952 and 2070 cm−1 vanish simultaneously [24], however, the peak at 1928 cm−1 showcases a four to five times faster absorption decay. All kinetics are plotted in ‘Number of Spectra’ to avoid misinterpretation of temporal information.
Figure 3B shows the increase of the 1978 and 1990 cm−1 contribution over time. In an earlier publication we followed oxidation from Fe(II)–CO to Fe(III)–CO as a result of H2O2 treatment via the 1990 cm−1 band [24]. However, upon closer examination it seems the initial product of oxidation is a band that fits best to 1978 cm−1 and whose kinetics can be monitored exponentially. Underlying this is a somewhat linear process that includes a non–redox shift to 1990 cm−1. In the current experiment this process was incomplete, which is why in fig. 3A a two–component Gaussian fit in the region of 1978/1990 cm−1 was used. The 1928 cm−1 peak was not found to be coupled with the 1952 and 2070 cm−1 contributions. Figure 3C depicts the diverging reaction kinetics. The new band at 1928 cm−1 (red slope) decreased four to five times faster than the Fe(II)–CO and Fe(II)–CN1 marker bands (blue and black slope, respectively). This hints at a mixed population in HypCDMC in contrast to the homogenous isolation that has been published for HypCDBL [17], [18].
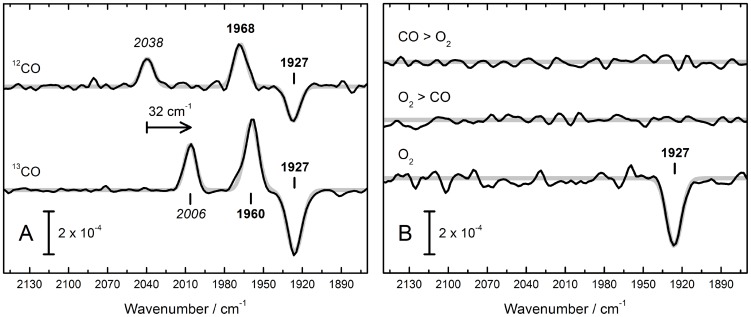
Semi–dry films of HypCD were treated with air (referred to as O2 henceforth), 12CO, and 13CO. While HypCDBL was insensitive to either O2 or CO, HypCDMC exhibited distinct changes. Figures 4A and B show the resulting difference spectra. Only the band at 1927 cm−1 reacted with CO and O2. Upon incubation with 12CO gas, a shift of 1927 to 1968 cm−1 was recorded (fig. 4A). Simultaneously, a pronounced peak at 2038 cm−1 appeared. A peak at 2038 cm−1 was observed earlier as a negative contribution in the difference spectrum of HypCDMC after H2O2 oxidation (fig. 3A). The 1968 cm−1 band was assigned as a contribution to the broad 1954 cm−1 peak of HypCDMC in fig. 2. In contrast, the peak centered at 2038 cm−1 was absent from the absolute spectrum. Binding of 13CO to HypCD gave rise to the expected isotope shift. A new peak appeared at 2006 cm−1, and the shift of 1927 to 1960 cm−1 was eight wavenumbers smaller than for the reaction with 12CO. A single negative peak appeared at 1927 cm−1 upon treatment with O2 (fig. 4B). This indicates a loss of absorbing molecules. Oxygen was not found to react with HypCDMC after CO treatment, and vice versa. It is important to note that no difference signals were observed for the peaks assigned to Fe(II)–CO (1954 cm−1), Fe(II)–CN1 (2072 cm−1), and Fe(II)–CN2 (2098 cm−1). Small changes in the cyanide region became visible only upon prolonged incubation under 1 mbar 13CO (see fig. S1). This effect was not observed with 12CO.
Figure 4. IR difference spectra of HypCDMC that show the effect of 12CO, 13CO, and O2.
(Fig. 4A) Binding of external 12CO (13CO) produces a new peak, 2038 cm−1 (2006 cm−1) and induces a shift of the original Fe–CO peak from 1927 to 1968 cm−1 (1960 cm−1). Note these spectra do not give rise to difference signals in the CN− region. (Fig. 4B) A strong negative contribution at 1927 cm−1 is observed. Double difference spectra depict on the influence of O2 on CO–soaked sample (CO>O2) and vice versa (O2>CO). No additional difference bands are observed. Note these spectra do not give rise to difference signals in the CN− region.
Discussion
Synthesis of CN− from carbamoyl–phosphate for the active site cofactor of [NiFe]–hydrogenases has been elucidated in ever–increasing detail since Reissmann and co–workers published their pioneering work in 2003 [21]. Crystal structures of the accessory proteins HypE and HypF have been resolved, both individually and in the relevant quaternary complexes, such as CDE [15], [20], [31], [32]. Recently, a specifically cyanated form of HypE has been crystallized after incubation with OCN− and ATP [23]. Moreover, the HypD enzyme was characterized as a central scaffold in [NiFe]–hydrogenase maturation [17], [18]. HypD binds a single iron ion as acceptor of the CN− ligands from cyanated HypE in vivo [16]. HypD carries a [4Fe–4S] cluster linked to an ‘active site’ iron ion within tunneling distance via one or two disulfide bridges [20]. HypD is discussed to be a redox enzyme that catalyzes the reductive transfer of CN− and CO to iron in an ATP–dependent reaction. The metabolic origin of CO is matter of ongoing debate [3] although prominent IR signatures on HypC and HypCD hint at iron–bound CO2 as a precursor of the CO ligand [14], [18], [24]. While it is reasonably clear what the source of the CN− ligands is, and strong circumstantial evidence for one possible source of CO has been suggested, precise details of how the 2∶1 ratio of CN− to CO is maintained remains unclear.
Isolation of HypCD from E. coli BL21(DE3) phypCDEF revealed an iron cofactor with two CN− ligands (CN1 at 2072 and CN2 at 2098 cm−1) and a single CO ligand (1955 cm−1). Strain BL21(DE3) naturally lacks hydrogenase activity because of deficiencies in nickel uptake and insertion [28], which presumably results in accumulation of the HypCD complex as the final acceptor of a pre–formed Fe–(CN)2CO cofactor [17], [18]. In this study, we isolated a mixed–state sample of HypCD from the hydrogenase–active E. coli wild–type strain MC4100. The increased spectral features probably result from the fact that hydrogenase maturation is unrestricted when compared with BL21(DE3). HypCDMC revealed an additional band at 1927 cm−1 and a shoulder of the main peak at 1954 cm−1, which fits best with 1968 cm−1. It is unlikely for this new state to represent an ‘off–pathway’ intermediate as trace amounts of the 1927 cm−1 contribution have been observed previously in difference spectra of a preparation derived from BL21(DE3) [24]; however this contribution barely accumulated in the hydrogenase–negative strain. The signal was attributed to a reduced population Fe(I)–CO, well in accord with a shift of ∼30 cm−1. In [NiFe]–hydrogenases, a band at 1922 cm−1 was assigned to the reduced, hydride–binding ‘SIR1’ state [33]. The reduced H–cluster of [FeFe]–hydrogenases includes a CO ligand that absorbs at 1916 cm−1 (Fed(I)–CO) [34]. Therefore, we assign the 1927 cm−1 band to Fe(I)–CO. We rule out the possibility of a second CO ligand to Fe(II)–(CN)2CO because the positions of the three original ligands are unchanged; an additional ligand on the central coordination metal site would result in a blue–shift of the spectral signature of the whole cofactor. No peaks indicative of a conceivable Fe(I)–CN− moiety were detected. However, these contributions might be lost in the combination band of H2O that partially masks iron–cyanide absorption.
Further support for this finding was yielded by a kinetic analysis of the effects of H2O2 treatment on the Fe–(CN)2CO cofactor. We have shown previously that H2O2 rapidly oxidizes the iron ion from Fe(II) to Fe(III) by Fenton chemistry [24]. While the overall effect on HypCDMC is not significantly different to what was reported for HypCDBL, the Fe(I)–CO (1927 cm−1) signature was found to react four to five times faster to chemical oxidation, thus being uncoupled from the other spectral contributions. The three diatomic ligands bound to Fe(II) shift with basically identical time constants. This comparison provides an explanation how the HypCDMC spectrum is a mixture of (at least) two redox states.
The kinetic analysis elucidated two further important findings. First, the initial product of H2O2 oxidation of Fe(II)–CO is not Fe(III)–CO at 1990 cm−1 as previously suggested [24] but rather results in a band at 1978 cm−1 (+24 cm−1). The latter peak forms first but shifts to 1990 cm−1 in a non–redox process fitting to a linear behavior. Radical chemistry associated with H2O2 oxidation, plus the instability of a Fe(III)–CO is very likely to cause structural changes in the active site environment. Infrared signatures of gaseous CO2 upon prolonged H2O2 incubation results from highly oxidized protein. The second finding is that the reduced–versus–oxidized difference spectrum revealed a (negative) contribution at 2038 cm−1 not visible in the absolute spectrum of HypCDMC, possibly due to a contribution of liquid water (which negate each other in the difference spectrum). We were able to assign this peak only after analysis of the effect of O2 and CO on HypCDMC.
The O2 sensitivity of many [NiFe]– and all [FeFe]–hydrogenases is well documented [8]–[10], [35]–[37]. It is also known that O2 not only interferes with active hydrogenases but inhibits cofactor maturation as well [5]. In order to gain comprehensive knowledge of O2 inhibition in vivo it is important to understand at which point in maturation the oxidant competes with cofactor ligation. Remarkably, the pre–formed Fe(II)–(CN)2CO moiety on HypCD and on HypD was found not to react with O2. We could show previously that HypD alone is able to coordinate the Fe–(CN)2CO cofactor [24]. Next to the three non–protein ligands we experimentally identified a cysteine residue as occupying one binding site of low–spin Fe(II) [18], [24]. Obviously, the non–reactivity of HypCDBL towards O2 and CO requires complete saturation of all binding sites.
Oxygen treatment of the HypCDMC mixed–state sample results in a loss of the 1927 cm−1 peak exclusively. No shift was detected in the other bands and we could not identify any new bands. The results strongly suggest that this 1927 cm−1 contribution (potentially a precursor Fe(I)–CO form of the cofactor) is released from HypCD in the presence of O2. Interestingly, when the HypCDMC film is treated with CO prior to contact with O2, no such loss is observed. It is tempting to suggest a protective CO inhibition against O2 damage, as it has been observed for [FeFe]–hydrogenases [36]. Carbon monoxide was not found to react with O2–soaked films, which argues in favor of a displacement of the Fe(I)–CO site upon incubation with O2.
The effect of CO on HypCDMC is even more illustrative. The band at 1927 cm−1 shifts to 1968 cm−1 and a new peak forms at 2038 cm−1. We interpret the latter peak to be due to exogenous CO at Fe(I), accompanied by a +41 cm−1 shift of the original CO peak (Fe(I)–(CO)2). This is reasonable for coupled vibrations. Treatment of HypCDMC with 13CO results in the same effect, however, the specific shifts are different and allow for definitive assignment of the 2038 cm−1 band to exogenous 12CO (2004 cm−1 for 13CO). Accordingly, the effect of 13CO binding to Fe(I)–CO is slightly smaller (8 cm−1) in comparison to 12CO. For both 12CO and 13CO, no changes were detected in the CN− region although intensive and prolonged incubation with 13CO partly replaces the natural CO ligand in Fe(II)–(CN)2CO (ligand scrambling [34]) and gives rise to iron–cyanide difference signals (see fig. S1). Re–examination of the absolute spectrum of HypCDMC reveals that both the 1968 cm−1 and 1927 cm−1 bands arise from the mixed–state sample. Obviously, HypCDMC includes not only an electron–rich intermediate Fe(I)–CO but a CO–inhibited species Fe(I)–(CO)2 as well. Self–poisoning of hydrogenase samples by CO is a frequent phenomenon referred to as ‘cannibalization’ [34] and explains why it is not surprising to find a fraction of CO–contaminated enzyme in HypCDMC. This interpretation is supported by the negative band at 2038 cm−1 in the H2O2 difference spectrum, which must have been present initially but was potentially masked by the H2O combination band.
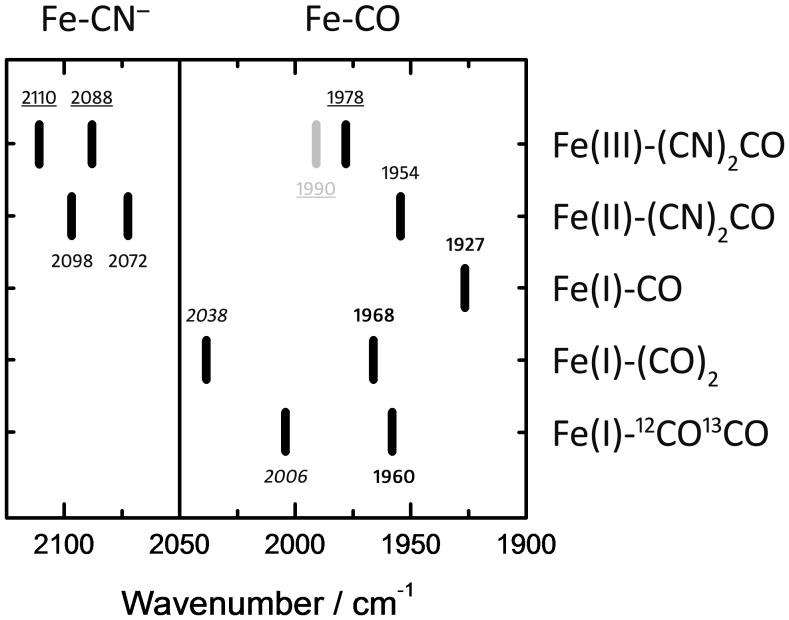
Conclusion
The novel population isolated from the HypCDMC mixed–state sample reacts with O2 and binds external CO. Steady–state and kinetic comparison with the characterized Fe(II)–(CN)2CO cofactor tentatively allows the assignment of the 1927 cm−1 contribution to Fe(I)–CO. Most importantly, no effect in the iron–cyanide region of the spectrum was observed upon treatment with O2, CO, or both. We could identify additional CO–binding species assigned to Fe(I)–(CO)2 (1968, 2038 cm−1) and Fe(I)–12CO13CO (1960, 2006 cm−1). No cyanide–binding intermediate was isolated, and CO2 was also absent from the sample. Figure 5 shows a plot of the identified HypCDMC redox states. While Fe(I)–(CO)2 might be a ‘contamination’ due to cannibalization or to CO–scrambling, Fe(I)–CO possibly represents the first of two inter–dependent steps in cofactor synthesis: The first step involves binding of CO to an Fe(I) ion on HypD, or binding of Fe–CO2 delivered by HypC [14] and reduction to Fe(I)–CO by a CO2 reductase–like mechanism, e.g. by HypD [38]; the second step results in binding of CN1 and CN2 to Fe(I)–CO as proposed by Watanabe and co–workers [15]. Our data potentially explains how externally added CO can be incorporated into the active site during in vivo biosynthesis but fails to exchange with bound CO in the mature active site [39], [40]. The same observation was made for O2; once Fe(II)–(CN)2CO is formed HypCD is not affected. These findings suggest that O2–sensitivity occurs exclusively at an earlier stage of maturation, e.g. immediately prior to addition of the CN– ligands and explains how cofactor synthesis is inhibited by O2 in vivo.
Figure 5. Conclusive comparison of the detected bands on HypCDMC.
Including Fe(II)–(CN)2CO (as–isolated), Fe(III)–(CN)2CO (oxidized by H2O2), Fe(I)–CO (reduced fraction), Fe(I)–(CO)2, and Fe(I)–12CO13CO. See discussion for details.
In a recent study Bürstel and co–workers have suggested that the binding of CN− ligands occurs prior to CO–ligation of the iron ion [17]. However, in that publication no iron–cyanide species devoid of CO was presented. We have identified CO bound to iron that lacks any CN− components, strongly suggesting that CO is the first diatomic ligand to be attached. This order of addition explains how the conserved 2∶1 ratio of CN∶CO is maintained in all [NiFe]–hydrogenases.
Supporting Information
Cyanide ligand scrambling upon prolonged incubation with 13CO. At 1 mbar and incubation times +1 h (room temperature, ambient light) 13CO not only binds to Fe(I)–CO but continuously replaces the ‘natural’ 12CO ligand of the Fe(II)–(CN)2CO cofactor whose spectrum serves as background in fig. S1. The black trace shows 12CO binding to a sample of HypCDMC as discussed in the main script. The red trace (13CO binding) gives rise to derivative–shaped signals in the Fe(II)–CN– region (here 2050–2100 cm−1). Both spectra were recorded under comparable conditions.
(PDF)
Acknowledgments
The authors are indebted to Joachim Heberle for discussion and support. Coordinates for the modified HypCD complex (fig. 1) were generously provided by Luis F. Pacios.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
RGS acknowledges the EU for EFRE funds and the DFG (Grant SA 494/3–2) for support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vignais PM, Billoud B (2007) Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev 107(10): 4206–4272. [DOI] [PubMed] [Google Scholar]
- 2. Tard C, Pickett CJ (2009) Structural and functional analogues of the active sites of the [Fe]–, [NiFe]–, and [FeFe]–hydrogenases. Chem. Rev 109(6): 2245–2274. [DOI] [PubMed] [Google Scholar]
- 3. Lubitz W, Ogata H, Ru O, Reijerse E (2014) Hydrogenases. Chem. Rev 114: 4081–4148. [DOI] [PubMed] [Google Scholar]
- 4. Hexter S V, Grey F, Happe T, Climent V, Armstrong FA (2012) Electrocatalytic mechanism of reversible hydrogen cycling by enzymes and distinctions between the major classes of hydrogenases. Proc. Natl. Acad. Sci. U. S. A 109(29): 11516–11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forzi L, Sawers RG (2007) Maturation of [NiFe]–hydrogenases in Escherichia coli . BioMetals 20(3–4): 565–578. [DOI] [PubMed] [Google Scholar]
- 6. Ogata H, Kellers P, Lubitz W (2010) The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni–A state). J. Mol. Biol 402(2): 428–444. [DOI] [PubMed] [Google Scholar]
- 7. Ogata H, Hirota S, Nakahara A, Komori H, Shibata N, et al. (2005) Activation process of [NiFe] hydrogenase elucidated by high–resolution X–ray analyses: conversion of the ready to the unready state. Structure 13(11): 1635–1642. [DOI] [PubMed] [Google Scholar]
- 8. Fritsch J, Scheerer P, Frielingsdorf S, Kroschinsky S, Friedrich B, et al. (2011) The crystal structure of an oxygen–tolerant hydrogenase uncovers a novel iron–sulphur centre. Nature 479: 249–252. [DOI] [PubMed] [Google Scholar]
- 9. Lukey MJ, Roessler MM, Parkin A, Evans RM, Davies RA, et al. (2011) Oxygen–Tolerant [NiFe]–Hydrogenases: The Individual and Collective Importance of Supernumerary Cysteines at the Proximal Fe–S Cluster. J. Am. Chem. Soc 133: 16881–16892. [DOI] [PubMed] [Google Scholar]
- 10. Fritsch J, Lenz O, Friedrich B (2013) Structure, function and biosynthesis of O2–tolerant hydrogenases. Nat. Rev 11(2): 106–114. [DOI] [PubMed] [Google Scholar]
- 11. Happe R, Roseboom W, Plerik A (1997) Biological Activation of Hydrogen. Nature 385: 126. [DOI] [PubMed] [Google Scholar]
- 12. Watanabe S, Sasaki D, Tominaga T, Miki K (2012) Structural basis of [NiFe] hydrogenase maturation by Hyp proteins. Biol. Chem 393(10): 1089–1100. [DOI] [PubMed] [Google Scholar]
- 13. Casalot L, Rousset M (2001) Maturation of the [NiFe] hydrogenases. Trends Microbiol 9(5): 228–237. [DOI] [PubMed] [Google Scholar]
- 14. Soboh B, Stripp ST, Bielak C, Lindenstrauß U, Braussemann M, et al. (2013) The [NiFe]–hydrogenase accessory chaperones HypC and HybG of Escherichia coli are iron– and carbon dioxide–binding proteins. FEBS Lett 587(16): 2512–2516. [DOI] [PubMed] [Google Scholar]
- 15. Watanabe S, Matsumi R, Atomi H, Imanaka T, Miki K (2012) Crystal Structures of the HypCD Complex and the HypCDE Ternary Complex: Transient Intermediate Complexes during [NiFe] Hydrogenase Maturation. Structure 20(12): 2124–2137. [DOI] [PubMed] [Google Scholar]
- 16. Blokesch M, Albracht SPJ, Matzanke BF, Drapal NM, Jacobi A, et al. (2004) The complex between hydrogenase–maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]–hydrogenases. J. Mol. Biol 344(1): 155–167. [DOI] [PubMed] [Google Scholar]
- 17. Bürstel I, Siebert E, Winter G, Hummel P, Zebger I, et al. (2012) A universal scaffold for synthesis of the Fe–(CN)2(CO) moiety of [NiFe] hydrogenase. J. Biol. Chem 287(46): 38845–38853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soboh B, Stripp ST, Muhr E, Granich C, Braussemann M, et al. (2012) [NiFe]–hydrogenase maturation: isolation of a HypC–HypD complex carrying diatomic CO and CN– ligands. FEBS Lett 586(21): 3882–3887. [DOI] [PubMed] [Google Scholar]
- 19. Albareda M, Palacios JM, Imperial J, Pacios LF (2013) Computational study of the Fe–(CN)2CO cofactor and its binding to HypC protein. J. Phys. Chem. B 117(43): 13523–13533. [DOI] [PubMed] [Google Scholar]
- 20. Watanabe S, Matsumi R, Arai T, Atomi H, Imanaka T, et al. (2007) Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling. Mol. . Cell 27(1): 29–40. [DOI] [PubMed] [Google Scholar]
- 21. Reissmann S, Hochleitner E, Wang H, Paschos A, Lottspeich F, et al. (2003) Taming of a poison: biosynthesis of the [NiFe]–hydrogenase cyanide ligands. Science 299(5609): 1067–1070. [DOI] [PubMed] [Google Scholar]
- 22. Paschos A, Bauer A, Zimmermann A, Zehelein E, Böck A (2002) HypF, a carbamoyl phosphate–converting enzyme involved in [NiFe] hydrogenase maturation. J. Biol. Chem 277(51): 49945–49951. [DOI] [PubMed] [Google Scholar]
- 23. Tominaga T, Watanabe S, Matsumi R, Atomi H, Imanaka T, et al. (2013) Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation. Proc. Natl. Acad. Sci. U. S. A 110(51): 20485–20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stripp ST, Soboh B, Lindenstrauss U, Braussemann M, Herzberg M, et al. (2013) HypD Is the Scaffold Protein for Fe–(CN)2CO Cofactor Assembly in [NiFe]–Hydrogenase Maturation. Biochemistry 52: 3289–3296. [DOI] [PubMed] [Google Scholar]
- 25. Casadaban MJ (1976) Regulation of the regulatory gene for the arabinose pathway, araC. J. Mol. Biol 104(3): 557–566. [DOI] [PubMed] [Google Scholar]
- 26. Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high–level expression of cloned genes. J. Mol. Biol 189(1): 113–130. [DOI] [PubMed] [Google Scholar]
- 27. Jacobi A, Rossmann R, Böck A (1992) The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli . Arch. Microbiol 58(6): 444–451. [DOI] [PubMed] [Google Scholar]
- 28. Pinske C, Sawers G (2011) Iron restriction induces preferential down–regulation of H2–consuming over H2–evolving reactions during fermentative growth of Escherichia coli . BMC Microbiol 11(196): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stadtman ER, Berlett BS (1991) Fenton Chemistry. J. Biol. Chem 266(26): 17201–17211. [PubMed] [Google Scholar]
- 30. Wink DA, Nims RW, Saavedra JE, Utermahlen WE, Ford PC (1994) The Fenton oxidation mechanism: reactivities of biologically relevant substrates with two oxidizing intermediates differ from those predicted for the hydroxyl radical. Proc. Natl. Acad. Sci. U. S. A 91(14): 6604–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shomura Y, Higuchi Y (2012) Structural basis for the reaction mechanism of S–carbamoylation of HypE by HypF in the maturation of [NiFe]–hydrogenases. J. Biol. Chem 287(34): 28409–28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tominaga T, Watanabe S, Matsumi R, Atomi H, Imanaka T, et al. (2012) Structure of the [NiFe]–hydrogenase maturation protein HypF from Thermococcus kodakarensis KOD1. Struct. Biol. Cryst. Commun 68(10): 1153–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shafaat HS, Rüdiger O, Ogata H, Lubitz W (2013) [NiFe] hydrogenases: a common active site for hydrogen metabolism under diverse conditions. Biochim. Biophys. . Acta 1827(8–9): 986–1002. [DOI] [PubMed] [Google Scholar]
- 34. Roseboom W, De Lacey AL, Fernandez VM, Hatchikian EC, Albracht SPJ (2006) The active site of the [FeFe]–hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO–ligand exchange as observed by infrared spectroscopy. J. Biol. Inorg. Chem 11(1): 102–118. [DOI] [PubMed] [Google Scholar]
- 35. Stripp ST, Goldet G, Brandmayr C, Sanganas O, Vincent KA, et al. (2009) How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. U. S. A 106(41): 17331–17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goldet G, Brandmayr C, Stripp ST, Happe T, Cavazza C, et al. (2009) Electrochemical kinetic investigations of the reactions of [FeFe]–hydrogenases with carbon monoxide and oxygen: comparing the importance of gas tunnels and active–site electronic/redox effects. J. Am. Chem. Soc 131(41): 14979–14989. [DOI] [PubMed] [Google Scholar]
- 37. Vincent KA, Parkin A, Lenz O, Albracht SPJ, Fontecilla–Camps JC, et al. (2005) Electrochemical definitions of O2 sensitivity and oxidative inactivation in hydrogenases. J. Am. Chem. Soc 127(51): 18179–18189. [DOI] [PubMed] [Google Scholar]
- 38. Jeoung JH, Dobbek H (2007) Carbon dioxide activation at the [NiFe]–cluster of anaerobic carbon monoxide dehydrogenase. Science 318(5855): 1461–1464. [DOI] [PubMed] [Google Scholar]
- 39. Forzi L, Hellwig P, Thauer RK, Sawers RG (2007) The CO and CN– ligands to the active site Fe in [NiFe]–hydrogenase of Escherichia coli have different metabolic origins. FEBS Lett 581(17): 3317–3321. [DOI] [PubMed] [Google Scholar]
- 40. Lenz O, Zebger I, Hamann J, Hildebrandt P, Friedrich B (2007) Carbamoylphosphate serves as the source of CN−, but not of the intrinsic CO in the active site of the regulatory [NiFe]–hydrogenase from Ralstonia eutropha . FEBS Lett 581(17): 3322–3326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cyanide ligand scrambling upon prolonged incubation with 13CO. At 1 mbar and incubation times +1 h (room temperature, ambient light) 13CO not only binds to Fe(I)–CO but continuously replaces the ‘natural’ 12CO ligand of the Fe(II)–(CN)2CO cofactor whose spectrum serves as background in fig. S1. The black trace shows 12CO binding to a sample of HypCDMC as discussed in the main script. The red trace (13CO binding) gives rise to derivative–shaped signals in the Fe(II)–CN– region (here 2050–2100 cm−1). Both spectra were recorded under comparable conditions.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.